Abstract
Protein kinases orchestrate signal transduction pathways involved in central nervous system functions ranging from neurodevelopment to synaptic transmission and plasticity. Abnormalities in kinase-mediated signaling are involved in the pathophysiology of neurological disorders, including neuropsychiatric disorders. Here, we expand on the hypothesis that kinase networks are dysregulated in schizophrenia. We investigated changes in serine/threonine kinase activity in cortical excitatory neurons differentiated from induced pluripotent stem cells (iPSCs) from a schizophrenia patient presenting with a 4-bp mutation in the disrupted in schizophrenia 1 (DISC1) gene and a corresponding control. Using kinome peptide arrays, we demonstrate large scale abnormalities in DISC1 cells, including a global depression of serine/threonine kinase activity, and changes in activity of kinases, including AMP-activated protein kinase (AMPK), extracellular signal-regulated kinases (ERK), and thousand-and-one amino acid (TAO) kinases. Using isogenic cell lines in which the DISC1 mutation is either introduced in the control cell line, or rescued in the schizophrenia cell line, we ascribe most of these changes to a direct effect of the presence of the DISC1 mutation. Investigating the gene expression signatures downstream of the DISC1 kinase network, and mapping them on perturbagen signatures obtained from the Library of Integrated Network-based Cellular Signatures (LINCS) database, allowed us to propose novel drug targets able to reverse the DISC1 kinase dysregulation gene expression signature. Altogether, our findings provide new insight into abnormalities of kinase networks in schizophrenia and suggest possible targets for disease intervention.
Graphical Abstract

iPSCs from a schizophrenia patient harboring a mutation in the DISC1 gene show large scale abnormalities in serine/threonine kinase activity.
1. Introduction
Schizophrenia is a debilitating neuropsychiatric disorder, of largely unknown pathophysiology. Patients affected display a complex symptomatology characterized by psychotic symptoms, such as hallucinations, delusions, and disorganization, cognitive deficits, and negative symptoms that together exert a profound impact on the quality of life1. The disease affects 1% of the worldwide population2, and presents with a significant genetic component with heritability estimates of 50–85%3.
Disrupted in schizophrenia 1 (DISC1) has emerged as a strong candidate gene underlying the risk for major mental disorders. DISC1 was originally identified in a large Scottish family in which the balanced reciprocal chromosomal translocation t(1;11) (q42.1;q14.3) disrupts this gene and segregates with major mental disorders, including schizophrenia, major depressive disorder, and bipolar disorder4. Subsequently, a rare mutation resulting in a 4 base-pair (bp) frameshift deletion in the C-terminus of DISC1 was identified in an American family (pedigree H) with schizophrenia and schizoaffective disorder5. In addition, variants and polymorphisms of DISC1 have been genetically associated with neuropsychiatric disorders including schizophrenia, major depressive disorder, bipolar disorder and autism, suggesting that DISC1 may underlie common endophenotypes associated with major mental disorders6. Animal and cell model studies support this notion, as DISC1 has a significant role in mediating central processes in the brain both during development and adulthood, including neurite outgrowth, neural migration, proliferation, and differentiation, neurogenesis, as well as synapse formation and regulation6,7. The precise mechanisms underlying these effects remain, however, incompletely understood, and may involve the function of DISC1 as intracellular scaffold protein, its postsynaptic localization, and/or its effects on intracellular signal transduction pathways6.
Protein kinases, including serine/threonine kinases, phosphorylate target proteins as a mechanism to fine tune signaling in complex biological pathways. Phosphorylation is one of the most wide-spread posttranslational modifications in eukaryotic cells and is involved in almost all aspects of cellular behavior, including metabolism, transcription, differentiation, apoptosis, and cytoskeletal regulation8. In the central nervous system, protein kinases regulate pathways that are important for synaptic transmission and plasticity, as well as circuit formation and refinement during development9,10. As such, dysregulation in kinase signaling can lead to synaptic impairment, and is often associated with neurological disorders where it contributes to the underlying pathogenic processes and functional impairment9,11. Protein kinase signaling has been classically studied by evaluating expression and phosphorylation level of individual kinase targets of a particular signaling network. However, intracellular signaling is complex, with many interconnected pathways and cross-talk between individual pathways12. In addition, the classical phosphoprotein approaches measure end-products of enzymatic reactions and do not provide direct information on protein kinase activity. Profiling of kinase activity on an “-omics” scale has been made possible by the development of kinome arrays, which are able to simultaneously detect activity changes in a large array of kinases present in the same sample13. Kinome array chips contain immobilized peptide substrates containing consensus phosphorylation sequences, covering a wide range of kinase specificities. Monitoring the phosphorylation levels at these reporter substrates after exposure to the kinases present in the sample allows the capture of information across different kinase families in a native signaling environment. Importantly, kinome arrays detect changes in kinase enzyme activity, and are unbiased towards particular signaling pathways, allowing for novel hypotheses on signal transduction pathways to be generated.
Using the kinome array platform, we recently described global changes in kinase signaling in the anterior cingulate cortex of chronic schizophrenia patients14. While this provided a unique view into the dysfunction of kinase networks in postmortem schizophrenia samples, much less is known regarding changes in kinase activity in schizophrenia patients in vivo. Recently, generation of patient-derived induced pluripotent stem cells (iPSCs) and their differentiation into disease-relevant cell types has provided a unique window into pathophysiological changes occurring in living human cells15. The use of iPSCs has emerged as a paradigm shift in modeling neurological disorders, allowing the recapitulation of cellular and molecular mechanisms in genetic contexts that faithfully resemble the human disease. This is especially suitable in the context of neurodevelopmental disorders, such as schizophrenia, as the use of iPSCs allows the recapitulation of pathological development of human tissue in a genetic context that is known to be permissive for reaching the disease state16.
In this study we extended our investigation into kinase signaling abnormalities in schizophrenia by analyzing the serine/threonine kinase activity changes in a human iPSC model of DISC1 schizophrenia. To do so, we employed kinome arrays to profile the kinase activity in cortical excitatory neurons differentiated from iPSCs of a patient presenting with schizophrenia and the DISC1 4-bp mutation17. We previously provided an in-depth characterization of this human iPSC model, which recapitulates features of the human pathology, including synaptic transmission defects, and gene expression changes mapping to pathways that include nervous system development, dendritic spine maintenance, and cytoskeletal organization18,19. Using gene editing, we tested whether the DISC1 4-bp mutation is required and/or sufficient for the identified kinome dysregulation profile, by creating isogenic cell lines in which the DISC1 mutation is either introduced in the control cell line, or rescued in the schizophrenia cell line. Doing so, we identified common nodes of kinase dysregulation that are both disease-driven and causally linked with the DISC1 mutation. Using an in silico approach, we further contrasted the kinase dysregulation signature of DISC1 cells to transcriptomic data obtained from postmortem schizophrenia samples, and generated a DISC1 kinase network dysregulation transcriptome profile using gene expression profiles obtained from the Library of Integrated Network-based Cellular Signatures (LINCS)20. Finally, using LINCS, we identified chemical perturbagens that may reverse the DISC1 kinase network gene expression signature. Our findings support the notion that kinase networks are dysfunctional in DISC1 schizophrenia and propose possible small molecules that can reverse the kinase dysregulation gene expression signature.
2. Materials and Methods
2.1. Experimental Design
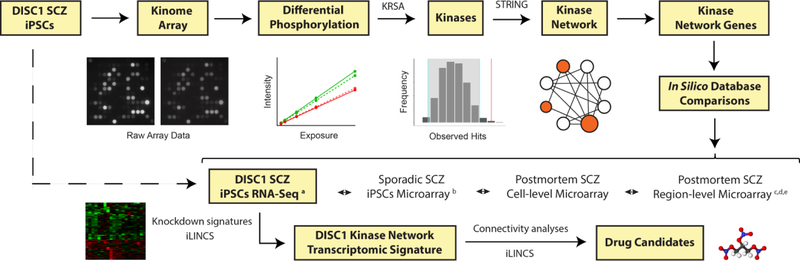
The experimental approach we followed is depicted in Fig. 1. Cortical neurons differentiated from iPSCs of a patient with schizophrenia and the DISC1 4-bp mutation (D2–1), and from iPSCs of a healthy sibling without the DISC1 mutation (C3–1), as well as isogenic cell lines in which the mutation was introduced in the control cell line (C3-M), or rescued in the schizophrenia cell line (D2-R), were subjected to kinome array analysis. All 4 samples (D2–1, C3–1, C3-M, D2-R) were prepared and run at the same time through the kinome array platform. Each sample was run in technical triplicate, and the results were averaged across the 3 identical kinome array runs.
Fig. 1.
Study workflow. By using the results of the kinome analysis, in combination with curated databases and published gene expression studies, we defined a DISC1 schizophrenia kinase dysregulation signature, and identified drug candidates that may reverse the disease gene expression signature. Peptides were initially selected based on difference in phosphorylation between groups, and the upstream kinases that most likely drive these effects identified via a random sampling statistical model. The emerging kinase hits were integrated in a larger kinase-to-kinase interaction network model. Expression of the kinase network genes was investigated in a published transcriptomic database of DISC1 schizophrenia iPSCs18. Kinase genes downregulated in DISC1 cells were used to generate a consensus gene expression signature. Knockdown signatures of the downregulated genes were retrieved from LINCS, aligned, and clustered, in order to identify common sets of gene expression changes downstream of the DISC1 kinase network. Performing connectivity analyses with perturbagen signatures from LINCS allowed us to further identify chemical perturbagens that reverse the gene expression signature of DISC1 schizophrenia as possible drug candidates targeting the kinase dysregulation network. Abbreviations: iLINCS integrative Library of Integrated Network-based Cellular Signatures, iPSCs induced pluripotent stem cells, KRSA kinase resampling analysis, SCZ schizophrenia, STRING Search Tool for Retrieval of Interacting Genes/Proteins. References: a – Wen et al. 201418, b – Brennand et al. 201130, c – Higgs et al. 200627, d – Roussos et al. 201228, e – Gandal et al. 201829.
2.2. Cell lines
Patient (D2–1) and control (C3–1) iPSCs and isogenic cell lines (D2-R and C3-M) were generated at Johns Hopkins School of Medicine as previously described18. All experiments were performed in compliance with the relevant laws and institutional guidelines. Patient D2–1 is part of a previously characterized American family, pedigree H5 presenting with schizophrenia and a 4-bp frameshift deletion at the DISC1 C-terminus. The control C3–1 is an unaffected sibling of D2–1 and does not carry the DISC1 mutation. The isogenic cell lines were generated by transcription activator-like effector nuclease (TALEN), either by correcting the 4-bp deletion in the D2–1 cell line (resulting in the cell line D2-R), or by introducing the 4-bp deletion in the C3–1 cell line (resulting in the cell line C3-M)18. Human iPSCs were differentiated into cortical neurons following the previously established protocol18. Briefly, hiPSCs colonies were detached from the feeder layer with 1 mg/ml collagenase treatment for 1 hour and suspended in embryonic body (EB) medium, consisting of FGF-2-free iPSC medium supplemented with 2 µM Dorsomorphin and 2 µM A-83, in non-treated polystyrene plates for 4 days with a daily medium change. After 4 days, EB medium was replaced by neural induction medium (NPC medium) consisting of DMEM/F12, N2 supplement, NEAA, 2 µg/ml heparin and 2 µM cyclopamine. The floating EBs were then transferred to matrigel-coated 6-well plates at day 7 to form neural tube-like rosettes. The attached rosettes were kept for 15 days with NPC medium change every other day. On day 22, the rosettes were picked mechanically and transferred to low attachment plates (Corning) to form neurospheres in NPC medium containing B27. The neurospheres were then dissociated with Accutase at 37°C for 10 minutes and placed onto Poly-D-Lysine/laminin-coated coverslips in the neuronal differentiation medium, consisting of Neurobasal medium supplemented with 2 mM L-glutamine, B27, cAMP (1 µM), L-Ascorbic Acid (200 ng/ml), BDNF (10 ng/ml) and GDNF (10 ng/ml).
2.3. Kinomics
2.3.1. Sample preparation
Cell pellets generated from cultures of C3–1, D2–1, C3-M, and D2-R were lysed on ice for 30 min using M-PER lysis buffer (ThermoFisher) containing 1:100 Halt Protease and Phosphatase Inhibitor Cocktail (ThermoFisher). Samples were centrifuged (14000 rpm, 10 min, 4°C), and the supernatants collected and assayed for total protein concentration (Pierce BCA Protein Assay Kit, ThermoFisher). Samples were diluted to 1 µg/µL and stored, across multiple aliquots, at −80°C. As freeze-thawing can result in loss of kinase activity21, frozen aliquots were used only once for kinase activity determination. Samples were run in triplicate on the kinome array, using 3 separate chips. Identical protein amounts were loaded for each condition.
2.3.2. Serine-threonine kinase activity profiling
Profiling of serine-threonine kinase activity was performed using the PamStation12 microarray (PamGene International) and STK (4-well) PamChips containing 144 consensus phosphopeptide sequences (3 of which are internal controls) per well, immobilized on porous ceramic membranes. Each PamChip well was blocked with 2% bovine serum albumin (BSA) before 2 µg of protein in the manufacturer’s kinase buffer (PamGene), 157 µM adenosine triphosphate (ATP), and FITC-labeled anti-phospho serine-threonine antibodies (PamGene) were added in each well. The homogenized samples containing the active kinases and assay mix were pumped through the wells to facilitate interaction between kinases in the sample and specific peptide substrates immobilized on the chip. The degree of phosphorylation per well was measured in real time using Evolve (PamGene) kinetic image capture software. The software captures FITC-labeled anti-phospho antibodies binding to each phosphorylated peptide substrate every 6 sec for 60 min. Peptide spot intensity was captured across multiple exposure times (10, 20, 50, 100, 200 ms) during post-wash, and the linear regression slope was calculated and used as the signal (i.e. peptide phosphorylation intensity) in comparative analyses. The signal ratio between pairs of samples (D2–1 vs. C3–1; C3-M vs. C3–1; and D2–1 vs. D2-R) was used to calculate fold change (FC) for each peptide. Peptides with a FC of at least 30% (i.e. FC > 1.30 or FC < 0.70) were considered changed in degree of phosphorylation. This threshold value was guided by previous reports suggesting that changes in kinase activity with similar (or smaller) orders of magnitude are sufficient to trigger biologically relevant changes14,22,23. Peptides that were undetectable or non-linear in the post-wash phase (i.e. the coefficient of determination R2 of the corresponding linear regression less than 0.90) were excluded from subsequent analyses. From the 141 peptides, 72 peptides were excluded under this criterion, leaving a total of 69 substrates used in the final analysis. Kinome arrays were run in triplicate, and the FC per peptide was averaged across the 3 replicates.
2.3.3. Heatmap and global phosphorylation plots
For generating the peptide phosphorylation heatmap and comparing the global phosphorylation levels across the 4 samples, the linear regression slope of each peptide was multiplied by 100 and log2 transformed14,23. The peptide phosphorylation heatmap was generated in R Software (R version 3.4.2, 2017 The R Foundation for Statistical Computing).
2.3.4. Waterfall plots and individual peptide phosphorylation curves
For each comparison considered (D2–1 vs. C3–1; C3-M vs. C3–1; and D2–1 vs. D2-R), FC values were calculated at individual reporter peptides and averaged across the 3 replicate chips. A graphical representation of the change in phosphorylation across reporter substrates is illustrated using waterfall plots, in which the reporter peptides are arranged on the y-axis in decreasing order of FC. In addition, post-wash phosphorylation curves are plotted at individual reporter peptides, using the peptide spot intensities captured during post-wash (10, 20, 50, 100, 200 ms exposure times), averaged across the 3 replicate chips.
2.3.5. Kinase resampling analysis
Using GPS 3.024 and Kinexus Phosphonet (Kinexus Bioinformatics) (http://www.phosphonet.ca), we identified protein kinases acting on phosphorylation sites within the array peptide sequences. These programs provide ranked predictions for serine-threonine kinases targeting putative phosphorylation sites in the peptide sequence. The top 5 kinases predicted by Kinexus and kinases with scores more than twice the prediction threshold for each phosphorylation site in GPS 3.0 were included as predicted kinases for each substrate. This list was complemented by kinases documented to act on the phosphorylation sites of the substrates extracted from public databases PhosphoELM (http://phospho.elm.eu.org), and PhosphoSite Plus (https://www.phosphosite.org). A frequency distribution of kinases predicted to act across the peptide substrates is shown in Supplementary Fig. S1.
To determine which upstream kinases are most likely to be important in the DISC1 signaling network, we performed random sampling analysis14,23. We generated data sets (n = 2000) where each data point includes randomly selected reporter peptide substrates from the kinome array. Each analysis included either 14 substrates, 20 substrates, or 8 substrates, matching the number of substrates changing in phosphorylation in D2–1 vs. C3–1, C3-M vs. C3–1, and D2–1 vs. D2-R respectively. We identified kinases predicted to target each phosphorylation site and calculated the frequency of each kinase for all 2000 data points in the data sets. From these we generated an expected distribution for each kinase. Means and standard deviations were calculated for each expected distribution. Kinases with observed frequencies falling outside two standard deviations from the expected mean (derived from randomly generated data points) were carried forward into our network analyses.
2.3.6. Kinase network modeling
Kinases implicated by the random sampling analyses were used to create a kinase signaling network. We generated a network model that represents the number of direct interactions between protein kinases identified from our random sampling analyses, as well as additional protein kinases that are connected with our array hits. For growing and connecting the kinase network we used the Search Tool for Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.org), selecting interactors that connect with the kinase proteins using the highest confidence interaction score. Because signaling networks may be amplified based on the number of interactions between kinases, we weighted our model by the number of interactions made by each kinase in the network. Visualization of the obtained kinase network model was performed using Cytoscape ver. 3.6.125.
2.3.7. Kinase network pathway analysis
To gain insight into the biological pathways affected downstream of the DISC1 kinase network, we employed Enrichr (http://amp.pharm.mssm.edu/Enrichr)26, using as input the 121 genes forming the extended kinase network (Supplementary Table S1). Results are based on the KEGG 2016 cell signaling pathway database and presented in order of statistical significance of association and number of genes enriched in each pathway.
2.4. In silico analyses
To gain insight into transcriptomic changes associated with our kinases of interest, we evaluated their expression in a previously published RNA-Seq database of gene expression changes in the same D2–1 vs. C3–1 cell lines18. In addition, to test whether the kinase dysregulation signature we identified in DISC1 schizophrenia would extrapolate to spontaneous schizophrenia cases, we also probed publicly available databases of gene expression changes in postmortem schizophrenia samples. The databases we queried are the Stanley Medical Research Institute (SMRI) Online Genomics Database27, the Mount Sinai School of Medicine microarray dataset28, and a recent meta-analysis of microarray studies29 (all region level). In addition, we investigated gene expression changes in a microarray database of neuronally differentiated human iPSCs derived from 4 schizophrenia patients and 4 controls30, and a recently generated microarray dataset of laser capture microdissected (LCM) pyramidal neurons from deep and superficial cortical layers of postmortem schizophrenia (unpublished data) (both cell level).
2.5. Bioinformatic analyses
2.5.1. Generation of a “seed profile”
For performing bioinformatic analyses, we selected a “seed gene profile” based on the kinase network model (Fig. 6), as well as changes in expression of the kinase network genes in D2–1 vs. C3–118. The seed profile contained 20 genes encoding for kinase families identified in the kinome array analysis and decreased in expression at transcript level in the DISC1 cells (Supplementary Table S7). This selection was unbiased, and guided only by the current analysis of kinomic and transcriptomic changes in DISC1 cells.
Fig. 6.
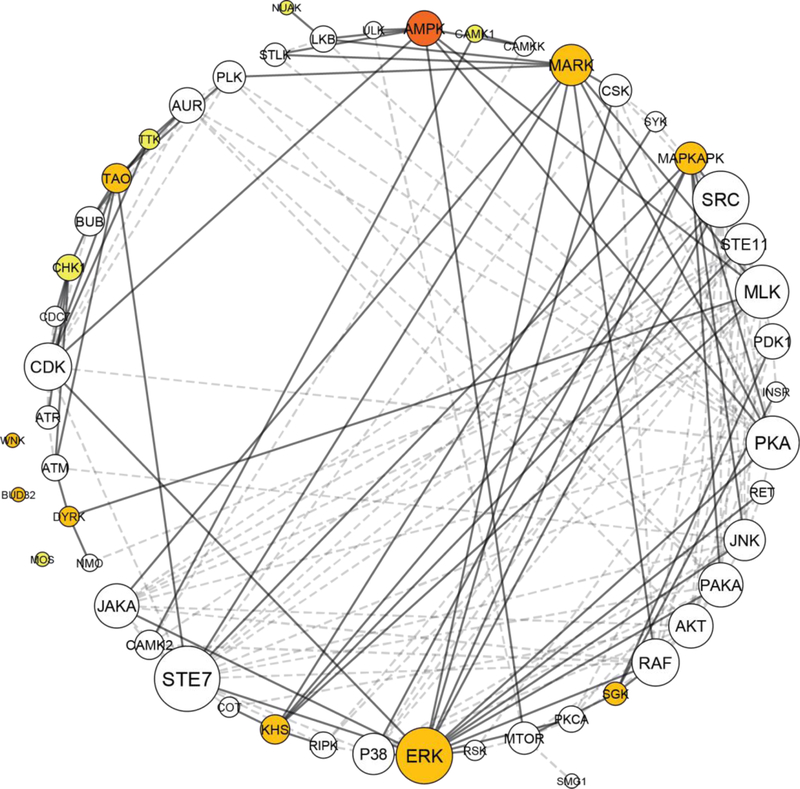
Kinase network dyregulation model of DISC1 schizophrenia. The kinase network was obtained by growing the kinome array hits with kinase interacting partners as identified using STRING. The kinome array hits are color coded, depending on the strength of association with DISC1 schizophrenia and the DISC1 mutation: dark orange indicates a kinome array hit identified in all 3 comparisons, orange a kinome array hit identified in 2/3 comparisons, and yellow a kinome array hit identified in 1/3 comparison (as depicted in the Venn diagram, Fig. 5). White circles represent indirect hits obtained after growing the network in STRING. Circle size corresponds to the number of interactions, with larger circles having more interactions. Thick lines represent interactions with a kinome array direct hit, while dashed lines represent interactions made between kinome array indirect hits. Kinase network model generated using Cytoscape ver. 3.6.1.
2.5.2. Generation of knockdown signatures for seed genes
Using integrative LINCS (iLINCS), an online platform for accessing and analyzing LINCS data (http://ilincs.org), we retrieved knockdown signatures for each seed gene individually. A gene knockdown signature is defined as the set of gene expression changes in the 978 landmark genes (L1000 genes31) when that gene is knocked down in a particular cell line. Seed genes often have multiple knockdown signatures from experiments done in different cell lines. We selected the knockdown signature from a vertebral-cancer of the prostate (VCAP) cell line, as it was the only cell line with knockdown signatures available for all seed genes. While VCAP is not a neuronal cell line, the downstream regulation of transcriptional profiles is often comparable across cell lines32, allowing the use of VCAP as a useful substrate for aligning gene expression signatures across different knockdown signatures.
2.5.3. iLINCS clustering analyses
In order to identify panels of correlated transcriptomic changes in our seed gene knockdown signatures, we used iLINCS to perform unsupervised clustering of the top 50 differentially expressed L1000 genes from our 20 seed gene knockdown signatures (maximum of 1000 L1000 genes). Due to gene overlap between top 50 differentially expressed L1000 genes in seed gene knockdown signatures, this resulted in the clustering of expression data for 601 of the L1000 genes. To generate the clustered heatmap, we used the interactive “gene clusters” function in iLINCS to cluster expression data from 601 differentially expressed genes using Pearson correlation coefficients. Using the y-axis dendrogram function, we selected genes that displayed similar changes in expression across the signatures. We refer to genes that are upregulated across all signatures as “panels of clustered upregulated genes” and genes that are downregulated across all signatures as “panels of clustered downregulated genes.” Panels of clustered upregulated genes and clustered downregulated gene were selected separately and the data were exported to Excel. These clustered gene panels were carried forward to separate Enrichr analyses.
2.5.4. Enrichr analyses of panels of clustered upregulated and downregulated genes from iLINCS clustering analysis
To investigate the biological pathways best describing our panels of upregulated and downregulated genes, we performed three separate Enrichr pathway analyses, using as input either the 40 genes forming the upregulated cluster (Supplementary Table S8), the 38 genes forming the downregulated cluster (Supplementary Table S9), or a combination of both clusters. Results are based on the KEGG 2016 cell signaling pathway database and presented in order of statistical significance of association and number of genes enriched in each pathway.
2.5.5. iLINCS connectivity analyses (drug discovery)
With the goal of identifying small molecules with inverse signatures, we probed iLINCS for chemical perturbagens that result in transcriptomic signatures that are highly discordant (anti-correlated as denoted by negative concordance values) with the kinase network transcriptional signature. In order to do so, we downloaded each individual knockdown gene signature from iLINCS and generated a kinase network transcriptional signature by averaging the log FC values across all seed genes. This average signature was re-uploaded on iLINCS as a new signature, and used to perform perturbagen analyses. We identified the top 20 perturbagens (sorted by concordance score), that are able to reverse the signature associated with the panel of upregulated genes, the panel of downregulated genes, simultaneously affect both the up- and downregulated genes, or anti-correlated with the complete signature.
3. Results
3.1. Decreased serine/threonine protein kinase activity in DISC1 schizophrenia
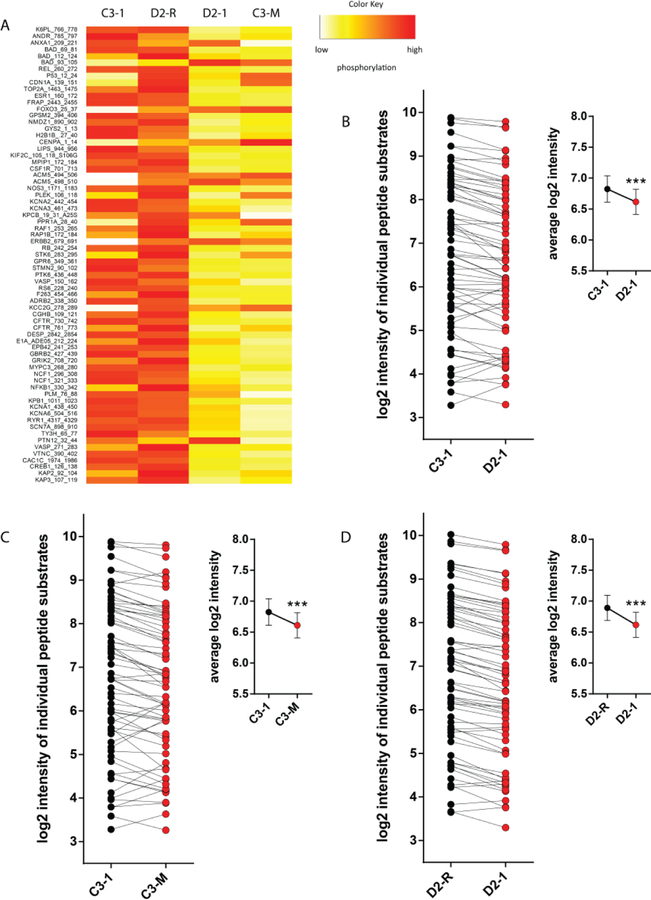
After excluding peptides that were un-detectable or did not increase in signal linearly with exposure time in the post-wash phase, 69 out of the total of 144 substrates on the PamChip kinome array were included in the final analysis. These 69 reported peptides, together with their average levels of phosphorylation across the 3 replicate chips, are depicted in heatmap form in Fig. 2A. We could detect a significant decrease in global phosphorylation levels in the schizophrenia cell line D2–1 compared to the control cell line C3–1 (Fig. 2B; p < 0.001). This decrease in peptide phosphorylation levels was robustly reproduced by introducing the DISC1 mutation in the control cell (C3-M vs. C3–1, Fig. 2C; p < 0.001), as well as rescuing the DISC1 mutation in the schizophrenia cell line (D2–1 vs. D2-R, Fig. 2D; p < 0.001). These findings suggest decreased overall serine/threonine protein kinase activity in DISC1 schizophrenia, causally linked with the DISC1 mutation.
Fig. 2.
Decreased serine/threonine protein kinase activity in DISC1 schizophrenia iPSC cultures. (A) Global phosphorylation heatmap, depicting the relative signal intensity at each reporter peptide for the 4 different cell lines. For ease of clarity, the heatmap is normalized per row to highlight relative changes at each individual peptide between the groups. Orange-red indicates high levels of phosphorylation and yellow-white indicates low levels of phosphorylation. Heatmap generated in R Software version 3.4.2. (B-D) Global phosphorylation plots, showing changes in degree of phosphorylation at each reporter peptide, as well as the average phosphorylation values (inset), when comparing C3–1 with D2–1 (B), C3–1 with C3-M (C), and D2-R with D2–1 (D). Data are presented as mean ± standard error of the mean and analyzed statistically using a paired Student’s t-test. *** p < 0.001.
3.2. Changes in phosphorylation at reporter substrates in DISC1 schizophrenia
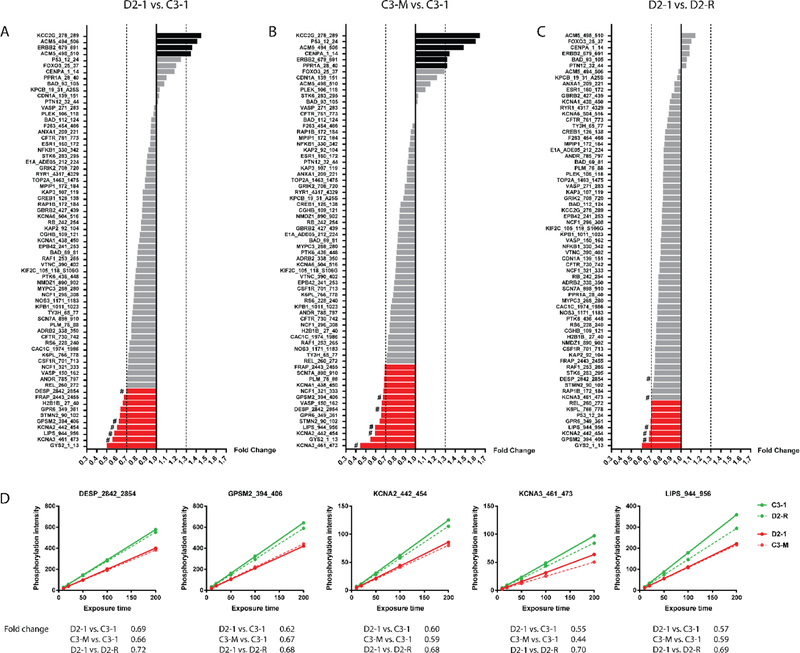
In order to identify substrates with robust changes in magnitude of phosphorylation, we restricted our selection of peptides using a fold change cutoff of ± 30%. This threshold value was guided by previous literature findings suggesting that changes in kinase activity with similar (or smaller) orders of magnitude are sufficient to trigger biologically relevant changes14,22,23. Using this threshold for peptide selection, we identified 14 peptides changed in phosphorylation in D2–1 vs. C3–1 (Fig. 3A), 20 peptides changed in phosphorylation in C3-M vs. C3–1 (Fig. 3B) and8 peptides changed in phosphorylation in D2–1 vs. D2-R (Fig. 3C). Examples of post-wash phosphorylation curves at AMPK-sensitive reporter peptides are indicated in Fig. 3D, together with the fold change values for each comparison considered. Phosphorylation at these substrates was consistently decreased in the DISC1 schizophrenia cell line (D2–1 vs. C3–1), with the pattern being reproduced in the control cell line with the DISC1 4-bp mutation introduced (C3-M vs. C3–1), and the effect rescued in the corrected DISC1 schizophrenia cell line (D2–1 vs. D2-R).
Fig. 3.
Altered kinase activity in DISC1 schizophrenia iPSC cultures. (A-C) Waterfall plots showing changes in degree of phosphorylation at reporter peptides for D2–1 vs. C3–1 (A), C3-M vs. C3–1 (B), and D2–1 vs. D2-R (C). Peptides with increased (FC > 1.30) or decreased phosphorylation (FC < 0.70) are highlighted in black and red, respectively. (D) Representative examples of post-wash phosphorylation curves at five AMPK-sensitive reporter peptides in D2–1 vs. C3–1, C3-M vs. C3–1 and D2–1 vs. D2-R (marked with # in A-C).
3.3. Changes in kinase activity in DISC1 schizophrenia
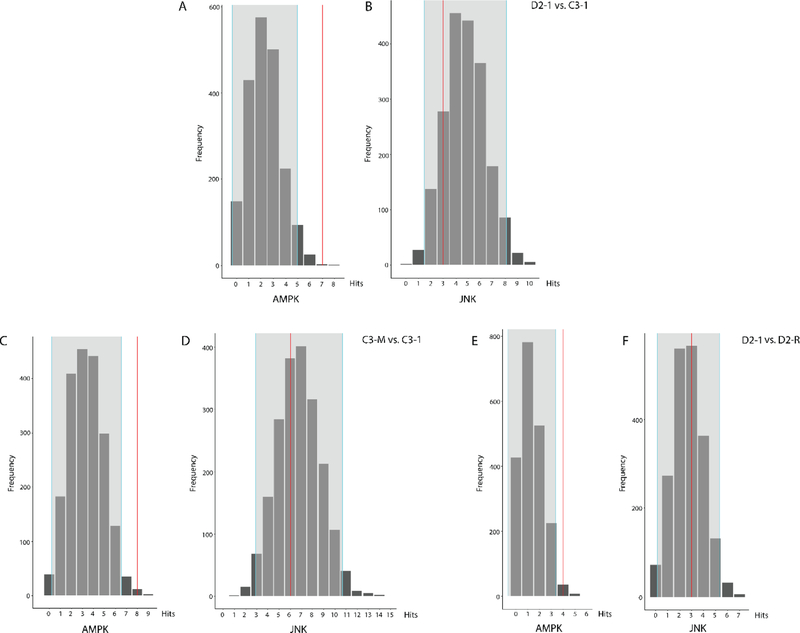
Using the peptide substrates with ± 1.30 fold change in phosphorylation, we performed kinase random sampling analysis to identify the upstream kinases that are most likely driving these effects14,23. This led to the identification of 9 different serine/threonine protein kinase families differentially represented in D2–1 vs. C3–1 (Table 1), 9 kinase families differentially represented in C3-M vs. C3–1 (Table 2), and 8 kinase families differentially represented in D2–1 vs. D2-R (Table 3). Fig. 4 shows examples of histograms reflecting the results of the random sampling analysis for a kinase found to be overrepresented in the 3 different comparisons – AMP-activated protein kinase (AMPK) (Fig. 4A, 4C, and 4E; for each comparison, AMPK was identified more than by random chance alone), as well as for a kinase which was not differentially represented – c-Jun N-terminal kinase (JNK) (Fig. 4B, 4D, 4F; for each comparison, JNK was identified as expected by random chance alone).
Table 1.
Predicted kinases and distributions for D2–1 vs. C3–1.
| Kinase | Observed Hits | Distribution Mean | Standard Deviation | Z-score | Confidence Interval |
|---|---|---|---|---|---|
| AMPK | 7 | 2.31 | 1.33 | 3.51 | −0.36 to 4.98 |
| SGK | 7 | 2.56 | 1.37 | 3.23 | −0.18 to 5.31 |
| KHS | 1 | 0.09 | 0.29 | 3.15 | −0.49 to 0.67 |
| TAO | 1 | 0.10 | 0.30 | 2.96 | −0.50 to 0.71 |
| WNK | 1 | 0.11 | 0.31 | 2.90 | −0.51 to 0.72 |
| DYRK | 1 | 5.36 | 1.67 | 2.61 | 2.02 to 8.70 |
| CAMK1 | 5 | 2.04 | 1.26 | 2.34 | −0.48 to 4.57 |
| ERK | 2 | 5.90 | 1.73 | 2.25 | 2.43 to 9.37 |
| MARK | 2 | 0.56 | 0.70 | 2.06 | −0.85 to 1.96 |
Table 2.
Predicted kinases and distributions for C3-M vs. C3–1.
| Kinase | Observed Hits | Distribution Mean | Standard Deviation | Z-score | Confidence Interval |
|---|---|---|---|---|---|
| AMPK | 8 | 3.38 | 1.58 | 2.93 | 0.22 to 6.53 |
| WNK | 1 | 0.13 | 0.33 | 2.60 | −0.54 to 0.80 |
| TAO | 1 | 0.14 | 0.34 | 2.51 | −0.55 to 0.82 |
| BUD32 | 1 | 0.14 | 0.35 | 2.44 | −0.56 to 0.85 |
| KHS | 1 | 0.15 | 0.36 | 2.35 | −0.57 to 0.87 |
| DYRK | 3 | 7.61 | 2.03 | 2.27 | 3.55 to 11.66 |
| ERK | 4 | 8.38 | 2.06 | 2.13 | 4.26 to 12.51 |
| SGK | 7 | 3.68 | 1.59 | 2.10 | 0.50 to 6.85 |
| MAPKAPK | 11 | 6.98 | 1.98 | 2.03 | 3.01 to 10.94 |
Table 3.
Predicted kinases and distributions for D2–1 vs. D2-R.
| Kinase | Observed Hits | Distribution Mean | Standard Deviation | Z-score | Confidence Interval |
|---|---|---|---|---|---|
| MARK | 3 | 0.35 | 0.57 | 4.69 | −0.78 to 1.48 |
| BUD32 | 1 | 0.05 | 0.22 | 4.34 | −0.39 to 0.49 |
| MOS | 1 | 0.06 | 0.23 | 4.12 | −0.40 to 0.51 |
| MAPKAPK | 7 | 2.78 | 1.28 | 3.30 | 0.22 to 5.34 |
| TTK | 1 | 0.10 | 0.30 | 3.00 | −0.51 to 0.70 |
| CHK1 | 3 | 0.73 | 0.79 | 2.88 | −0.85 to 2.31 |
| NUAK | 1 | 0.11 | 0.32 | 2.79 | −0.54 to 0.75 |
| AMPK | 4 | 1.34 | 1.01 | 2.64 | −0.68 to 3.36 |
Fig. 4.
Observed frequency of selected kinases relative to expected random sampling distribution in D2–1 vs. C3–1 (A, B), C3-M vs. C3–1 (C, D), and D2–1 vs. D2-R (E, F). Examples are shown for a kinase identified in the reporter peptides more than by random chance alone (AMPK; A, C, D), as well as for a kinase identified as expected by random chance alone (JNK; B, D, F). The distribution is derived from 2000 randomly sampled same-size subsets from the kinome array. Gray areas indicate ± 2 standard deviations from the expected distribution mean.
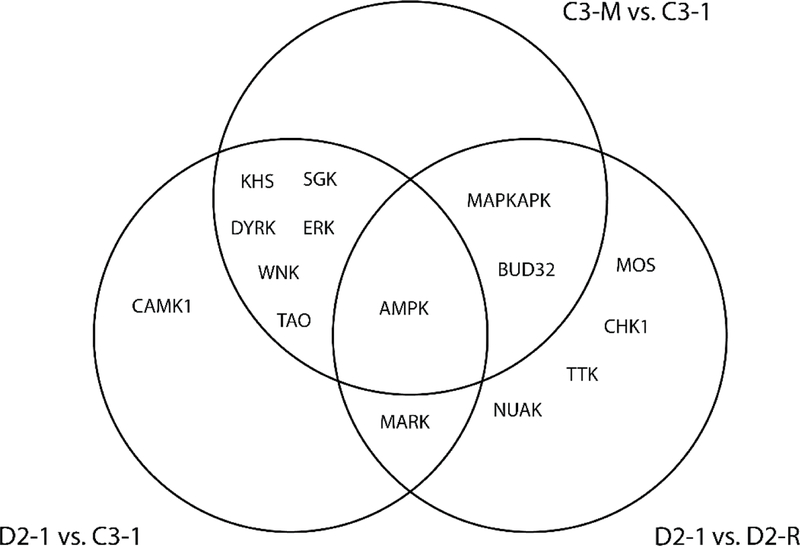
By making an overlap of the kinases identified for the 3 different comparisons (D2–1 vs. C3–1, C3-M vs. C3–1, and D2–1 vs. D2-R), we identified AMPK as a point of convergence, being both disease-linked, as well as strongly related to the DISC1 mutation (Fig. 5). In addition, kinase members of extracellular signal-regulated kinases (ERK), thousand-and-one amino acid (TAO) kinases, KHS, WNK, SGK, and dual-specificity tyrosine phosphorylation-regulated kinases (DYRK) were differentially represented both in DISC1 cells as well as in the control cell line where the DISC1 mutation was introduced (C3-M), MAP/microtubule affinity-regulating kinase (MARK) was differentially represented in both DISC1 cells and the schizophrenia cell line where the DISC1 mutation was rescued (D2-R), while CAMK1 was identified as a schizophrenia kinase not overlapping with either C3-M or D2-R. In addition, NUAK, TTK, CHK1, and MOS were uniquely identified in the rescue cell line (D2-R) compared to the parent schizophrenia cell line (D2–1).
Fig. 5.
Venn diagram highlighting overlap between kinases identified for each of the indicated comparisons.
3.4. Kinase network model of DISC1 schizophrenia
Kinases do not act in isolation, and may be best visualized as part of an interacting network regulating complex biological pathways12. In order to generate a kinase network model that would better reflect kinase network dysregulations characteristic of DISC1 schizophrenia, we connected our initial kinase hits from the kinome array (Tables 1–3) with kinase families to which they are known to interact with using the STRING database. In addition, as kinase networks may be amplified depending on the number of interactions, we weighted the network by the number of connections made by each individual kinase family. The extended kinase network, with the associated kinase interactions, is depicted in Fig. 6. In addition to AMPK, this analysis revealed additional kinase families that act as nodes of regulation of the DISC1 network, including members of AKT, mammalian target of rapamycin (mTOR), STE7, cyclin-dependent kinases (CDK), SRC, protein kinase A (PKA), and class A p21-activated protein kinase (PAKA) families.
Enrichr cell pathway analysis performed using as input the 121 genes that form the extended kinase network (Supplementary Table S1), revealed “MAPK signaling pathway”, “FoxO signaling pathway”, “Neurotrophin signaling pathway”, “Insulin signaling pathway”, “mTOR signaling pathway”, and “AMPK signaling pathway” as top cellular pathways associated with the DISC1 network (Supplementary Table S2).
3.5. In silico analysis of kinase network gene expression changes in DISC1 cells
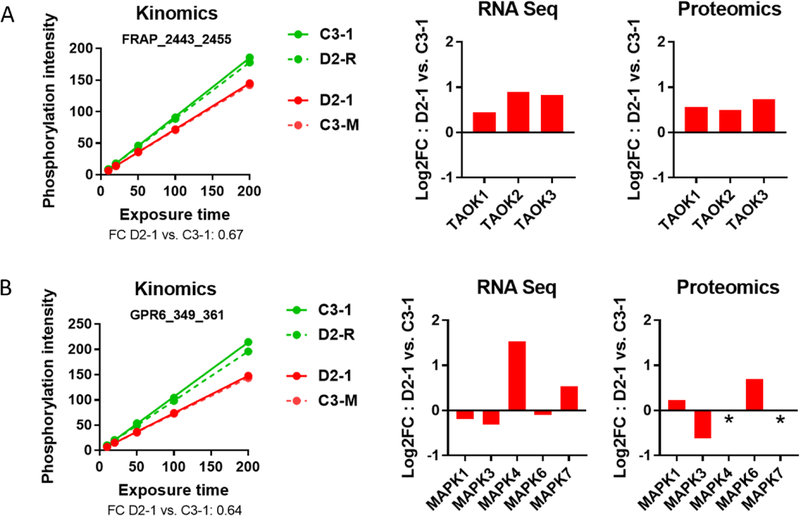
In order to identify whether the changes in kinase activity are linked with transcriptional changes in the corresponding genes, we evaluated the expression of the kinase network genes (Supplementary Table S1), in a previously published RNA-Seq database of transcriptional changes in D2–1 vs. C3–118. This revealed a complex transcriptional signature with a large subset of genes (73 of the 121 kinase network genes; 60.3%) showing changes in expression in DISC1 cells at a ± 0.3 log2 FC threshold (Supplementary Tables S3 and S4). Changes at mRNA level were supported at protein level, with overlap in kinase changes between the RNA-Seq results and a proteomic database of protein expression changes in D2–1 vs. C3–1 (Supplementary Tables S3 and S4) (unpublished observations). These findings support the kinase dysregulation profile we identified using the kinome array, as changes in kinase activity would predict perturbation of the corresponding kinase gene/protein expression. For example, the TAO family of kinases show decreased kinase activity as evaluated on the peptide reporter substrate in the kinome array (FRAP_2443_2455), and increased mRNA and protein expression levels of its corresponding kinase members (TAOK1–3) (Fig. 7). This particular case reveals that changes in kinase activity can be associated transcriptional changes of the corresponding genes possibly as a result of negative feedback regulation.
Fig. 7.
Comparison of kinome array changes with mRNA and protein expression changes for TAO and ERK family of kinases in D2–1 vs. C3–1 cells. A) Post-wash phosphorylation curve for FRAP_2443_2455, a peptide substrate for the TAO family of kinases, and mRNA and protein expression changes of individual members of TAO (TAOK1, TAOK2, TAOK3) in D2–1 vs. C3–1. B) Post-wash phosphorylation curve for GPR6_349_361, a peptide substrate for the ERK family of kinases, and mRNA and protein expression changes of individual members of ERK (MAPK1, MAPK3, MAPK4, MAPK6, MAPK7) in D2–1 vs. C3–1. * no peptide fragments identified in LC-MS/MS.
3.6. In silico analysis of kinase network gene expression changes in postmortem schizophrenia databases
In order to contrast whether the transcriptional changes we detect in the DISC1 kinase genes are consistent with profiles identified in spontaneous cases of schizophrenia, we performed in silico analyses using cell-level or region-level postmortem schizophrenia datasets. First, we analyzed expression of the corresponding genes in 2 cell-level databases. To do so, we probed a recently generated microarray dataset of transcriptional changes in LCM captured pyramidal neurons from deep or superficial layers of the dorsolateral prefrontal cortex (DLPFC) of postmortem schizophrenia (unpublished data), as well as a publicly available microarray database of gene expression changes in an iPSC model of schizophrenia30. In parallel, we used publicly available datasets of gene expression changes in schizophrenia to investigate whether similar changes are mirrored at the region-level. Our findings revealed a heterogeneous kinase expression profile, with partly overlapping patterns of gene expression changes between the different datasets (Supplementary Tables S5 and S6). Although the findings were not always consistent, changes in expression of at least one of the members of AMPK, CAMK1, CAMK2, CAMKK, DYRK, JAKA, JNK, KHS, MAPKAPK, PAKA, SGK, STE7, TAO, and WNK kinase families were transcriptionally changed across at least 4/7 schizophrenia databases, suggesting a more wide-spread kinase dysregulation signature in schizophrenia. At the same time, the gene expression profiles of DISC1 iPSCs and iPSCs generated from spontaneous schizophrenia cases30 were only partially overlapping, hinting at a unique DISC1 transcriptional program driven by the DISC1 mutation.
3.7. iLINCS analyses
In order to identify transcriptional profiles acting downstream of the DISC1 kinase network, we employed iLINCS. iLINCS is an online platform for accessing the LINCS database20, a reference library of cell-based perturbation-response signatures containing data on expression of 978 landmark genes (L1000 genes) following knockdown of genes in different cell lines. This allows the description of transcriptional signatures which are associated with a gene of interest, as expression of the 978 landmark genes can provide insight into changes contained within the entire transcriptome31.
3.7.1. Generation of a DISC1 schizophrenia kinase dysregulation profile
We first generated a DISC1 schizophrenia kinase dysregulation “seed gene” profile, using a combination of our findings from the kinome array (Fig. 6), as well as results from a previous study of transcriptomic changes in D2–1 vs. C3–118. We defined the seed gene profile by the set of genes that encode kinase family members differentially active in our kinome array (based on a ± 1.3 FC in peptide phosphorylation level) and decreased in expression at transcript level in the DISC1 cells (based on a ± 0.3 log2 FC in gene expression level). This revealed a subset of 20 seed genes (Supplementary Table S7), for which knockdown signatures can be accessed from iLINCS. Identifying a consensus transcriptomic signature across all knockdown signatures allowed us to converge on a common set of targets changing downstream of the DISC1 schizophrenia kinase network.
3.7.2. iLINCS clustering analyses
After identifying knockdown signatures for each of the 20 seed genes in iLINCS, we performed unsupervised clustering of the top 50 differentially expressed L1000 genes to identify panels of genes that show similar changes in expression across all signatures. This identified a panel of 40 consistently upregulated genes and a panel of 38 consistently downregulated genes across all knockdown signatures (Supplementary Fig. S2) (see Supplementary Tables S8 and S9 for the corresponding list of genes).
Enrichr analyses identified “Endocytosis”, “TGFβ signaling pathway”, “MAPK signaling pathway”, “FoxO signaling pathway”, and “Calcium signaling pathway” as cell pathways enriched in the panel of upregulated genes (Supplementary Table S10), and “Cell cycle”, “p53 signaling pathway”, and “Pyrimidine metabolism” as cell pathways enriched in the panel of downregulated genes (Supplementary Table S11). Pathway analysis using as input a combination of both clusters of up- and downregulated genes identified “Cell cycle”, “FoxO signaling pathway”, “MAPK signaling pathway, and “Endocytosis” as top enriched cellular pathways (Supplementary Table S12).
3.7.3. iLINCS drug discovery analyses
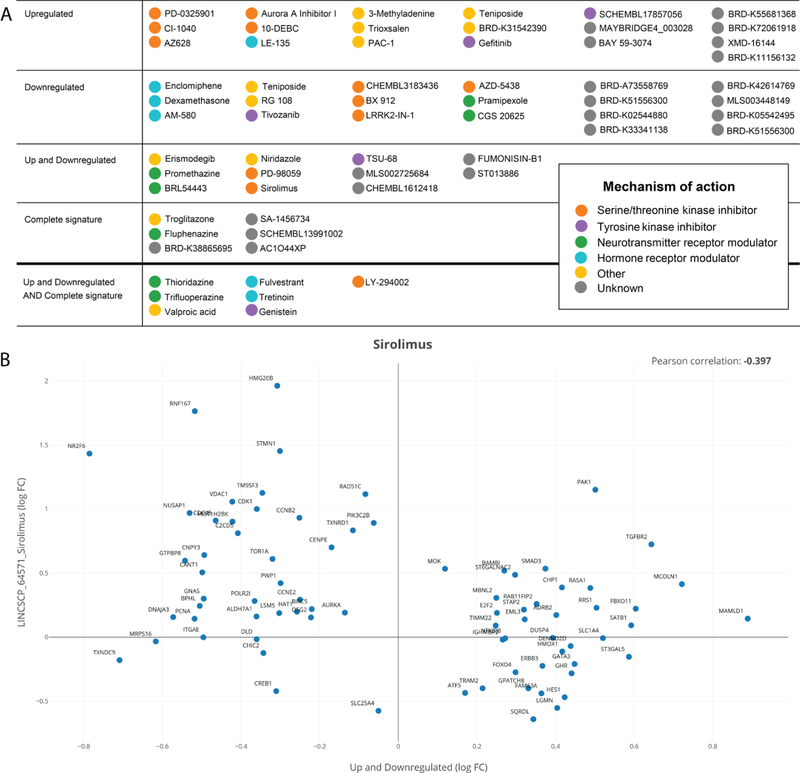
In order to identify perturbagens that are able to reverse the DISC1 schizophrenia kinase network dysregulation signature, we queried iLINCS for the top 20 chemical perturbagens that produce signatures that are highly discordant (anti-correlated) with the transcriptional signature averaged across all seed gene knockdowns. We identified drugs that specifically target the panel of clustered upregulated genes (Supplementary Table S13), the panel of clustered downregulated genes (Supplementary Table S14), simultaneously affect both the up- and downregulated panels (Supplementary Table S15), or are anti-correlated with the complete signature (Supplementary Table S16). An overview of the perturbagens identified, clustered by mechanism of action, is shown in Fig 8. This list of drugs includes MEK inhibitors (PD-0325901, PD-98059, CI-1040), mTOR inhibitors (Sirolimus, LY-294002), protein tyrosine kinase inhibitors (Genistein, Gefitinib), as well as non-kinase modulators, such as estrogen receptor antagonists (Enclomiphene, Fulvestrant), peroxisome proliferator-activated receptor (PPAR) agonists (Troglitazone), and histone deacetylase (HDAC) inhibitors (Valproic acid).
Fig. 8.
iLINCS drug discovery analysis for identifying perturbagens targeting the DISC1 kinase network. (A) Perturbagens predicted to reverse the signature associated with the panel of upregulated genes (drug set 1, first row), the panel of downregulated genes (drug set 2, second row), simultaneously affect the up- and downregulated genes (drug set 3, third row), or target the complete signature (drug set 4, fourth row). Also depicted are perturbagens common to drug sets 3 and 4, listed separately in fifth row. For each category, drugs are clustered by mechanism of action. For a more detailed description of the iLINCS perturbagen results, including concordance scores and p-values, see Supplementary Tables S13–S16. (B) Example of perturbagen concordance plot generated in iLINCS for Sirolimus (LINCSCP_64571) comparing it to the signature defined by the up- and downregulated gene clusters (drug set 3). Each dot represents an individual gene, plotted as log FC values in the perturbagen signature (y axis) vs. the DISC1 kinase network signature (x axis).
4. Discussion
In this study, we provide evidence of kinase network dysregulation in cortical neurons differentiated from iPSCs obtained from a patient presenting with schizophrenia and the 4-bp DISC1 mutation. We show that the overall serine-threonine kinase activity is depressed in patient-derived cells, and this effect can be attributed to the DISC1 mutation. In addition, we identified changes in activity of a wide range of kinase families in the DISC1 cells, including AMPK, ERK, TAO, KHS, DYRK, SGK, WNK, MARK, and CAMK1. Using RNAseq and shotgun LCMS for confirmatory studies, we found changes in expression levels for specific isoforms/subunits of our kinome array hits, including AMPK, KHS, DYRK, ERK, TAO and CAMK1. Integrating the kinome array hits within a larger interaction network highlighted kinase-to-kinase interactions between our array hits and additional kinase families as nodes of network regulation, including members of AKT, mTOR, PAKA, SRC, and STE7. Finally, using the DISC1 schizophrenia kinase dysregulation network model, we interrogated the LINCS database to identify perturbagens that reverse the disease gene expression signature as possible drug interventions targeting the DISC1 kinase network.
Accumulating evidence suggests a role for DISC1 in key processes during early brain development and synapse formation and regulation, including synaptogenesis, neurite outgrowth, neuronal migration and neurogenesis7. However, the precise signaling mechanisms via which DISC1 regulates these processes remain incompletely understood. Changes in serine/threonine kinase signaling have been proposed to mediate some of the downstream effects of DISC1 in the central nervous system. For instance, DISC1 regulates morphogenesis and dendritic development of newborn neurons in the adult brain via the AKT-mTOR pathway33. In addition, DISC1 regulates proliferation of adult neural progenitor cells during embryonic brain development via inhibition of GSK3β34, while data obtained from model systems support an involvement of DISC1 in modulating the activity of kinase families including ERK35,36, PI3K37, and PAKA38. These findings provide an intriguing framework in which changes in serine/threonine kinase signaling mediates effects of DISC1 on brain development and synaptic transmission. However, the full complement of kinome changes downstream of DISC1 has yet to be established.
Our initial observation revealed a small, but consistent, decrease in global peptide phosphorylation across most of the reporter peptides in the DISC1 schizophrenia cells (D2–1) when compared to the control cells (C3–1). Interestingly, this effect was fully mimicked by introducing the DISC1 mutation in the control cell line (C3-M), and restored by correcting the DISC1 mutation in the disease cell line (D2-R), suggesting that the decrease in global phosphorylation levels we observed in the schizophrenia cell line is an effect of the DISC1 mutation. Protein phosphorylation levels are influenced by a carefully regulated balance between protein kinase and protein phosphatase activities. Given that the samples were lysed in the presence of phosphatase inhibitors, our findings support a decrease in kinase activity, rather than an increase in phosphatase activity, in DISC1 cells. As the reporter peptides of the kinome array map to 82 different kinase families (Supplementary Fig. S1), and given that our findings extended across different reporter peptides, these results suggest a more global depression of serine/threonine kinase activity in DISC1 cells. This surprising result, not previously identified in postmortem tissue of chronic schizophrenia patients14,39, might be a particular effect of the DISC1 4-bp mutation, and/or be cell-type specific. While postmortem tissue contains a complex mixture of cell types and extracellular matrix, the DISC1 cells are 90% glutamatergic neurons18, suggesting that the kinase dysregulation signature we identified is mainly driven by changes in neurons. As kinase activity is critical for a wide range of functions including neuronal development and synaptic communication9,10, the global decrease in kinase activity we identified in DISC1 cells may impact a large number of physiological processes, and drive some of the phenotypic differences and functional deficits previously described in this cell line18,19.
Our kinome array identified kinase families in DISC1 schizophrenia cells that were shared with C3-M (ERK, TAO, KHS, WNK, DYRK and SGK), D2-R (MARK), or were uniquely associated with the DISC1 schizophrenia signature (CAMK1). Furthermore, a series of kinase families were causally associated with the DISC1 mutation (MAPKAPK), or were specifically induced by rescuing the DISC1 mutation in the schizophrenia cell line (CHK1, TTK, and NUAK). Interestingly, AMPK emerged as the single strongest hit in our array, overlapping with all 3 comparisons (Fig. 5). While some of the kinases we identified as part of the extended kinase network have been previously implicated in schizophrenia and/or linked with regulation by DISC1 (such as ERK, AKT, mTOR, and PAKA) in agreement with our findings, many of the additional associations are new. These include members of the AMPK, TAO, MARK, NUAK, MAPKAPK, and CHK1 kinase families, which, to our knowledge, have not been previously related to DISC1 function or identified downstream of the DISC1 4-bp mutation. One of the newly identified kinases, AMPK, in particular, was found to have the clearest association, being both schizophrenia-linked as well as directly related to the DISC1 mutation. AMPK is a serine/threonine kinase that acts as a central regulator of cellular energy homeostasis in peripheral tissues as well as the central nervous system40. The activity of AMPK is regulated by the cellular levels of AMP and Ca2+, via the main upstream activators liver kinase B1 (LKB1) and calmodulin-dependent protein kinase kinase β (CaMKKβ). AMPK is activated in conditions of energy deficits and metabolic stress, and acts to restore energy balance by diminishing energy consuming pathways and increasing alternate catabolic pathways that produce ATP41. AMPK is highly expressed in the brain where it plays an important role in regulating processes such as neural polarization42, synaptic transmission and plasticity43,44, and neuronal membrane excitability45,46. In addition, AMPK activity is critically required for proper neuronal development, including axon formation, axon and dendritic growth and dendritic arborization, under conditions of metabolic stress47,48.
Accumulating evidence points to the presence of bioenergetic abnormalities in schizophrenia, including evidence of mitochondrial dysfunction49, and deficits in glucose metabolism, the lactate shuttle, bioenergetic coupling, and ATP metabolism, suggesting that synaptic abnormalities are accompanied by widespread metabolic dysfunction that contributes to disease pathogenesis50. In addition, recent findings provide support for a role for DISC1 in modulating bioenergetic balance in neurons51. DISC1 predominantly localizes to mitochondria52, modulates axonal trafficking of mitochondria53,54, and is essential for oxidative phosphorylation55. Moreover, a reduction in DISC1 function was found to trigger mitochondrial dysfunction, as evidenced by decreases in mitochondrial NADH dehydrogenase activities and ATP production, and perturbed mitochondrial Ca2+ buffering56. It is tempting to speculate that the DISC1 4-bp mutation, leading to strongly decreased levels of DISC118, triggers a state of metabolic stress that determines changes in AMPK activity. Alternatively, due to its function as a sensor of AMP:ATP ratios, changes in activity of AMPK might result from the global decrease in serine/threonine kinase activity that leads to decreased consumption of ATP (as phosphate-donating group) in DISC1 cells. In either case, changes in AMPK could, in turn, impact neuronal function, possibly via downstream mTOR and/or AKT signaling pathways33,57, and explain some of the functional and phenotypic deficits observed in DISC1 loss of function models. While changes in AMPK activity have not been as yet linked with the pathophysiology of schizophrenia, our findings provide the first suggestions that AMPK might be implicated in schizophrenia downstream of the DISC1 4-bp mutation. Further studies, including in patients with spontaneous cases of schizophrenia, are warranted to investigate AMPK as a possible hub integrating genetic risk and metabolic disturbances with aberrant processes neurodevelopment and synaptic plasticity in this disorder.
Using the extended DISC1 kinase interaction model (Fig. 6), in conjunction with a previously published study of transcriptomic changes in the same DISC1 cells18, we aimed to identify the gene expression signature downstream of the DISC1 kinase network. In order to do so, we interrogated LINCS, a large database containing information on the expression of 978 landmark genes (L1000 genes) following knockdown of genes or application of chemical perturbagens in various cell lines20. Using the 20 kinases that were decreased in expression in DISC1 cells as input (Supplementary Table S7), we aligned their corresponding knockdown signatures in iLINCS to identify clusters of up-regulated and down-regulated gene expression modules (Supplementary Fig. S2). Using the generated DISC1 kinase dysregulation signature, we queried iLINCS for perturbagens that may act to reverse the disease gene expression signature. Mapping transcriptomic effects of drugs to disease-specific gene expression signatures has emerged as a novel and powerful methodology in drug discovery58. A recent large-scale computational study in models of cancer demonstrated the feasibility of using LINCS for identifying perturbagens able to reverse the disease gene expression signature as method to generate new therapeutic leads59. Using a similar approach, we screened the LINCS database for chemical perturbagens that are anti-correlated with the DISC1 kinase network dysregulation signature. This revealed a series of candidate drug targets, including MEK inhibitors, mTOR inhibitors, protein tyrosine kinase inhibitors, as well as non-kinase modulators, such as estrogen receptor antagonists, PPAR receptor agonists, and HDAC inhibitors (Fig. 8). We propose that some of these drug candidates might be interesting to pursue further as a means of providing therapeutic benefit. Interestingly, mTOR inhibition is an efficient strategy to reverse the cognitive and affective deficits observed after knocking down DISC1 in adult-born dentate gyrus neurons in vivo57. In addition, changes in mTOR signaling have been suggested to play an etiological role in the pathology of schizophrenia60. Given that mTOR represents one of the main effector pathways via which AMPK connects bioenergetic fluctuations to changes in synaptic transmission43,44,61 and neurodevelopment47,48, we propose that targeting the AMPK-mTOR signaling axis might represent a novel therapeutic approach in schizophrenia.
To augment the discovery-based focus of this kinome array study, we performed confirmatory RNAseq and LCMS studies using two of the cell lines, D2–1 and C3–1. We chose this approach to provide the most comprehensive data for the complex array of kinase families implicated by our kinome array studies. Our confirmatory expression studies show changes in mRNA and/or protein levels for nearly all of the hits from out study, including AMPK, KHS, DYRK, ERK, TAO, and CAMK1. As indicated in the Supplementary Table S3, many of these kinase families have a complex array of subunits and/or isoforms. Future work in cell culture model systems is needed to establish which of these individual kinase isoforms is contributing to changes in activity seen at the network level. The data for TAO kinase highlight the challenges of such studies. We found decreased activity at a peptide mapping to TAO kinase, with increases in mRNA and protein for all TAO isoforms (Fig. 7), suggesting a compensatory response to diminished TAO activity. We also found an increase in mRNA for one of the AMPK subunits (PRKGA2), while our LCMS study did not yield any detectable AMPK subunit peptide fragments (Supplementary Table S3). In general, our findings of disturbed gene and protein expression are in line with the kinase activity differences identified, as changes in enzyme activity are predicted to be associated either causally or consequentially with changes in gene/protein expression.
While the discovery-based platform we employed allows for a high degree of novelty and generation of unforeseen hypothesis on a network scale, it is not amenable to traditional statistical analyses. The variation of Monte Carlo simulation we have used, while being a well-accepted Bayesian statistical approach for data analysis, does not inherently include false discovery corrections. As such, further evaluation of the currently identified targets will be an important step, and future work will need to include direct assessment of kinase activity levels in postmortem schizophrenia brain and models of schizophrenia.
In conclusion, we provide the first comprehensive evaluation of signal transduction changes mediated by serine/threonine kinases in a human iPSC model of DISC1 schizophrenia. Kinome profiling revealed a global depression of serine/threonine kinase activity, and identified, amongst others, members of the AMPK, ERK, and TAO kinase families as differentially active in DISC1 schizophrenia iPSCs. Using an extended kinase interaction model, we provide transcriptional programs that may act downstream of the affected kinases, and identify perturbagens that are able to reverse the disease gene expression signature. Altogether, our findings provide new insight into abnormalities in kinase signaling in schizophrenia and suggest possible targets for disease intervention.
Supplementary Material
Acknowledgments
This work was supported by the following grants: R.E.M.: R01 MH107487 and R21 MH107916; J.H.M.: RC1 MH088752; Z.W.: NIH (U19 AI131130; R01 NS107505), Edward Mallinckrodt, Jr. Foundation, and Emory ADRC (P50 AG025688). We kindly thank Dr. Joshua Anderson of the University of Alabama Birmingham Kinome core for his technical assistance.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Kahn RS et al. Schizophrenia. Nature reviews. Disease primers 1, 15067, doi: 10.1038/nrdp.2015.67 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Bhugra D The global prevalence of schizophrenia. PLoS medicine 2, e151; quiz e175, doi: 10.1371/journal.pmed.0020151 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardno AG & Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. American journal of medical genetics 97, 12–17 (2000). [PubMed] [Google Scholar]
- 4.Blackwood DH et al. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. American journal of human genetics 69, 428–433 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sachs NA et al. A frameshift mutation in Disrupted in Schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Molecular psychiatry 10, 758–764, doi: 10.1038/sj.mp.4001667 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Brandon NJ & Sawa A Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nature reviews. Neuroscience 12, 707–722, doi: 10.1038/nrn3120 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ & Millar JK The DISC locus in psychiatric illness. Molecular psychiatry 13, 36–64, doi: 10.1038/sj.mp.4002106 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Manning G, Whyte DB, Martinez R, Hunter T & Sudarsanam S The protein kinase complement of the human genome. Science 298, 1912–1934, doi: 10.1126/science.1075762 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Baltussen LL, Rosianu F & Ultanir SK Kinases in synaptic development and neurological diseases. Progress in neuro-psychopharmacology & biological psychiatry 84, 343–352, doi: 10.1016/j.pnpbp.2017.12.006 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Giese KP & Mizuno K The roles of protein kinases in learning and memory. Learning & memory 20, 540–552, doi: 10.1101/lm.028449.112 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Chico LK, Van Eldik LJ & Watterson DM Targeting protein kinases in central nervous system disorders. Nature reviews. Drug discovery 8, 892–909, doi: 10.1038/nrd2999 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao Z, Petschnigg J, Ketteler R & Stagljar I Application guide for omics approaches to cell signaling. Nature chemical biology 11, 387–397, doi: 10.1038/nchembio.1809 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Arsenault R, Griebel P & Napper S Peptide arrays for kinome analysis: new opportunities and remaining challenges. Proteomics 11, 4595–4609, doi: 10.1002/pmic.201100296 (2011). [DOI] [PubMed] [Google Scholar]
- 14.McGuire JL et al. Abnormalities of signal transduction networks in chronic schizophrenia. NPJ schizophrenia 3, 30, doi: 10.1038/s41537-017-0032-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen Z, Christian KM, Song H & Ming GL Modeling psychiatric disorders with patient-derived iPSCs. Current opinion in neurobiology 36, 118–127, doi: 10.1016/j.conb.2015.11.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen Z Modeling neurodevelopmental and psychiatric diseases with human iPSCs. Journal of neuroscience research 95, 1097–1109, doi: 10.1002/jnr.24031 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Chiang CH et al. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Molecular psychiatry 16, 358–360, doi: 10.1038/mp.2011.13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen Z et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515, 414–418, doi: 10.1038/nature13716 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao L et al. Disrupted-in-Schizophrenia-1 (DISC1) protein disturbs neural function in multiple disease-risk pathways. Human molecular genetics 26, 2634–2648, doi: 10.1093/hmg/ddx147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keenan AB et al. The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell systems 6, 13–24, doi: 10.1016/j.cels.2017.11.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilhorst R et al. Peptide microarrays for profiling of serine/threonine kinase activity of recombinant kinases and lysates of cells and tissue samples. Methods in molecular biology 977, 259–271, doi: 10.1007/978-1-62703-284-1_21 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Appuhamy JA et al. Effects of AMP-activated protein kinase (AMPK) signaling and essential amino acids on mammalian target of rapamycin (mTOR) signaling and protein synthesis rates in mammary cells. Journal of dairy science 97, 419–429, doi: 10.3168/jds.2013-7189 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Dorsett CR et al. Traumatic Brain Injury Induces Alterations in Cortical Glutamate Uptake without a Reduction in Glutamate Transporter-1 Protein Expression. Journal of neurotrauma 34, 220–234, doi: 10.1089/neu.2015.4372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue Y et al. GPS-SNO: computational prediction of protein S-nitrosylation sites with a modified GPS algorithm. PloS one 5, e11290, doi: 10.1371/journal.pone.0011290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cline MS et al. Integration of biological networks and gene expression data using Cytoscape. Nature protocols 2, 2366–2382, doi: 10.1038/nprot.2007.324 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen EY et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC bioinformatics 14, 128, doi: 10.1186/1471-2105-14-128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgs BW, Elashoff M, Richman S & Barci B An online database for brain disease research. BMC genomics 7, 70, doi: 10.1186/1471-2164-7-70 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roussos P, Katsel P, Davis KL, Siever LJ & Haroutunian V A system-level transcriptomic analysis of schizophrenia using postmortem brain tissue samples. Archives of general psychiatry 69, 1205–1213, doi: 10.1001/archgenpsychiatry.2012.704 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Gandal MJ et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359, 693–697, doi: 10.1126/science.aad6469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brennand KJ et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225, doi: 10.1038/nature09915 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437–1452 e1417, doi: 10.1016/j.cell.2017.10.049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iorio F, Rittman T, Ge H, Menden M & Saez-Rodriguez J Transcriptional data: a new gateway to drug repositioning? Drug discovery today 18, 350–357, doi: 10.1016/j.drudis.2012.07.014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JY et al. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 63, 761–773, doi: 10.1016/j.neuron.2009.08.008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao Y et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell 136, 1017–1031, doi: 10.1016/j.cell.2008.12.044 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashimoto R et al. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Human molecular genetics 15, 3024–3033, doi: 10.1093/hmg/ddl244 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Wang S et al. DISC1 regulates astrogenesis in the embryonic brain via modulation of RAS/MEK/ERK signaling through RASSF7. Development 143, 2732–2740, doi: 10.1242/dev.133066 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Seoane C, Ramos A, Korth C & Requena JR DISC1 regulates expression of the neurotrophin VGF through the PI3K/AKT/CREB pathway. Journal of neurochemistry 135, 598–605, doi: 10.1111/jnc.13258 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Hayashi-Takagi A et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nature neuroscience 13, 327–332, doi: 10.1038/nn.2487 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGuire JL et al. Altered serine/threonine kinase activity in schizophrenia. Brain research 1568, 42–54, doi: 10.1016/j.brainres.2014.04.029 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez M, Nogueiras R, Tena-Sempere M & Dieguez C Hypothalamic AMPK: a canonical regulator of whole-body energy balance. Nature reviews. Endocrinology 12, 421–432, doi: 10.1038/nrendo.2016.67 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Hardie DG AMPK--sensing energy while talking to other signaling pathways. Cell metabolism 20, 939–952, doi: 10.1016/j.cmet.2014.09.013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amato S et al. AMP-activated protein kinase regulates neuronal polarization by interfering with PI 3-kinase localization. Science 332, 247–251, doi: 10.1126/science.1201678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potter WB et al. Metabolic regulation of neuronal plasticity by the energy sensor AMPK. PloS one 5, e8996, doi: 10.1371/journal.pone.0008996 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han Y et al. AMPK Signaling in the Dorsal Hippocampus Negatively Regulates Contextual Fear Memory Formation. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 41, 1849–1864, doi: 10.1038/npp.2015.355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuramoto N et al. Phospho-dependent functional modulation of GABA(B) receptors by the metabolic sensor AMP-dependent protein kinase. Neuron 53, 233–247, doi: 10.1016/j.neuron.2006.12.015 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikematsu N et al. Phosphorylation of the voltage-gated potassium channel Kv2.1 by AMP-activated protein kinase regulates membrane excitability. Proceedings of the National Academy of Sciences of the United States of America 108, 18132–18137, doi: 10.1073/pnas.1106201108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramamurthy S, Chang E, Cao Y, Zhu J & Ronnett GV AMPK activation regulates neuronal structure in developing hippocampal neurons. Neuroscience 259, 13–24, doi: 10.1016/j.neuroscience.2013.11.048 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Williams T, Courchet J, Viollet B, Brenman JE & Polleux F AMP-activated protein kinase (AMPK) activity is not required for neuronal development but regulates axogenesis during metabolic stress. Proceedings of the National Academy of Sciences of the United States of America 108, 5849–5854, doi: 10.1073/pnas.1013660108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ben-Shachar D Mitochondrial dysfunction in schizophrenia: a possible linkage to dopamine. Journal of neurochemistry 83, 1241–1251 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Sullivan CR, O’Donovan SM, McCullumsmith RE & Ramsey A Defects in Bioenergetic Coupling in Schizophrenia. Biological psychiatry 83, 739–750, doi: 10.1016/j.biopsych.2017.10.014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norkett R, Modi S & Kittler JT Mitochondrial roles of the psychiatric disease risk factor DISC1. Schizophrenia research 187, 47–54, doi: 10.1016/j.schres.2016.12.025 (2017). [DOI] [PubMed] [Google Scholar]
- 52.James R et al. Disrupted in Schizophrenia 1 (DISC1) is a multicompartmentalized protein that predominantly localizes to mitochondria. Molecular and cellular neurosciences 26, 112–122, doi: 10.1016/j.mcn.2004.01.013 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Atkin TA, MacAskill AF, Brandon NJ & Kittler JT Disrupted in Schizophrenia-1 regulates intracellular trafficking of mitochondria in neurons. Molecular psychiatry 16, 122–124, 121, doi: 10.1038/mp.2010.110 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Ogawa F et al. DISC1 complexes with TRAK1 and Miro1 to modulate anterograde axonal mitochondrial trafficking. Human molecular genetics 23, 906–919, doi: 10.1093/hmg/ddt485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pinero-Martos E et al. Disrupted in schizophrenia 1 (DISC1) is a constituent of the mammalian mitochondrial contact site and cristae organizing system (MICOS) complex, and is essential for oxidative phosphorylation. Human molecular genetics 25, 4157–4169, doi: 10.1093/hmg/ddw250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park YU et al. Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin. Proceedings of the National Academy of Sciences of the United States of America 107, 17785–17790, doi: 10.1073/pnas.1004361107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou M et al. mTOR Inhibition ameliorates cognitive and affective deficits caused by Disc1 knockdown in adult-born dentate granule neurons. Neuron 77, 647–654, doi: 10.1016/j.neuron.2012.12.033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sirota M et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Science translational medicine 3, 96ra77, doi: 10.1126/scitranslmed.3001318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen B et al. Reversal of cancer gene expression correlates with drug efficacy and reveals therapeutic targets. Nature communications 8, 16022, doi: 10.1038/ncomms16022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gururajan A & van den Buuse M Is the mTOR-signalling cascade disrupted in Schizophrenia? Journal of neurochemistry 129, 377–387, doi: 10.1111/jnc.12622 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Gao XJ et al. AMPK signaling in the nucleus accumbens core mediates cue-induced reinstatement of cocaine seeking. Scientific reports 7, 1038, doi: 10.1038/s41598-017-01043-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.