Abstract
The transcriptome of a cell dictates its unique cell-type biology.. We used single-cell RNA sequencing to determine the transcriptomes for essentially every cell type of a complete animal: the regenerative planarian Schmidtea mediterranea. Planarians contain a diverse array of cell types, possess lineage progenitors for differentiated cells (including pluripotent stem cells), and constitutively express positional information, making them ideal for this undertaking. We generated data for 66,783 cells, defining transcriptomes for known and many previously unknown planarian cell types and for putative transition states between stem and differentiated cells. We also uncovered regionally expressed genes in muscle, which harbors positional information. Identifying the transcriptomes for potentially all cell types for many organisms should be readily attainable and is a powerful new approach to metazoan biology.
One Sentence Summary
Single-cell RNA sequencing identifies the transcriptomes for most-to-all cell types of the planarian Schmidtea mediterranea.
The complete sequence of animal genomes, such as that of C. elegans reported in 1998 and humans in 2001, has had an immeasurable impact on research (1-3). Whereas the genome sequence of an organism contains the information for its development and physiology, the transcriptomes (the sets of actively transcribed genes) of the cell types in an organism define how the genome is utilized for the unique functions of its cells. Recent advances in RNA sequencing of individual cells have greatly enhanced the ability to determine cell-type transcriptomes (4, 5), and single-cell RNA sequencing (SCS) of thousands of cells has become readily achievable (6). For example, the transcriptomes of most cell types of complete C. elegans L2 larvae and numerous mouse cells were recently reported with this approach (7, 8). We reasoned that it might be possible, given these advances, to determine the transcriptomes of essentially every cell type of a complete adult organism possessing an unknown number of cell types.
Multicellular organisms can have many millions of cells and hundreds of different cell types, and the cellular composition of organisms varies dramatically over the course of development. This complexity has historically made the identification of all cell types, much less their transcriptomes, for most multicellular organisms an extreme challenge. The planarian Schmidtea mediterranea is an attractive case study organism for which to generate the transcriptomes for all cells in an animal. Planarians are famous for their ability to regenerate essentially any missing body part and possess a complex body plan containing many characterized cell types (9, 10). Despite this complexity, with an average planarian possessing ~105-106 cells (11), planarians are smaller with simpler anatomy than humans and many other model systems such as mice. Planarians are also easily dissociated into single-cell suspensions, allowing potential characterization of all cells. Because some planarian cell types, such as glia (12, 13), have only recently been defined with molecular markers, it is probable that undescribed planarian cell types exist. The combination of known and potentially unknown cell types is attractive for developing approaches that can apply to diverse organisms with varying amounts of available cell type information. Planarians possess a population of proliferative cells called neoblasts that contain pluripotent stem cells, enabling their ability to regenerate and replace aged cells in tissue turnover (14). Neoblasts are the only cycling somatic cells and the source of all new tissue. Neoblasts contain multiple classes of specialized cells, with transcription factors expressed to specify cell fate (15, 16). Because of the constant turnover of planarian tissues, essentially all stages of all cell lineages from pluripotent stem cell to differentiated cell are anticipated to be present in the adult (9, 17). Planarians also constitutively and regionally express dozens of genes that have roles in positional information (18). These genes, referred to as positional control genes (PCGs), are expressed in a complex spatial map spanning anterior-posterior (AP), medial-lateral (ML), and dorsal-ventral (DV) axes (18), and their expression is largely restricted to muscle (19). PCGs are hypothesized to constitute instructions for the maintenance and regeneration of the body plan. Because of these features, comprehensive SCS at a single time point (the adult) could allow transcriptome identification for all differentiated cell types, lineage precursors for these cells, and the patterning information guiding new cell production and organization. To capture this information in most organisms would require sampling the adult and many transient stages of embryogenesis.
Single-cell RNA sequencing of 50,562 planarian cells
Planarians have a complex internal anatomy including a brain, ventral nerve cords, peripheral nervous system, epidermis, intestine, muscle, an excretory system (the protonephridia), and a centrally located pharynx (10). These major tissues are composed of multiple different cell types that, together with other gland and accessory cells, comprise the planarian anatomy.
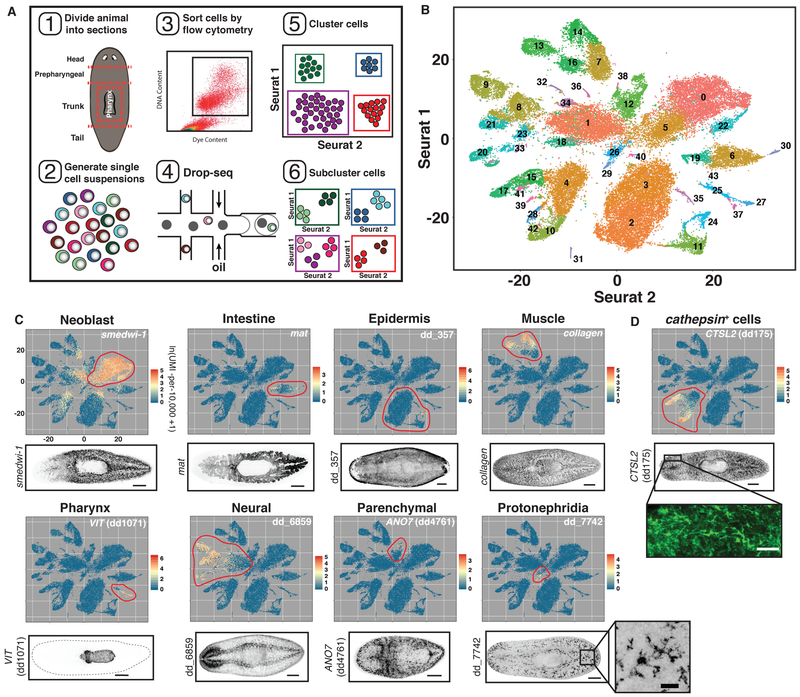
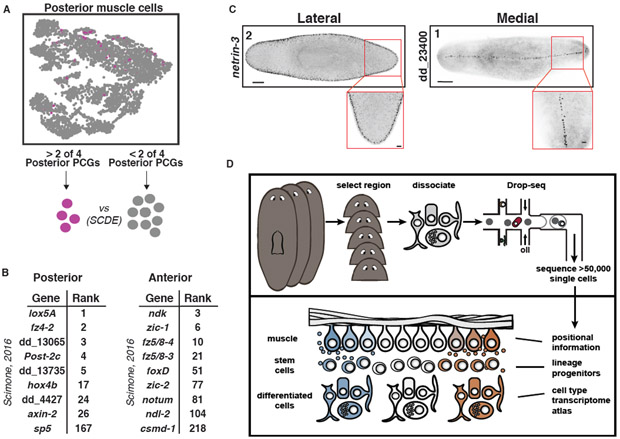
To detect planarian cell types and states in an unbiased manner, including rare cell types, we used the SCS method Drop-seq (6) to determine the transcriptomes for 50,562 individual cells from adults (Fig 1A, Fig. S1A, Table S1). Planarians contain 105106 cells (11), and yet some cell types are extremely rare, such as the ~100 photoreceptor neurons of eyes (20). Given such rarity, sequencing random cells from entire animals might not reach cell type saturation with even 105 cells sequenced. Therefore, we divided animals into five sections (head, prepharyngeal region, trunk with pharynx removed, tail, and the pharynx itself) and cells from each region were dissociated, sorted by flow cytometry, and sequenced (Fig. 1A, Fig S1A, Table S1). Sequences were aligned to a previously assembled transcriptome (21). We targeted cell type saturation by assessing coverage of known, rare cell types during iterative rounds of cell isolation and sequencing in a region-by-region approach. In total, 25 separate Drop-seq runs were completed, yielding cells with an average of 3,020 unique molecular identifiers (UMIs) and 1,404 genes on average (~13% of the estimated detection limit) (Fig. S1A-C, Table S1, Methods).
Fig. 1. Drop-seq of 50,562 planarian cells.
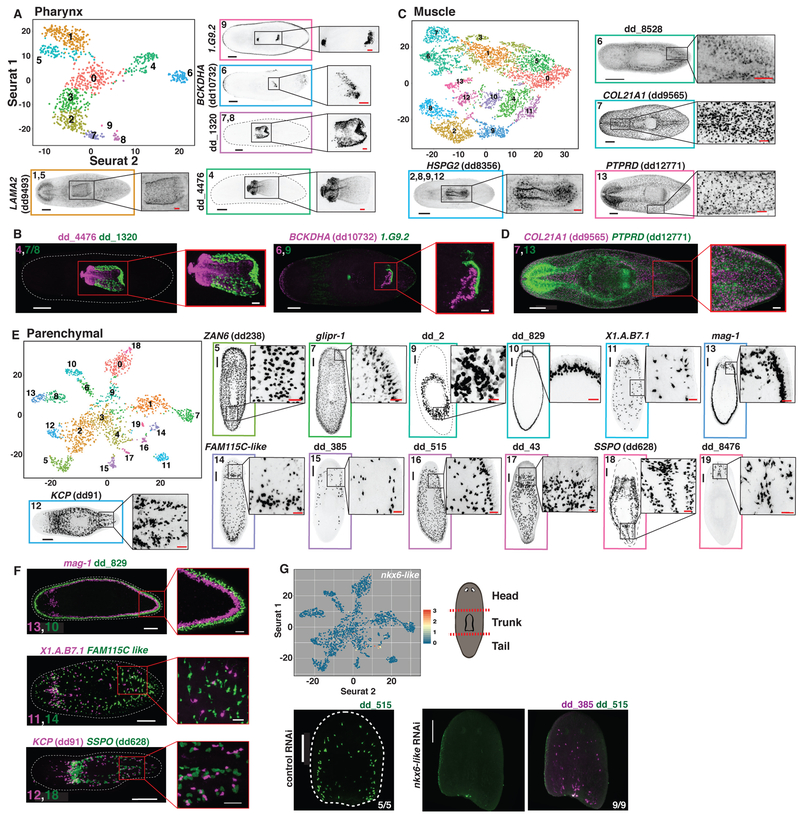
(A) Schematic illustrating the workflow used to isolate and cluster single cells. (B) t-SNE representation of 44 clusters generated from the data. (C and D) Top: t-SNE plots, colored by gene expression for highly enriched genes from 9 planarian tissue classes (Red to yellow to blue indicates high to medium to low expression). Red circles denote clusters assigned to that tissue class. Bottom: FISH images for tissue-enriched genes. Scale bars: whole-animal images, 200 μm; insets, 50 μm.
Genes with high variance and expression across cells were used to generate informative principal components using Seurat (6, 22). Cells were clustered using Seurat into 44 distinct major clusters using a graph-based clustering approach and were visualized by applying t-distributed stochastic neighbor embedding on transcriptomes (t-SNE; Methods) (Fig. 1B, Fig. S1D). Cells from different regions were largely interspersed in the t-SNE plots, except for cells from the pharynx, which contains many unique cell types (Fig. S2A). Cell doublets were scarce within the data and did not affect clustering results (Fig. S2B-D). To determine the identity of each cluster, cluster-specific genes were identified using a receiver operating characteristic curve (ROCC) analysis and a likelihood ratio test (LRT) test based on zero-inflated data (Table S2) (23). Expression of established cell-type markers within each cluster and fluorescent in situ hybridization (FISH) with cluster-specific markers enabled cluster assignment to one of eight previously identified planarian tissue classes: protonephridia, neural, epidermis, intestine, pharynx, muscle, neoblast, and parenchymal (Fig.1C). The parenchymal class was previously termed “parapharyngeal” because of localization of some enriched markers around the pharynx (24). However, most cell populations within this class exhibit broader localization in the planarian parenchyma. Unexpectedly, we also identified a ninth group of clusters marked by CTSL2 (dd175) expression (Fig. 1D). CTSL2 (dd175) FISH revealed cells with long processes distributed broadly. We designated this group of clusters the cathepsin+ class. Hierarchical clustering of a subset of 5,000 cells by Euclidean distance, independently of Seurat, recapitulated assignment of cells into these 9 tissue classes (Fig. S3).
Clusters comprising the major planarian tissue classes were generally heterogeneous in terms of gene expression. For example, neural clusters contained a large number of known neuronal cell types, suggesting that multiple distinct cell types could be identified within each major cluster (Fig. S4) Therefore, we systematically subclustered each major cluster group (Figs. 2, 3, 4, 5, 6; Methods), identifying >150 subclusters, and determined genes with enriched expression in cells of each subcluster (Table S2). Subclustering proved a powerful approach to defining the collection of cell types that constituted each major cluster and identified candidate transition states between stem cells and differentiated cells.
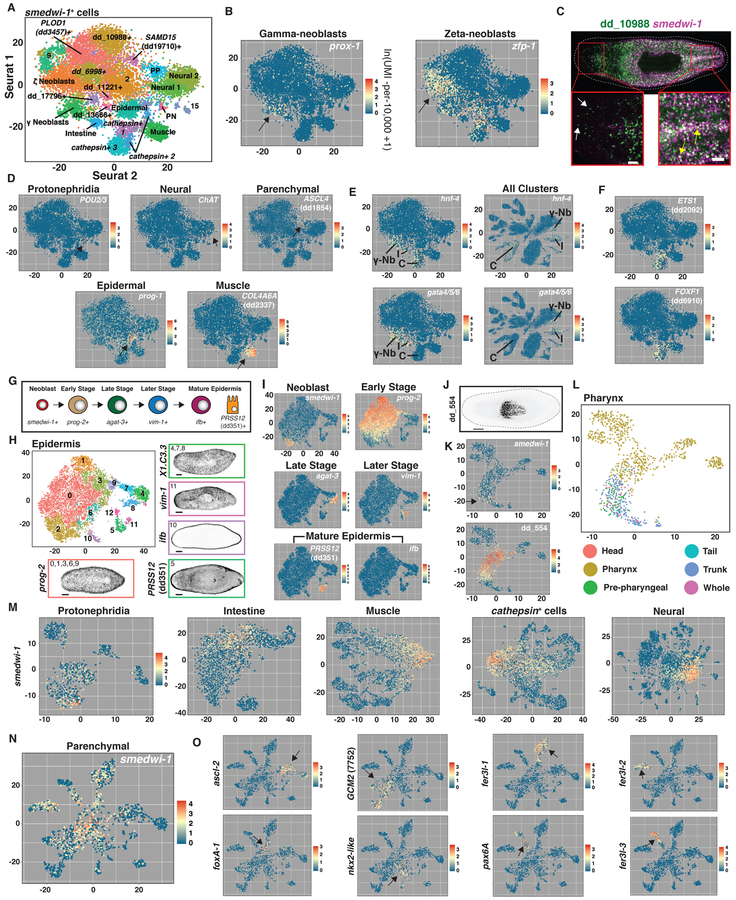
Fig. 2. Subclustering identifies neoblast subpopulations.
(A) t-SNE representation of 22 clusters generated from subclustering cells with smedwi-1 expression ≥ 2.5 (ln(UMI-per−10,000+1)). Identity of numbered clusters unknown. PP, parenchymal; PN, protonephridia. Intestine cluster indicated by lower expression of smedwi-1 and enriched gata4/5/6 and hnf4 expression. (B) smedwi-1+ t-SNE plots colored by prox-1 and zfp-1 expression. (C) Double FISH image for dd_10988 and smedwi-1. Yellow arrows: coexpression. White arrows: no co-expression. (D) smedwi-1+ t-SNE plots colored by expression of differentiated tissue-enriched genes. Arrows: gene expression sites. (E) Left: smedwi-1+ t-SNE plots colored by gata4/5/6 and hnf-4 expression. Right: All cluster t-SNE plots colored by gata4/5/6 and hnf-4 expression. C, cathepsin+ cells; I, Intestine; γ-Nb, γ Neoblasts. (F) smedwi-1+ t-SNE plots colored by ETS1 (dd2092) and FOXF1 (dd6910) expression. (G) Epidermal cell maturation stages. (H) t-SNE representation of epidermal subclusters. FISH images labeled by their associated cluster(s) are shown. (I) Epidermal t-SNE plots colored by epidermal-lineage marker expression from (G). (J) dd_554 FISH. (K) Pharynx t-SNE plots colored by smedwi-1 and dd_554 expression. (L) Pharynx t-SNE plot colored by the body region from which each cell was isolated. (M and N) t-SNE plots generated by subclustering cells identified as (M) protonephridia, intestine, muscle, cathepsin+ cells, neural, and (N) parenchymal. t-SNE plots colored by smedwi-1 expression. (O) Parenchymal t-SNE plots colored by expression of 8 transcription factor-encoding genes enriched in (N). Arrows: gene expression sites. Scale bars: whole-animal images, 200 μm; insets, 50 μm.
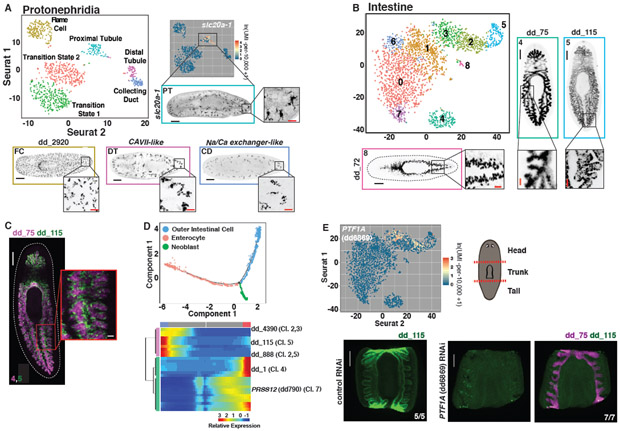
Fig. 3. Subclustering of tissues reveals transcriptomes for known and novel cell populations.
(A) t-SNE representation of the protonephridial subcluster. FISH images are labeled by their associated cluster. (B) t-SNE representation of intestinal subclusters. (C) Double FISH images of genes enriched in separate intestinal subclusters. Numbers indicate the associated subcluster for each marker. (D) Top: Cell trajectory of enterocyte and outer intestinal cell lineages produced by Monocle. Cells colored by identity. Bottom: Heat map of branch dependent genes (q-value < 1E-145) across cells plotted in pseudotime. Cells, columns; Genes, rows. Beginning of pseudotime at center of heatmap. “Cl.” annotation indicates a log-fold enrichment ≥ 1 of the gene in that intestine Seurat cluster. (E) Top left: Intestine t-SNE plot colored by expression of PTF1A (dd6869). Top right: Illustration of cutting scheme used to generate fragments. Bottom: dd_ 115 and dd_75 FISH of control and PTF1A (dd6869) RNAi animals. Animals cut and fixed 23 days following the start of dsRNA feedings. Scale bars: whole-animal/fragment images, 200 μm; insets 50 μm.
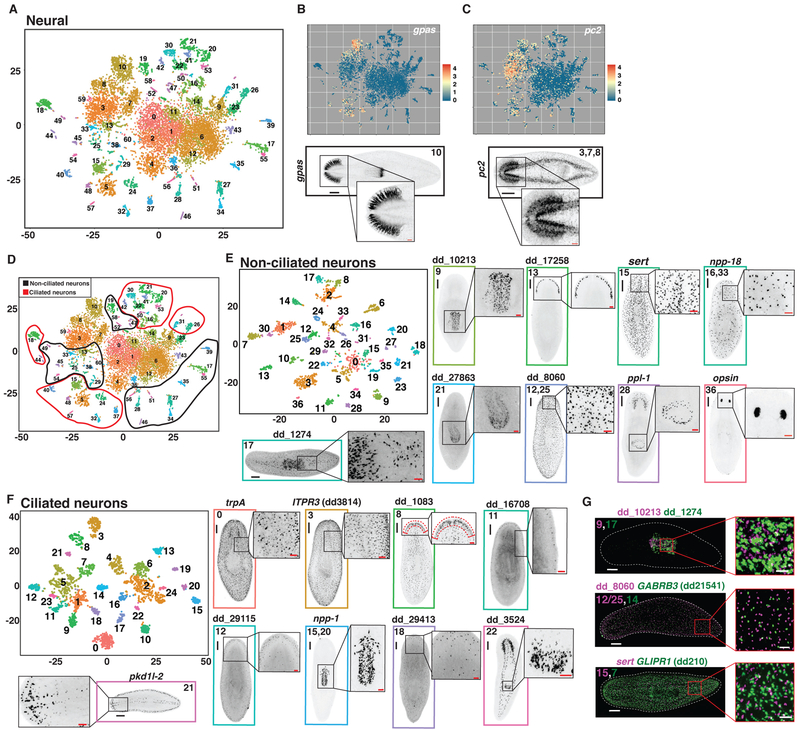
Fig. 4. Subclustering of neural cells reveals known and novel cell populations.
(A) t-SNE representation of the neural subcluster. (B and C) Top: t-SNE plots colored by expression of (B) gpas and (C) pc-2. Bottom: FISH for (B) gpas and (C) pc-2 labeled with the associated neural subcluster. (D) t-SNE plot in (A) overlaid with circles indicating the ascribed identity of each subcluster as ciliated or non-ciliated. (E and F) t-SNE representation of subclustered cells identified as (E) non-ciliated or (F) ciliated in (D). (G) Double FISH images of 3 sets of non-ciliated neuron genes enriched in separate subclusters. Numbers indicate the associated non-ciliated neuron subcluster(s) for each marker. Scale bars: whole-animal images, 200 μm; insets 50 μm.
Fig. 5. Tissue subclustering identifies cell populations of poorly characterized tissues.
(A) t-SNE representation of the pharynx subcluster. FISH images are labeled by their associated cluster(s). (B) Double FISH images of pharynx markers enriched in separate subclusters. Numbers indicate the associated pharynx subcluster(s) for each marker. (C) t-SNE representation of the muscle subcluster. (D) Double FISH images of 2 muscle markers enriched in separate subclusters. Numbers indicate the associated muscle subcluster for each marker. (E) t-SNE representation of the parenchymal subcluster. (F) Double FISH images of 3 sets of parenchymal markers enriched in separate subclusters. Numbers indicate the associated parenchymal subcluster for each marker. (G) Top left: Parenchymal t-SNE plot colored by expression of nkx6-like. Top right: Illustration of cutting scheme used to generate fragments. Bottom: dd_515 and dd_385 FISH of control and nkx6-like RNAi animals. Animals cut and fixed 23 days following the start of dsRNA feedings. Scale bars: whole-animal/fragment images, 200 μm; insets, 50 μm.
Fig. 6. Tissue subclustering reveals a previously unidentified class of cells.
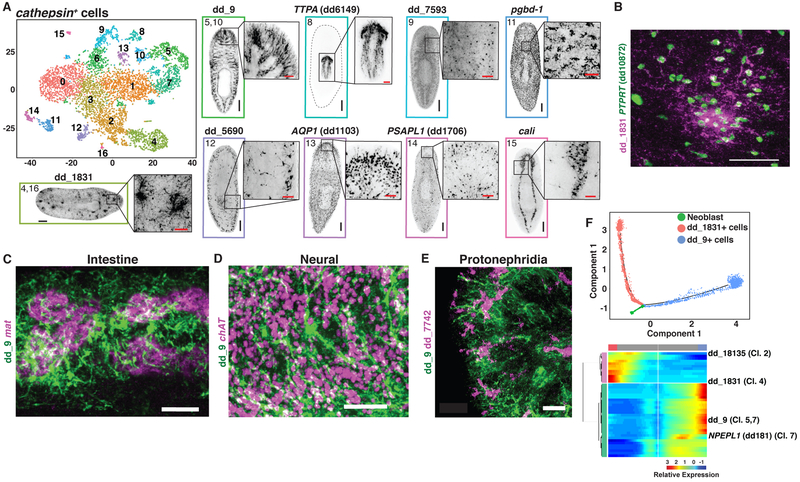
(A) t-SNE representation of the cathepsin+ cell subcluster. FISH images are labeled by their associated cluster(s). Images associated with subclusters 5/10 and 8 are single slices in the animal. All other images are maximum intensity projections. (B) Double FISH for 2 cathepsin+ cell markers enriched in the same subclusters, 4 and 16. (C-E) FISH for dd_9 and (C) mat, (D) ChAT, and (E) dd_7742. (F) Top: Cell trajectory of dd_1831+ and dd_9+ cathepsin+ cell lineages produced by Monocle. Cells colored by identity. Bottom: Heat map of branch dependent genes (q-value < 1E-175) across cells plotted in pseudotime. Cells, columns; Genes, rows. Beginning of pseudotime at center of heatmap. “Cl.” annotation indicates a log-fold enrichment ≥ 1 of the gene in that cathepsin+ cell Seurat cluster. Scale bars: whole-animal images, 200 μm; insets and B-E, 50 μm.
Progenitors in planarian cell lineages
Neoblasts are abundant and express canonical marker genes such as smedwi-1 (25), vasa (26), and bruli (27) (Fig. 1C, Fig. S5A,B). Neoblasts are cycling cells and consequently show enrichment in expression of S/G2/M cell cycle markers (Fig. S5C). To identify the transcriptomes of potential neoblast subpopulations, 12,212 cells with smedwi-1 expression ≥ 2.5 (ln(UMI-per−10,000+1)) were selected in silico and subclustered (Fig. 2A, Fig. S6A; Methods). Resulting clusters on the left of the plot were enriched in S/G2/M cell cycle markers (Fig. S6B). These clusters included the previously characterized major specialized neoblast classes, including gamma-neoblasts (intestine progenitors) and zeta-neoblasts (epidermis progenitors) (28) (Fig. 2B). A number of other subclusters were also identified, including one marked by expression of the contig dd_10988 (S6C,D). FISH confirmed that dd_10988 was expressed in a neoblast subset, as well as in a number of smedwi-1− cells (Fig. 2C).
The large number of subclustered neoblasts facilitated transcriptome determination for candidate progenitors for many planarian tissues. Clusters to the right of the plot were marked by a G1/G0 cell cycle status and displayed expression of various tissue markers (Fig. 2D, Fig. S6B). These included a population defined by expression of POU2/3, a marker for protonephridia specialized neoblasts (29), and a number of subclusters expressing markers also expressed in specific differentiated tissues or their post-mitotic precursors, such as ChAT for the nervous system, prog-1 for the epidermis, ASCL4 (dd1854) for parenchymal cells, and COL4A6A (dd2337) for muscle (Fig. 2D, S7A). Expression of these markers in smedwi-1+ cells suggests these cells could be transition states for those lineages. Several markers enriched in the dd_10988+ subcluster, including dd_10988, were also expressed in cells of the two smedwi-1+ neural subclusters, as well as in neural cells of the initial clustering (Fig. S6C, S7B,C), suggesting the dd_10988+ subcluster is enriched in neural progenitors. Likewise, many markers enriched in the PLOD1 (dd3457)+ subcluster were also expressed in the smedwi-1+ muscle subcluster, suggesting the PLOD1 (dd3457)+ subcluster is enriched in muscle progenitors (Fig. S7D). prox-1, hnf-4, nkx2.2, and gata4/5/6 encode transcription factors expressed in intestinal progenitors (28) and in this data all four genes were expressed in gamma neoblasts (Fig. 2B,E, Fig. S7E), with hnf-4, nkx2.2, and gata4/5/6 also expressed in intestinal clusters (Fig. 2E, Fig. S7F). hnf-4, but not prox-1, nkx2.2, and gata4/5/6, was also expressed in a smedwi-1+ cell cluster enriched in CTSL2 (dd175) expression (the cathepsin+ cell marker) and in differentiated cathepsin+ cells (Fig. 2E, Fig. S7G). The additional transcription factor-encoding genes ETS1 (dd2092) and FOXF1 (dd6910) were expressed with hnf-4 in these cells and also displayed similar expression patterns to CTSL2 (dd175) in the animal (Fig. 2F, Fig. S8A,B) and have recently been shown to regulate the planarian pigment cell lineage (30). Pigment cells clustered within the cathepsin+ cell class in our data (see below). By FISH, hnf-4 was indeed co-expressed with nkx2.2 and gata4/5/6 in the intestine, but was also coexpressed with cathepsin+ cell markers (Fig. S8C-E), suggesting that hnf4 is expressed in two distinct lineages. These data demonstrate the utility of this approach for identifying potentially novel neoblast progenitor populations and the transcription factors that define them.
Some planarian neoblasts called clonogenic neoblasts (or “cNeoblasts”) display pluripotency in clonal assays and are hypothesized to generate all lineage-committed neoblast subpopulations (14). We selected cells expressing high levels of smedwi-1 but that excluded zeta- and gamma-neoblasts (including subclusters 2, 9, dd_10988+, dd_6998+, dd_17796+, SAMD15 (dd19710)+, dd_11221+, dd_13666+, and PLOD1 (dd3457)+) and subclustered this set of neoblasts in isolation (Fig. S9A,B). A remnant zeta-neoblast population (Clusters 4 and 6), as well as protonephridia progenitors (Cluster 10) and the putative neural (Clusters 2 and 5) and muscle (Clusters 1 and 9) progenitor populations described above in the smedwi-1+ cell subclustering were identified (S9C; Table S2). Clusters 0, 3, 7, and 8 were largely devoid of specifically enriched markers (Table S2). It is therefore possible that cNeoblasts are defined by an absence of any tissue-specific markers as opposed to the unique expression of specific genes.
When all cells were clustered together, numerous smedwi-1+ cells were present regionally within each of the other eight major planarian tissue clusters (Fig. 1C). We reasoned that these smedwi-1+ cells could represent progenitors for the cell types within each associated tissue cluster. We therefore examined these smedwi-1+ cells after taking each tissue class in isolation and subclustering the data.
The planarian epidermis contains ciliated and non-ciliated cells as well as dorsal-ventral boundary epidermis (10, 28, 31) and the lineage from zeta-neoblasts to epidermal cells is well characterized (31-33) (Fig. 2G). SCS reveals gene expression transitions during neoblast epidermal differentiation (31); subclustering 11,021 epidermal lineage cells (Fig. 1C) produced subclusters associated with each epidermal lineage stage (Fig. 2H, Fig. S10A,B). Plotting gene expression onto this tSNE map showed a continuous progression from zeta-neoblast to differentiated cells (Fig. 2I, Fig. S10C).
The gene dd_554 (SmedASXL_059179 in (34)) is expressed in candidate pharynx progenitors (34) (smedwi-1+ cells at the pharynx base) and in smedwi-1− cells within the pharynx (34) (Fig. 2J, Fig. S11). Subclustering the 1,083 non-muscle, non-neuronal pharynx cluster cells (Fig. 1C) revealed that smedwi-1+ cells sequenced from non pharynx midbody tissue clustered with pharynx cells, despite not being part of the pharynx itself (Fig. 2K,L). Because the pharynx lacks neoblasts, pharynx-specialized neoblasts must be outside of the pharynx. This clustering of neoblasts with pharynx cells clearly demonstrates the ability of SCS data clustering to associate lineage precursors with differentiated cells. Similarly, many dd_554+ cells sequenced from outside of the pharynx clustered with pharynx cells (Fig. 2K,L). Plotting smedwi-1 /dd_554 expression onto pharyngeal subclusters revealed a progression from smedwi-1+ cells isolated outside the pharynx to dd_554+ cells isolated outside the pharynx to dd_554+ cells isolated inside the pharynx to pharyngeal cells (Fig. 2K,L). These epidermis and pharynx examples demonstrate how precursor stages within cell lineages can be identified from subclustering cells within a major tissue class. Because planarians constantly generate new differentiated cells for essentially all tissue types (17, 20), transcriptomes for lineage precursors for essentially every cell type in the body could in principle be studied with this approach.
Cell lineages for many planarian cell types are largely uncharacterized. Following tissue-type subclustering, smedwi-1+ cells were present with expression high locally in resultant t-SNE plots; expression gradually declined in level in cells across subclusters (Fig. 2M,N). These smedwi-1+ cells, similar to the epidermis and pharynx cases, could represent transition states between pluripotent neoblasts and differentiated cells for the various cells of the protonephridia, intestine, muscle, nervous system, parenchymal, and cathepsin+ cells (Fig. 2M,N). The smedwi-1+ cells found within subclusters of the major tissue type classes generally displayed enriched expression of at least one transcription factor. For example, smedwi-1 expression was high within cells at the center of the parenchymal cell t-SNE plot and displayed a graded decrease projecting in all directions into seven major parenchymal subclusters (Fig. 2N). Each projection was associated with enriched expression of one or more distinct transcription factors, identifying candidate transcription factors associated with the specification of different parenchymal cell types (Fig. 2O).
Subclustering cells by tissue type uncovers rare cell types
The protonephridia is the planarian excretory and osmoregulatory system, and is comprised of flame cells for filtering fluids, proximal and distal tubule cells, and a collecting duct (29, 35, 36). The protonephridia is a model tissue for studying organ regeneration and the evolution of kidney-like excretory systems. Subclustering of 890 protonephridia cells (Fig. 1C) identified each known protonephridia cell type as a separate subcluster, revealing the complete transcriptomes of these cells (Fig. 3A, Fig. S12A-C). Furthermore, two protonephridia subclusters with smedwi-1+ cells were identified (Fig. 2M). One was enriched in flame cell gene expression (e.g., dd_2920) and the other in a proximal tubule marker (dd_10830), suggesting they might be flame and tubule cell precursors, respectively (Fig. S12D,E).
Less is known regarding the full complement of cell types in other planarian tissues. Ultrastructural studies suggested that the planarian intestine contains two cell types, absorptive enterocytes and secretory goblet cells (37, 38). Subclustering of 3,025 intestinal cells (Fig. 1C) revealed 3 distinct cell populations (Clusters 4, 5, and 8) (Fig. 3B, Fig. S13A,B). FISH with subcluster-enriched markers (Table S2, Methods) revealed distinct intestine components. Cluster 4 represented an inner intestine cell layer (Fig. 3C) and was enriched for absorptive enterocyte markers (39). Cluster 8 cells were largely present within the primary intestine branches, resembling the pattern of goblet cells (40). A third group (cluster 5) represented an outer intestine cell layer, and displayed a strikingly different set of enriched genes from clusters 4/8 (Fig. 3C, Table S2). In addition to these three main intestine components, clusters representing putative transition states were also identified. Clusters 1, 3, and 6 included many smedwi-1+ cells (Fig. 2M). Genes with enriched expression in clusters 0 and 7 displayed expression spanning into the enterocyte cluster (cluster 4), suggesting these might be enterocyte transition states (Fig. S14A). Genes with enriched expression in clusters 2 and 3 displayed expression spanning into the outer gut cluster (cluster 5) and might reflect transition or variant states of these cells (Fig. S14B). The Monocle toolkit can be used to predict cellular transitions in lineages (41) and was used to build single cell trajectories for the enterocyte and outer gut cell lineages, closely recapitulating the candidate transition states identified by Seurat (Fig. 3D, Fig. S15, Table S3).
Several transcription factors required for the specification of various planarian cell types have been identified with RNAi and gene expression studies. Because of constant tissue turnover, RNAi of transcription factor-encoding genes expressed in specific classes of specialized neoblasts in adult planarians can lead to steady depletion of the cell type generated by that specialized neoblast class (29, 42, 43). The transcriptomes identified here generate a resource of enriched gene expression for different cell types, including transcription factor-encoding genes. Accordingly, inhibition of the transcription factorencoding PTF1A (dd_6869) gene, which had enriched expression in candidate transition states for the outer intestine cluster, strongly reduced this cell population while not affecting absorptive enterocytes of the intestine (Fig. 3E).
The nervous system displays by far the greatest known cell-type composition complexity of the major planarian tissues. By subclustering 11,907 neuronal cells (Fig. 1C), we identified 61 distinct subclusters representing a diversity of cell types and states (Fig. 4A, Fig. S16A). Twelve subclusters had high smedwi-1 expression, suggesting they are neuronal precursors (Fig. S16B). Cluster 10 contained cells of the brain branches, as determined by expression of gpas (44) and pds (45) (Fig. 4B, Fig. S16C). Three subclusters (Clusters 3, 7, and 8) were defined by expression of pc2 (encoding a neuropeptide-processing proprotein convertase), as well as an assortment of markers for rare neuron classes in the cephalic ganglia and ventral nerve cords (Fig. 4C, Fig S16D,E). We also sequenced an additional 7,766 cells from the brain region to expand the number of cells in these clusters (Fig. S16F-H). In addition to these large clusters, there existed a number of smaller, compact, and well-separated subclusters. These could be further divided into ciliated and non-ciliated neurons based upon the expression of rootletin (dd_6573), which encodes a ciliary rootlet component (Fig. 4D, Fig. S17A). Because of further heterogeneity within these clusters (e.g., opsin+ presumptive photoreceptors were present together, but not as a separate cluster), data from these two cell sets (ciliated, not ciliated) were each taken in isolation for further subclustering. This yielded 37 non-ciliated neuron subclusters (Fig. 4E, Fig. S17B, S18A,B, S19-21B) and 25 putatively ciliated neuron clusters (Fig. 4F, Fig. S17B, S22A,B, S23B). We assessed the localization of cells associated with 46 of 62 of these subclusters by FISH using subcluster-specific markers. The observed cell types had a wide range of patterns including rare cell types such as photoreceptor neurons (Fig. 4E,F, Fig. S18-23). Many genes had enriched expression in multiple clusters; the distribution of neural cell types they represented was defined by a combinatorial set of markers (Fig. S18-23). A number of identified cell types from different subclusters displayed similar localization patterns. FISH demonstrated no overlap in subcluster-specific markers, however, consistent with the SCS data (Fig. 4G). For several neural subtypes, we found smedwi-1+ candidate precursor cells. Four non-ciliated neuron subclusters (1, 2, 4, and 12) and a single ciliated neuron subcluster (1) were enriched in smedwi-1 expression (Fig. S24A). Non-ciliated neuron subcluster 4 also expressed gata4/5/6, as did six smedwi-1− clusters (Clusters 14,16/33, 24, 26, and 32) that radiated out from central smedwi-1+ cells, raising the possibility that these smedwi-1+ cells constitute precursors for these populations (Fig. S24B).
The pharynx is a muscular tube used for feeding and defecation (10). It is contained within an epithelial cavity, and connects to the intestine at its anterior end via an esophagus. Pharyngeal muscle cells and pharyngeal neurons clustered together with the other muscle cells and neurons of the body (figs. S25A, S26A). Other pharynx-associated cells, including cells from isolated pharynges and surrounding tissue, constituted the other major pharynx clusters. These non-neural, non-muscle pharynx and pharynx-associated cells (Fig. 1C, n=1,083 cells) were subclustered and FISH was performed on cluster-enriched markers (Fig. 5A, Fig. S25B,C). Subclusters included pharyngeal-cavity epithelium cells (Clusters 7 and 8), the epithelial pharynx lining (Clusters 1 and 5), the mouth and esophagus (Cluster 9), cells near the pharynx opening (Cluster 6), and cells that constitute the connection to the planarian body (Cluster 4). FISH confirmed non-overlapping expression patterns for markers of tested separate cell populations (Fig. 5B).
Planarian muscle expresses collagen, in addition to canonical muscle genes such as troponin and tropomyosin (19). Muscle exists in a subepidermal body wall layer, in the pharynx, surrounding the intestine, and in a DV domain (46). Subclustering 5,014 muscle cells (Fig. 1C) revealed seven smedwi-1/+ candidate precursor subclusters (Clusters 0, 1, 3, 4, 5, 10, and 11) (Fig. 2M), as well as subclusters containing body wall muscle (Cluster 7), pharyngeal muscle (Cluster 2, 8, 9, and 12) (Fig. S26A), a population of muscle cells enriched around the intestine (Cluster 6), and an unidentified population (Cluster 13) (Fig. 5C,Fig. S26B,C). Markers for body wall muscle (Cluster 7) and Cluster 13 were expressed in non-overlapping cells by FISH (Fig. 5D).
Whereas some molecular characterization existed for the seven broad planarian tissue classes previously mentioned, very little is known regarding the cellular composition of the two remaining classes. The parenchymal class (Fig. 1C) (24) was highly heterogeneous, with subclustering of 2,120 cells identifying many distinct cell populations (Fig. 5E, Fig. S27A,B, S28B). In addition to eight smedwi-1+ putative precursor subclusters (Clusters 0, 1, 2, 3, 4, 6, 8, and most of 9) (Fig. 2N), parenchymalcell subclustering revealed 13 well-separated differentiated cell subclusters. FISH showed that each of these differentiated cell populations were present as scattered cells, presumably within a mesenchymal tissue layer called the parenchyma that surrounds major planarian organs (10). Previous morphological studies determined that the parenchyma is composed of multiple gland cells, neoblasts, and “fixed parenchymal cells” characterized through histological and electron microscopy studies as a likely phagocytic cell with long cellular processes filling most of the parenchymal space (10, 47, 48). Some identified parenchymal subclusters appeared to be gland cells, displaying processes extending to the epidermis, defining transcriptomes for these cells. Candidate gland cell types included two that were exclusively dorsal (Clusters 16 and 17), two exclusively lateral (Clusters 10 and 13, including marginal adhesive gland cells and an unknown cell population), four present both dorsally and ventrally (Clusters 7, 11, 14, and 15), and one present ventrally near the brain (Cluster 19). Three subclusters (Clusters 5, 12, and 18) contained cells with similar patterns to planarian neoblasts, but were not neoblasts. Finally, a single subcluster contained large cells surrounding the pharynx (small group of cluster 9 cells) and were enriched for expression of previously identified metalloprotease-encoding genes (49). Three pairs of parenchymal subclusters (six subclusters total) were confirmed to exist in non-overlapping populations by FISH (Fig. 5F).
The transcription factor-encoding gene nkx6-like was expressed in a parenchymal cell population marked by dd_515. Inhibition of nkx6-like ablated dd_515 cells, while not affecting a distinct, non-enriched parenchymal cell population marked by dd_385 (Fig. 5G). These results further highlight the potential to utilize the data to ablate many specific cell types in the animal.
The final major class of cells, the cathepsin+ group, contained 7,034 cells (Fig. 1D). This group of clusters contained recently described glia and pigment cells (12, 13, 50). Subclustering of cathepsin+ cells identified 4 subclusters expressing smedwi-1 that represented putative precursor cells (Clusters 0, 1,3, and 6) (Fig. 2M), a glial subcluster (Cluster 15), and two pigment cell populations (Clusters 11 and 14), identifying transcriptomes for these cell types (Fig. 6A, Fig. S29A,B, S30B). Eight cathepsin+ subclusters represented previously unidentified cell populations. FISH revealed striking, elaborate morphologies for most of these cells, involving long processes and unique distributions (Fig. 6A, Fig. S29B, S30B). Cells from subclusters 5 and 10 were spread throughout the planarian body, with long processes filling substantial parenchyma space. Subcluster 8 represented cells specific to the pharynx. Subcluster 9 cells were scattered throughout the animal. Subclusters 4 and 16 identified cells with dense aggregated foci of elaborate processes at scattered locations throughout the animal that lacked definitive positions - an unusual and unanticipated cell-type distribution. FISH identified markers labeling cell bodies of these cells, revealing that the aggregates were comprised of many cells (Fig. 6B). Subclusters 12 and 13 also exhibited processes with visible cell bodies. Subcluster 12 cells were largely subepidermal. The most elaborate of these newly identified cells (subclusters 5 and 10) were excluded from the intestine and brain, but had processes around the branches of the intestine and protonephridia and interspersed within the cephalic ganglia (Fig. 6C-E, Fig. S31A,B). FISH confirmed non-overlapping expression patterns for two tested subclusters (Fig. S31C).
Subcluster 7 of the cathepsin+ group of cells was enriched in expression of genes with expression spanning into clusters 5 and 10 (Fig. S32A). Similarly, expression of cluster 2 marker genes spanned into clusters 4 and 16 (Fig. S32B). These cells might reflect transition or variant states of cells for clusters 5/10 and 4/16, respectively. SMEDWI-1 protein perdures in neoblast progeny following loss of smedwi-1 mRNA, allowing detection of newly produced neoblast progeny (27). MAP3K5 (dd_4849)+ cells, which were predicted to be expressed in cells transitioning from the smedwi-1+ state in the cathepsin+ cell plot, were SMEDWI-1+ / smedwi-1−, supporting the interpretation that these cells are progenitors in the cathepsin+ cell lineage (Fig. S32C,D). The Monocle toolkit was also used to build single cell trajectories for these clusters, with data closely recapitulating the transition states identified by Seurat (Fig. 6F, Fig. S33, Table S3).
A near complete discovery of planarian cell type transcriptomes
The number of cell types identified in this study vastly exceeded prior planarian SCS data (24). Within the neuronal subclusters, a 17 cell subcluster represented photoreceptor neurons (Fig. 4E), which are present at ~100 total cells in a medium-sized (~2-3mm) animal (20). Therefore, our data should have readily included unknown cell types as rare as photoreceptor neurons. Similarly, an average-sized planarian has ~60 cintillo+ neurons and our data included 10 cintillo+ neurons (Fig. S34A). These cells were grouped within a larger subcluster (cluster 3) of non-ciliated neurons (Fig. S34B), suggesting that even further subclustering of this "subcluster 3" could reveal additional distinct cell types. Indeed, cintillo+ cells emerged as a unique cluster from such additional (fourth tier) subclustering of original data (Fig. S34C, Table S2). Esophagus cells, connecting pharynx to intestine, clustered with mouth cells in the pharynx subclustering data (Fig. S34D). ~50 of these cells exist in an average-sized animal and 3 such cells were present in the data (Fig. S34A,D). Several known rare cell types did not separate into individual clusters, though most could still be identified in the data, suggesting the data is largely saturated for rare cell types. These include anterior pole cells, which function as an anterior organizer (51-53), notum+ neurons in the brain (54), and posterior pole cells (45), which are among the rarest known cell types in the animal, with only ~10 each present in an average animal (Fig. S34A). Five anterior pole cells, 10 notum+ neurons, and one posterior pole cell were identified in the data (Fig. S34E-G). Similarly ~25 ovo+ eye progenitors (42) and ~90 nanos+ germ cells (55) are present in an average animal (Fig. S34A). Two eye progenitors and 19 germ cells were identified in the data (Fig. S34H,I). In addition to the asexual strain of Schmidtea mediterranea used in this study, a sexual strain of cross-fertilizing hermaphrodites exists. We sequenced 8,455 cells from this strain, adding sexual strain cells to this resource as well, including 7 yolk and 7 testes cells in addition to the 19 germ cells described above (Fig. S35A-D). Further sequencing of sexual cell types could be a target for future studies. Together, our data indicate that we have essentially reached saturation for determining the cell type transcriptomes of asexual planarians.
Discovery of novel patterning genes
Planarians constitutively express dozens of genes associated with patterning (PCGs) in complex spatial patterns across body axes (18). PCGs are almost exclusively expressed in muscle (19). AP axis PCGs are well established, including with muscle SCS (56). Muscle cells did not subcluster based on their anatomical positions (Fig. 7A, Fig. S36A- C). However, we reasoned that expression of known PCGs could ascribe locations to muscle cells in the data. Because of variability in the expression of any one PCG, muscle cell regional identity was determined based on expression of at least two PCGs. For example, posterior muscle cells were identified by co-expression of at least 2/4 posterior PCGs wnt11-1, wnt11-2, fz4-1, and wntP-2, yielding 163 cells (Fig. 7A, S36D). Differential expression analysis using the algorithm SCDE (57) was performed on these 163 cells against the 4,851 other muscle cells (Fig. 7A, Table S4). Strikingly, nine of the differentially expressed genes were identified by Scimone et al. (56) as posterior-enriched; eight of these were within the top 26 genes identified by differential expression analysis (Fig. 7B, hypergeometric p = 2.75E-9). A similar analysis on 837 anterior muscle cells was also performed (Fig. S36A,D, Table S4). Nine of the differentially expressed genes were identified by Scimone et al. (56); four of the genes were within the top 25 genes identified by SCDE (Fig. 7B, hypergeometric p = 5.80E-5). We also applied this approach to the less well-studied ML axis. 62 lateral muscle cells were identified and FISH with 15 of the top genes identified seven with lateral muscle expression (Fig. 7C, Fig. S36B,D-G (58, 59)). 90 medial muscle cells were isolated and the top ranked gene displayed a striking thin stripe of expression down the dorsal midline (Fig. 7C, Fig. S36C,D,H). Together, these results demonstrate the power of deep SCS for identifying regional gene expression, such as that involved in patterning, in adult animal tissues.
Fig. 7. Identification of new regionally expressed genes in muscle.
(A) Top: t-SNE plot colored by muscle cells positive for expression ≥ 0.5 (ln(UMI-per−10,000+1)) of 2 of the 4 posterior PCGs wnt11-1, wnt11-2, fz-4, and wntP-2. Positive cells, pink; negative cells, grey. Bottom: Transcriptomes for posterior muscle cells were compared to all other muscle cells by SCDE. (B) List of differentially expressed genes in (left) posterior and (right) anterior muscle cells that were identified in Scimone et al. (56). Rank indicates the rank of the gene in our analysis. (C) FISH images of one (left) lateral and one (right) medial expressed gene ranked highly in this analysis (59). Number indicates gene rank in the list generated by SCDE. Scale bars: whole-mount images, 200 μm; insets, 50 μm. (D) Illustration highlighting the capacity of the dataset to identify almost all cell types in the planarian, as well specialized neoblast progenitors, and novel patterning information from the adult animal.
Discussion
RNA sequencing of >50,000 cells (in total, 66,783 cells were sequenced) of the planarian S. mediterranea allowed the identification of transcriptomes for most-to-all cell types of an adult animal. This includes transcriptomes for cell types present as rarely as 10 cells in an animal with 105 - 106 cells, strongly suggesting we have reached near saturation. Sequencing of different body regions, and assessment of rare cell-type coverage in an iterative process enabled reaching this saturation level. Some cell types might have escaped detection if they are exceptionally rare or hard to dissociate from the animal. Our data did indicate some cell types were preferentially recovered based on the abundance of that cell type by FISH, while others were less represented (Fig. S37A,B). In particular, prog-1+ epidermal progenitor cells were highly over-represented in the data compared to their prevalence in the animal, perhaps because their small size made their isolation easier (Fig. S37A). Absent prog-1+ cells, most other cell types analyzed were represented similarly to their relative abundance in the animal (Fig. S37B). Regardless of differences in ease of dissociation between cell types, we nonetheless recovered data from all known cell types assessed. Not every known rare cell type emerged as a separable cluster - i.e., these cells were sometimes embedded within a larger cluster. In some instances, further rounds of subclustering based upon such knowledge resulted in splitting of subclusters into additional subclusters. Therefore, further subclustering analyses and even deeper sequencing will likely continue to enhance the capacity to computationally isolate rare cell types from other clusters. Nonetheless, the transcriptomes for such rare cell types are present in our data and can be studied by searching for the desired cells. Another challenge with assessing saturation of cell type sequencing is ambiguity with the term cell type. Gene expression heterogeneity exists within well-defined clusters and could reflect differences ranging from technical sampling error to cell type state differences to robust differences in biological function. Further in vivo morphological and functional studies with identified cell clusters, further computational analyses, and ever more sequencing data can continue to refine the knowledge of biologically significant cell-type differences.
Cell types have been previously identified largely through morphological descriptions and perhaps a few marker genes. Determining cell-type transcriptomes with large-scale SCS is a powerful new approach to defining the cell-type constitution of a tissue, organ, or even a complete animal. In our study, we identified a large number of previously uncharacterized planarian cell populations across multiple tissues. This included multiple cell populations (in the cathepsin+ group) previously undescribed at the molecular level. One cell population, defined by dd_9 expression, had long processes filling parenchymal space and surrounding, but excluded from, other planarian tissues. This pattern is reminiscent of “fixed parenchymal cells”, a largely uncharacterized cell population described by histology and electron microscopy (EM) (48). Previous EM work suggested that fixed parenchymal cells are likely phagocytic, with clearly observed lysosomes. dd_9+ cells highly expressed genes encoding a variety of digestive enzymes and endocytosis proteins, providing further support for this hypothesis (Table S2). The biology of these cathepsin+ cells and all the other diverse cell types identified in this work can now be studied in depth utilizing identified transcriptomes and the tools of planarian biology research. For instance, we show for two case studies above that RNAi of a gene encoding a transcription factor with enriched expression in a candidate cell lineage leads to ablation of the predicted differentiated cell.
Generating transcriptomes for most-to-all cell types in an animal will be invaluable for studying gene function and the biology and evolution of a large range of important cell types. Because of their phylogenetic position within the Spiralian superphylum (60), major cell types found across diverse bilaterians (e.g., shared between humans and Drosophila, C. elegans, molluscs, and/or annelids, etc.) should have been present in the last common ancestor of planarians and humans. As such, studying the transcriptomes and associated genes with cell type-enriched expression in this dataset can allow characterization of the gene function underlying the biology of these cells.
Planarian biology presents many features that made this organism attractive for comprehensive SCS. Planarians are a model for studying numerous important problems in regeneration, stem cell biology, patterning, and evolution. At a single timepoint - the adult - there exist progenitors for essentially all cell types and the patterning information for guiding new cell type production. We identified the transcriptomes of numerous candidate transition states in lineages from pluripotent stem cell to diverse differentiated cell types. Furthermore, we utilized the data to identify novel regionally expressed genes in planarian muscle (the site of patterning gene expression). Together, these results illustrate the capacity of our dataset to define cell type transcriptomes, identify lineage transition states, and ascertain novel patterning information, all from a single timepoint (Fig. 7D). Much like the genome of an animal, we propose this atlas-like dataset of cell-type transcriptomes can serve as a resource fueling an immense amount of research, not only in planarians, but in other bilaterians with similar cell types. To facilitate such study, we developed an online resource that generates cluster expression data, for any gene, across all clusters and subclusters (digiworm.wi.mit.edu). Case study model organisms proved valuable testing grounds for developing approaches to complete genome sequencing - these planarian SCS data demonstrate an approach to near complete cell-type transcriptome identification that could be applied broadly to diverse organisms with varying degrees of information about cell-type composition. The remarkable ability of single-cell RNA sequencing to reach near-to-complete saturation of transcriptome identification for the cell types of a complete animal is a new and powerful approach for describing the anatomy of complete organisms at the molecular level.
Supplementary Material
Acknowledgements
We thank M.L. Scimone, C. McQuestion, and K.D. Atabay for their help in the targeted dissociation of tissue from the planarian brain. We thank all Reddien Lab members for valuable comments and discussion. We thank E.Z. Macosko, M. Goldman, and S.A. McCarroll for making protocols available.
Funding: We acknowledge NIH (R01GM080639) support. P.W.R. is an Investigator of the Howard Hughes Medical Institute and an associate member of the Broad Institute of Harvard and MIT. We thank the Eleanor Schwartz Charitable Foundation for support.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: All raw and processed data files associated with this study have been deposited to Gene Expression Omnibus (GEO) under the accession number GSE111764.
References
- 1.The C elegans Sequencing Consortium, Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282, 2012–2018 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian D, Rogers Wyman, J., Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X, The sequence of the human genome. Science 291, 1304–1351 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I, Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, Schwartz S, Fowler B, Weaver S, Wang J, Wang X, Ding R, Raychowdhury R, Friedman N, Hacohen N, Park H, May AP, Regev A, Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510, 363–369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, Adey A, Waterston RH, Trapnell C, Shendure J, Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, Huang D, Xu Y, Huang W, Jiang M, Jiang X, Mao J, Chen Y, Lu C, Xie J, Fang Q, Wang Y, Yue R, Li T, Huang H, Orkin SH, Yuan GC, Chen M, Guo G, Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 172, 1091–1107 e1017 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Reddien PW, Sánchez Alvarado A, Fundamentals of planarian regeneration. Ann. Rev. Cell Dev. Bio. 20, 725–757 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Hyman LH, The Invertebrates: Platyhelminthes and Rhynchocoela The acoelomate bilateria. (McGraw-Hill Book Company Inc., New York, 1951), vol. II. [Google Scholar]
- 11.Baguñà J, Romero R, Quantitative analysis of cell types during growth, degrowth and regeneration in the planarians Dugesia mediterranea and Dugesia tigrina. Hydrobiologia 84, 181–194 (1981). [Google Scholar]
- 12.Wang IE, Lapan SW, Scimone ML, Clandinin TR, Reddien PW, Hedgehog signaling regulates gene expression in planarian glia. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts-Galbraith RH, Brubacher JL, Newmark PA, A functional genomics screen in planarians reveals regulators of whole-brain regeneration. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner DE, Wang IE, Reddien PW, Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scimone ML, Kravarik KM, Lapan SW, Reddien PW, Neoblast Specialization in Regeneration of the Planarian Schmidtea mediterranea. Stem Cell Reports 3, 339–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddien PW, Specialized progenitors and regeneration. Development 140, 951–957 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newmark P, Sánchez Alvarado A, Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol 220, 142–153 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Reddien PW, Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet 27, 277–285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witchley JN, Mayer M, Wagner DE, Owen JH, Reddien PW, Muscle cells provide instructions for planarian regeneration. Cell Rep 4, 633–641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LoCascio SA, Lapan SW, Reddien PW, Eye Absence Does Not Regulate Planarian Stem Cells during Eye Regeneration. Dev Cell 40, 381–391 e383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu SY, Selck C, Friedrich B, Lutz R, Vila-Farre M, Dahl A, Brandl H, Lakshmanaperumal N, Henry I, Rink JC, Reactivating head regrowth in a regeneration-deficient planarian species. Nature 500, 81–84 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Satija R, Farrell JA, Gennert D, Schier AF, Regev A, Spatial reconstruction of single-cell gene expression data. Nature Biotechnology 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, Gottardo R, Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29, 461–467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wurtzel O, Cote LE, Poirier A, Satija R, Regev A, Reddien PW, A Generic and Cell-Type-Specific Wound Response Precedes Regeneration in Planarians. Dev Cell 35, 632–645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A, SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Wagner DE, Ho JJ, Reddien PW, Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell 10, 299–311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo T, Peters AH, Newmark PA, A Bruno-like gene is required for stem cell maintenance in planarians. Dev Cell 11, 159–169 (2006). [DOI] [PubMed] [Google Scholar]
- 28.van Wolfswinkel JC, Wagner DE, Reddien PW, Single-Cell Analysis Reveals Functionally Distinct Classes within the Planarian Stem Cell Compartment. Cell Stem Cell 15, 326–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scimone ML, Srivastava M, Bell GW, Reddien PW, A regulatory program for excretory system regeneration in planarians. Development 138, 4387–4398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He X, Lindsay-Mosher N, Li Y, Molinaro AM, Pellettieri J, Pearson BJ, FOX and ETS family transcription factors regulate the pigment cell lineage in planarians. Development 144, 4540–4551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wurtzel O, Oderberg IM, Reddien PW, Planarian Epidermal Stem Cells Respond to Positional Cues to Promote Cell-Type Diversity. Dev Cell 40, 491–504 e495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eisenhoffer GT, Kang H, Sánchez Alvarado A, Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 3, 327–339 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu KC, Cheng LC, T. K. V. H, Lange JJ, McKinney SA, Seidel CW, Sánchez Alvarado A, Egr-5 is a post-mitotic regulator of planarian epidermal differentiation. eLife 4, e10501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu SJ, Hallows SE, Currie KW, Xu C, Pearson BJ, A mex3 homolog is required for differentiation during planarian stem cell lineage development. eLife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vu HT, Rink JC, McKinney SA, McClain M, Lakshmanaperumal N, Alexander R, Sánchez Alvarado A, Stem cells and fluid flow drive cyst formation in an invertebrate excretory organ. eLife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rink JC, Vu HT, Sánchez Alvarado A, The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development 138, 3769–3780 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willier BH, Hyman LH, Rifenburgh SA, A histochemical study of intracellular digestion in triclad flatworms. J. Morph 40, 299–340 (1925). [Google Scholar]
- 38.Ishii S, Electron microscopic observations on the Planarian tissues II. The intestine. Fukushima J. Med. Sci. 12, 67–87 (1965). [PubMed] [Google Scholar]
- 39.Forsthoefel DJ, James NP, Escobar DJ, Stary JM, Vieira AP, Waters FA, Newmark PA, An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Dev Cell 23, 691–704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zayas RM, Cebria F, Guo T, Feng J, Newmark PA, The use of lectins as markers for differentiated secretory cells in planarians. Dev Dyn 239, 2888–2897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL, The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lapan SW, Reddien PW, Transcriptome Analysis of the Planarian Eye Identifies ovo as a Specific Regulator of Eye Regeneration. Cell Rep 2, 294–307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapan SW, Reddien PW, dlx and sp6-9 control optic cup regeneration in a prototypic eye. PLoS Genet 7, e1002226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cebrià F, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Agata K, Dissecting planarian central nervous system regeneration by the expression of neural-specific genes. Development, Growth & Differentiation 44, 135–146 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Petersen CP, Reddien PW, Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319, 327–330 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Cebrià F, Planarian Body-Wall Muscle: Regeneration and Function beyond a Simple Skeletal Support. Frontiers in Cell and Developmental Biology 4, 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedersen KJ, Some features of the fine structure and histochemistry of planarian subepidermal gland cells. Z. Zellf 50, 121–142 (1959). [Google Scholar]
- 48.Pedersen KJ, Studies on the nature of planarian connective tissue. Zeitschrift fur Zellforschung 53, 569–608 (1961). [Google Scholar]
- 49.Newmark PA, Reddien PW, Cebria F, Sánchez Alvarado A, Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc Natl Acad Sci 100, 11861–11865 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stubenhaus BM, Dustin JP, Neverett ER, Beaudry MS, Nadeau LE, Burk-McCoy E, He X, Pearson BJ, Pellettieri J, Light-induced depigmentation in planarians models the pathophysiology of acute porphyrias. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scimone ML, Lapan SW, Reddien PW, A forkhead transcription factor is wound-induced at the planarian midline and required for anterior pole regeneration. PLoS Genetics 10, e1003999 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogg MC, Owlarn S, Perez Rico YA, Xie J, Suzuki Y, Gentile L, Wu W, Bartscherer K, Stem cell-dependent formation of a functional anterior regeneration pole in planarians requires Zic and Forkhead transcription factors. Developmental Biology 390, 136–148 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Petersen CP, Reddien PW, Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332, 852–855 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hill EM, Petersen CP, Wnt/Notum spatial feedback inhibition controls neoblast differentiation to regulate reversible growth of the planarian brain. Development 142, 4217–4229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Zayas RM, Guo T, Newmark PA, nanos function is essential for development and regeneration of planarian germ cells. Proc Natl Acad Sci 104, 5901–5906 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scimone ML, Cote LE, Rogers T, Reddien PW, Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kharchenko PV, Silberstein L, Scadden DT, Bayesian approach to single cell differential expression analysis. Nature Methods 11, 740–742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barberán S, Martín-Durán JM, Cebrià F, Evolution of the EGFR pathway in Metazoa and its diversification in the planarian Schmidtea mediterranea. Scientific Reports 6, 28071 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scimone ML, Cote LE, Reddien PW, Orthogonal muscle fibres have different instructive roles in planarian regeneration. Nature 551, 623–628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laumer CE, Bekkouche N, Kerbl A, Goetz F, Neves RC, Sorensen MV, Kristensen RM, Hejnol A, Dunn CW, Giribet G, Worsaae K, Spiralian phylogeny informs the evolution of microscopic lineages. Curr Biol 25, 2000–2006 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL, BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rouhana L, Vieira AP, Roberts-Galbraith RH, Newmark PA, PRMT5 and the role of symmetrical dimethylarginine in chromatoid bodies of planarian stem cells. Development 139, 1083–1094 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayashi T, Asami M, Higuchi S, Shibata N, Agata K, Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Development, Growth & Differentiation 48, 371–380 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin JR, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CG, Kazer SW, Gaillard A, Kolb KE, Villani AC, Johannessen CM, Andreev AY, Van Allen EM, Bertagnolli M, Sorger PK, Sullivan RJ, Flaherty KT, Frederick DT, Jane-Valbuena J, Yoon CH, Rozenblatt-Rosen O, Shalek AK, Regev A, Garraway LA, Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.King RS, Newmark PA, In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC developmental biology 13, 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rouhana L, Weiss JA, Forsthoefel DJ, Lee H, King RS, Inoue T, Shibata N, Agata K, Newmark PA, RNA interference by feeding in vitro-synthesized double-stranded RNA to planarians: methodology and dynamics. Dev Dyn 242, 718–730 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Sánchez Alvarado A, Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol 347, 24–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petersen CP, Reddien PW, A wound-induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci 106, 17061–17066 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gurley KA, Rink JC, Sánchez Alvarado A, Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319, 323–327 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rink JC, Gurley KA, Elliott SA, Sánchez Alvarado A, Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science 326, 1406–1410 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Felix DA, Aboobaker AA, The TALE class homeobox gene Smed-prep defines the anterior compartment for head regeneration. PLoS Genetics 6, e1000915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Molina MD, Neto A, Maeso I, Gomez-Skarmeta JL, Saló E, Cebria F, Noggin and noggin-like genes control dorsoventral axis regeneration in planarians. Curr Biol 21, 300–305 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Gavino MA, Reddien PW, A Bmp/Admp regulatory circuit controls maintenance and regeneration of dorsal-ventral polarity in planarians. Curr Biol 21, 294–299 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adell T, Saló E, Boutros M, Bartscherer K, Smed-Evi/Wntless is required for beta-catenin-dependent and -independent processes during planarian regeneration. Development 136, 905–910 (2009). [DOI] [PubMed] [Google Scholar]
- 75.Molina MD, Saló E, Cebrià F, Expression pattern of the expanded noggin gene family in the planarian Schmidtea mediterranea. Gene Expr Patterns 9, 246–253 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Cebrià F, Newmark PA, Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development 134, 833–837 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Orii H, Kato K, A. K., Watanabe K, Molecular cloning of bone morphogenetic protein (BMP) gene from the planarian Dugesia japonica. Zool. Science 15, 871–877 (1998). [Google Scholar]
- 78.Reddien PW, Bermange AL, Kicza AM, Sánchez Alvarado A, BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134, 4043–4051 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Molina MD, Saló E, Cebria F, The BMP pathway is essential for respecification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev Biol 311, 79–94 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Cebrià F, Newmark PA, Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 132, 3691–3703 (2005). [DOI] [PubMed] [Google Scholar]
- 81.Nishimura K, Kitamura Y, Inoue T, Umesono Y, Yoshimoto K, Takeuchi K, Taniguchi T, Agata K, Identification and distribution of tryptophan hydroxylase (TPH)-positive neurons in the planarian Dugesia japonica. Neuroscience Research 59, 101–106 (2007). [DOI] [PubMed] [Google Scholar]
- 82.Sánchez Alvarado A, Newmark PA, Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci 96, 5049–5054 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nishimura K, Kitamura Y, Inoue T, Umesono Y, Sano S, Yoshimoto K, Inden M, Takata K, Taniguchi T, Shimohama S, Agata K, Reconstruction of dopaminergic neural network and locomotion function in planarian regenerates. Developmental Neurobiology 67, 1059–1078 (2007). [DOI] [PubMed] [Google Scholar]
- 84.Collins JJ 3rd, Hou X, Romanova EV, Lambrus BG, Miller CM, Saberi A, Sweedler JV, Newmark PA, Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol 8, e1000509 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nishimura K, Kitamura Y, Inoue T, Umesono Y, Yoshimoto K, Taniguchi T, Agata K, Characterization of tyramine beta-hydroxylase in planarian Dugesia japonica: cloning and expression. Neurochemistry International 53, 184–192 (2008). [DOI] [PubMed] [Google Scholar]
- 86.Nishimura K, Kitamura Y, Umesono Y, Takeuchi K, Takata K, Taniguchi T, Agata K, Identification of glutamic acid decarboxylase gene and distribution of GABAergic nervous system in the planarian Dugesia japonica. Neuroscience 153, 1103–1114 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Azimzadeh J, Wong ML, Downhour DM, Sanchez Alvarado A, Marshall WF, Centrosome loss in the evolution of planarians. Science 335, 461–463 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wenemoser D, Reddien PW, Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol 344, 979–991 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nishimura K, Kitamura Y, Taniguchi T, Agata K, Analysis of motor function modulated by cholinergic neurons in planarian Dugesia japonica. Neuroscience 168, 18–30 (2010). [DOI] [PubMed] [Google Scholar]
- 90.Oviedo NJ, Newmark PA, Sánchez Alvarado A, Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev Dyn 226, 326–333 (2003). [DOI] [PubMed] [Google Scholar]
- 91.Adler CE, Seidel CW, McKinney SA, Sánchez Alvarado A, Selective amputation of the pharynx identifies a FoxA-dependent regeneration program in planaria. eLife 3, e02238 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vasquez-Doorman C, Petersen CP, zic-1 Expression in planarian neoblasts after injury controls anterior pole regeneration. PLoS Genetics 10, e1004452 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marz M, Seebeck F, Bartscherer K, A Pitx transcription factor controls the establishment and maintenance of the serotonergic lineage in planarians. Development 140, 4499–4509 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.