Abstract
Background & objectives:
Bacterial biofilms a multi-layered defence, comprise extracellular DNA (eDNA) and proteins, protect bacteria from harmful environment and nutrient limitation and utilize the mutual benefits within a community. Bacterial biofilms also defend bacteria from harsh environments such as antibiotic treatment. This leads to poor antibiotic penetration, slow growth, adaptive stress responses, and formation of persister cells. This study was done to determine the relation of antibiotic resistance deciphered by the biofilms in Lactobacillus plantarum, a lactic acid bacteria (LAB) with probiotic significance.
Methods:
The gentamicin-resistant L. plantarum isolates were allowed to form biofilms and subjected to DNase I and proteinase K treatment. The optical density (OD) values were recorded for the biofilm assay and the cell count for the number of viable cells was taken for the control and the test samples. Percentage reduction was calculated based on the difference between the initial and final OD for both the parameters.
Results:
The biofilm assay revealed that the native L. plantarum isolates which were phenotypically susceptible, possessed the ability to form biofilms. The OD values were significantly decreased in comparison to the biofilm-forming control culture when these were treated with DNase I and proteinase K.
Interpretation & conclusions:
The study revealed that the biofilms formed by L. plantarum comprised of eDNA and proteins which was evidenced by the reduction in OD values and percentage in comparison to the control upon DNase I and proteinase K treatment. This indicates that the eDNA and biofilm matrix proteins are vital constituents of biofilms and may carry significant risk when coupled with antibiotic resistance.
Keywords: Antibiotic resistance, biofilms, extracellular DNA, lactic acid bacteria, nutrient limitation, persister cells
Lactic acid bacteria (LAB) are extensively researched for their probiotic utility owing to the beneficial properties. These are generally regarded as safe hence widely used in the food industry1. The genus Lactobacillus pertaining to the group of LAB is commonly used as starter and non-starter cultures in the dairy and fermented food industry. However, these have been reported to exhibit variability in nature which is a leading concern for food spoilage2. The main cause of spoilage is the formation of biofilms on the raw material3 or on the processing units during food processing4. Biofilms consist of microorganisms attached to a surface, implanted in a matrix of extracellular polymers5. In food processors, the extracellular matrix protects cells from processing and disinfectant treatments. In addition, biofilm-forming ability of Lactobacillus species could lead to treatment failures as a few species, viz. L. acidophilus,6, L. gasseri7, L. rhamnosus8, L. leichmanii9 and Pediococcus pentosaceus10, have been reported to cause clinical infections, such as endocarditis6, septic urinary infection7, chest infection8, bacteremia9, abscess6, peritonitis6, endometritis11 and septicaemia10.
The biofilm-forming ability of LAB varies from strain to strain and is known to shield the bacteria against disinfectants and organic acids during food processes3,12. The extracellular matrix in Lactobacillus plantarum comprises proteinaceous materials such as subunits of flagella as well as pili, cell surface adhesins, extracellular proteins, proteins of outer membrane vesicles and extracellular DNA (eDNA)5. The strain-to-strain variation in biofilm formation depends on differences in the eDNA levels owing to the altered lysis behaviour13. eDNA is a by-product of cell lysis and was stated as a significant structural constituent of the Enterococcus faecalis biofilm matrix. The release of eDNA has been described as fratricide which includes gelatinase-mediated cell death14.
Biofilms have been proposed to play a major role in deciphering antibiotic resistance, especially aminoglycosides15. The existence of a close microbial niche within biofilms and varying genetic responses are caused due to the density-dependent mechanism. This may aid the adaptive tolerance to antibiotics and transmission of resistance genes16. Biofilm-associated bacteria are less susceptible to antimicrobials than free-living or planktonic cells17. In certain instances eDNA has been reported to induce antimicrobial resistance in pathogenic bacteria. In Pseudomonas aeruginosa, eDNA regulates Pho PQ and PmrAB system and induces antimicrobial resistance18. It alters lipid polysaccharides and lowers outer membrane permeability to gentamicin in Salmonella enterica serovar Typhimurium19. In a certain case, a lower inner membrane proton motive force due to acidic pH induces drug resistance in P. aeruginosa20. Biofilm formation in Streptococcus pneumoniae21, Staphylococcus aureus22, Listeria monocytogenes23 and L. plantarum24 constitutes eDNA; however, the mechanism of resistance to antibiotics is not known.
Cells within the biofilms experience harsh growth conditions. Tolerance to antibiotics may be seen as a phenotypic shift in its nature when cells adapt to a sessile life style25. The diversity and metabolic state of cells present in a biofilm play an important role in deciphering antibiotic resistance. Generally, persistent cells being more resistant to antibiotics play a major role in supporting and maintaining the biofilm community26,27.
Biofilm matrix of LAB is poorly understood. Since most species of LAB are non-motile, the process of biofilm formation in the latter may be different from motile bacteria, thereby prompting the necessity to understand its mechanisms. In this study, the relation of antibiotic resistance deciphered by the biofilms and its components such as eDNA and proteins in bacteria has been described.
Material & Methods
L. plantarum MCC2774, L. plantarum CSG-8, L. plantarum CSG-21, L. plantarum C26a and L. plantarum MCC3011 isolated from chicken sausages were subjected to minimum inhibitory concentration (MIC) determination28. These were also found to possess the bi-functional aminoglycoside resistance gene15. The native cultures were grown and maintained in LAB susceptibility medium (LSM) and were used in this study. The study was conducted in the department of Microbiology and Fermentation Technology, Central Food Technological Research Institute, Mysuru, India. L. rhamnosus GG (LRGG) was used as a positive control as described earlier29 for the evaluation of biofilm formation and was obtained from the National institute of Nutrition (NIN), Hyderabad. It was subjected to MIC determination to ensure its susceptibility.
DNase I and proteinase K treatment of biofilm-forming L. plantarum cultures: The role of eDNA and proteins in biofilm formation was evaluated as described earlier13. L. plantarum isolates (n=5) were inoculated in LSM containing polystyrene cell culture plates (Tarsons, Bangalore) with 4 μg/ml of gentamicin (HiMedia, Mumbai) and allowed to grow till 48 h at 37°C. LRGG was used as a positive control. Phosphate-buffered saline (PBS) was used to wash and remove the unadhered cells. To test the presence of eDNA and proteins, DNase I (100 μg/ml) and proteinase K (10 μg/ml) (Genei, Bangalore) were added separately to the wells containing biofilm-formed adhered cells. The cells were incubated at 37°C for one hour and subsequently washed in PBS to remove traces of the DNase I and proteinase K.
Fluorescent microscopy: The DNase I- and proteinase K-treated cells of MCC3011 and LRGG were subjected to fluorescence microscopy. Before this, the cells were washed in PBS to remove traces of the DNase I and proteinase K. To visualize the live and dead cells, ethidium bromide (EtBr)/acridine orange (AcOr) dye (HiMedia) were used as described earlier13 to stain the test culture, MCC 3011 as well as control, LRGG cells and immediately viewed under fluorescent filters (3) with ×100 magnification (BX51TRF, Olympus, Japan). Images were captured with a Digital Olympus camera.
Assessment of the reduction in optical density (OD) values and cell enumeration: Crystal violet (CV, 0.1%) (HiMedia) was added to the microtitre plates which were kept for incubation at room temperature for 30 min. These were washed with 225 μL PBS thrice followed by the addition of 70 per cent ethanol to solubilize the dye. The optical density (OD) at 595 nm (ELISA Spec, Thermo Scientific, Finland) was recorded and the cell count was taken by serial dilution and pour plating in De Man, Rogosa and Sharpe (MRS) agar in triplicates. The percentage reduction in the CV staining and the culturable cells was calculated by the formula:
Percentage of reduction = Control OD595nm − Test595nm/Control OD595nm × 100.
The statistical analysis was carried out by one-way analysis of variance (ANOVA).
Results
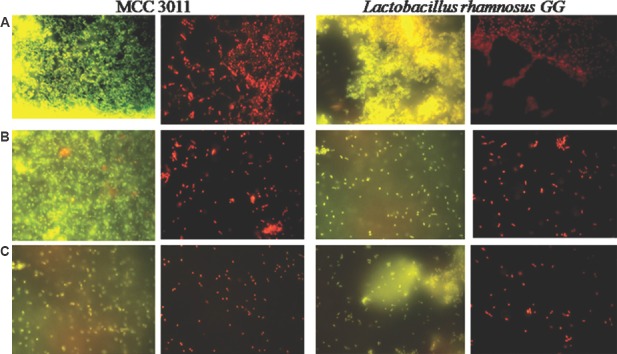
Role of eDNA and proteins on biofilm formation: To assess the function of eDNA in biofilm formation, mature biofilms in L. plantarum were subjected to DNase I and proteinase K treatment. In the present study, fluorescent images of the test culture, MCC 3011, as well as control, LRGG cells (Figure A), were observed to possess significantly dense population of both live and dead cells which accounted for the presence of matured biofilms. However, a considerable decrease in the cell density was evident in the DNase I-treated (Figure B) and proteinase K-treated (Figure C) cells. The decrease in the number of cells was also evidenced from the cell counts which ranged from 107 to 108 in the control cultures, while it reduced to 106-107 in cells subjected to DNase I and proteinase K treatment (Table). The OD values and log cfu/ml were observed to decrease upon addition of DNase I and proteinase K. L. plantarum MCC3011 and LRGG were found to form considerable quantity of biofilms. The reduction in the OD values due to the addition of proteinase K was more prominent than DNase I in all the isolates.
Figure.
Fluorescent images of Lactobacillus rhamnosus GG (control) and Lactobacillus plantarum MCC3011 (Test) (A) 48 h mature biofilms in phosphate buffered saline, (B) DNase I-treated and incubated for one hour, (C) proteinase K-treated and incubated for one hour. The cells viewed under fluorescent filters (3) with ×100 magnification and scale (2 μ).
Table.
Per cent reduction in biofilm after DNase I and proteinase K treatment for Lactobacillus plantarum cultures
| L. plantarum cultures | Initial OD | Final OD | Per cent reduction in CV staining | Initial log cfu/ml | Final log cfu/ml | Per cent reduction in cell count | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| DNase I | Proteinase K | DNase I | Proteinase K | DNase I | Proteinase K | DNase I | Proteinase K | |||
| CSG-8 | 0.64 | 0.34 | 0.30 | 46.4±0.03d | 52.3±0.1B | 8.18 | 7.15 | 7.40 | 23.7±0.21c | 21.8±0.28C |
| CSSG-21 | 0.53 | 0.33 | 0.24 | 36.5±0.02c | 54.2±0.08B | 8.68 | 7.15 | 7 | 29.2±0.21c | 30.9±0D |
| C26a | 0.39 | 0.35 | 0.20 | 10.2±0.03b | 47.3±0.01A | 8.46 | 7.71 | 7.73 | 8.8±0.03a | 8.5±0.08A |
| MCC3011 | 0.83 | 0.76 | 0.33 | 7.9±0.08a | 59.6±0.05B | 8.5 | 7.51 | 7.04 | 11.6±0.11b | 17.1±0.00B |
| MCC 2774 | 0.63 | 0.56 | 0.30 | 12±0.04b | 51.6±0.03B | 8.60 | 7.23 | 7.70 | 15.9±0.12b | 10.5±0.13A |
| LRGG | 0.94 | 0.46 | 0.29 | 50.9±0.03d | 69.2±0.01C | 8.56 | 7.10 | 7.40 | 28.7±0.18c | 25.2±0.14C |
The control biofilm by individual control was taken as 100 per cent (SD=±), The percentage of reduction has been calculated with respect to the control. The difference in the superscript for treatment with DNase I (a, b, c, d) and Proteinase K (A, B, C, D) in the given columns are significantly different (P<0.05). L. plantarum, Lactobacillus plantarum; OD, optical density; cfu, colony-forming unit; CV, crystal violet; SD, standard deviation; LRGG, Lactobacillus rhamnosus GG
Percentage reduction in the control LRGG was found to be the maximum when treated with DNase I (50.9%) and proteinase K (69.2%). On DNase I treatment, the highest reduction in eDNA was observed in CSG-8 (46.4%) followed by CSG-21 (36.5%). C26a and MCC 2774 were found to have only 10.2 and 12 per cent reduction, respectively. The least reduction (7.9%) was found in MCC 3011 (Table). This showed that C26a, MCC 2774 and MCC3011 produced less eDNA in comparison to CSG-8 and CSG-21. The percentage reduction in cell counts of all the five isolates was found to relate with the percentage reduction in OD values upon DNase I treatment.
The percentage reduction in the proteinase K-treated cells was significant and was in the same range. The reduced cell counts accounted to the increase in the number of dead cells owing to proteinase K treatment. The reduction values depicted in Table showed that the proteinase K treatment had high effect on the biofilm degradation.
The highest reduction in CV staining after DNase I treatment was observed for CSG-8 although less than the control and the least for MCC3011. Among the L. plantarum cultures treated with proteinase K, the highest reduction was found in MCC3011 (59.6%) and the least for C26a (47.3%) which corresponded with the percentage reduction in cell enumeration (Table).
Fluorescence microscopy revealed the presence of dead bacteria in biofilms. This explained the reduction of cell counts in DNase I- and proteinase K-treated cells. The role of eDNA was evident by fluorescent microscopy with EtBr/AcOr staining. The fluorescent microscopy images displayed the LRGG and the MCC3011 which comprised the live cells and the mature biofilm-stained orange indicated the presence of dead bacteria along with scattered live bacteria (Figure A). Figure B and C showed cells treated with DNase I and proteinase K, respectively. MCC3011 was observed to be less affected by DNase I treatment although the presence of eDNA was evident.
Discussion
This study was an attempt to evaluate the role of eDNA in biofilm formation which might aid in exhibiting phenotypic resistance. The strain-to-strain variation in exhibiting phenotypic resistance as well as biofilm formation may be explained owing to variation in levels of eDNA, as a probable consequence in lysis behaviour13. The CV assay depicts the quantitative figures of total biofilm formed. The reductions in the OD values upon DNase I and proteinase K treatment indicate the role of eDNA and proteins in biofilms formed by L. plantarum isolates. Similar patterns of reduction of OD values in L. plantarum isolates have been observed earlier13,23. Despite individual differences, the biofilm formation was affected by DNase I treatment. The contribution of eDNA and proteinase K as a component of the biofilm matrix has been shown earlier for other species including S. pneumoniae21, S. aureus22, L. monocytogenes23 and L. plantarum LM324.
In the present study, the percentage reduction due to proteinase K treatment was higher in comparison to eDNA in all the L. plantarum isolates. In line with our results, it has been stated that the biofilm formation and its spread are also synchronized by several proteins30. It was also demonstrated that biofilms were immensely susceptible to the administration of proteinase K, indicating an imperative role of proteins on surface colonization30. It was interesting to observe the occurrence of adhesive molecules on the membrane of probiotic bacteria.
CV may bind to dead cells which contribute to the L. plantarum biofilm matrix. This was supported by fluorescence microscopy which was evident by reduced cell counts, indicating an increase in the number of dead cells. Similar results were observed by researchers in earlier studies13,23. The existence of eDNA in the biofilm matrix of L. plantarum was further proved by DNase treatment of the mature biofilms displaying a considerable reduction in both CV staining and number of culturable cells for LRGG, CSG-8 and CSG-21.
Biofilms are known to be encased in an extracellular matrix of DNA, bacterial polysaccharides and proteins. Embedded on an extracellular mass, the bacterial colony tends to display a 1000-fold resistance to antibiotics when compared to its planktonic strains18. Extracellular DNA, a constituent of biofilms, was observed to induce antibiotic resistance and to provide a structural framework to the biofilm18.
In a study conducted in P. aeruginosa the resistance to antibiotics was reasoned to be due to an unknown function of DNA which was able to attach and sequester cations, including magnesium, from the neighbouring surroundings18. Precisely, the eDNA was detected to increase resistance against cationic antimicrobial peptides, such as aminoglycosides but not to fluoroquinolones or β-lactams. In another study, as bacterial eDNA builds up, there is equivalent generation of acidic domains within the biofilms20. Acidic pH disrupts the inner membrane proton motive force of anaerobic bacteria, hindering the uptake of positively charged aminoglycosides. These findings show evidence of a novel role for DNA in biofilms as well as identify cation chelation by DNA as a formerly unidentified mechanism, which can elucidate the amplified resistance of biofilms to antimicrobial agents.
The function of eDNA and proteins biofilm-mediated aminoglycoside resistance in L. plantarum are not yet known. However, assorted functions, such as, being a major structural constituent, a source of energy and nutrition, or a gene pool for horizontal gene transfer within naturally competent bacteria, can be a possibility. In this study, variations in the adaptive and phenotypic biofilm formation posed a limitation to the understanding of the molecular mechanisms. Moreover, CV assays may not be reliable owing to its lack of sensitivity and specificity.
The interest in biofilm research is due to the stability of these complex structures and their inherent ability to resist clearance by antimicrobials. There is a possibility of biofilm-mediated antibiotic resistance in Lactobacillus species, especially in clinical settings. This prompts the need of understanding the mechanisms that underlie antibiotic resistance mediated by biofilms in Lactobacillus species. The study indicates the possible risks associated with biofilms in terms of antibiotic resistance in clinical settings.
Acknowledgment:
Authors acknowledge the Director, CSIR-CFTRI, Mysuru, for facilities and permission to publish this study (PMC approval No. PMC/2017-18/313). The first author (JG) acknowledges University Grants Commission, New Delhi, for Maulana Azad National Fellowship.
Footnotes
Financial support & sponsorship: The financial support for the work was provided by the Indian Council of Medical Research, New Delhi (Project No. AMR/24/2011-ECD-I).
Conflicts of Interest: None.
References
- 1.Leroy F, De Vuyst L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol. 2004;15:67–78. [Google Scholar]
- 2.Sanders JW, Oomes SJ, Membré JM, Wegkamp A, Wels M. Biodiversity of spoilage lactobacilli: Phenotypic characterisation. Food Microbiol. 2015;45:34–44. doi: 10.1016/j.fm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Kubota H, Senda S, Nomura N, Tokuda H, Uchiyama H. Biofilm formation by lactic acid bacteria and resistance to environmental stress. J Biosci Bioeng. 2008;106:381–6. doi: 10.1263/jbb.106.381. [DOI] [PubMed] [Google Scholar]
- 4.Somers EB, Johnson ME, Wong AC. Biofilm formation and contamination of cheese by nonstarter lactic acid bacteria in the dairy environment. J Dairy Sci. 2001;84:1926–36. doi: 10.3168/jds.S0022-0302(01)74634-6. [DOI] [PubMed] [Google Scholar]
- 5.Watnick P, Kolter R. Biofilm, city of microbes. J Bacteriol. 2000;182:2675–9. doi: 10.1128/jb.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayer AS, Chow AW, Betts D, Guze LB. Lactobacillemia – Report of nine cases. Important clinical and therapeutic considerations. Am J Med. 1978;64:808–13. doi: 10.1016/0002-9343(78)90521-1. [DOI] [PubMed] [Google Scholar]
- 7.Dickgiesser U, Weiss N, Fritsche D. Lactobacillus gasseri as the cause of septic urinary infection. Infection. 1984;12:14–6. doi: 10.1007/BF01641017. [DOI] [PubMed] [Google Scholar]
- 8.Rahman M. Chest infection caused by Lactobacillus casei ss rhamnosus. Br Med J (Clin Res Ed) 1982;284:471–2. doi: 10.1136/bmj.284.6314.471-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharpe ME, Hill LR, Lapage SP. Pathogenic lactobacilli. J Med Microbiol. 1973;6:281–6. doi: 10.1099/00222615-6-3-281. [DOI] [PubMed] [Google Scholar]
- 10.Corcoran GD, Gibbons N, Mulvihill TE. Septicaemia caused by Pediococcus pentosaceus: A new opportunistic pathogen. J Infect. 1991;23:179–82. doi: 10.1016/0163-4453(91)92190-g. [DOI] [PubMed] [Google Scholar]
- 11.Aguirre M, Collins MD. Lactic acid bacteria and human clinical infection. J Appl Bacteriol. 1993;75:95–107. doi: 10.1111/j.1365-2672.1993.tb02753.x. [DOI] [PubMed] [Google Scholar]
- 12.Kubota H, Senda S, Tokuda H, Uchiyama H, Nomura N. Stress resistance of biofilm and planktonic Lactobacillus plantarum subsp. plantarum JCM 1149. Food Microbiol. 2009;26:592–7. doi: 10.1016/j.fm.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Fernández Ramírez MD, Smid EJ, Abee T, Nierop Groot MN. Characterisation of biofilms formed by Lactobacillus plantarum WCFS1 and food spoilage isolates. Int J Food Microbiol. 2015;207:23–9. doi: 10.1016/j.ijfoodmicro.2015.04.030. [DOI] [PubMed] [Google Scholar]
- 14.Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol Microbiol. 2009;72:1022–36. doi: 10.1111/j.1365-2958.2009.06703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George J, Halami PM. Sub-inhibitory concentrations of gentamicin triggers the expression of Aac(6’)Ie-aph(2″)Ia, chaperons and biofilm related genes in Lactobacillus plantarum MCC 3011. Res Microbiol. 2017;168:722–31. doi: 10.1016/j.resmic.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Jayaraman A, Wood TK. Bacterial quorum sensing: Signals, circuits, and implications for biofilms and disease. Annu Rev Biomed Eng. 2008;10:145–67. doi: 10.1146/annurev.bioeng.10.061807.160536. [DOI] [PubMed] [Google Scholar]
- 17.Algburi A, Comito N, Kashtanov D, Dicks LM, Chikindas ML. Control of biofilm formation: Antibiotics and beyond. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.02508-16. pii: e02508-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murata T, Tseng W, Guina T, Miller SI, Nikaido H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar Typhimurium. J Bacteriol. 2007;189:7213–22. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilton M, Charron-Mazenod L, Moore R, Lewenza S. Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60:544–53. doi: 10.1128/AAC.01650-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, Mershon WJ, et al. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 2008;8:173. doi: 10.1186/1471-2180-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harmsen M, Lappann M, Knøchel S, Molin S. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl Environ Microbiol. 2010;76:2271–9. doi: 10.1128/AEM.02361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muscariello L, Marino C, Capri U, Vastano V, Marasco R, Sacco M. CcpA and three newly identified proteins are involved in biofilm development in Lactobacillus plantarum. J Basic Microbiol. 2013;53:62–71. doi: 10.1002/jobm.201100456. [DOI] [PubMed] [Google Scholar]
- 25.Ghannoum M, O’Toole GA. Washington, DC: ASM Press; 2004. Microbial biofilms. [Google Scholar]
- 26.Vega NM, Gore J. Collective antibiotic resistance: Mechanisms and implications. Curr Opin Microbiol. 2014;21:28–34. doi: 10.1016/j.mib.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–8. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 28.Jaimee G, Halami PM. High level aminoglycoside resistance in Enterococcus, Pediococcus and Lactobacillus species from farm animals and commercial meat products. Ann Microbiol. 2016;66:101–10. [Google Scholar]
- 29.Lebeer S, Verhoeven TL, Perea Vélez M, Vanderleyden J, De Keersmaecker SC. Impact of environmental and genetic factors on biofilm formation by the probiotic strain lactobacillus rhamnosus GG. Appl Environ Microbiol. 2007;73:6768–75. doi: 10.1128/AEM.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simões M, Bennett RN, Rosa EA. Understanding antimicrobial activities of phytochemicals against multidrug resistant bacteria and biofilms. Nat Prod Rep. 2009;26:746–57. doi: 10.1039/b821648g. [DOI] [PubMed] [Google Scholar]