Abstract
The guanine nucleotide exchange factor (GEF) Son of Sevenless (SOS) is a key Ras activator that is autoinhibited in the cytosol and activates upon membrane recruitment. Autoinhibition release involves structural rearrangements of the protein at the membrane and thus introduces a delay between initial recruitment and activation. In this study, we designed a single-molecule assay to resolve the time between initial receptor-mediated membrane recruitment and the initiation of GEF activity of individual SOS molecules on microarrays of Ras-functionalized supported membranes. The rise-and-fall shape of the measured SOS activation time distribution and the long mean time scale to activation (~50 seconds) establish a basis for kinetic proofreading in the receptor-mediated activation of Ras. We further demonstrate that this kinetic proofreading is modulated by the LAT (linker for activation of T cells)-Grb2-SOS phosphotyrosine-driven phase transition at the membrane.
A key step in the mitogen-activated protein kinase (MAPK) pathway is the activation of the membrane-anchored Ras guanosine triphosphatase (GTPase) by guanine nucleotide exchange factors (GEFs), including Son of Sevenless (SOS) (1–5). SOS, which is an important Ras GEF in T cell receptor (TCR) and epidermal growth factor receptor (EGFR) signaling, is recruited via Grb2 to activated receptors or scaffold proteins on the membrane, where it subsequently activates Ras (5). The influence of SOS dynamics in MAPK signaling is underscored by the relatively low SOS protein copy number in the EGFR-MAPK pathway (6). In addition to its GEF activity, SOS serves as a cross-linking molecule that, by interacting with two Grb2 molecules, forms linkages between receptor or scaffold molecules (7, 8). In the case of the scaffold protein linker for activation of T cells (LAT), multivalency of the phosphorylated tyrosine (pY) sites on LAT, which serve as Grb2 binding sites, enables the networked assembly of LAT-Grb2-SOS into a condensed two-dimensional gel or liquid phase on the membrane surface (9–13). Although similar protein assembly-mediated phase transitions are emerging as a general theme in several different signaling systems (14,15), little is known about how they regulate signaling.
We performed a detailed analysis of the molecular mechanism by which SOS autoinhibition is released after receptor-mediated recruitment to the membrane. We reconstituted purified full-length SOS (SOSFL) along with Grb2 on supported membranes containing phosphorylated LAT and Ras to mimic the signaling geometry of the inner leaflet of the T cell plasma membrane. By developing a single-molecule activation assay in a membrane microarray format (16,17), we resolved the timing process from the initial membrane association to the initiation of GEF activity for each individual SOS molecule. The results reveal a long mean time delay (~50 s) between initial recruitment and the activation of SOS GEF activity. Additionally, the delay time distribution—exhibiting the distinctive rise and fall of a gamma distribution—reveals the existence of rate-limiting kinetic intermediates in the activation of SOS. These features of SOS autoinhibition release, along with the system’s being intrinsically out of equilibrium, provide the basic requirements for a kinetic proofreading mechanism (18) in the activation of Ras (10). In such a mechanism, shortdwelling SOS molecules at the membrane may fail to achieve release of autoinhibition and thus fail to activate any Ras, whereas long-dwelling SOS molecules would exhibit disproportionately higher probabilities of activation. Such a screening process may be especially important for SOS because, once activated, SOS remains trapped at the membrane, where it can processively activate hundreds of Ras molecules (16,19,20).
Lastly, we experimentally demonstrate that the LAT-Grb2-SOS pY-mediated phase transition, which has been shown to extend SOS dwell times (10), strongly modulates the amount of Ras activated by a group of SOS molecules. These observations suggest a mechanism by which protein assembly phase transitions, such as LAT-Grb2-SOS, modulate downstream signaling activity via kinetic discrimination or proofreading processes.
Receptor-mediated activation of SOS is tightly regulated (3, 5, 10, 20–24). The catalytic core of SOS, consisting of the Ras exchanger motif and a CDC25 domain, is flanked by a C-terminal proline-rich (PR) domain and N-terminal autoinhibitory domains consisting of the histone fold, Dbl homology, and Pleckstrin homology domains (Fig. 1A). Mutations in the N-terminal domains of SOS can lead to the dysregulation of autoinhibition and the development of Noonan syndrome disorder (25). The PR domain binds the Src homology 3 (SH3) domains of Grb2, whereas the SH2 domain of Grb2 interacts with pY residues on membrane receptors such as LAT or EGFR, thereby localizing SOS onto membrane surfaces in response to receptor activation (4,26). Upon membrane recruitment, interactions of the SOS N-terminal domains with anionic lipids, such as phosphatidylinositol 4,5-bisphosphate (PIP2), facilitate the release of autoinhibition (21, 27). The catalytic domains of SOS contain two binding sites for Ras: catalytic and allosteric (28). With Ras engaged in the allosteric site, SOS remains on membranes for extended periods of time (>1 min) and enters a highly processive mode in which it catalyzes hundreds of Ras guanosine triphosphate (GTP)-loading events (16, 19, 20). Although there is growing understanding that a multitude of molecular interactions are critical for efficient SOS activation on membranes (5), real-time monitoring of receptor- mediated SOS activation has been inaccessible, in part because of challenges in SOS purification and the development of a membrane assay capable of resolving such features.
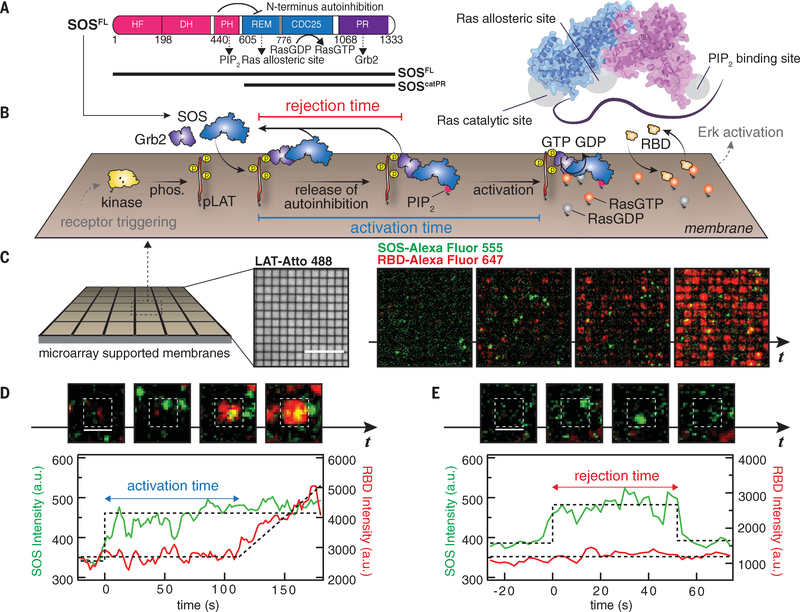
Fig. 1. Single-molecule activation assay of SOSFL on supported membranes.
(A) Domain architecture (left) and a cartoon model (right) of SOSFL The crystal structure was rendered with Protein Data Bank entry 3KSY. HF, histone fold; DH, Dbl homology domain; PH, Pleckstrin homology domain; REM, Ras exchanger motif. Numbers indicate amino acid positions. (B) Schematic of the reconstitution of Grb2-mediated SOS activation. LAT phosphorylated by membrane-bound kinase Hck (pLAT) and Ras preloaded with GDP were decorated on the supported membranes corralled by 1-μm by 1-μm or 2-μm by 2-μm chromium grids. The injection of Grb2, SOSFL-Alexa Flour 555, RBD-K65E-Alexa Fluor 647, and GTP into the solution triggered SOS recruitment, subsequent release of autoinhibition, and activation. Receptor triggering and downstream activation (in gray) are shown to illustrate the signaling pathway, and not included in the experiments. phos., phosphorylation. (C) Snapshot images of SOS (green) and RBD (red) recruitment in microarray supported membranes. Scale bar, 10 μm. t, time. (D) Definition of activation time. (E) Definition of rejection time. [(D) and (E)] Scale bars, 2 μm. a.u., arbitrary units.
We developed a single-molecule activation assay on supported membranes to resolve the timing of SOS activation from membrane recruitment to the initiation of nucleotide turnover in Ras (Fig. 1B). Supported membranes consisting primarily of DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) lipids doped with 2% PIP2, 2% MCC-DOPE {1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidomethyl)cyclohexane-carboxamide]}, and 4% Ni2+-NTA-DOGS {1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl]} were deposited on a glass substrate by vesicle fusion. Cytoplasmic domains of the Src kinase Hck and the adaptor protein LAT, both with six-histidine (His6) tags, were tethered to the supported membrane through His6 interactions with nickel-chelating lipids (29). Although Hck is not an efficient kinase for the phosphorylation of LAT (30), it phosphorylates LAT to completion, given sufficient time. H-Ras molecules were covalently attached to membranes via maleimide chemistry with the MCC-DOPE lipids (31). All of these proteins reconstituted on supported membranes were laterally mobile, with typical densities of <100, ~800, and ~500 μm−2 for Hck, LAT, and Ras, respectively. The LAT densities in the reconstitution are comparable to those measured in cells (32). Although Ras local densities in cells can vary widely, we used densities similar to those of Ras clusters (>1000 μm−2) in cells (33, 34). LAT was fully phosphorylated by Hck before the addition of SOS, as monitored by the recruitment of fluorescent Grb2 (10). SOSFL was prepared by intein ligation (35) of the N-terminal flanked catalytic domain and the C-terminal PR domain of human SOS1. By adding ~1 nM SOSFL and 20 nM Grb2 in solution [the concentrations are comparable to recent quantifications of protein abundance in the EGFR-MAPK pathway (6); dissociation constant Kd ~ 10 μM (36)], single-molecule membrane recruitment of SOS via Grb2 can be clearly resolved by total internal reflection fluorescence (TIRF) microscopy. If Grb2 was not included, we observed ~90% lower SOS recruitment. On membranes, SOS catalyzes the nucleotide exchange of Ras [preloaded with guanosine diphosphate (GDP)] with GTP in solution. Ras-GTP levels are read out via the rapidly reversible binding of a fluorescently labeled Ras-binding domain (RBD) (40 nM in solution). The RBD used in this study is a mutant [Lys65→Glu (K65E)] derived from Raf-1; it has faster binding kinetics than the wild-type RBD (37), providing real-time (~1-s) Ras-GTP density measurements to precisely identify the moment of SOS activation.
We resolve the activity of single SOSFL molecule recruitment events by localizing the reactions into small corrals separating patches of fluid membranes (16) (Fig. 1C). This strategy allows precise assignment of the amount and timing of Ras activation to individual SOS recruitment events. In practice, this was achieved by corralling the supported membranes with 1-μm by 1-μm or 2-μm by 2-μm grid arrays of chemically inert chromium barriers, which are prefabricated onto the underlying substrate before membrane deposition (17). The chromium barriers are typically ~10 nm in height, and although they are effective barriers to the mixing of membrane-associated components, they pose essentially no barrier to the flow of molecules in the adjoining solution (fig. S1). Both LAT and Ras showed small variations (<10%) across different arrays (fig. S1). This membrane microarray strategy also facilitates simultaneous sampling of large numbers (~1000) of nearly identical membrane corrals, all in contact with the same solution.
The simultaneous imaging of SOSFL-Alexa Fluor 555 and RBD-K65E-Alexa Fluor 647 at a frame rate of 0.5 Hz recorded when and where SOS recruitment and activation occurred (Fig. 1, C to E). Once activated, SOS processively catalyzes nucleotide exchange in Ras at a relatively rapid rate, providing a distinctly discernable transition between inactive and active states of SOS (see fluorescence intensity traces in Fig. 1D). Additionally, SOS tends to stay active for an extended period of time (16,19). Specifically, we parsed out corrals with a single SOS recruitment event and then measured the time to the onset of RBD recruitment, indicating the activation of SOS. The SOS recruitment-activation trajectories can be classified into two types: recruitment of SOS followed by activation (Fig. 1D) and recruitment and dissociation of SOS without activation (Fig. 1E). The first type of trajectory reveals the time interval between initial membrane engagement and the release of SOS autoinhibition, enabling GEF activity, which we define as the activation time. The latter trajectory indicates the membrane dwell times for SOS molecules that unbind from the membrane before activating, which we define as the rejection time. The shape of the activation time distribution is especially informative. The existence of rate-limiting intermediates, through which SOS must pass en route to activation, will create a distinctive rise-and-fall shape (gamma distribution), whereas an exponential activation time distribution will indicate that autoinhibition release follows a first-order process. The implication of this kinetic detail is that kinetic proofreading mechanisms, under which long-dwelling molecules are disproportionately more likely to activate, can operate only in the first case, with rate-limiting kinetic intermediates (10,18).
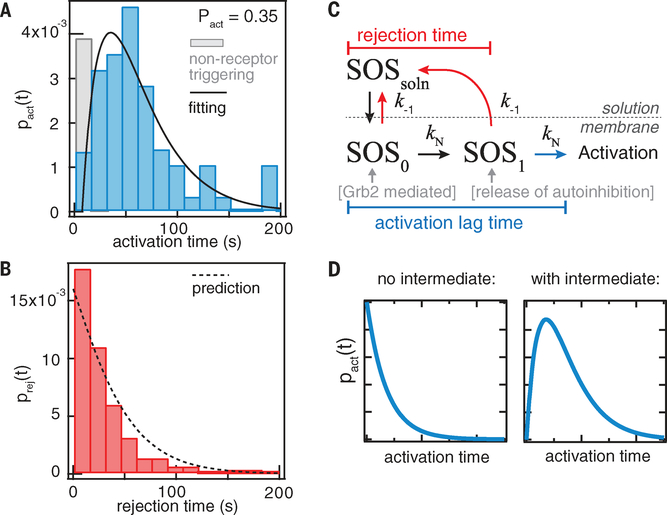
A single SOS recruitment event results in either activation or rejection. By collecting ensembles of these single-molecule recruitment- activation trajectories, we compiled histograms of the activation and rejection time distributions, which also reveal the relative probabilities of these two outcomes (Fig. 2, A and B). The activation time distribution exhibits the rise- and-fall shape of a gamma distribution, with a rather long mean delay of 55 s and a standard deviation of 44 s (Fig. 2A). The time resolution of detectable activation in the current experiment is <2 s, evident because RBD-K56E binding to Ras-GTP has fast binding kinetics (dwell time, ~100 ms) and some trajectories show rapid RBD recruitment (<2 s) after SOS binding to membranes (fig. S3). The rejection time distribution is roughly exponential in shape, with a mean of 30 s and a standard deviation of 29 s (Fig. 2B). The shapes of these distributions reveal underlying mechanistic features of SOS regulation.
Fig. 2. Activation time distribution of SOS.
(A and B) Histograms of the activation time and rejection time of Grb2-mediated SOSFL recruitment from the single-molecule activation assay. The solid line is fitting to the model in (C); the fitted values are kN = 0.02 s−1 and k−1 = 0.016 s−1. The dashed line represents the prediction by the model using fitted values from the activation time distribution. (C) A simple model of SOSFL activation. kN denotes the transition rate constants for the kinetic intermediates, and k−1 represents the dissociation rate constants from membranes. SOSsoln, SOS in solution. (D) Without a kinetic intermediate (N = 0), the activation time distribution is an exponential distribution peaked at t = 0. In contrast, the existence of at least one intermediate produces the characteristic rise-and-decay shape for the activation time distribution.
To quantitatively analyze these measured probability distributions, we construct a simple kinetic model in which the activation of SOS on membranes is described by the competing kinetics between activation and dissociation from membrane surfaces (Fig. 2C) (for derivations, see materials and methods) (10, 38, 39). In this model, SOS binds to membranes initially in an autoinhibition state (SOSo), and the release of autoinhibition is achieved after overcoming a series of kinetic intermediates. Alternatively, SOS may unbind from the membrane, going back into solution, where it eventually relaxes back to its native auto- inhibited state (SOS0). Given N number of kinetic intermediate(s) in the activation pathway, the activation time distribution has the form
where pN(t) is the activation time distribution for a linear multistep process, arising from the convolution of (N + 1) single Poisson steps with rate constants ki and k−1 is the dissociation rate constant. The overall probability of SOS activation, Pact, depends on both the transition rate (ki) and the unbinding rate (k−1) and is given by . A similar expression defines the overall probability of rejection, Prej, in terms of the rejection time distribution, prej(t), and the integrated sum of these two distributions normalizes to one.
In a case with a single rate-limiting step or multiple steps with similar rates, pN(t) approaches a gamma distribution, . This approximation provides a useful intuition about the shape of the activation time distribution: The existence of intermediate(s) predicts a rise-and-fall shape to the distribution (Fig. 2D). In the alternative case, without any kinetic intermediates (N = 0), the activation time distribution is strictly exponential (Fig. 2D). The observed data (Fig. 2A) are consistent with the first case, indicating that SOS activation on membranes involves progressing through at least one rate- limiting kinetic intermediate, with a low transition rate (kN) of 0.02 s−1. In practice, the kinetic observation will be dominated by the slowest kinetic step for a unidirectional activation mechanism (20). Thus, we used N = 1 to analyze the distributions, i.e., . If this model is sufficient to describe SOS activation, the fitted values from the activation time distribution should, with no additional fît parameters, predict the rejection time distribution (Fig. 2B), which is given by
where , the upper incomplete gamma function. In the case of N =1, . The experimentally measured rejection time distribution is well described by the prediction based on parameters measured from the activation time distribution (Fig. 2B). The activation time distribution of SOS is indicative of an activation mechanism dominated by a single rate-limiting kinetic intermediate. This model does not exhaust all possible routes of activation but delineates the main pathway. In addition, it is possible that SOS dissociation from membranes is a multistep process, but it is likely much faster than the activation time scale such that the data are well described by the simple model.
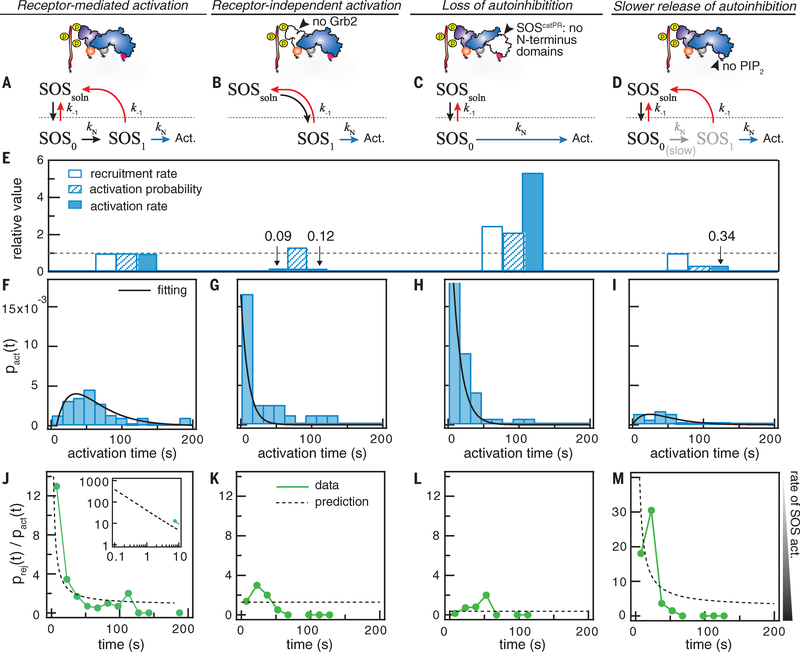
The activation time of SOS is determined, in part, by the release of autoinhibition of the catalytic domains by the N-terminal domains. We compared Grb2-mediated SOS recruitment and activation with three other orthogonal activation profiles (Fig. 3 and fig. S2): (i) receptor- independent activation without Grb2 (Fig. 3B), (ii) activation without N-terminal autoinhibition using the construct SOScatPR (Figs. 1A and 3C), and (iii) hindered activation without PIP2 (Fig. 3D). Although the recruitment rate of SOS is markedly enhanced by Grb2 (Fig. 3E), these three cases exhibit fundamental differences in the activation time compared with Grb2-dependent activation (Fig. 3, F to I). Cases (i) and (ii) result in single-step activation (i.e., the activation time distribution is exponentially shaped) over the experimental time scale of 1 to 200 s. Without Grb2, spontaneous activation of SOSFL is (rarely) observed as a result of SOS apparently fluctuating into an active configuration before interacting with the membrane. In this state, SOSFL binds to Ras directly and begins to processively activate Ras without the need to pass through the kinetic intermediate. This detail is revealed by the exponential shape of the activation time distribution for these spontaneous activation events. Before successful purification of SOSFL, these are the only activation events that have been observed in prior studies (16, 20). The SOScatPR construct lacks N-terminal autoinhibition and also apparently lacks the kinetic intermediate, as revealed by its exponentially shaped distribution (Fig. 3, G and H). Case (iii), lacking PIP2 in the membrane, exemplifies the scenario where the activation pathway on the membrane is hindered, as revealed by a decreased activation rate, yet the kinetic bottleneck is retained, as revealed by the rise-and-fall shape of the activation time distribution (Fig. 3I).
Fig. 3. The regulation of autoinhibition defines the activation timing of SOS and its kinetic proofreading capability.
The top schematics show the experimental design (details are in fig. S2). (A to D) The kinetic models for Grb2-mediated SOSFL activation, SOSFL activation without Grb2, SOScatPR activation, and SOSFL activation without PIP2. Act., activation. (E) The relative recruitment rate, activation probability, and activation rate for each condition. Activation rate = recruitment rate × activation probability. The Grb2-mediated recruitment rate of SOSFL was 3 × 10−3 s−1 μm−2. The dashed line is at 1. (F to I) The activation time distribution for each condition. Black lines are fitting to the models in (A) to (D). (F) is the same data from Fig. 2A. (J to M) The ratio between rejection and activation counts as a function of SOS dwell time. The inset in (J) shows the model’s extrapolation for very short dwelling SOS. Dashed lines are predictions by the models in (A) to (D).
Together, these data provide a molecular description of the autoinhibition release process of SOSFL: The first recruited state (SOSo) corresponds to Grb2-mediated recruitment while under the protection of autoinhibition. The slowest intermediate (SOS) involves PIP2-mediated release of autoinhibition (Figs. 2C and 3, A to D). Finally, fully activated SOS presumably involves locking Ras into the allosteric pocket (16, 23), enabling the processive catalysis.
A multistep activation process, such as we have observed for SOSFL, can, in principle, lead to longer-dwelling molecules having disproportion- ally higher activation rates (10)—this is classically known as kinetic proofreading (18). To test for this behavior experimentally, we examined the ratio prej(t)/pact(t) as a function of SOS dwell time from the corral experiments. This analysis compares the rate (probability per unit of time) of SOS activation for different dwell times (Fig. 3, J to M). The kinetic intermediate defined by autoinhibition release greatly enhances the proofreading capability, as evident from the decaying functional form of the prej(t)/pact(t) curve for SOSFL (Fig. 3, J and M). By contrast, a weakly discriminating process will have a near-constant curve, as observed for SOScatPR molecules lacking kinetic intermediates (Fig. 3, K and L). These data provide evidence that SOSFL activity is capable of being modulated by a kinetic proofreading mechanism.
With a proofreading mechanism at play, the rate of Ras activation depends disproportionately on the membrane dwell times of SOS. Given that the individual pY-Grb2 interactions exhibit fast kinetics [mean dwell time, ~0.1 s (10)] compared with the mean activation time of SOS measured in this study (~55 s), we speculate that monovalent recruitment of SOS to the membrane by Grb2 is unlikely to lead to Ras activation. However, SOS dwell time on the membrane can be greatly elongated when SOS interacts with more than one Grb2 molecule. Single-molecule tracking of SOS shows that the initial stable recruitment of SOS (dwell time > 1 s) is mediated primarily by Grb2 instead of PIP2 or Ras (figs. S4 and S5). Furthermore, mobility analysis indicates that stably recruited SOS is interacting with at least two LAT molecules via multiple Grb2 molecules (fig. S6).
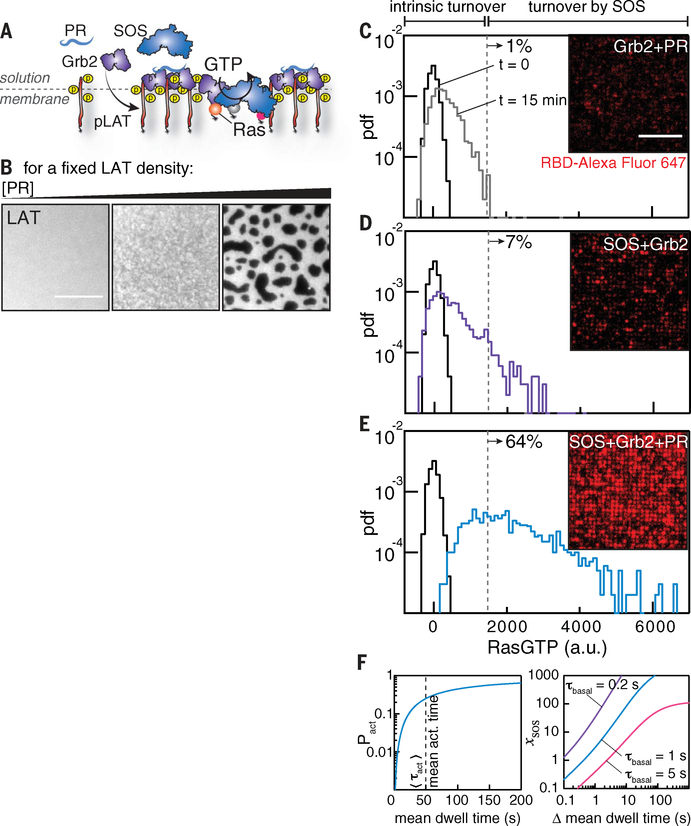
Recent studies have vividly demonstrated that LAT-Grb2-SOS can undergo a phase transition to form a condensed two-dimensional liquid or gel state consisting of a networked molecular assembly on the membrane surface (9–13). A distinctive feature of these condensed phases is that multivalent interaction of SOS within the network can markedly elongate SOS dwell time on the membrane (10). To explore the effect of this protein assembly network on Ras activation, we added the PR domain of SOS, which has no intrinsic catalytic activity but allows formation of the assembly, into the corral experiments (Fig. 4, A and B). If the PR domain acted solely as a competitor, titrating it would decrease Ras activation by outcompeting the fixed concentration of SOSFL. Instead, we observed that the rate of Ras activation increased substantially when the PR domain was added (50 nM) (Fig. 4, C to E), suggesting that LAT assemblies promote Ras activation. Notably, this enhanced activation by the PR domain was absent at low LAT densities, where assemblies cannot form; titrating the PR domains to a higher concentration (≥100 nM in our experiments) also abolished this enhancement by outcompeting SOSFL (fig. S7). Furthermore, the twofold increase in SOSFL recruitment with the PR domain is insufficient to explain the eightfold increase in Ras activation (Fig. 4, C to E).
Fig. 4. Molecular assembly enhances Ras activation by SOS.
(A and B) LAT assemblies are tuned by the addition of Grb2 and the PR domain of SOS, which enables multivalent cross-linking but has no intrinsic catalytic activity. Images represent LAT-Alexa Flour 555 after the addition of Grb2 and the PR domain. (C to E) Snapshot images of RBD after 15 min of reaction time. The histograms show the RBD intensities per corral of the images. The dashed lines are guidelines for the maximum extent of intrinsic nucleotide turnover. The percentages show the number of corrals with turnover above the level of the dashed line. Scale bar, 10 μm. pdf, probability density function. (F) Calculation of the probability of SOS activation as a function of Grb2-mediated mean dwell time (left) and the increase in SOS activity as a function of the change in mean dwell time (right). The change in mean dwell time is defined as .
To quantitatively account for the effect of Grb2-mediated SOS dwell time at the membrane on the probability of SOS activation, we first evaluated the activation probability of SOS per recruitment event, is intrinsically a function of the Grb2-mediated SOS dwell time, as longer-dwelling SOS molecules have a higher likelihood of activation before unbinding. The functional dependence of Pact on the mean dwell time is plotted in Fig. 4F, on the basis of the measured activation time distribution of SOS (Fig. 2) (N =1 intermediate). This calculation reveals that Pact is highly sensitive to the SOS dwell time when the mean activation time is substantially longer than the mean dwell time, which is the case in our reconstitution and likely in live cells as well (14). Next, we evaluated the fold increase in SOS activity per molecule, xSOS (the definition is provided in materials and methods), as a function of change in the mean dwell time. Figure 4F enables a direct comparison of SOS activation probabilities in the unassembled and assembled LAT configurations in Fig. 4, D and E. Single-molecule analysis reveals that the fraction of long-dwelling species of SOS increases with the addition of the PR domain (and consequent LAT assembly) in this experiment (fig. S8), although the degree of molecular assembly is not as large as that observed in a fully condensed phase (10). The roughly fourfold net increase in Ras activation per SOS molecule (after accounting for recruitment) is comparable to the estimation on the basis of mean dwell time changes (from ~4 to 6 s). Thus, relatively small changes in the SOS dwell time lead to large changes in the probability of activation because the system is operating under conditions in which the overall probability of activation is low (e.g., the mean dwell time is substantially shorter than the mean activation time). Monovalent recruitment interactions between SOS and phosphorylated LAT via Grb2 are brief (<1 s) (10) and will thus have an exceptionally low probability of successful SOS activation. In the fully condensed state, the LAT-Grb2-SOS assembly can elongate SOS dwell times by more than an order of magnitude (10), which is even more than that achieved in the PR domain experiments presented here. Collectively, these data suggest that LAT-Grb2-SOS assembly can exert a powerful influence over SOS activation by modulating the SOS dwell time at the membrane.
In resting T cells, LAT typically exhibits some basal level of phosphorylation (40). A proofreading mechanism in SOS activation can prevent the spontaneous membrane localization of SOS from activating Ras and thus prevent upstream basal pY activity from activating certain downstream pathways (fig. S9). This kinetic proofreading at the level of SOS is different from kinetic proofreading at the TCR, which has been proposed to contribute to antigen discrimination on the basis of peptide major histocompatibility complex-TCR binding kinetics (41).
The type of kinetic proofreading identified here in the activation of Ras by SOS reveals a potentially broad mechanism by which membrane recruitment kinetics can be used to modulate downstream signaling activity. Many membrane-associated signaling molecules activate in a multistep fashion—for example, Vav (GEF for Rho family GTPase) (42), Raf (43), class I protein kinase C-α (44), phosphoinositide 3-kinases (45), and inducible T cell kinase (46). The initiation of actin nucleation by the Arp2/3 complex is also a slow multistep process. Notably, the actin regulator neuron Wiskott-Aldrich syndrome protein (N-WASP) and the Arp2/3 complex have recently been shown to exhibit dwell time-dependent activity modulation by nephrin-Nck-N-WASP condensation phase transition, in a mechanism highly parallel to that described here for Ras and SOS (15,47). Hence, we speculate that the timing and proofreading mechanism observed in this study is not exclusive to SOS but is a common theme in signal regulation at the membrane.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Rosen and L. Case for their insightful comments and for communicating results before publication. We thank H.-K. Chiang for assistance in the pilot experiments. We thank S. Hansen, M. Triplet, and other members in the Groves lab and Kuriyan lab for helpful discussion.
Funding: This work was supported by National Institutes of Health (NIH) National Cancer Institute (NCI) Physical Sciences in Oncology Network (PS-ON) project 1-U01CA202241 and by the Novo Nordisk Foundation Challenge Program under the Center for Geometrically Engineered Cellular Systems. Additional support was provided by NIH grant PO1 A1091580.
Footnotes
Competing interests: None declared.
Data and materials availability: All data necessary to evaluate the conclusions in the paper are available in the paper or the supplementary materials.
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/363/6431/1098/suppl/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S9
References (48–54)
Movie S1
REFERENCES AND NOTES
- 1.Samelson LE, Annu. Rev. Immunol 20, 371–394 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J, Cell 103, 211–225 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Sondermann H et al. , Cell 119, 393–405 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Gale NW, Kaplan S, Lowenstein EJ, Schlessinger J, Bar-Sagi D, Nature 363, 88–92 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Bandaru P, Kondo Y, Kuriyan J, Cold Spring Harb. Perspect. Med 9, a031534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi T et al. , Sci. Signal 9, rs6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houtman JCD et al. , Nat Struct Mol. Biol 13, 798–805 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Kortum RL et al. , Sci. Signal 6, ra99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nag A, Monine MI, Faeder JR, Goldstein B, Biophys. J 96, 2604–2623 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang WYC et al. , Proc. Natl. Acad. Sci. U.S.A 113, 8218–8223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su X et al. , Science 352, 595–599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang WYC, Chiang HK, Groves JT, Biophys. J 113, 1807–1813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang WYC, Ditlev JA, Chiang HK, Rosen MK, Groves JT, J. Am. Chem. Soc 139, 18009–18015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer BJ, Yu J, J. Mol. Biol 430, 4547–4556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banjade S, Rosen MK, eLife 3, e04123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iversen L et al. , Science 345, 50–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groves JT, Ulman N, Boxer SG, Science 275, 651–653 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Hopfield JJ, Proc. Natl. Acad. Sci. U.S.A 71, 4135–4139 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YK et al. , Nat. Commun 8, 15061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen SM et al. , Nat. Struct. Mol. Biol 23, 838–846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gureasko J et al. , Proc. Natl. Acad. Sci. U.S.A 107, 3430–3435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groves JT, Kuriyan J, Nat. Struct. Mol. Biol 17, 659–665 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gureasko J et al. , Nat. Struct. Mol. Biol 15, 452–461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yadav KK, Bar-Sagi D, Proc. Natl. Acad. Sci. U.S.A 107, 3436–3440 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tartaglia M et al. , Nat. Genet 39, 75–79 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Houtman JCD et al. , Biochemistry 43, 4170–4178 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D, Nat. Cell Biol 9, 707–712 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Margarit SM et al. , Cell 112, 685–695 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Nye JA, Groves JT, Langmuir 24, 4145–4149 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Shah NH et al. , eLife 5, e20105 (2016).27700984 [Google Scholar]
- 31.Lin WC et al. , Proc. Natl. Acad. Sci. U.S.A 111, 2996–3001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williamson DJ et al. , Nat. Immunol 12, 655–662 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Gurry T, Kahramanogullari O, Endres RG, PLOS ONE 4, e6148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho KJ, Hancock JF, Small GTPases 4, 57–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens AJ et al. , J. Am. Chem. Soc 138, 2162–2165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonald CB et al. , Biochemistry 51, 2122–2135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaitner BK et al. , J. Biol. Chem 272, 29927–29933 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Moffitt JR, Chemla YR, Bustamante C, Methods Enzymol. 475, 221–257 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Kou SC, Cherayil BJ, Min W, English BP, Xie XS, J. Phys. Chem. B 109, 19068–19081 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Lin J, Weiss A, J. Biol. Chem 276, 29588–29595 (2001). [DOI] [PubMed] [Google Scholar]
- 41.McKeithan TW, Proc. Natl. Acad. Sci. U.S.A 92, 5042–5046 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner M, Billadeau DD, Nat. Rev. Immunol 2, 476–486 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Cutler RE Jr., Stephens RM, Saracino MR, Morrison DK, Proc. Natl. Acad. Sci. U.S.A 95, 9214–9219 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziemba BP et al. , Biochemistry 53, 1697–1713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vadas O, Burke JE, Zhang X, Berndt A, Williams RL, Sci. Signal 4, re2 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Andreotti AH, Schwartzberg PL, Joseph RE, Berg LJ, Cold Spring Harb. Perspect. Biol 2, a002287 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Case LB, Zhang X, Ditlev JA, Rosen MK, Science 363, 1093–1097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.