Abstract
Context
Primary ovarian insufficiency (POI) encompasses a spectrum of premature menopause, including both primary and secondary amenorrhea. For 75% to 90% of individuals with hypergonadotropic hypogonadism presenting as POI, the molecular etiology is unknown. Common etiologies include chromosomal abnormalities, environmental factors, and congenital disorders affecting ovarian development and function, as well as syndromic and nonsyndromic single gene disorders suggesting POI represents a complex trait.
Objective
To characterize the contribution of known disease genes to POI and identify molecular etiologies and biological underpinnings of POI.
Design, Setting, and Participants
We applied exome sequencing (ES) and family-based genomics to 42 affected female individuals from 36 unrelated Turkish families, including 31 with reported parental consanguinity.
Results
This analysis identified likely damaging, potentially contributing variants and molecular diagnoses in 16 families (44%), including 11 families with likely damaging variants in known genes and five families with predicted deleterious variants in disease genes (IGSF10, MND1, MRPS22, and SOHLH1) not previously associated with POI. Of the 16 families, 2 (13%) had evidence for potentially pathogenic variants at more than one locus. Absence of heterozygosity consistent with identity-by-descent mediated recessive disease burden contributes to molecular diagnosis in 15 of 16 (94%) families. GeneMatcher allowed identification of additional families from diverse genetic backgrounds.
Conclusions
ES analysis of a POI cohort further characterized locus heterogeneity, reaffirmed the association of genes integral to meiotic recombination, demonstrated the likely contribution of genes involved in hypothalamic development, and documented multilocus pathogenic variation suggesting the potential for oligogenic inheritance contributing to the development of POI.
Exome sequencing was used to investigate genes and mutational mechanisms contributing to primary ovarian insufficiency, and to gain insights into disease biology and underlying pathophysiology.
Hypergonadotropic hypogonadism (HH) is a developmental disorder characterized in females by ovarian dysgenesis with a clinical presentation ranging from delayed to arrested puberty associated with primary or secondary amenorrhea, elevated gonadotropins, and low sex steroid hormones. Additional clinical features of HH include delayed bone age on bone scanning, possible short stature, undetectable or hypoplastic ovaries on pelvic ultrasound, and absent or delayed development of secondary sex characteristics. Without proper diagnosis and treatment, clinical sequelae such as infertility, short stature, pubertal underdevelopment, osteoporosis, and early onset menopause are possible, especially in those with a more severe clinical phenotype (1, 2). Additionally, molecular diagnoses in severe disease presentations, as evidenced by ovarian failure, likely represent dramatic perturbations of pathways underlying common ovarian physiology.
Primary ovarian insufficiency (POI) is a general term used to describe depletion or dysfunction of ovarian follicles with cessation of menses before age 40, and HH is one type of POI. Although not characterized in younger ages, the incidence of POI in women ages 18 to 25 is ∼1:10,000 and the frequency increases with age (3). Cytogenetic causes of POI, the most common being Turner syndrome, have been well described (4). Female carriers of premutation repeat expansions in FMR1, one of the most common causes of inherited intellectual disability, have a 16% to 25% risk of developing POI (5). More recently, pathogenic variants have been described in a number of genes such as NOBOX, FIGLA, BMP15, and GDF9 with pleiotropic syndromic phenotypes that include POI. These disease gene studies reveal that 46,XX POI is a genetically complex disease (6–13). Evidence suggests that additional disease genes, and potential gene by environmental effects remain to be elucidated. Up to 90% of nonsyndromic premature ovarian failure cases are estimated to be idiopathic, with ∼30% having an affected first degree relative, supporting a potential underlying genetic etiological basis (14). These observations in patients are further supported by animal model studies that have uncovered a wealth of candidate disease genes associated with ovarian dysgenesis (15–19). Recent studies have continued to demonstrate human disease genes in association with POI, such as SOHLH1 and NUP107, in a small number of families, further illustrating the genetic heterogeneity of this condition (1, 2, 20–22).
In this study, we applied exome sequencing (ES) and family-based genomics in a cohort of 36 unrelated families with POI for whom clinical phenotyping and molecular studies had not previously yielded a molecular diagnosis. We identified variants in known genes, propose candidate genes in association with POI, and provide evidence for aggregation of multilocus pathogenic variation driven by transmission genetics and identity-by-descent.
Methods
Patients
The institutional review board at Baylor College of Medicine approved this study. All study participants (family members and patients) were evaluated and referred by one or more pediatrician(s), endocrinologist(s), and clinical geneticist(s), and provided informed consent (institutional review board protocol number: H-29697) prior to enrollment in the study.
Patients with elevated gonadotropin levels (FSH > 40 mIU/mL) who presented with a POI phenotype, including primary/secondary amenorrhea and small or invisible ovaries on imaging, were recruited for study by endocrinologists and/or clinical geneticists at Marmara University School of Medicine, Istinye University, Bezmialem University, and Kanuni Sultan Suleyman Training and Research Hospital in Istanbul, Turkey and at Sami Ulus Children’s Hospital in Ankara, Turkey (23). Patients with chromosomal abnormalities, except a patient with a maternally inherited, presumably balanced translocation, were excluded.
Genomic DNA was extracted from venous blood using previously described methods (20). Samples from 27 families underwent proband only ES, whereas samples from 10 families underwent trio (proband + parents) or quad (proband + unaffected sib + parents) ES study design.
Exome sequencing
ES was performed by the Baylor College of Medicine Human Genome Sequencing Center through the Baylor Hopkins Center for Mendelian Genomics (BHCMG) research initiative. Exome capture was implemented using Nimblegen reagents and a custom-designed capture platform, VCRome2.1, optimized for experimental sequencing and rare variant sequence analysis. ES postsequence processing of raw data was performed using the Mercury pipeline, available via DNANexus (http://blog.dnanexus.com/2013-10-22-run-mercury-variant-calling-pipleine/) (24). The ATLAS2 variant calling method and Sequence Alignment/Map were used to call variants, followed by a customized Cassandra annotation pipeline, which is based on Annotation of Genetic Variants. Raw sequencing data were parsed for rare variants using bioinformatics algorithms set to specific variant allele frequency controls. Insertions/deletions (indels) had to pass through quality controls and were parsed for minor allele frequency (MAF) <2% using the Thousand Genomes Project (1000GP) database to be reported (25). However, indels reported as variants in the Human Gene Mutation Database only had to have a MAF <5% in 1000GP to be retained (26). Single nucleotide variants also had to pass quality controls and were parsed for rare variants in 1000GP, National Heart Lung and Blood Institute GO Exome Sequencing Project (esp5400) African, and esp5400 European databases for initial inclusion (27). If a single nucleotide variant had already been reported as a disease-associated variant in Human Gene Mutation Database, or had been defined as a dbSNP clinical variant, parsing criteria were less stringent.
After quality control and parsing, variant analysis and family-based genomics were performed within the BHCMG, retaining variants with a minimum variant read count >4; MAF ≤0.001 in the local BHCMG database, gnomAD, esp5400, and 1000GP; Atherosclerosis Risk in Communities MAF of ≤2.74 × 10−4; conservation score using phyloP greater than 0; and variant type with synonymous and noncoding variants given lower priority (28–31). Variants thought to parsimoniously contribute to pathogenicity were orthogonally validated and segregated within each family by dideoxy Sanger sequencing of PCR amplicons (32).
Custom microarray
A custom 4 × 180k tiling-path oligonucleotide microarray was designed to interrogate a 120-kb segment surrounding the MND1 gene mapping to chromosome 4q31.3. The high resolution array consisted of 60-mer oligonucleotide probes that were designed to span chr4:154,265,801 to 154,386,247 (National Center for Biotechnology Information build 37) using the Agilent SureDesign website (https://earray.chem.agilent.com/suredesign). Probes had an average distribution of one interrogating oligonucleotide probe per 135 bp within MND1 and one probe per 520 bp for the regions flanking MND1. Coriell Institute sex-matched controls were used (GM10851C-male; GM15510-female) and comparative genomic hybridization was carried out as described (33). Copy number variants (CNVs) were identified after comparative genomic hybridization using the normalized log2 ratio average of Cy5/Cy3. Agilent Feature Extraction software was used to process scanned array images, and Agilent Genomic Workbench (version 7.0.4.0) was used to analyze extracted files. An average log2 ratio threshold of −1 was used to call heterozygous deletion, and an average log2 ratio threshold of −4 was used to call homozygous deletion.
Breakpoint junction PCR assay
Inward facing primers flanking the deleted region of MND1 were designed based on apparent breakpoint junctions revealed by chromosomal microarray analysis (23). Internal primers were also generated to amplify exon 6 of MND1, which was predicted to be deleted by HMZDelFinder (23, 34). Standard PCR amplification and gel electrophoresis were used to confirm zygosity of the deletion (23).
Identification of absence of heterozygosity regions
BafCalculator (35) (https://github.com/BCM-Lupskilab/BafCalculator) was used to identify regions of absence of heterozygosity (AOH) from exome variant data as follows: first ES data were used to calculate B-allele frequency data by comparing the ratio of variant to total reads; then, the Circular Binary Segmentation algorithm (36) was used to delineate regions of AOH using calculated B-allele frequency. Coefficient of consanguinity was calculated using the GENESIS package hosted on Bioconductor (37).
Collaborative candidate match and modeling software
Candidate genes in which variants segregated with disease were submitted to GeneMatcher, and genes for which successful matches were identified were included for cohort-wide lower-stringency reanalysis (38, 39). The cBioPortal Mutation Mapper was used to map and visualize variant changes within protein functional domains (40, 41). GeneMANIA was used to interrogate gene and protein interaction networks for genes proposed to be involved in an oligogenic inheritance model (42).
Screening for previously reported candidate genes
We screened all 42 exomes for rare variants in POI candidate genes identified by previous animal model and human cohort studies (23). Parsing for rare variants was performed as described above. Combined Annotation Dependent Depletion score was not used as a strict cut-off for pathogenic variants but was considered as a suggestive feature when considering which variants likely contributed to pathogenicity (43). Likewise, loss-of-function intolerance was not used as a strict parameter for variant/gene consideration, so that gain-of-function could also be considered. All Mendelian modes of inheritance were explored for the family-based genomics analysis: autosomal dominant (AD), autosomal recessive (AR), and X-linked (XL). Variants were also analyzed by putative mode of trait inheritance. Biallelic variants, considered potentially responsible when exploring Mendelian expectations for an AR disease trait segregation pattern, were parsed using lower stringency in terms of allelic frequency, from ≤0.001 to ≤0.005 in the BHCMG, 1000GP, esp5400, and gnomAD databases. Potentially causative alleles were subsequently examined for homozygous frequency in populations. These additional analysis steps were performed to include variants that may have a higher heterozygous allele frequency, but a low or absent homozygous allele frequency in population databases. Genes with variant hits were analyzed across BHCMG cohorts to identify additional cases, and CNV analysis was performed using XHMM (44) and HMZDelFinder (34) ES-based CNV callers. Additionally, the anonymized Baylor Genetics clinical sequencing database was queried for CNV encompassing reported candidates.
Results
We performed ES on 42 affected females with POI from 36 families (Fig. 1) (23). This analysis identified a molecular diagnosis or potential molecular diagnosis in 16 families (44%), including 11 families for whom a likely damaging variant in a known gene was identified and five families for whom a likely damaging variant in a potential disease gene was identified (23). Homozygous predicted deleterious variants in SOHLH1 (Online Mendelian Inheritance in Man [OMIM] 617690) and MRPS22 (OMIM 618117) were reported as disease gene discoveries in this POI cohort (20, 45). Of the 16 families in which evidence for a molecular diagnosis was concluded, 2 (13%) harbored potentially pathogenic variants at more than one locus: one family with variants in three phenotype associated disease genes, and one family with variants in one candidate and one known disease gene.
Figure 1.
Pedigrees for all families studied by ES. Filled shapes represent affected individuals. Family ID is written above each pedigree. Filled in arrows designate the proband. Patient ID is written underneath probands in each family.
Variants in known genes
Rare likely damaging variants in eight known disease genes associated with POI were found as a monogenic cause of POI in 10 families (Table 1). In all but three of these genes, TG (OMIM 274700), GALT (OMIM 230400), and one of three MCM9 variants (OMIM 616185), the identified variants were distinct compared with those previously reported (46). For 9/10 (90%) of these families, homozygous predicted pathogenic variants were found within a region of AOH ranging in size from 2 to 20 Mb. Molecular diagnoses involved genes with a known nonsyndromic disease association in eight families, including premature ovarian failure (NOBOX, OMIM 611548; MCM8 OMIM 612885), ovarian dysgenesis (PSMC3IP, OMIM 614324), and preimplantation embryonic lethality (PADI6, OMIM 617234; Fig. 2). Notably, variant segregation in the family HOU 2581, with proband BAB6996 having documented HH and POI, demonstrated a male sibling homozygous for the identified variant. This pattern could be consistent with a sex-limited trait expressed only in females.
Table 1.
Summary of Variants Found in Known Disease Genes
| Gene | Proband | Hg19 Coordinates | vR/tR | Variant/Protein Change | Zyg | AOH Block Around Gene (Mb) | Total AOH (Mb) | Previously Reported Variant | Linked OMIM Disease | FSH (mIU/mL) | LH (mIU/mL) | Estradiol (pg/mL) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C3 a | BAB7008 | chr19:6709705 | 49/49 | c.1835C>T;p.T612M | Hom | 10 | 535 | No | 613779 | 84 | 24 | 15 |
| GALT | BAB7662 | chr9:34649520 | 35/35 | c.1018G>T; pE340* | Hom | 7 | 328 | Yes | 230400 | 73 | 18 | <10 |
| MCM8 | BAB7100 | chr20:5932750 chr20:5953777 | 27/60 40/81 | c.89A>C;p.K30T c.1330A>G;p.I444V | Comp Het | — | 275 | No | 612885 | 80 | 14 | 45 |
| MCM8 | BAB7675 | chr20:5948131 | 125/125 | c.925C>T; p.R309* | Hom | 2 | 160 | No | 612885 | 160 | 33 | 3.2 |
| MCM9 | BAB9435 | chr6:119245203 | 37/37 | c.394C>T;p.R132* | Hom | 8 | 465 | Yes | 616185 | 77 | 19 | 5 |
| MCM9 | BAB10068 | chr6:119252669 | 76/76 | c.220C>T;p.R74* | Hom | 20 | 351 | No | 616185 | 91 | 33 | 5 |
| NOBOX | BAB6985 | chr7:144096926 | 50/51 | c.1078C>T;p.R360* | Hom | 3 | 114 | No | 611548 | 110 | 35 | Undetectable |
| PADI6 | BAB6996 | chr1:17707625 | 102/102 | c.519G>C;p.E173D | Hom | 5 | 381 | No | 617234 | 62 | 29 | 27 |
| PSMC3IP | BAB7116 | chr17:40725025 | 114/114 | c.377delA;p.E126Gfs*30 | Hom | 3 | 256 | No | 614324 | 123 | 48 | <12 |
| TG a | BAB7094 | chr8:133900662 | 202/202 | c.2610G>T;p.Q870H | Hom | 10 | 405 | Yes | 274700, 608175 | 56 | 22 | <12 |
Abbreviations: Comp Het, compound heterozygous; Hom, homozygous; OMIM, Online Mendelian Inheritance in Man; tR, total reads; vR, variant reads; Zyg, zygosity.
C3 and TG are included due to the association of autoimmune disease with diminished ovarian reserve.
Figure 2.
Pedigrees and segregation of identified variants for families in which candidate and previously described variants were found in known disease genes. Filled shapes represent affected individuals. Family ID is written above each pedigree. Name of the genes and respective variants (red) are written below the pedigrees. Filled in arrows designate the proband; unfilled arrows designate males with the same genotype without the sex-limited POI phenotype. AOH maps for probands are shown below the relevant pedigrees. Vertical red line delineates the gene of interest, and regions shaded gray represent regions of AOH. Size of AOH region encompassing identified variant is indicated in red font at the top of each AOH plot.
In three families, the identified variants involved genes associated with systemic disease: thyroid dyshormonogenesis (TG, OMIM 274700), C3 deficiency (C3, OMIM 613779), and Galactosemia (GALT, OMIM 230400).
In the case of BAB7094, a diagnosis of Hashimoto Thyroiditis subclinical hypothyroidism was documented [TSH = 11.9 μIU/mL (0.34 to 5.6 μIU/mL), fT4 = 10.99 ng/dL (0.61 to 1.12 ng/dL)], and ultrasonography had demonstrated hyperechoic nodules suspicious for thyroiditis. The diagnosis of C3 deficiency in BAB7008 was further supported by the finding of a low serum C3 (C3 = 0.71 g/L, N = 0.75 to 1.4 g/L). BAB7008 was not reported to have recurrent infections or renal dysfunction, and her only other clinical diagnosis was hypertension.
A homozygous nonsense variant in GALT was found in an individual with nonsyndromic HH from a consanguineous pedigree (HOU 2771). Sanger sequencing confirmed segregation of potentially pathogenic variants from heterozygous parents (Fig. 2; Table 1). Clinical testing of the patient after sequencing and pathogenic rare variant identification confirmed reduced serum GALT activity (GALT activity = 0.3 µmol/mL, N = 4.0 to 13.0 µmol/mL). Clinical follow-up revealed phenotypic features consistent with galactosemia: milk avoidance, mild intellectual disability, and bilateral cataracts with mild opacification. Although both initial and repeat testing yielded negative urine reducing substances, gas chromatography was positive for galactose enrichment. This suggests that the patient’s diagnosis of galactosemia presenting as HH was missed by current screening standards but was identified by ES.
HH candidate genes
IGSF10
We identified two unrelated Turkish probands in consanguineous families with homozygous IGSF10 variants that segregated with disease. In BAB7110, who has a phenotype of POI and GH deficiency without detectable anatomical variation in the hypophysis on MRI, a predicted deleterious homozygous (c.2237C>G; p.P746R) variant was found by ES and confirmed by Sanger sequencing (Fig. 3; Table 2). Both unaffected parents and one unaffected sister were heterozygous for the variant, and one brother and another unaffected sister did not have the identified variant (Fig. 3). The variant was found within a region of AOH of ∼23 Mb in size, with total genome-wide AOH of ∼269 Mb consistent with consanguineous parents who were known second degree cousins. Although the gnomAD database frequency of the variant allele is relatively high (6.831 × 10−3; 1932 alleles), only 10 individuals are homozygous for this variant. Five of these are male and five are female, aged 45 to 65 years; thus, the frequency for finding a homozygote with a potential sex influenced trait in gnomAD is 0.007%.
Figure 3.
Pedigrees and segregation of identified variants for families in which variants were found in candidate disease genes. Filled shapes represent affected individuals. Family ID is written above each pedigree. Name of the genes and base changes (red) are written below the pedigrees. Filled in arrows designate the proband. AOH maps for probands and affected siblings are shown below the relevant pedigrees. Vertical red line delineates the gene of interest, and regions shaded gray represent regions of AOH. Size of AOH region encompassing identified variant is indicated in red font at the top of each AOH plot.
Table 2.
Summary of Variants Found in Candidate Disease Genes
| Gene | Proband | Hg19 Coordinates | vR/tR | Variant/Protein Change | Zyg | AOH Block Around Gene (Mb) | Total AOH (Mb) | FSH (mIU/mL) | LH (mIU/mL) | Estradiol (pg/mL) |
|---|---|---|---|---|---|---|---|---|---|---|
| IGSF10 | BAB7110 | chr3:151165532 | 110/110 | c.2237C>G;p.P746R | Hom | 23 | 269 | 111 | 39 | 46 |
| IGSF10 | HHMI00103.97 | chr3:151171554 | — | c.346A>G; p.K116E | Hom | — | — | 43 | 27 | 16 |
| IGSF10 | Erasmus MC | chr3: 151154500 | 11/28 | c.7849delG; p.A2617Qfs*6 | Het | — | — | 6 | 4 | <5 |
| MND1 | BAB9440 BAB9444 | chr4:154325119-chr4:154316483 | HMZDel Finder | 8.636Kb Deletion | Hom | 65 | 493 | 73 | 18 | <10 |
| 71 | 525 | 97 | 38 | <10 | ||||||
| MRPS22 a | BAB7669 | chr3:139067066 | 69/69 | c.404G>A; p.R135Q* | Hom | 23 | 211 | 99 | 20 | 0.01 |
| SOHLH1 a | BAB4619 | chr9:138589450 | 40/41 | c.705delT; p.K236Rfs*3 | Hom | 7 | 333 | 99 | 25 | <5 |
| BAB4620 | 87/88 | 9 | 199 | 109 | 25 | 42 |
Abbreviations: Het, heterozygous; Hom, homozygous; tR, total reads; vR, variant reads; Zyg, zygosity.
aPrevious candidate disease genes found in cohort.
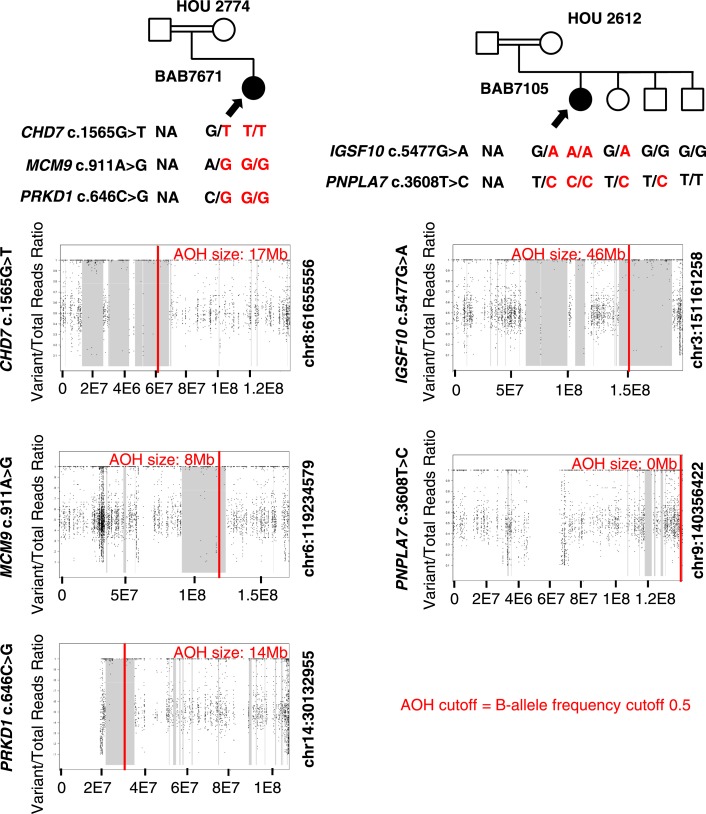
We do not know whether any of these female individuals had infertility or delayed puberty phenotypes. In our own database, one maternal sample of a proband with neuronal migration disorder is homozygous for the same variant in IGSF10 and has no known history of infertility (purportedly conceived naturally) or hypogonadism, suggesting the possibility of incomplete penetrance of some IGSF10 alleles. In BAB7105, who has a phenotype of nonsyndromic HH, a homozygous missense variant in IGSF10 (c.5477G>A; p.G1862D) was identified (Fig. 4; Table 3). No homozygotes for this allele are present in gnomAD, and in the BHCMG database only BAB7105 is homozygous for the variant. An additional homozygous predicted pathogenic variant in PNPLA7 may further contribute to the phenotype in this case (see below for dual molecular diagnosis cases).
Figure 4.
Pedigrees, segregation of identified variants, and AOH plots for families in which variants were thought to be consistent with oligogenic inheritance. Filled shapes represent affected individuals. Family ID is written above each pedigree. Name of the genes and base changes (red) are written below the pedigrees. Filled in arrows designate the proband. AOH maps for probands and affected siblings are shown below the relevant pedigrees. Vertical red line delineates the gene of interest, and regions shaded gray represent regions of AOH. Size of AOH region gene is within and is shown in red at the top of the AOH plot.
Table 3.
Summary of Variants Found in Families With a Potential Oligogenic Inheritance
| Gene | Proband | Hg19 Coordinates | vR/tR | Variant/Protein Change | Zyg | AOH Block Around Gene (Mb) | Total AOH (Mb) | Linked OMIM Disease | FSH (mIU/mL) | LH (mIU/mL) | Estradiol (pg/mL) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CHD7 | BAB7671 | chr8:61655556 | 80/80 | c.1565G>T;p.G522V | Hom | 17 | 351 | 214800, 612370 | 47 | 23 | 17 |
| MCM9 | BAB7671 | chr6:119234579 | 56/56 | c911A>G;p.N304S | Hom | 8 | 351 | 616185 | 47 | 23 | 17 |
| PRKD1 | BAB7671 | chr14:30132955 | 108/108 | c.646C>G;p.R216G | Hom | 14 | 351 | 617364 | 47 | 23 | 17 |
| IGSF10 | BAB7105 | chr3:151161258 | 57/57 | c.5477G>A;p.G1862D | Hom | 46 | 608 | — | 126 | 34 | 20 |
| PNPLA7 | BAB7105 | chr9:140356422 | 15/15 | c.3608T>C;p.I1203T | Hom | NA | 608 | — | 126 | 34 | 20 |
Abbreviations: Hom, homozygous; OMIM, Online Mendelian Inheritance in Man; tR, total reads; vR, variant reads; Zyg, zygosity.
A third family was identified by a collaborating group at the Center for the Study of Rare Hereditary Diseases in Milan, Italy. The proband, HH-MI00103.97, is affected with nonsyndromic HH and harbors a homozygous variant in IGSF10 (c.346A>G; p.K116E). Both parents are heterozygous for the variant, and an unaffected sister did not inherit the allele. The variant is not present in the 1000G database and has a gnomAD allele frequency of 2.794 × 10−5 with no homozygotes (Fig. 3; Table 2). No individuals within the BHCMG database are homozygous for the allele. The variant is predicted to be deleterious by 6/7 in silico prediction algorithms (23, 47–53).
GeneMatcher (39, 40) facilitated identification of a fourth family with a frameshift variant in IGSF10 from the Erasmus MC, University Medical Center Rotterdam. The proband is a female who presented to the genetics department at 20 years of age with primary amenorrhea, hyposmia, and findings of hypogonadism with normal gonadotropin levels that distinguish her case from Kallmann syndrome. ES revealed a c.7849delG; p.A2617Qfs*6 variant in IGSF10, which is maternally inherited. The variant is not present in gnomAD or within the BHCMG database (23). Maternal inheritance suggests this allele may lead to incomplete penetrance of the phenotype. Although ES was not performed on the maternal sample, chromatogram peaks were not suggestive of maternal germline mosaicism.
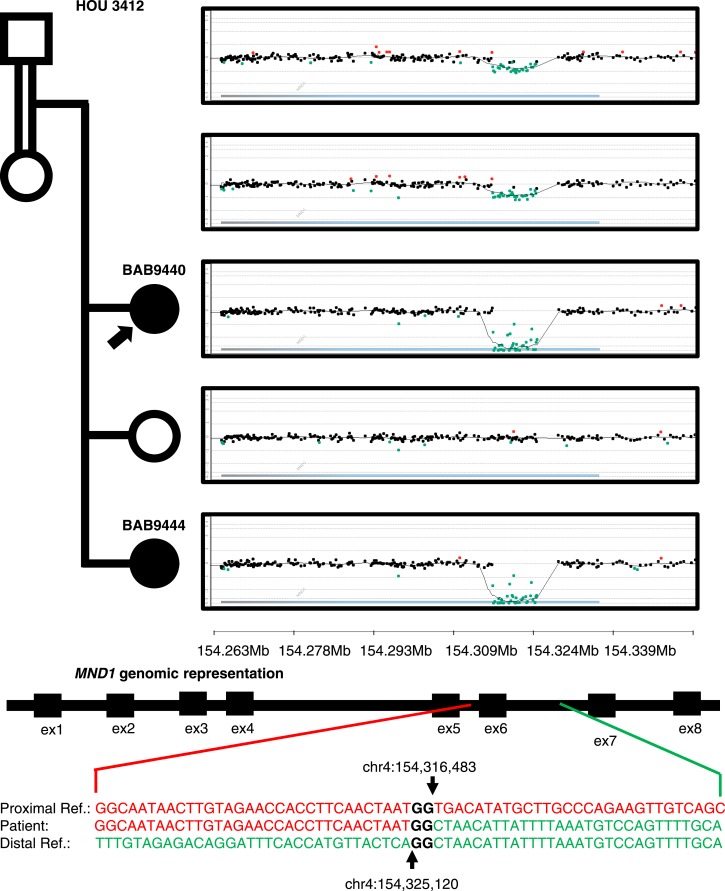
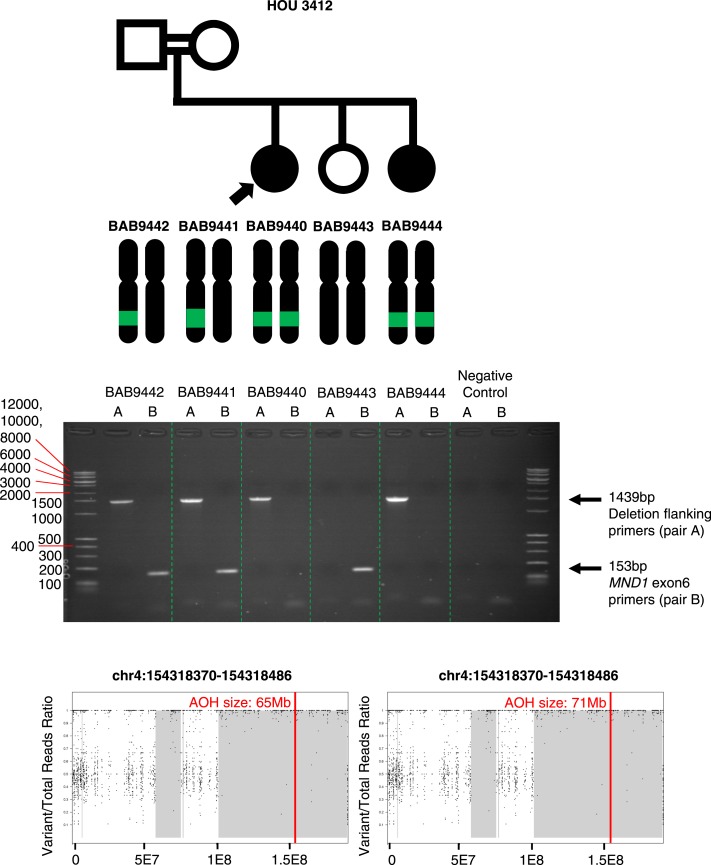
MND1
Two sisters with a diagnosis of HH, BAB9440 and BAB9444, had homozygous deletion of exon 6 (out of eight exons) of MND1 encompassing 116 bp as called on ES data by HMZDelFinder (34). A custom microarray targeted to the region revealed an 8.6-kb genomic deletion inherited from heterozygous parents (Fig. 5). Sequencing of the breakpoint junction via PCR amplification using inward-facing primers flanking the deletion breakpoints revealed 2-bp microhomology and enabled us to further refine the deletion breakpoint coordinates (chr4:154,316,483 to 154,325,120; Fig. 6). The proximal breakpoint did not map within a repeat structure such as a low-copy repeat or a repetitive element. However, the distal breakpoint coincides with an Alu element, AluJb. The lack of a pair of repeats at both breakpoints and the presence of 2-bp microhomology at the junction together suggest nonhomologous recombination underlying the formation of the observed MND1 deletion. Possible mechanisms include nonhomologous end joining, microhomology-mediated end joining, or fork stalling and template switching/microhomology-mediated break-induced replication, FoSTeS/MMBIR (54, 55).
Figure 5.
Pedigree, segregation of identified MND1 deletion, and custom microarrays for BAB9440, BAB9441, BAB9442, BAB9443, and BAB9444. Filled in shapes represent affected individuals. Family ID is written above the pedigree. Filled in arrows designate the proband. Breakpoint junction sequence and coordinates are shown at bottom. Proximal reference sequence in green and distal reference sequence in red.
Figure 6.
PCR assay results for primers designed to amplify the MND1 deletion (expected size 1439 bp) and for primers designed to amplify exon 6 of MND1 (expected size 153 bp). AOH maps for probands and affected siblings are shown at bottom. Vertical red line delineates the gene of interest, and regions shaded gray represent regions of AOH. Size of AOH region encompassing identified variant is shown in red font at the top of the AOH plot.
MND1 is integral to meiotic recombination and interacts with the encoded protein of a known HH associated gene, PSMC3IP (56). MND1 deletion is within a genomic region of AOH of size 65 Mb in BAB9440 and 71 Mb in BAB9444 (Fig. 6). Total AOH for BAB9440 and BAB9444 was 493 Mb (coefficient of consanguinity = 0.13) and 525 Mb (coefficient of consanguinity = 0.13), respectively. The coefficient of consanguinity is consistent with a reported family history of consanguinity.
Dual molecular diagnosis and potential oligogenic inheritance
CHD7, MCM9, and PRKD1
Homozygous, predicted deleterious variants in CHD7 (c.1565G>T; p.G522V) and MCM9 (c.911A>G; p.N304S), two genes previously associated with hypogonadism, and a homozygous variant in PRKD1 (c.646C>G; p.R216G), a previously described candidate gene for obesity (57), were found in patient BAB7671 with HH and obesity (body mass index, 35). Consistent with reported parental consanguinity, all three variants are located within regions of AOH (Fig. 4; Table 3). Total AOH was ∼351 Mb (coefficient of consanguinity = 0.024), with variants in CHD7, MCM9, and PRKD1 each identified within regions of AOH measuring 17, 8, and 14 Mb, respectively. These results suggest a mutational burden and multiple molecular diagnosis in this individual. We propose that both CHD7 and MCM9 variants contribute to the HH phenotype and that the PRKD1 variant contributes to the obesity phenotype.
IGSF10 and PNPLA7
Homozygous missense variants in IGSF10 (c.5477G>A; p.G1862D), described above, and in PNPLA7 (c.3608T>C; p.I1203T) were found by ES in a patient with nonsyndromic HH from a consanguineous pedigree (HOU 2612). IGSF10, but not PNPLA7, likely damaging variants were found in a region of AOH (total AOH ∼608 Mb; coefficient of consanguinity = 0.042). The region of AOH containing variants in IGSF10 was ∼46 Mb in size. Although paternal DNA was not available, Sanger study of maternal DNA, as well as that of an unaffected sister, revealed that they were heterozygous for all three variants. One brother harbored both PNPLA7 variants but did not carry the c.5477G>A IGSF10 variant. A second unaffected brother was wild type for both PNPLA7 and IGSF10 alleles (Fig. 4; Table 3).
Discussion
ES reveals genes potentially contributing to HH
One challenge to the family-based genomics approach in nonsyndromic 46,XX POI has been the limited number of families with more than one affected relative available for study. Access to consanguineous families with a relatively uniform POI phenotype as a potential AR trait enables candidate disease gene identification through a homozygous variant analysis approach. This cohort represents a unique opportunity to elucidate genes and variants that lead to a POI phenotype as a means to garner further insights into disease biology. These findings may further our understanding of ovarian dysgenesis disease biology and provide families the opportunity for molecular diagnosis, informing both disease biology and potentially medical management. Molecular diagnosis also allows for identification of at-risk relatives.
In 16/36 (44%) of the families studied, a potential disease gene contributing to the observed clinical phenotype was identified by ES. Three additional families with rare variants in candidate genes (MRPS22 and IGSF10) were identified through GeneMatcher (38, 39) and direct collaboration. Discoveries included variants in six candidate disease genes, eight known or associated POI disease genes, one gene previously associated with menstrual irregularity, and one candidate gene associated with obesity. Four genes (IGSF10, MCM8, MCM9, and MRPS22) were implicated in more than one family. In two families (13% of molecularly diagnosed cases), variants in more than one gene were proposed to likely contribute to pathogenesis of the patient’s phenotype. ES in this cohort provided additional human genomic evidence to support candidate disease genes identified in animal models, particularly for SOHLH1 and PADI6 (58–60). In our cohort, 5/42 (12%) patients had a diagnosis of Hashimoto thyroiditis. Of patients diagnosed with Hashimoto thyroiditis, four have a potential molecular diagnosis. Notably, only one has variants in an autoimmune susceptibility gene (TG), whereas the others have variants proposed to affect ovarian development through a non-autoimmune mechanism (SOHLH, IGSF10, PSMC3IP). This includes a sibling-pair (BAB4619 and BAB4620) with discordant phenotypes in regard to Hashimoto thyroiditis (23). The possibility that POI phenotypes in these patients are influenced by antiovarian antibodies cannot be ruled out.
ES enables a dissection of phenotypic differences resulting from unique variants in the same gene. For example, a heterozygous c.1076-1G>C variant in TG that causes skipping of exon 9 was recently reported to be causative of Hashimoto thyroiditis in a four-generation family with eight affected individuals. The variant was suggested to be almost completely penetrant, with only one unaffected individual carrying the heterozygous change (61). Here we describe a biallelic missense variant of TG in a patient with both Hashimoto thyroiditis and HH. However, the occurrence of both monoallelic and biallelic variants in genes other than TG has been reported in association with similar or distinct phenotypes (62–64).
The utility of ES in further dissecting POI phenotypes has also been demonstrated through the diagnosis of galactosemia in BAB7662, who was initially characterized as having nonsyndromic HH. After a putatively damaging variant in GALT was found, it prompted evaluation of serum galactose levels and GALT enzymatic function—both of which were abnormal. Further clinical evaluation identified a history of milk avoidance, mild intellectual disability, and bilateral cataracts with mild opacification, consistent with galactosemia. In this subject, a diagnosis of galactosemia presented as HH. However, screening for galactosemia via urinary reducing substance testing was negative. ES results helped the physicians to uncover an enzymatic dysfunction, which was not clinically recognizable prior to ES.
We propose that identified genes and variants in the present HH cohort contribute to POI in an allele-dependent manner, highlighting the common molecular etiologies underlying ovarian development and maintenance. Trait manifestation is thus impacted by the presence and degree of perturbation of contributory molecular pathways.
Common molecular mechanisms of ovarian development
In the current study, likely genetic contributors to ovarian development and maintenance were elucidated. Known genes MCM8, MCM9, and PSMC3IP are thought to contribute to oogenesis through their role in DNA damage repair and meiosis-specific recombination (22, 65, 66). We propose homozygous deletion in a candidate gene, MND1, affects oogenesis through disruption of the same processes (Fig. 7a). MND1 and PSMC3IP are interacting partners in processes integral to DNA repair via homologous recombination, and mediate the function of the meiosis-specific recombination protein DMC1 (67, 68). Homozygous deletion in MND1 was associated with an early presentation of HH in BAB9440 and BAB9444, at ages 15.9 and 10.8 years. However, heterozygous deletion in the mother of BAB9440 and BAB9444 may be associated with her presentation of delayed puberty and early menopause at age 35 years (23). The deletion in MND1 is a single-exon dropout, a variant allele that may be missed by clinical arrays due to lower-density coverage of the region. Moreover, heterozygous CNV at a locus, including exon dropout alleles, are challenging to identify from ES data. Particularly, read depth is potentially distorted due to the experimental design of capture sequencing that depends on hybridization kinetics whose efficiency is partially determined by guanine/cytosine content. Although intragenic duplication in PDE8A has been reported in association with Turner syndrome, this is the first report of intragenic deletion in association with a POI phenotype to our knowledge (69). Intragenic deletions have been implicated as pathogenic alleles in a diverse spectrum of phenotypes (BBS9; Bardet-Biedl-Syndrome, DMD, GRID2, SNX14; brain malformation; RIPPLY1, Heterotaxy; DOCK8, immunodeficiency; CNTNAP2, intellectual disability; WWOX, epileptic encephalopathy; TANGO2, rhabdomyolysis) (34, 70, 71) and HMZDelFinder continues to be a useful tool in intragenic CNV detection. Predicting genes that may undergo genomic instability due to Alu-Alu mediated rearrangements (AAMR) (72) resulting in exon dropout may facilitate the molecular diagnosis of heterozygous deletion alleles. MND1 is above the suggested 0.6 AAMR risk score cutoff, suggesting that it may be in a region of relative genomic instability due to AAMR (23).
Figure 7.
Proposed functional models for known and candidate genes in POI. (a) Development of the primary oocyte to mature ovum. (b) Development of the pituitary gland and hypothalamic-pituitary synapses.
In addition to more ubiquitous processes such as DNA repair, proper and timely development of the hypothalamic-pituitary axis is thought to affect germ cell development (73). IGSF10 has been proposed to play a role in the development of the gonadotropin axis, based on observations of aberrant migration of GnRH neurons in zebrafish models of IGSF10 knockdown and the association of rare missense variants segregating in an AD pattern with delayed puberty in 10 families (74). Here, we report four unrelated families with biallelic missense or monoallelic nonsense variants in IGSF10 and a phenotype of POI. IGSF10 missense variants in individuals with POI are unique from the variants described in families with delayed puberty in type of missense change and in segregation pattern (AD vs AR). In the three families in whom biallelic missense variants in IGSF10 are thought to contribute to the proband’s phenotype, clinical features were consistent with HH (Tables 2 and 3) (23). In the case from Erasmus MC, a maternally inherited single base-pair deletion is associated with a phenotype that is Kallmann syndrome-like, including delayed puberty, primary amenorrhea, hypogonadism, and anosmia, but with normal gonadotropin levels. This allele is predicted to cause a change in five amino acids of the C-terminus along with a truncation of two amino acids. This variant is not reported in gnomAD, and we propose it is associated with this patient’s Kallmann syndrome-like phenotype with variable penetrance. This supports the possibility of allele-specific phenotypes for IGSF10 and expands on the previously described association of IGSF10 with delayed puberty (74). Additionally, in those cases with variants in IGSF10, an additional disorder of neuronal or pituitary development was more likely. Two cases with variants in IGSF10, BAB7110 and the Erasmus MC case, had an additional phenotype of growth hormone deficiency and reduced olfaction, respectively (23).
Two of the known genes presented, TG and C3, have a known autoimmune-mediated impact on ovarian development through effects on follicular maintenance and ovarian reserve. Taken together, molecular diagnosis in our cohort suggest that regulators of ovarian and hypothalamic development, as well as genes with autoimmune-mediated impact on ovarian reserve, contribute to the development of POI (Fig. 7b).
MRPS22, a known cause of combined oxidative phosphorylation deficiency 5 (OMIM 611719), additionally represents a possible effector in ovarian physiology. Heterozygous MRPS22 null mouse breeding populations produce no homozygous offspring, suggesting embryonic lethality. However, germ cell specific knockout of MRPS22 in Drosophila demonstrates that MRPS22 has a cell autonomous effect on ovarian development (45). Although defects in mitochondrial oxidative phosphorylation and transcription were not detected, the non-cell autonomous role that subtle mitochondrial defects may play should be considered (45). Another gene implicated in the mitochondrial disease combined oxidative phosphorylation deficiency 7 (OMIM 613559), C12orf65, has also been associated with HH in a family that includes two affected siblings (75). The importance of mitochondrial function to ovarian physiology is further linked by the mitochondrial interacting protein ATAD3A, which funnels cholesterol for steroidogenesis and has been shown to be important for Leydig cell steroidogenesis via mitochondrial membrane association (62, 76, 77). An association between mitochondrial deficiency and a POI phenotype would also be advantageous from an evolutionary perspective, as the egg is the sole contributor of mitochondrial function during human development. Genes involved in mitochondrial pathology are therefore excellent candidates for POI phenotypes. Additionally, those patients with mitochondrial related molecular diagnosis may have an additional variant associated neurologic phenotype, as BAB7669 was noted to have both HH and neuropathy (23). PNPLA7 is an insulin-regulated lysophospholipase shown to be important for energy metabolism, and for which polymorphisms have been associated with menstrual irregularity (78, 79). We suggest that its role in energy metabolism may impact ovarian reserve, perhaps through mechanisms overlapping with mitochondrial dysregulation.
Allele frequency interpretation in sex-limited disease
In this study, three families (HOU 2611, HOU 2776, and HOU 2581) illustrate a pattern of inheritance consistent with a sex-limited disease trait. The first carries likely damaging pathogenic variant in a known HH disease gene, MCM8 in both a proband (BAB7100) and her father. Although POI is a sex-limited trait, MCM8 variants have been reported to cause gonadal deficiency in males as well (65). However, the degree of perturbation of function caused by specific variants may affect sexes in a differential manner. MCM8 and MCM9 function together to perform integral roles in meiosis and DNA repair (80), and egg production in females is uniquely susceptible to perturbation of meiosis (Fig. 7b). Phenotypic variation between sexes is also relevant for the family of BAB7675, in which an unaffected younger brother harbors the same homozygous MCM8 variant. The brother, age 13 and Tanner stage III, does not have evidence of hypogonadism, gonadotropin levels are normal and appropriate for pubertal stage and age, and dehydroepiandrosterone sulfate levels are low—similar to the gonadotropin/dehydroepiandrosterone sulfate levels in the affected proband. It is possible that the effect of these genes on gonadal development is purely through hormonal regulation, and that the differing hormonal milieus required for female and male development ultimately lead to the sex-specific phenotypic effects of deleterious alleles.
The genetic loci affecting sex-limited traits have unique evolutionary characteristics in terms of frequency of observed deleterious variants (81, 82). In the family HOU 2581, both the proband (BAB6996) and an unaffected brother (BAB6999) have the homozygous c.519G>C variant in PADI6. However, functional data have shown that this is a sex-limited phenotype with a functional role in oocyte cytoplasmic lattices in model systems. Here, we use human data to demonstrate a similar sex-limited phenotype in association with the identified genotype, with one unaffected brother having the same homozygous variant (58, 59, 83). Cases like these, for which rare, sex-limited deleterious variants come together in a clan exemplify how these alleles may be introduced and continue to propagate in a population through the nonaffected sex. Further study is warranted to better define which loci are under sex-limited selection and to what extent. The potential molecular diagnoses in family HOU 2581 suggests that the stringent allele-frequency cutoffs used for genomic analysis may be too high when analyzing sex-limited phenotypes. Variant functional assessment algorithms also consider allele frequency when determining whether an allele is predicted to be deleterious, and these may need adjustment to reflect sex-specific differences in allele frequency.
Potential oligogenic inheritance and complex allele contributions to pathophysiology of genetic disease
Multilocus variation has increasingly explained blended phenotypic outcomes in disease cohorts (35, 84, 85). Mutational burden has been shown to contribute to complex phenotypic presentations including distal symmetric polyneuropathy (86). A retrospective study of 7374 consecutive unrelated patients referred for clinical ES identified 101 cases (4.9% of cases for which ES was diagnostic) with molecular diagnoses resulting from pathogenic variants at two or more distinct disease-associated loci (35, 84, 85). The clinical variability of POI has contributed to the hypothesis that ovarian development is also a clinical phenotype in which multilocus variation may explain apparent phenotypic variability (35).
AOH regions in the consanguineous families studied are hypothesized by the Clan Genomics model to harbor unique, disease-causing rare variants that have been rapidly brought to homozygosity instead of cleared by selection due to consanguineous marriage (87). Just as this could occur for a single disease locus, homozygosity of rare variants at more than one locus can occur (35, 85). In our proposed oligogenic families, homozygous variants in four of five genes (80%) are located in regions of AOH, demonstrating an AOH-mediated recessive disease burden in these cases (35). One of these families, HOU 2774, involved mutational burden and dual molecular diagnosis of two known-associated genes for POI and a candidate gene for obesity in a patient with HH and obesity. All identified variants in these oligogenic families were homozygous, one with AR + AR variants and one with AR + AR + AR variants. Consistent with the Clan Genomics hypothesis, AOH-mediated recessive disease burden contributes to 15 of 16 (94%) molecular diagnoses in this cohort (87).
Conclusion
Ovarian development is complex, as is the maintenance of its physiological functions during germ cell production. The presence of direct and indirect effectors of ovarian development function shows that a single pathway does not sufficiently explain ovarian physiology. However, ES of POI cohorts can elucidate molecular candidates involved in female gonadal development and decades of physiological function required for germ cell production. Identifying variants causative of POI provides patients with a molecular diagnosis and accurate counseling regarding recurrence risk. Furthermore, this molecular diagnosis can guide expectant management. The continual elucidation of disease genes in POI allows for a deeper understanding of the molecular physiology of ovarian development and function, and the relative contributions of multilocus variation and AOH-mediated recessive disease burden to POI.
Acknowledgments
Financial Support: This study was supported in part by the National Human Genome Research Institute (NHGRI) and National Heart Lung and Blood Institute (NHBLI) to the Baylor-Hopkins Center for Mendelian Genomics (BHCMG; UM1 HG006542 to J.R.L.); NHGRI grant to Baylor College of Medicine Human Genome Sequencing Center (U54HG003273 to R.A.G.), and National Institute of Neurologic Disorders and Stroke (NINDS; R35NS105078 to J.R.L.). J.E.P. was supported by NHGRI K08 HG008986. A.J. was supported in part by the Howard Hughes Medical Research Fellows Program and the Baylor College of Medicine Medical Scientist Training Program. D.P. is supported by Clinical Research Training Scholarship in Neuromuscular Disease partnered by the American Academy of Neurology (AAN), American Brain Foundation (ABF), and Muscle Study Group (MSG). T.S.B. was supported by the Netherlands Organization for Scientific Research (ZonMW Veni, Grant 91617021) and by an NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation.
Current Affiliation: Y. Bayram’s current affiliation is the Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, New York.
Disclosure Summary: J.R.L. has stock ownership in 23andMe and is a paid consultant for Regeneron Pharmaceuticals. J.R.L. is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical exome sequencing offered at Baylor Genetics (MGL; http://www.bcm.edu/geneticlabs/); J.R.L. is a member of the SAB of Baylor Genetics. The remaining authors have nothing to disclose.
Glossary
Abbreviations:
- 1000GP
Thousand Genomes Project
- AAMR
Alu-Alu mediated rearrangement
- AD
autosomal dominant
- AOH
absence of heterozygosity
- AR
autosomal recessive
- BHCMG
Baylor Hopkins Center for Mendelian Genomics
- CNV
copy number variant
- ES
exome sequencing
- HH
hypergonadotropic hypogonadism
- MAF
minor allele frequency
- OMIM
Online Mendelian Inheritance in Man
- POI
primary ovarian insufficiency
References and Notes
- 1. Deligeoroglou E, Athanasopoulos N, Tsimaris P, Dimopoulos KD, Vrachnis N, Creatsas G. Evaluation and management of adolescent amenorrhea. Ann N Y Acad Sci. 2010;1205(1):23–32. [DOI] [PubMed] [Google Scholar]
- 2. Shelling AN. Premature ovarian failure. Reproduction. 2010;140(5):633–641. [DOI] [PubMed] [Google Scholar]
- 3. Franić-Ivanišević M, Franić D, Ivović M, Tančić-Gajić M, Marina L, Barac M, Vujović S. Genetic etiology of primary premature ovarian insufficiency. Acta Clin Croat. 2016;55(4):629–635. [DOI] [PubMed] [Google Scholar]
- 4. Turner HH. A syndrome of infantilism, congenital webbed neck, and cubitus valgus. Endocrinology. 1938;23(5):566–574. [PubMed] [Google Scholar]
- 5. Allingham-Hawkins DJ, Brown CA, Babul R, Chitayat D, Krekewich K, Humphries T, Ray PN, Teshima IE. Tissue-specific methylation differences and cognitive function in fragile X premutation females. Am J Med Genet. 1996;64(2):329–333. [DOI] [PubMed] [Google Scholar]
- 6. Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocr Rev. 2009;30(6):624–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qin Y, Choi Y, Zhao H, Simpson JL, Chen Z-J, Rajkovic A. NOBOX homeobox mutation causes premature ovarian failure. Am J Hum Genet. 2007;81(3):576–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao H, Qin Y, Kovanci E, Simpson JL, Chen ZJ, Rajkovic A. Analyses of GDF9 mutation in 100 Chinese women with premature ovarian failure. Fertil Steril. 2007;88(5):1474–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kovanci E, Rohozinski J, Simpson JL, Heard MJ, Bishop CE, Carson SA. Growth differentiating factor-9 mutations may be associated with premature ovarian failure. Fertil Steril. 2007;87(1):143–146. [DOI] [PubMed] [Google Scholar]
- 10. Laissue P, Christin-Maitre S, Touraine P, Kuttenn F, Ritvos O, Aittomaki K, Bourcigaux N, Jacquesson L, Bouchard P, Frydman R, Dewailly D, Reyss AC, Jeffery L, Bachelot A, Massin N, Fellous M, Veitia RA. Mutations and sequence variants in GDF9 and BMP15 in patients with premature ovarian failure. Eur J Endocrinol. 2006;154(5):739–744. [DOI] [PubMed] [Google Scholar]
- 11. Rossetti R, Di Pasquale E, Marozzi A, Bione S, Toniolo D, Grammatico P, Nelson LM, Beck-Peccoz P, Persani L. BMP15 mutations associated with primary ovarian insufficiency cause a defective production of bioactive protein. Hum Mutat. 2009;30(5):804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao H, Chen ZJ, Qin Y, Shi Y, Wang S, Choi Y, Simpson JL, Rajkovic A. Transcription factor FIGLA is mutated in patients with premature ovarian failure. Am J Hum Genet. 2008;82(6):1342–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shiina H, Matsumoto T, Sato T, Igarashi K, Miyamoto J, Takemasa S, Sakari M, Takada I, Nakamura T, Metzger D, Chambon P, Kanno J, Yoshikawa H, Kato S. Premature ovarian failure in androgen receptor-deficient mice. Proc Natl Acad Sci USA. 2006;103(1):224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chapman C, Cree L, Shelling AN. The genetics of premature ovarian failure: current perspectives. Int J Womens Health. 2015;7:799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bilgin EM, Kovanci E. Genetics of premature ovarian failure. Curr Opin Obstet Gynecol. 2015;27(3):167–174. [DOI] [PubMed] [Google Scholar]
- 16. Fonseca DJ, Patiño LC, Suárez YC, de Jesús Rodríguez A, Mateus HE, Jiménez KM, Ortega-Recalde O, Díaz-Yamal I, Laissue P. Next generation sequencing in women affected by nonsyndromic premature ovarian failure displays new potential causative genes and mutations. Fertil Steril. 2015;104(1):154–62.e2. [DOI] [PubMed] [Google Scholar]
- 17. Laissue P. Aetiological coding sequence variants in non-syndromic premature ovarian failure: From genetic linkage analysis to next generation sequencing. Mol Cell Endocrinol. 2015;411:243–257. [DOI] [PubMed] [Google Scholar]
- 18. Pangas SA, Choi Y, Ballow DJ, Zhao Y, Westphal H, Matzuk MM, Rajkovic A. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci USA. 2006;103(21):8090–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qin Y, Jiao X, Simpson JL, Chen ZJ. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum Reprod Update. 2015;21(6):787–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bayram Y, Gulsuner S, Guran T, Abaci A, Yesil G, Gulsuner HU, Atay Z, Pierce SB, Gambin T, Lee M, Turan S, Bober E, Atik MM, Walsh T, Karaca E, Pehlivan D, Jhangiani SN, Muzny D, Bereket A, Buyukgebiz A, Boerwinkle E, Gibbs RA, King MC, Lupski JR. Homozygous loss-of-function mutations in SOHLH1 in patients with nonsyndromic hypergonadotropic hypogonadism. J Clin Endocrinol Metab. 2015;100(5):E808–E814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weinberg-Shukron A, Renbaum P, Kalifa R, Zeligson S, Ben-Neriah Z, Dreifuss A, Abu-Rayyan A, Maatuk N, Fardian N, Rekler D, Kanaan M, Samson AO, Levy-Lahad E, Gerlitz O, Zangen D. A mutation in the nucleoporin-107 gene causes XX gonadal dysgenesis. J Clin Invest. 2015;125(11):4295–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiao X, Ke H, Qin Y, Chen ZJ. Molecular genetics of premature ovarian insufficiency. Trends Endocrinol Metab. 2018;29(11):795–807. [DOI] [PubMed] [Google Scholar]
- 23. Jolly A, Bayram Y, Turan S, Aycan Z, Tos T, Abali ZY, Hacihamdioglu B, Coban Akdemir ZH, Hijazi H, Bas S, Atay Z, Guran T, Abali S, Bas F, Darendeliler F, Colombo R, Barakat TS, Rinne T, White JJ, Yesil G, Gezdirici A, Gulec EY, Karaca E, Pehlivan D, Jhangiani SN, Muzny DM, Poyrazoglu S, Bereket A, Gibbs RA, Posey JE, Lupski JR. Data from: Exome sequencing of a primary ovarian insufficiency cohort reveals common molecular etiologies for a spectrum of disease. Figshare. Deposited 29 January2019. 10.6084/m9.figshare.7647215. [DOI] [PMC free article] [PubMed]
- 24. Reid JG, Carroll A, Veeraraghavan N, Dahdouli M, Sundquist A, English A, Bainbridge M, White S, Salerno W, Buhay C, Yu F, Muzny D, Daly R, Duyk G, Gibbs RA, Boerwinkle E. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA, Donnelly P, Egholm M, Flicek P, Gabriel SB, Gibbs RA, Knoppers BM; 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136(6):665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. NHBLI. NHBLI GO Sequencing Projecting (ESP). Available at: http://evs.gs.washing.edu/EVS. Accessed 22 October 2018.
- 28. Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, Harrell TM, McMillin MJ, Wiszniewski W, Gambin T, Coban Akdemir ZH, Doheny K, Scott AF, Avramopoulos D, Chakravarti A, Hoover-Fong J, Mathews D, Witmer PD, Ling H, Hetrick K, Watkins L, Patterson KE, Reinier F, Blue E, Muzny D, Kircher M, Bilguvar K, López-Giráldez F, Sutton VR, Tabor HK, Leal SM, Gunel M, Mane S, Gibbs RA, Boerwinkle E, Hamosh A, Shendure J, Lupski JR, Lifton RP, Valle D, Nickerson DA, Bamshad MJ; Centers for Mendelian Genomics. The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am J Hum Genet. 2015;97(2):199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG; Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Virani SS, Brautbar A, Davis BC, Nambi V, Hoogeveen RC, Sharrett AR, Coresh J, Mosley TH, Morrisett JD, Catellier DJ, Folsom AR, Boerwinkle E, Ballantyne CM. Associations between lipoprotein(a) levels and cardiovascular outcomes in black and white subjects: the Atherosclerosis Risk in Communities (ARIC) Study. Circulation. 2012;125(2):241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20(1):110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74(12):5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carvalho CMB, Zhang F, Liu P, Patel A, Sahoo T, Bacino CA, Shaw C, Peacock S, Pursley A, Tavyev YJ, Ramocki MB, Nawara M, Obersztyn E, Vianna-Morgante AM, Stankiewicz P, Zoghbi HY, Cheung SW, Lupski JR. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum Mol Genet. 2009;18(12):2188–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gambin T, Akdemir ZC, Yuan B, Gu S, Chiang T, Carvalho CMB, Shaw C, Jhangiani S, Boone PM, Eldomery MK, Karaca E, Bayram Y, Stray-Pedersen A, Muzny D, Charng WL, Bahrambeigi V, Belmont JW, Boerwinkle E, Beaudet AL, Gibbs RA, Lupski JR. Homozygous and hemizygous CNV detection from exome sequencing data in a Mendelian disease cohort. Nucleic Acids Res. 2017;45(4):1633–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karaca E, Posey JE, Coban Akdemir Z, Pehlivan D, Harel T, Jhangiani SN, Bayram Y, Song X, Bahrambeigi V, Yuregir OO, Bozdogan S, Yesil G, Isikay S, Muzny D, Gibbs RA, Lupski JR. Phenotypic expansion illuminates multilocus pathogenic variation. Genet Med. 2018;20(12):1528–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. [DOI] [PubMed] [Google Scholar]
- 37. Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12(2):115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat. 2015;36(4):425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue, Suppl 2):W214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fromer M, Purcell SM. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet. 2014;81(Suppl 81):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen A, Tiosano D, Guran T, Baris HN, Bayram Y, Mory A, Shapiro-Kulnane L, Hodges CA, Akdemir ZC, Turan S, Jhangiani SN, van den Akker F, Hoppel CL, Salz HK, Lupski JR, Buchner DA. Mutations in the mitochondrial ribosomal protein MRPS22 lead to primary ovarian insufficiency. Hum Mol Genet. 2018;27(11):1913–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Corral J, Martín C, Pérez R, Sánchez I, Mories MT, San Millan JL, Miralles JM, González-Sarmiento R. Thyroglobulin gene point mutation associated with non-endemic simple goitre. Lancet. 1993;341(8843):462–464. [DOI] [PubMed] [Google Scholar]
- 47. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7(Suppl 76):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012; 40(Web Server issue, W1)W452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Firoozi H, Rahimpour Z. Condition assessment of power transformers- logical process of decision making to successful diagnosis. Int. Rev. Model. Simulations. 2012;5(4):1486–1490. [Google Scholar]
- 51. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(10):e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leskovec J, Sosič R. SNAP: a general purpose network analysis and graph mining library. ACM Trans Intell Syst Technol. 2016;8(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bendl J, Musil M, Štourač J, Zendulka J, Damborský J, Brezovský J. PredictSNP2: A unified platform for accurately evaluating SNP effects by exploiting the different characteristics of variants in distinct genomic regions. PLOS Comput Biol. 2016;12(5):e1004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5(1):e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee JA, Carvalho CMB, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131(7):1235–1247. [DOI] [PubMed] [Google Scholar]
- 56. Zhao W, Sung P. Significance of ligand interactions involving Hop2-Mnd1 and the RAD51 and DMC1 recombinases in homologous DNA repair and XX ovarian dysgenesis. Nucleic Acids Res. 2015;43(8):4055–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Williams MJ, Almén MS, Fredriksson R, Schiöth HB. What model organisms and interactomics can reveal about the genetics of human obesity. Cell Mol Life Sci. 2012;69(22):3819–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Esposito G, Vitale AM, Leijten FPJ, Strik AM, Koonen-Reemst AMCB, Yurttas P, Robben TJAA, Coonrod S, Gossen JA. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol. 2007;273(1-2):25–31. [DOI] [PubMed] [Google Scholar]
- 59. Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, Coonrod SA. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135(15):2627–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kan R, Yurttas P, Kim B, Jin M, Wo L, Lee B, Gosden R, Coonrod SA. Regulation of mouse oocyte microtubule and organelle dynamics by PADI6 and the cytoplasmic lattices. Dev Biol. 2011;350(2):311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lo MS, Towne M, VanNoy GE, Brownstein CA, Lane AA, Chatila TA, Agrawal PB. Monogenic Hashimoto thyroiditis associated with a variant in the thyroglobulin (TG) gene. J Autoimmun. 2018;86:116–119. [DOI] [PubMed] [Google Scholar]
- 62. Harel T, Yoon WH, Garone C, Gu S, Coban-Akdemir Z, Eldomery MK, Posey JE, Jhangiani SN, Rosenfeld JA, Cho MT, Fox S, Withers M, Brooks SM, Chiang T, Duraine L, Erdin S, Yuan B, Shao Y, Moussallem E, Lamperti C, Donati MA, Smith JD, McLaughlin HM, Eng CM, Walkiewicz M, Xia F, Pippucci T, Magini P, Seri M, Zeviani M, Hirano M, Hunter JV, Srour M, Zanigni S, Lewis RA, Muzny DM, Lotze TE, Boerwinkle E, Gibbs RA, Hickey SE, Graham BH, Yang Y, Buhas D, Martin DM, Potocki L, Graziano C, Bellen HJ, Lupski JR; Baylor-Hopkins Center for Mendelian Genomics, University of Washington Center for Mendelian Genomics. Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am J Hum Genet. 2016;99(4):831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Harel T, Yesil G, Bayram Y, Coban-Akdemir Z, Charng WL, Karaca E, Al Asmari A, Eldomery MK, Hunter JV, Jhangiani SN, Rosenfeld JA, Pehlivan D, El-Hattab AW, Saleh MA, LeDuc CA, Muzny D, Boerwinkle E, Gibbs RA, Chung WK, Yang Y, Belmont JW, Lupski JR; Baylor-Hopkins Center for Mendelian Genomics. Monoallelic and biallelic variants in EMC1 identified in individuals with global developmental delay, hypotonia, scoliosis, and cerebellar atrophy. Am J Hum Genet. 2016;98(3):562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rainger J, Pehlivan D, Johansson S, Bengani H, Sanchez-Pulido L, Williamson KA, Ture M, Barker H, Rosendahl K, Spranger J, Horn D, Meynert A, Floyd JAB, Prescott T, Anderson CA, Rainger JK, Karaca E, Gonzaga-Jauregui C, Jhangiani S, Muzny DM, Seawright A, Soares DC, Kharbanda M, Murday V, Finch A, Gibbs RA, van Heyningen V, Taylor MS, Yakut T, Knappskog PM, Hurles ME, Ponting CP, Lupski JR, Houge G, FitzPatrick DR; UK10K, Baylor-Hopkins Center for Mendelian Genomics. Monoallelic and biallelic mutations in MAB21L2 cause a spectrum of major eye malformations. Am J Hum Genet. 2014;94(6):915–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tenenbaum-Rakover Y, Weinberg-Shukron A, Renbaum P, Lobel O, Eideh H, Gulsuner S, Dahary D, Abu-Rayyan A, Kanaan M, Levy-Lahad E, Bercovich D, Zangen D. Minichromosome maintenance complex component 8 (MCM8) gene mutations result in primary gonadal failure. J Med Genet. 2015;52(6):391–399. [DOI] [PubMed] [Google Scholar]
- 66. Dondik Y, Lei Z, Gaskins J, Pagidas K. Minichromosome maintenance complex component 8 and 9 gene expression in the menstrual cycle and unexplained primary ovarian insufficiency. J Assist Reprod Genet. 2019;36(1):57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zierhut C, Berlinger M, Rupp C, Shinohara A, Klein F. Mnd1 is required for meiotic interhomolog repair. Curr Biol. 2004;14(9):752–762. [DOI] [PubMed] [Google Scholar]
- 68. Zhao W, Saro D, Hammel M, Kwon Y, Xu Y, Rambo RP, Williams GJ, Chi P, Lu L, Pezza RJ, Camerini-Otero RD, Tainer JA, Wang HW, Sung P. Mechanistic insights into the role of Hop2-Mnd1 in meiotic homologous DNA pairing. Nucleic Acids Res. 2014;42(2):906–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Castronovo C, Rossetti R, Rusconi D, Recalcati MP, Cacciatore C, Beccaria E, Calcaterra V, Invernizzi P, Larizza D, Finelli P, Persani L. Gene dosage as a relevant mechanism contributing to the determination of ovarian function in Turner syndrome. Hum Reprod. 2014;29(2):368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lalani SR, Liu P, Rosenfeld JA, Watkin LB, Chiang T, Leduc MS, Zhu W, Ding Y, Pan S, Vetrini F, Miyake CY, Shinawi M, Gambin T, Eldomery MK, Akdemir ZHC, Emrick L, Wilnai Y, Schelley S, Koenig MK, Memon N, Farach LS, Coe BP, Azamian M, Hernandez P, Zapata G, Jhangiani SN, Muzny DM, Lotze T, Clark G, Wilfong A, Northrup H, Adesina A, Bacino CA, Scaglia F, Bonnen PE, Crosson J, Duis J, Maegawa GHB, Coman D, Inwood A, McGill J, Boerwinkle E, Graham B, Beaudet A, Eng CM, Hanchard NA, Xia F, Orange JS, Gibbs RA, Lupski JR, Yang Y. Recurrent muscle weakness with rhabdomyolysis, metabolic crises, and cardiac arrhythmia due to bi-allelic TANGO2 mutations. Am J Hum Genet. 2016;98(2):347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Posey JE, Rosenfeld JA, James RA, Bainbridge M, Niu Z, Wang X, Dhar S, Wiszniewski W, Akdemir ZHC, Gambin T, Xia F, Person RE, Walkiewicz M, Shaw CA, Sutton VR, Beaudet AL, Muzny D, Eng CM, Yang Y, Gibbs RA, Lupski JR, Boerwinkle E, Plon SE. Molecular diagnostic experience of whole-exome sequencing in adult patients. Genet Med. 2016;18(7):678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Song X, Beck CR, Du R, Campbell IM, Coban-Akdemir Z, Gu S, Breman AM, Stankiewicz P, Ira G, Shaw CA, Lupski JR. Predicting human genes susceptible to genomic instability associated with Alu/Alu-mediated rearrangements. Genome Res. 2018;28(8):1228–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. MacColl G, Quinton R, Bouloux PMG. GnRH neuronal development: insights into hypogonadotrophic hypogonadism. Trends Endocrinol Metab. 2002;13(3):112–118. [DOI] [PubMed] [Google Scholar]
- 74. Howard SR, Guasti L, Ruiz-Babot G, Mancini A, David A, Storr HL, Metherell LA, Sternberg MJ, Cabrera CP, Warren HR, Barnes MR, Quinton R, de Roux N, Young J, Guiochon-Mantel A, Wehkalampi K, André V, Gothilf Y, Cariboni A, Dunkel L. IGSF10 mutations dysregulate gonadotropin-releasing hormone neuronal migration resulting in delayed puberty. EMBO Mol Med. 2016;8(6):626–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Karaca E, Harel T, Pehlivan D, Jhangiani SN, Gambin T, Coban Akdemir Z, Gonzaga-Jauregui C, Erdin S, Bayram Y, Campbell IM, Hunter JV, Atik MM, Van Esch H, Yuan B, Wiszniewski W, Isikay S, Yesil G, Yuregir OO, Tug Bozdogan S, Aslan H, Aydin H, Tos T, Aksoy A, De Vivo DC, Jain P, Geckinli BB, Sezer O, Gul D, Durmaz B, Cogulu O, Ozkinay F, Topcu V, Candan S, Cebi AH, Ikbal M, Yilmaz Gulec E, Gezdirici A, Koparir E, Ekici F, Coskun S, Cicek S, Karaer K, Koparir A, Duz MB, Kirat E, Fenercioglu E, Ulucan H, Seven M, Guran T, Elcioglu N, Yildirim MS, Aktas D, Alikaşifoğlu M, Ture M, Yakut T, Overton JD, Yuksel A, Ozen M, Muzny DM, Adams DR, Boerwinkle E, Chung WK, Gibbs RA, Lupski JR. Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron. 2015;88(3):499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Issop L, Fan J, Lee S, Rone MB, Basu K, Mui J, Papadopoulos V. Mitochondria-associated membrane formation in hormone-stimulated Leydig cell steroidogenesis: role of ATAD3. Endocrinology. 2015;156(1):334–345. [DOI] [PubMed] [Google Scholar]
- 77. Rone MB, Midzak AS, Issop L, Rammouz G, Jagannathan S, Fan J, Ye X, Blonder J, Veenstra T, Papadopoulos V. Identification of a dynamic mitochondrial protein complex driving cholesterol import, trafficking, and metabolism to steroid hormones. Mol Endocrinol. 2012;26(11):1868–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kienesberger PC, Lass A, Preiss-Landl K, Wolinski H, Kohlwein SD, Zimmermann R, Zechner R. Identification of an insulin-regulated lysophospholipase with homology to neuropathy target esterase. J Biol Chem. 2008;283(9):5908–5917. [DOI] [PubMed] [Google Scholar]
- 79. Su Y, Kong GL, Su YL, Zhou Y, Lv LF, Wang Q, Huang BP, Zheng RZ, Li QZ, Yuan HJ, Zhao ZG. Correlation analysis of the PNPLA7 gene polymorphism and susceptibility to menstrual disorder. Genet Mol Res. 2015;14(1):1733–1740. [DOI] [PubMed] [Google Scholar]
- 80. Park J, Long DT, Lee KY, Abbas T, Shibata E, Negishi M, Luo Y, Schimenti JC, Gambus A, Walter JC, Dutta A. The MCM8-MCM9 complex promotes RAD51 recruitment at DNA damage sites to facilitate homologous recombination. Mol Cell Biol. 2013;33(8):1632–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Reinhold K. Evolutionary genetics of sex-limited traits under fluctuating selection. J Evol Biol. 1999;12(5):897–902. [Google Scholar]
- 82. Connallon T, Jordan CY. Accumulation of deleterious mutations near sexually antagonistic genes. G3 (Bethesda). 2016;6(8):2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. White J, Mazzeu JF, Hoischen A, Jhangiani SN, Gambin T, Alcino MC, Penney S, Saraiva JM, Hove H, Skovby F, Kayserili H, Estrella E, Vulto-van Silfhout AT, Steehouwer M, Muzny DM, Sutton VR, Gibbs RA, Lupski JR, Brunner HG, van Bon BWM, Carvalho CMB; Baylor-Hopkins Center for Mendelian Genomics. DVL1 frameshift mutations clustering in the penultimate exon cause autosomal-dominant Robinow syndrome. Am J Hum Genet. 2015;96(4):612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pehlivan D, Beck CR, Okamoto Y, Harel T, Akdemir ZHC, Jhangiani SN, Withers MA, Goksungur MT, Carvalho CMB, Czesnik D, Gonzaga-Jauregui C, Wiszniewski W, Muzny DM, Gibbs RA, Rautenstrauss B, Sereda MW, Lupski JR. The role of combined SNV and CNV burden in patients with distal symmetric polyneuropathy. Genet Med. 2016;18(5):443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Posey JE, Harel T, Liu P, Rosenfeld JA, James RA, Coban Akdemir ZH, Walkiewicz M, Bi W, Xiao R, Ding Y, Xia F, Beaudet AL, Muzny DM, Gibbs RA, Boerwinkle E, Eng CM, Sutton VR, Shaw CA, Plon SE, Yang Y, Lupski JR. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376(1):21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gonzaga-Jauregui C, Harel T, Gambin T, Kousi M, Griffin LB, Francescatto L, Ozes B, Karaca E, Jhangiani SN, Bainbridge MN, Lawson KS, Pehlivan D, Okamoto Y, Withers M, Mancias P, Slavotinek A, Reitnauer PJ, Goksungur MT, Shy M, Crawford TO, Koenig M, Willer J, Flores BN, Pediaditrakis I, Us O, Wiszniewski W, Parman Y, Antonellis A, Muzny DM, Katsanis N, Battaloglu E, Boerwinkle E, Gibbs RA, Lupski JR; Baylor-Hopkins Center for Mendelian Genomics. Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Reports. 2015;12(7):1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147(1):32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]