Abstract
We recently pioneered several analyses of small-molecule sensitivity data collected from large-scale perturbation of hundreds of cancer cell lines with hundreds of small molecules, with cell viability measured as a readout of compound sensitivity. We performed these studies using cancer cell lines previously annotated with cellular, genomic, and basal gene-expression features. By combining small-molecule sensitivity data with these other datasets, we identified new candidate biomarkers of sensitivity, gained insights into small-molecule mechanisms of action, and proposed candidate hypotheses for cancer dependencies (including candidate combination therapies). Nevertheless, given the size of these datasets, we expect that many connections between cellular features and small-molecule sensitivity remain under-explored. In this chapter, we provide a step-by-step account of foundational data-analysis methods underlying our published studies, including working MATLAB code applied to our own public datasets. These procedures will allow others to repeat analyses of our data with new parameters, in additional contexts, and to adapt our procedures to their own datasets.
Keywords: computational biology, chemical biology, pharmacogenomics, biomarkers, cancer dependencies, combination therapy, public datasets, data sharing, reproducibility
1. Introduction
In this chapter, we provide a hands-on MATLAB walk-through of foundational data-analysis procedures underlying the results in the Cancer Therapeutics Response Portal (CTRP; https://portals.broadinstitute.org/ctrp/), which provides access to data and visualizations stemming from small-molecule sensitivity profiling of cancer cell lines [1–3]. When combined with large-scale genomic and transcriptomic characterization of cancer cell lines, such as those from the Cancer Cell Line Encyclopedia (CCLE) [4] and the Genomics of Drug Sensitivity in Cancer (GDSC) [5–7], small-molecule sensitivity data have been used to identify new candidate cancer dependencies [1–3,8–14]. Their availability has also stimulated development of foundational analysis methods to find novel cancer dependencies in these or similar data, such as those emerging from genetic perturbation experiments in cancer cell lines [15–18].
Foundational analysis methods for CTRP include creating cell-line sensitivity profiles [1–3] based on area-under-curve (AUC) values computed from concentration-response curves; enrichment analysis for cellular (lineage or mutational) features among cell lines sensitive to small molecules [1,2]; and cross-correlation analysis of small-molecule sensitivity with basal gene-expression patterns [3]. The data underlying CTRP have also been used in the development of several additional methods that integrate prior knowledge, harness new datasets, or result in more complex models. For example, we developed annotated cluster multidimensional enrichment (ACME) analysis [2], a novel method to combine clustering of small-molecule sensitivity data with prior knowledge (e.g., about compound targets or cellular pathways) to formulate new cancer-dependency hypotheses.
The emergence of these datasets has excited the computational biology community, as there is a need for novel method development to mine these data and uncover novel connections. Other recent methods that include a component of small-molecule sensitivity analysis include DAISY [19], DiSCoVER [20], EDDY [21], OncoGPS [22], and RWEN [23]; extensions to The Cancer Proteome Atlas (TCPA) [24,25] and GDSC itself [26]; and a number of other related studies [27–32]. This rapid proliferation of tools, methods, and comparative analyses has also sparked an important line of critical reflection about the reproducibility of findings and their concordance across different data sources with different experimental designs [33–36]. These developments evince an appetite in the computational biology community to use small-molecule sensitivity profiling data in diverse and creative ways.
Accordingly, we present this chapter to carefully describe the foundational analyses of CTRP, showcase a MATLAB reference implementation of them, and share tips, caveats, and other considerations that we encountered during their development. We cover three procedures in detail:
Procedure 1: preparation of concentration-response areas-under-curve (AUCs) as a measure of small-molecule sensitivity [1–3]
Procedure 2: enrichment analysis of mutation features among cell lines of a particular lineage sensitive to individual compounds [1,2]
Procedure 3: correlation analysis of basal gene-expression levels to compound sensitivity across panels of cell lines from a lineage [3]
In each case, the procedure selects at random an appropriate subset of available public data and is followed by a basic visualization, interpretable by users without a strong computational background, that illustrates the essential output, including statistical and quality-control measures we use to prioritize the most important connections. We focus on datasets that best illustrate the core steps in the analysis and that best highlight important tips and caveats. Our goal here is to present step-by-step instruction in the core methods, including working code, advice, and notes on the process, rather than highlight specific findings of potential biological interest. Indeed, our choice of including a random component into each analysis subset selection means not only that specific output figures will differ from run to run, but also that the code in principle could highlight a previously unappreciated biological connection. We strive to present the workflow in a manner that is logically modular, so that studies of reproducibility (by removing the random component), extensions to larger data subsets (at the cost of run-time), and the addition of other perturbations (e.g., new compounds, genetic perturbations) or alternative cellular features (e.g., proteomic, metabolomic, or epigenomic features), are straightforward for a user with baseline familiarity in MATLAB.
2. Materials
Conceptually, the analyses presented in the chapter are general enough to be carried out in any programming language. In our experience, scientific computing software, such as R (R Foundation for Statistical Computing; Vienna, Austria) or MATLAB (MathWorks, Inc.; Natick, MA), provides the appropriate balance of flexibility and the provision of built-in functions for common data manipulations. Therefore, to minimize environment configuration, and focus the discussion on the core principles of the analyses, we have based this narrative on a modular MATLAB reference implementation designed to operate on publicly available data and metadata files.
2.1. Supplementary ZIP file
The code to accompany this chapter is provided as a supplementary ZIP file (implement.zip) containing the main script (master.m) and 3 subservient procedural scripts (code\scr folder), plus all custom functions necessary to execute the procedures (code\fun folder and subfolders). The ZIP file also contains a pre-configured directory structure, with folders to house input data (data folder), input metadata (meta folder), and output tables (out folder). The supplementary ZIP file can be downloaded from GitHub (https://github.com/remontoire-pac/ctrp-reference; see Note 1). The ZIP file should be unpacked using a standard utility and placed in the local computing environment in a location that can be added to the MATLAB search path (see Note 2).
2.2. MATLAB Environment
The implementation described in this chapter was initially developed on a Dell OptiPlex 9020 (Intel i7–4790 CPU @ 2×3.60GHz, 32.0 GB RAM) running 64-bit Windows 7 Enterprise, Service Pack 1, and MATLAB 2014b. Development was completed, and main testing performed, on a virtual machine (Intel Xeon CPU E5–2695 v4 @ 2×2.10GHz, 32.0 GB RAM) running Windows Server 2016 Datacenter and MATLAB 2018a. Development was initiated in MATLAB 2014b, and this is the earliest version of MATLAB that can execute the complete set of procedures as written, due to the addition of fishertest.m in that release. The procedures require the MATLAB Statistics and Machine Learning Toolbox (called the Statistics Toolbox in MATLAB 2014b), along with the author-provided custom functions. A successful independent set of tests was completed on a Microsoft Surface Pro 4 (Intel i7–6650 CPU @ 2×2.20GHz, 16 GB RAM) running 64-bit Windows 10 Pro and MATLAB 2017b. In all 3 of these environments, the entire code package runs in 5—10 minutes and creates 3 figures, with some time variation per instantiation depending on the random component of data subset selection for analysis (see Note 3).
2.3. Downloading Public Datasets
All data and metadata required to run the analyses presented in this chapter are available for free public download from the National Cancer Institute (NCI) Office of Cancer Genomics (OCG) and were originally produced in our laboratories as part of work supported by the Cancer Target Discovery and Development (CTD2) Network of research centers (see Note 4). To complete the download and deployment of the necessary input files for analysis, the following steps are sufficient:
Navigate a web browser to the OCG FTP server link ftp://caftpd.nci.nih.gov/pub/OCG-DCC/CTD2/Broad/.
Navigate down to the directory CTRPv2.0_2015_ctd2_ExpandedDataset and download the ZIP file CTRPv2.0_2015_ctd2_ExpandedDataset.zip.
Extract the file v20.data.per_cpd_pre_qc.txt and place it in the local data folder (created by unpacking the Supplementary ZIP file implement.zip in the local environment; see Section 2.1).
Extract the files v20.meta.per_cell_line.txt, v20.meta.per_compound.txt, and v20.meta.per_experiment.txt, and place them in the local meta folder.
Return to the parent FTP directory, navigate down to the directory CTRPv2.2_2015_pub_CancerDisc_5_1210, and download the ZIP file CTRPv2.2_2015_pub_CancerDisc_5_1210.zip.
Extract the files v22.data.auc_sensitivities.txt, v22.anno.ccl_anno_features.txt, and v22.anno.ccl_mut_features.txt, and place them in the local data folder.
Extract the file v22.meta.per_compound.txt and place it in the local meta folder.
Return to the parent FTP directory, navigate down to the directory CTRPv2.1_2016_pub_NatChemBiol_12_109, and download the ZIP file CTRPv2.1_2016_pub_NatChemBiol_12_109.zip.
Extract the files v21.data.auc_sensitivities.txt and v21.data.gex_avg_log2.txt, and place them in the local data folder.
Extract the files v21.meta.gex_features.txt and v21.meta.per_compound.txt, and place them in the local meta folder.
3. Methods
Each of the following procedures features a specific analysis that assumes the user has downloaded and unpacked the author-provided files (see Section 2.1), configured an appropriate MATLAB environment (see Section 2.2), and downloaded the relevant public datasets (see Section 2.3).
3.1. Sensitivity Calculation from Small-Molecule Concentration-Response Data
Read input data and experiment metadata as DataFrames to prepare for curve-fitting analysis (master.m, lines 21–23; see Note 5).
Walk-through (Step 3) or run (skip to Step 21) the first subsidiary script (procedure1CurveFit.m) to perform curve-fit analysis (master.m, line 25; see Note 6).
Index input DataFrame with unique combinations of compound and experiment identifiers (procedure1CurveFit.m, lines 1—2; see Note 7).
Reconcile relationships between experiment identifiers and cell lines (procedure1CurveFit.m, lines 4—6; see Note 8).
Define a set of curves to be fit (procedure1CurveFit.m, lines 8–10; see Note 9).
Create an empty MATLAB structure to hold the curve-fitting results (procedure1CurveFit.m, lines 12—13).
Define concentration limits for area-under-curve (AUC) numeric integration (procedure1CurveFit.m, lines 15—17; see Note 10).
Initialize curve-fit parameters and options for non-linear fits (procedure1CurveFit.m, lines 19—22; see Note 11).
Get data points for an individual curve fit and AUC integration (procedure1CurveFit.m, lines 28—34; see Note 12).
Seed curve-fit parameters by guessing log2(EC50) from the data (procedure1CurveFit.m, lines 36—42; see Note 13).
Handle data censoring depending on the value of a pre-computed quality-control type (procedure1CurveFit.m, lines 44—72; see Note 14).
Decide whether enough data points remain to perform a curve fit (procedure1CurveFit.m, lines 75—97; see Note 15).
Provisionally fit a 3-parameter sigmoid curve to data points (procedure1CurveFit.m, lines 99—103).
Conditionally fit a 2-parameter sigmoid curve depending on apparent log2(EC50) of the provisional 3-parameter fit (procedure1CurveFit.m, lines 105–112; see Note 16).
Append the current curve-fit results to the growing MATLAB structure (procedure1CurveFit.m, lines 116–134; see Note 17).
Create output DataFrames for fitted-curve per-point and per-curve data for all fit curves (procedure1CurveFit.m, lines 138—141).
Iterate over curve-fit results structure to build DataFrames for quality control and output (procedure1CurveFit.m, lines 143—218; see Note 18).
Define post curve-fit quality-control filters and apply to DataFrames (procedure1CurveFit.m, lines 220—241; see Note 19).
Resolve experiment identifiers to cell-line identifiers (procedure1CurveFit.m, lines 243—245; see Note 20).
Write output files for downstream analysis and interpretation (procedure1CurveFit.m, lines 247—249).
Read additional metadata containing compound and cell-line information for interpretation and for figure labels (master.m, lines 27—29; see Note 21).
Select suitable curves to illustrate concentration-response curve fit and extract data (master.m, lines 31—43; see Note 22).
Plot both original data and predicted curves with error bars to produce a variant of Figure 1 (master.m, lines 45—62; see Note 23).
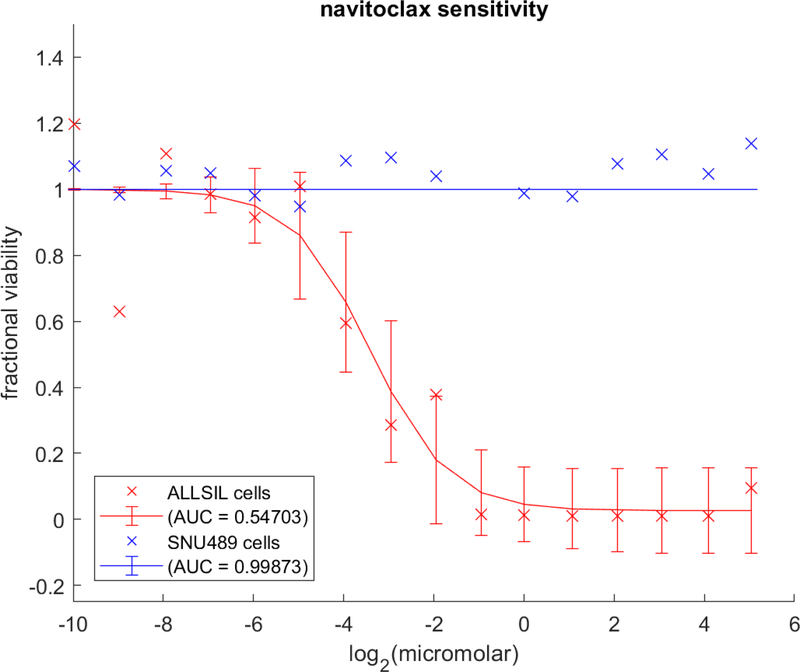
Figure 1:
Representative curve-fit visualization showing the differential sensitivity of two cell lines. Two plots are presented for each of two cell lines, one more sensitive (red) and one less sensitive (blue), to navitoclax, a compound annotated in the Cancer Therapeutics Response Portal (CTRP; http://portals.broadinstitute.org/ctrp) as an inhibitor of BCL2, BCL-xL, and BCL-W. Unconnected crosses represent the original data and are labeled in the MATLAB figure legend with the cell-line name. Line plots with error bars represent the corresponding fit curves and are labeled in the MATLAB figure legend with the computed area-under-curve (AUC).
3.2. Mutation Enrichment among Small-Molecule Sensitive Cell Lines
Read input data and metadata as DataFrames to prepare for enrichment analysis (master.m, lines 66—69; see Note 24).
Walk-through (Step 3) or run (skip to Step 16) the second subsidiary script (procedure2Enrichment.m) to perform enrichment analysis (master.m, line 71; see Note 25).
Index input DataFrame with unique combinations of compound and experiment identifiers (procedure2Enrichment.m, lines 1—2; see Note 26).
Verify that indices for rows and columns match compounds and cell lines, respectively (procedure2Enrichment.m, lines 4—6).
Create a matrix of sensitivity values for compounds by cell lines (procedure2Enrichment.m, lines 8—12; see Note 27).
Create an indicator (i.e., binary) matrix of cell lineage and histology features by cell lines (procedure2Enrichment.m, lines 15—18; see Note 28).
Define a subset of cell lines to be tested and restrict compound data to this subset (procedure2Enrichment.m, lines 20—25; see Note 29).
Create an indicator (i.e., binary) matrix of cellular mutation features by cell lines and restrict this matrix to the same cell-line subset (procedure2Enrichment.m, lines 27—34; see Note 30).
Restrict analysis to mutation features with an appropriate number of examples (procedure2Enrichment.m, lines 36—39; see Note 31).
Define a set of compounds to be tested (procedure2Enrichment.m, lines 41—43; see Note 32).
Create an empty DataFrame to hold enrichment results (procedure2Enrichment.m, lines 45—48; see Note 33).
For a given compound to be tested, verify that enough cell lines were examined, and compute raw enrichment output using Fisher’s exact tests (procedure2Enrichment.m, lines 52—58; see Note 34).
For the current compound, record cell lineage, compound, and mutation feature labels, and append labeled results to the growing result DataFrame (procedure2Enrichment.m, lines 60—66; see Note 35).
When all compounds are tested (by iterating over Steps 12—13), define statistical and quality-control filters for all enrichment results (procedure2Enrichment.m, lines 70—77; see Note 36).
Apply filters to raw enrichment results and write output files for downstream analysis and interpretation (procedure2Enrichment.m, lines 79—83; see Note 37).
Read additional metadata containing compound information for interpretation and for figure labels (master.m, lines 73—74; see Note 38).
Select suitable results to illustrate enrichment analysis and extract data (master.m, lines 76—90; see Note 39).
Perform one-sided T-test as an additional statistical annotation for boxplots (master.m, lines 92—93).
Create labels for visualizations using appropriate metadata (master.m, lines 95—98; see Note 40).
Plot enrichment results as both heatmap and boxplot representations to produce a variant of Figure 2 (master.m, lines 100—122; see Note 41).
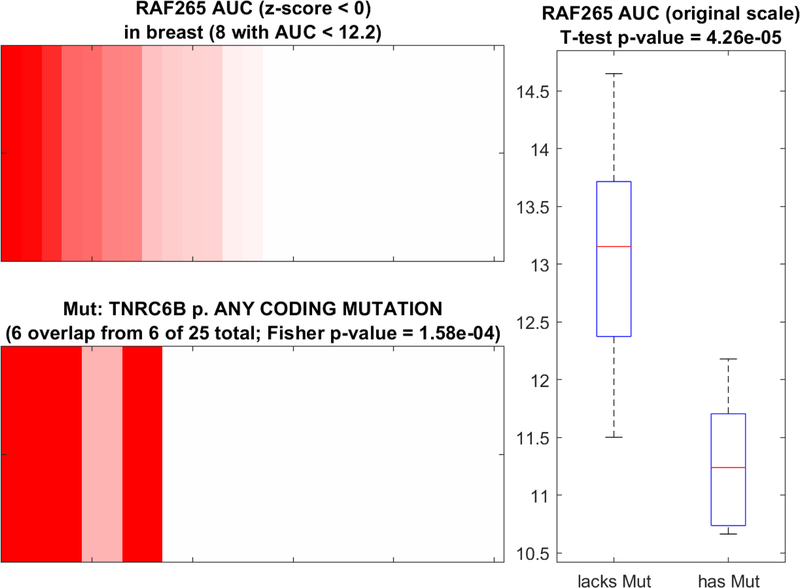
Figure 2:
Representative visualization of enrichment analysis for a single compound tested in multiple cell lines of the same type. In this case, 25 breast-derived cancer lines were tested with RAF265 (annotated in CTRP as an inhibitor of VEGFR2 and BRAF), and then sorted by area-under concentration-response curve (AUC) in the top left panel (increasing red color represents lower AUCs below the mean and therefore more sensitivity). Enrichment analysis resulted in an optimal cutoff of AUC < 12.2 which corresponds to 8 total cell lines in the bottom left panel, of which 6 carry a coding mutation in TNRC6B (red = has mutation; pink = lacks mutation). These were the only 6 TNRC6B mutants in this subset of 25 breast cancer-derived cell lines. The right panel depicts an alternative representation (box-whisker plot) and statistical analysis (t-test) of the same information, showing the relative distribution of AUC values for cell lines with or without coding mutations in TNRC6B.
3.3. Correlation of Small-Molecule Sensitivity with Basal Gene-Expression
Read input data as DataFrames to prepare for correlation analysis (master.m, lines 126—128; see Note 42).
Walk-through (Step 3) or run (skip to Step 17) the third subsidiary script (procedure3Correlation.m) to perform correlation analysis (master.m, line 130; see Note 43).
Index input DataFrame with unique combinations of compound and cell-line identifiers (procedure3Correlation.m, lines 1—2; see Note 44).
Create a matrix of sensitivity values for compounds by cell lines (procedure3Correlation.m, lines 4—8; see Note 45).
Index gene-expression DataFrame with unique combinations of gene and cell-line identifiers (procedure3Correlation.m, lines 10—11; see Note 46).
Create a matrix of gene-expression features by cell lines (procedure3Correlation.m, lines 13—17; see Note 47).
Define a subset of cell lines to be tested (procedure3Correlation.m, lines 19—22; see Note 48).
Restrict cell lines considered to those with both sensitivity and expression data (procedure3Correlation.m, lines 24—27; see Note 49).
Restrict genes considered to those with adequate dynamic range (procedure3Correlation.m, lines 29—33; see Note 50).
Restrict compounds considered to those with differentially sensitive cell lines (procedure3Correlation.m, lines 35—37; see Note 51).
Define a set of compounds to be tested (procedure3Correlation.m, lines 39—40; see Note 52).
Compute raw correlation output using normalized Pearson correlation coefficients (procedure3Correlation.m, lines 42—43; see Note 53).
Compute p-values and index to statistically filter the output correlations (procedure3Correlation.m, lines 45—48; see Note 54).
Create a results DataFrame and append cell lineage labels, plus compound and gene identifiers (procedure3Correlation.m, lines 50—57; see Note 55).
Append p-values, correlation z-scores, correlation coefficients, and the numbers of participating cell lines to results DataFrame (procedure3Correlation.m, lines 59—63; see Note 56).
Define and apply correlation quality-control filters, then write output DataFrame (procedure3Correlation.m, lines 65—86; see Note 57).
Read additional metadata containing gene information for interpretation and for figure labels (master.m, lines 132–133).
Select suitable correlation results to illustrate correlation analysis and extract data (master.m, lines 135—142; see Note 58).
Create labels for visualizations using appropriate metadata (master.m, lines 144—147; see Note 59).
Plot correlation results as a scatterplot to produce a variant of Figure 3 (master.m, lines 149—161; see Note 60).
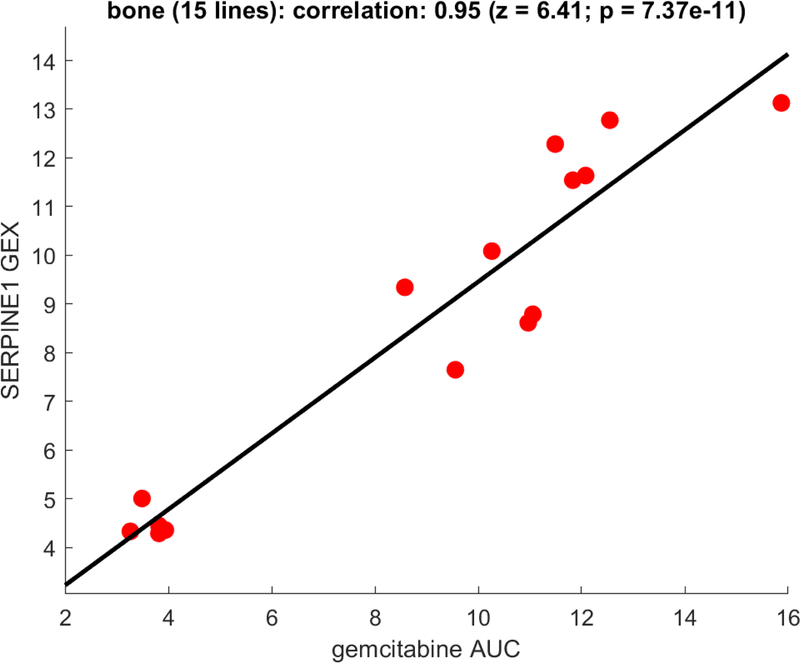
Figure 3:
Representative visualization of correlation analysis for a single compound tested in multiple cell lines of the same type. In this case, 15 bone-derived cancer lines were tested with gemcitabine (annotated in CTRP as an inhibitor of CMPK1, RRM1, TYMS). Sensitivity to gemcitabine (low AUC) is correlated with low expression of SERPINE1 in these cell lines, and each of the AUC and gene-expression distributions exhibit good dynamic ranges as described in the text (see also Note 57).
4. Notes
We anticipate that the GitHub repository may grow over time, possibly including code updates, additional procedures, and other information. However, we will keep the original version corresponding exactly to this chapter available indefinitely.
To keep the distribution file size small, we include the complete directory structure, but do not redistribute the source data and metadata. Rather, we include instructions for downloading the data and metadata from the National Cancer Institute (see also Section 2.3).
The demonstration code uses judiciously-sized subsets of compounds for each procedure to keep total run-time down while still providing a complete analysis. Our recent production dataset [2,3] includes 481 small molecules, and we imagine the provided code could be easily modified to perform a global analysis of the complete dataset.
The National Cancer Institute (NCI) has supported multiple Cancer Target Discovery and Development (CTD2) Centers nationwide in the United States through several rounds of funding with an evolving mission directed at improving cancer patient outcomes with basic research activities. The Cancer Therapeutics Response Portal (CTRP) is one flagship project resulting from the Broad Institute’s Chemical Biology and Therapeutics Science program participating in the NCI-funded CTD2 effort.
Throughout these procedures, we use a special type of MATLAB structure called a DataFrame, which was developed by Hyman Carrel in one of our laboratories (PAC) over a decade ago, inspired by data frames in R. DataFrames are MATLAB structures with one or more fields, constrained to each contain column vectors of equal length, but which collectively may mix numeric and text data types. In more modern releases of MATLAB, the utility of DataFrames has been essentially supplanted by the new MATLAB table variable type.
On first use, we recommend simply running the subsidiary script from within master.m, and skipping to Step 21. Doing so will ensure the user can get to Figure 1 more quickly and validate that procedure1CurveFit.m runs to completion in their environment. Detailed exploration of the inner workings of procedure1CurveFit.m (Steps 3—20) can be saved for later exploration.
In several steps, we make use of a special indexing function for DataFrames (DFindex.m) that allows for rapid conversion of tabular data to matrices and without requiring complete data or that tabular data be pre-sorted. We recommend studying the documentation within DFindex.m (and other DataFrame functions) to learn how it operates in detail.
During our cell-sensitivity profiling studies, we envisioned profiling data acquisition as a matrix of tests representing compounds by cell lines. As described in the relevant publications [2,3], however, the reality was less tidy. Checking the identity of cell lines by single-nucleotide-polymorphism (SNP) fingerprinting [16] revealed that sample-handling issues had resulted in a small fraction of intended cell lines being omitted, while others were inadvertently tested twice (or three times in one case). An important consequence of these practical considerations for data analysis is that the relationship of an experiment (a specific cell-line sample exposed to a compound collection) to a cell line (an abstract entity annotated with prior information about lineage, mutation, or basal gene expression) is not one-to-one. The public metadata reflects these details, and the exhibition code accounts for them.
In the demonstration, we select a single compound at random and fit all curves available for that compound. The code could easily be modified to select a specific compound of the user’s choice, and we recommend such a modification as a first step in customizing the analysis. More aggressive modifications might include studying multiple compounds, a single cell line across all compounds, or all possible curves in the dataset. However, such modifications will also require modification of the visualization code for Figure 1, since the current visualization code for curve-fitting expects a single compound.
In our earlier studies [1–3], we set limits of integration for area-under-curve (AUC) that were based on the concentrations tested for each compound individually, making the comparison of AUC values across compounds potentially problematic. In this chapter, consistent with our current best practices, we define a single set of integration limits across all compounds in the dataset, normalized from 0 (complete killing) to 1 (equivalent to untreated controls).
We seed 3 of the 4 possible sigmoid curve parameters here; the concentration parameter, log2(EC50), is seeded later, using the response data to improve the initial guess.
Within the for loop, data for each curve are collected and processed in a set of temporary variables that are reset with each loop iteration. Data kept for output are stored in a growing MATLAB structure before the loop ends.
We seed the concentration parameter, log2(EC50), at this stage, using the response data to improve the initial guess by choosing either the lowest concentration at which 50% cell killing is achieved, or the median percent killing if 50% cell killing is not achieved at any concentration.
In practice, we observed a number of different issues with data quality in our experiments, and therefore defined in data pre-processing [2] some quality-control measures (“QC-types”). Most curves either stayed flat (no compound effect) or relatively smoothly descended from no cell killing at low concentration to maximal cell killing at high concentration (QC-type 0). We observed cases where the top one (QC-type 1) or top two (QC-type 2) concentrations returned to the “no effect” baseline after observing concentration-dependent cell killing at lower concentrations. These aberrant data points are likely due to compound precipitation in the assay plate and were therefore omitted. We also observed cases where fluctuations in the data were more complex, presumably due to liquid-handling and other plate-reader artifacts (QC-type 3). In these cases, we used standard methods to censor individual data points (see the author-included function cooksdist.m for details).
In practice, we only fit curves with at least five data points passing pre-fit quality control. We regard this as a permissive choice, particularly for 16-point concentration-response experiments.
While curve-fits whose right asymptote is between 1 (no killing) and 0 (complete killing) are relatively common and may reflect a biological distinction between cytostatic effects of a compound and true cell killing, we noted a curve-fit failure mode where the predicted lower asymptote was strongly negative, which is not meaningful. This situation occurs when the predicted log2(EC50) is higher than the highest concentration tested. In such cases, we re-fit the curve with the lower asymptote constrained to zero.
To accommodate the fact that we eventually want two outputs, one with per-curve information and one with per-point information, we use an intermediate MATLAB structure to accumulate curve-fitting results and prepare the two desired outputs in a separate step.
We create two output DataFrames simultaneously. The first is for per-curve data, which is created directly by looping over the intermediate MATLAB structure, and accounts both for missing curves and whether a 3-parameter or 2-parameter curve is reported. The second is for per-point data and is created by appending to a growing DataFrame with each turn of the loop since the number of points to be included at each turn is not known in advance.
During our studies [2,3], we scanned thousands of concentration-response curves and identified multiple modes of failure. While these problematic curves represented a relatively small fraction, they fell into categories that we were able to trap computationally and exclude. Both the in-code documentation and our prior reports [2] detail the specific failure modes. These steps also illustrate the use of DFkeeprow.m, which applies the typical MATLAB logical or linear indexing to DataFrames.
To allow connection to cell-line metadata (e.g., the cell-line name), we reconcile the experiment number with the cell-line identifier at this stage (see also Note 8).
Each of our prior studies [1–3] uses a different subset of cell lines from the Cancer Cell Line Encyclopedia [4], and they also consider overlapping but not identical sets of compounds. To aid in reconciliation between datasets, we use global identifiers master_cpd_id (for compounds) and master_ccl_id (for cell lines) that have a shared meaning across all CTRP datasets.
To illustrate differential sensitivity, we choose a cell line among the top 5% of responders (sensitive) and a cell line near the median responder. These choices could easily be modified to display, for example, the most and least responsive cell lines.
The figure code is deliberately included in the calling script master.m for transparency and to allow facile modification by users without disrupting the scripts that do the calculations. Users are encouraged to further customize the appearance of figures according to their preferences.
In this procedure, two types of categorical variable are introduced, one describing the provenance of cancer cell lines as context to understand their sensitivity (primary site or lineage, histology terms, and other demographic information), and the other describing their mutational status.
On first use, we recommend simply running the subsidiary script from within master.m, and skipping to Step 16. Doing so will ensure the user can get to Figure 2 more quickly and validate that procedure2Enrichment.m runs to completion in their environment. Detailed exploration of the inner workings of procedure2Enrichment.m (Steps 3—15) can be saved for later exploration.
The original study [2] uses consecutive internal indices for compounds and cell lines in addition to the master identifiers (see also Note 21).
We anticipate missing data in the matrix of sensitivity scores by first seeding an appropriately-sized matrix with NaN (not a number) values, then filling in known values in a for loop over the indexed DataFrame.
In the present procedure, we use lineage and histology information about cell lines as a context feature to pick a subset of cell lines to study. We note that one could as easily check for enrichment of a single cell lineage versus all other lineages by treating lineage as a feature analogous to the way mutations are treated in the reference code.
We select a lineage, histology, or demographic term with at least 16 representative cell lines, but fewer than 25% of all cell lines, for illustration purposes. The user can modify these choices to expand the set of terms available or modify the code to specify a lineage of interest.
In the present procedure, we use mutation feature information as the primary type of feature for enrichment analysis, but we note that one could as easily use mutations for context (as we do here with lineage and histology information) to derive new and potentially interesting groups of cell lines, e.g., for enrichment or correlation analyses.
We choose among mutation features with at least 3 representative cell lines, but fewer than 50% of all cell lines, for illustration purposes. The user can modify these choices to expand the set of features available or modify the code to specify a mutation of interest.
We choose 12 random compounds, strictly to keep demonstration run-times low. Increasing the number of compounds, including choosing specific subsets of a user’s interest, is an obvious and recommended starting point for user customization.
The DataFrame to hold enrichment results will be grown by appending new rows because the number of rows to be appended will not, in general, be known in advance. Therefore, each field in the DataFrame is defined in advance and populated with an empty array.
The primary enrichment analysis at this stage is to perform many Fisher’s exact tests both to detect the optimal AUC cutoff for each compound and to iterate over candidate mutation features. We implement this feature using the custom function sensenfex.m. Importantly, this function avoids redundant calculations by first building an array of unique 2×2 contingency tables and tracking their relationship to metadata indices. We recommend studying the documentation within sensenfex.m to learn how it operates in detail.
The first output variable from sensenfex.m allows direct appending of mutational feature labels to the growing DataFrame along with the compound and lineage under consideration.
Experience has shown that statistical significance of an enrichment result is necessary but not sufficient to warrant continued biological interest. Accordingly, we filter on several other parameters, such as the minimum AUC (the compound must reliably kill at least one cell line), the enrichment confidence (fraction of mutant cell lines killed by the compound), the enrichment purity (fraction of sensitive cell lines harboring the mutation), and the enrichment overlap (at least two mutant cell lines must be sensitive). To ensure that the code produces at least one output for visualization, the enrichment with the best p-value is retained, even if it fails all the other criteria.
This step illustrates a relatively simple use of DFkeeprow.m to apply an accumulated set of filters (see also Note 19). To see output corresponding to those results passing each filter, a user could call DFkeeprow.m using each of the separate components (procedure2Enrichment.m, lines 71—76) in turn.
To use the consecutive compound index applied to the data from the original study [2], this procedure leverages the metadata file specific to that study (see also Note 26).
With the extensive pre-filtering of enrichment results based on confidence, purity, and overlap, the selection of data for visualization simply takes the most statistically significant result remaining after applying the filters. However, multiple parameters besides the p-value are retrieved from the results record for use in the visualization.
We note that human-readable context names and cell-line features are procured for use in the visualization directly from the result table, while compound names are procured from the master metadata. In general, we prefer a discipline where each human-readable string is stored exactly once, and database-like identifiers are used to represent data as far into a procedure as possible (e.g., until needed for visualization). We deliberately employed a mixed strategy here for illustration purposes.
The figure code is deliberately included in the calling script master.m (see also Note 23).
In this procedure, new numeric data are imported for AUCs, as well as for basal gene-expression values corresponding to our prior study of their cross-correlations [3]. However, we re-use the compound metadata from Procedure 1 and the cellular provenance information (lineage, histology, demographic) data from Procedure 2. Thus, if Procedure 3 is run in isolation, users should still load all data and metadata files specified in master.m.
On first use, we recommend simply running the subsidiary script from within master.m, and skipping to Step 17. Doing so will ensure the user can get to Figure 3 more quickly and validate that procedure3Correlation.m runs to completion in their environment. Detailed exploration of the inner workings of procedure3Correlation.m (Steps 3—16) can be saved for later exploration.
Unlike Procedure 2, here we use the master identifiers for compounds and cell lines (see also Note 21 and Note 26).
Again, we seed an appropriately-sized matrix with NaN (not a number) values to anticipate missing data (see also Note 27).
Indexing a DataFrame of gene-expression scores by cell lines works just as it does for AUCs by cell lines, but instead using a unique numeric identifier for gene names (see also Note 7).
As complete coverage of all cell lines with gene-expression data is not guaranteed, we start by seeding an appropriately-sized matrix with NaN (not a number) values (see also Note 27 and Note 45).
We select a lineage (primary site) with at least 16 representative cell lines for illustration purposes. The user can modify this choice to expand the set of terms available or modify the code to specify a lineage of interest.
Though our correlation procedure can handle missing values, we save some computation time by eliminating in advance those cell lines that have either no expression data or no sensitivity data. This step has the added benefit, as implemented, of aligning our sensitivity and gene-expression matrices so their columns correspond to the same cell-line identities as each other, in the same order.
An important idea in correlation analysis is that a gene whose expression correlates with small-molecule sensitivity has sufficient dynamic range to qualify as a potentially useful biomarker. Strong correlations with low effect sizes are less interesting. In practice, we save computation time by ruling out genes with low dynamic ranges in advance of computing correlations, but after the set of cell lines under consideration is known.
An important idea in correlation analysis is that compounds under consideration evince differential sensitivity across a set of cell lines, related to the idea of the “therapeutic window” between efficacy and toxicity. In practice, we save computation time by ruling out compounds with low dynamic ranges (or with little killing at all) in advance of computing correlations, but after the set of cell lines under consideration is known.
We choose 12 random compounds, strictly to keep demonstration run-times low (see also Note 32).
The primary correlation analysis at this stage is to perform many pairwise correlations between compound sensitivities and gene-expression levels. While we do take advantage of MATLAB’s powerful built-in pairwise similarity infrastructure, we note that accounting for missing values requires that we normalize correlation coefficients using Fisher’s z-transformation [37] to account for different numbers of cell lines participating in different comparisons. We implement these steps using the custom function nanpw2fishz.m and custom distance measure nanpwcor.m. We recommend studying the documentation within these two functions to learn how they operate in detail.
Since the output of nanpw2fishz.m is still a (potentially large) matrix of sensitivity-expression cross-correlations, we perform initial basic statistical filtering here, separately from quality-control filtering conducted downstream (in contrast to Procedure 2, where we performed both together).
We deliberately refrain from resolving compound and gene identifiers to human-readable names at this stage (see also Note 40).
For use in later visualizations, we record several (non-independent) expressions of the correlation, including the number of cell lines involved, the raw correlation coefficient, the correlation z-score from Fisher’s z-transformation [37], and a p-value derived from the Fisher’s z-transformation.
Experience has shown that statistical significance of a correlation result is necessary but not sufficient to warrant continued biological interest. Accordingly, we apply several additional filters on results to be output, including a minimum of 8 involved cell lines in the reference code (a relatively arbitrary value that is easy to modify). Most importantly, we have previously noticed many cases where a single cell line is responsible for the dynamic range of either sensitivity or gene-expression levels, and we are wary of investing much energy on such results even if their nominal p-values appear satisfactory. For nominally significant results, therefore, we censor the most extreme-valued cell line at each end of both the sensitivity and gene-expression distributions, then re-check whether the dynamic range of each vector satisfies our original criteria (see also Note 50 and Note 51). To ensure that the code produces at least one output for visualization, the correlation with the best p-value is retained, even if it fails these additional criteria.
With the extensive pre-filtering of correlation results based on dynamic-range considerations, the selection of data for visualization simply takes the largest raw (absolute) correlation coefficient result remaining after applying the filters (see also Note 39).
We note that human-readable context names are procured for use in the visualization directly from the result table in this case, while compound and gene names are procured from the master metadata (see also Note 40 and Note 55).
The figure code is deliberately included in the calling script master.m (see also Note 23).
Acknowledgements
Development of the code presented in the chapter was supported by the National Cancer Institute (NCI) through the Cancer Target Discovery and Development (CTD2) Network (grant numbers U01CA176152 and U01CA217848). The authors are grateful to Shubhroz Gill, Brittany Petros, and Bridget Wagner for helpful discussions on the manuscript.
Contributor Information
Matthew G. Rees, Cancer Biology Program, Broad Institute of Harvard and MIT; 415 Main Street, Cambridge, MA 02142.
Brinton Seashore-Ludlow, Department of Oncology-Pathology, Karolinska Institutet; Z1:00, Karolinska Universitetssjukhuset Solna 171 76 Stockholm, Sweden.
Paul A. Clemons, Chemical Biology and Therapeutics Science Program, Broad Institute of Harvard and MIT; 415 Main Street, Cambridge, MA 02142..
5. References
- 1.Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, Ebright RY, Stewart ML, Ito D, Wang S, Bracha AL, Liefeld T, Wawer M, Gilbert JC, Wilson AJ, Stransky N, Kryukov GV, Dancik V, Barretina J, Garraway LA, Hon CS, Munoz B, Bittker JA, Stockwell BR, Khabele D, Stern AM, Clemons PA, Shamji AF, Schreiber SL (2013) An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154 (5):1151–1161. doi: 10.1016/j.cell.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, Jones V, Bodycombe NE, Soule CK, Gould J, Alexander B, Li A, Montgomery P, Wawer MJ, Kuru N, Kotz JD, Hon CS, Munoz B, Liefeld T, Dancik V, Bittker JA, Palmer M, Bradner JE, Shamji AF, Clemons PA, Schreiber SL (2015) Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer Discov 5 (11):1210–1223. doi: 10.1158/2159-8290.CD-15-0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, Javaid S, Coletti ME, Jones VL, Bodycombe NE, Soule CK, Alexander B, Li A, Montgomery P, Kotz JD, Hon CS, Munoz B, Liefeld T, Dancik V, Haber DA, Clish CB, Bittker JA, Palmer M, Wagner BK, Clemons PA, Shamji AF, Schreiber SL (2016) Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol 12 (2):109–116. doi: 10.1038/nchembio.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr., de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483 (7391):603–607. doi: 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O’Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH (2012) Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483 (7391):570–575. doi: 10.1038/nature11005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, Ramaswamy S, Futreal PA, Haber DA, Stratton MR, Benes C, McDermott U, Garnett MJ (2013) Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 41 (Database issue):D955–961. doi: 10.1093/nar/gks1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Goncalves E, Barthorpe S, Lightfoot H, Cokelaer T, Greninger P, van Dyk E, Chang H, de Silva H, Heyn H, Deng X, Egan RK, Liu Q, Mironenko T, Mitropoulos X, Richardson L, Wang J, Zhang T, Moran S, Sayols S, Soleimani M, Tamborero D, Lopez-Bigas N, Ross-Macdonald P, Esteller M, Gray NS, Haber DA, Stratton MR, Benes CH, Wessels LFA, Saez-Rodriguez J, McDermott U, Garnett MJ (2016) A Landscape of Pharmacogenomic Interactions in Cancer. Cell 166 (3):740–754. doi: 10.1016/j.cell.2016.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams DJ, Ito D, Rees MG, Seashore-Ludlow B, Puyang X, Ramos AH, Cheah JH, Clemons PA, Warmuth M, Zhu P, Shamji AF, Schreiber SL (2014) NAMPT is the cellular target of STF-31-like small-molecule probes. ACS Chem Biol 9 (10):2247–2254. doi: 10.1021/cb500347p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aldrich LN, Kuo SY, Castoreno AB, Goel G, Kuballa P, Rees MG, Seashore-Ludlow BA, Cheah JH, Latorre IJ, Schreiber SL, Shamji AF, Xavier RJ (2015) Discovery of a Small-Molecule Probe for V-ATPase Function. J Am Chem Soc 137 (16):5563–5568. doi: 10.1021/jacs.5b02150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart ML, Tamayo P, Wilson AJ, Wang S, Chang YM, Kim JW, Khabele D, Shamji AF, Schreiber SL (2015) KRAS Genomic Status Predicts the Sensitivity of Ovarian Cancer Cells to Decitabine. Cancer Res 75 (14):2897–2906. doi: 10.1158/0008-5472.CAN-14-2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Waal L, Lewis TA, Rees MG, Tsherniak A, Wu X, Choi PS, Gechijian L, Hartigan C, Faloon PW, Hickey MJ, Tolliday N, Carr SA, Clemons PA, Munoz B, Wagner BK, Shamji AF, Koehler AN, Schenone M, Burgin AB, Schreiber SL, Greulich H, Meyerson M (2016) Identification of cancer-cytotoxic modulators of PDE3A by predictive chemogenomics. Nat Chem Biol 12 (2):102–108. doi: 10.1038/nchembio.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han T, Goralski M, Gaskill N, Capota E, Kim J, Ting TC, Xie Y, Williams NS, Nijhawan D (2017) Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356 (6336). doi: 10.1126/science.aal3755 [DOI] [PubMed] [Google Scholar]
- 13.Herold N, Rudd SG, Sanjiv K, Kutzner J, Bladh J, Paulin CBJ, Helleday T, Henter JI, Schaller T (2017) SAMHD1 protects cancer cells from various nucleoside-based antimetabolites. Cell Cycle 16 (11):1029–1038. doi: 10.1080/15384101.2017.1314407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, Kaffenberger SD, Eaton JK, Shimada K, Aguirre AJ, Viswanathan SR, Chattopadhyay S, Tamayo P, Yang WS, Rees MG, Chen S, Boskovic ZV, Javaid S, Huang C, Wu X, Tseng YY, Roider EM, Gao D, Cleary JM, Wolpin BM, Mesirov JP, Haber DA, Engelman JA, Boehm JS, Kotz JD, Hon CS, Chen Y, Hahn WC, Levesque MP, Doench JG, Berens ME, Shamji AF, Clemons PA, Stockwell BR, Schreiber SL (2017) Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547 (7664):453–457. doi: 10.1038/nature23007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, Jiang G, Hsiao J, Mermel CH, Getz G, Barretina J, Gopal S, Tamayo P, Gould J, Tsherniak A, Stransky N, Luo B, Ren Y, Drapkin R, Bhatia SN, Mesirov JP, Garraway LA, Meyerson M, Lander ES, Root DE, Hahn WC (2011) Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A 108 (30):12372–12377. doi: 10.1073/pnas.1109363108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, East-Seletsky A, Ali LD, Gerath WF, Pantel SE, Lizotte PH, Jiang G, Hsiao J, Tsherniak A, Dwinell E, Aoyama S, Okamoto M, Harrington W, Gelfand E, Green TM, Tomko MJ, Gopal S, Wong TC, Li H, Howell S, Stransky N, Liefeld T, Jang D, Bistline J, Hill Meyers B, Armstrong SA, Anderson KC, Stegmaier K, Reich M, Pellman D, Boehm JS, Mesirov JP, Golub TR, Root DE, Hahn WC (2014) Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data 1:140035. doi: 10.1038/sdata.2014.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, Meyers RM, Ali L, Goodale A, Lee Y, Jiang G, Hsiao J, Gerath WFJ, Howell S, Merkel E, Ghandi M, Garraway LA, Root DE, Golub TR, Boehm JS, Hahn WC (2017) Defining a Cancer Dependency Map. Cell 170 (3):564–576 e516. doi: 10.1016/j.cell.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald ER 3rd, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, Belur D, Castelletti D, Frias E, Gampa K, Golji J, Kao I, Li L, Megel P, Perkins TA, Ramadan N, Ruddy DA, Silver SJ, Sovath S, Stump M, Weber O, Widmer R, Yu J, Yu K, Yue Y, Abramowski D, Ackley E, Barrett R, Berger J, Bernard JL, Billig R, Brachmann SM, Buxton F, Caothien R, Caushi JX, Chung FS, Cortes-Cros M, deBeaumont RS, Delaunay C, Desplat A, Duong W, Dwoske DA, Eldridge RS, Farsidjani A, Feng F, Feng J, Flemming D, Forrester W, Galli GG, Gao Z, Gauter F, Gibaja V, Haas K, Hattenberger M, Hood T, Hurov KE, Jagani Z, Jenal M, Johnson JA, Jones MD, Kapoor A, Korn J, Liu J, Liu Q, Liu S, Liu Y, Loo AT, Macchi KJ, Martin T, McAllister G, Meyer A, Molle S, Pagliarini, Phadke, Repko, Schouwey, Shanahan, Shen Q, Stamm C, Stephan C, Stucke VM, Tiedt R, Varadarajan M, Venkatesan K, Vitari AC, Wallroth M, Weiler J, Zhang J, Mickanin C, Myer VE, Porter JA, Lai A, Bitter H, Lees E, Keen N, Kauffmann A, Stegmeier F, Hofmann F, Schmelzle T, Sellers WR (2017) Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell 170 (3):577–592 e510. doi: 10.1016/j.cell.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 19.Jerby-Arnon L, Pfetzer N, Waldman YY, McGarry L, James D, Shanks E, Seashore-Ludlow B, Weinstock A, Geiger T, Clemons PA, Gottlieb E, Ruppin E (2014) Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell 158 (5):1199–1209. doi: 10.1016/j.cell.2014.07.027 [DOI] [PubMed] [Google Scholar]
- 20.Hanaford AR, Archer TC, Price A, Kahlert UD, Maciaczyk J, Nikkhah G, Kim JW, Ehrenberger T, Clemons PA, Dancik V, Seashore-Ludlow B, Viswanathan V, Stewart ML, Rees MG, Shamji A, Schreiber S, Fraenkel E, Pomeroy SL, Mesirov JP, Tamayo P, Eberhart CG, Raabe EH (2016) DiSCoVERing Innovative Therapies for Rare Tumors: Combining Genetically Accurate Disease Models with In Silico Analysis to Identify Novel Therapeutic Targets. Clin Cancer Res 22 (15):3903–3914. doi: 10.1158/1078-0432.CCR-15-3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Speyer G, Mahendra D, Tran HJ, Kiefer J, Schreiber SL, Clemons PA, Dhruv H, Berens M, Kim S (2017) Differential Pathway Dependency Discovery Associated with Drug Response across Cancer Cell Lines. Pac Symp Biocomput 22:497–508. doi: 10.1142/9789813207813_0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JW, Abudayyeh OO, Yeerna H, Yeang CH, Stewart M, Jenkins RW, Kitajima S, Konieczkowski DJ, Medetgul-Ernar K, Cavazos T, Mah C, Ting S, Van Allen EM, Cohen O, McDermott J, Damato E, Aguirre AJ, Liang J, Liberzon A, Alexe G, Doench J, Ghandi M, Vazquez F, Weir BA, Tsherniak A, Subramanian A, Meneses-Cime K, Park J, Clemons P, Garraway LA, Thomas D, Boehm JS, Barbie DA, Hahn WC, Mesirov JP, Tamayo P (2017) Decomposing Oncogenic Transcriptional Signatures to Generate Maps of Divergent Cellular States. Cell Syst 5 (2):105–118 e109. doi: 10.1016/j.cels.2017.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basu A, Mitra R, Liu H, Schreiber SL, Clemons PA (2018) RWEN: Response-Weighted Elastic Net For Prediction of Chemosensitivity of Cancer Cell Lines. Bioinformatics:bty199-bty199. doi: 10.1093/bioinformatics/bty199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, Yang JY, Broom BM, Verhaak RG, Kane DW, Wakefield C, Weinstein JN, Mills GB, Liang H (2013) TCPA: a resource for cancer functional proteomics data. Nat Methods 10 (11):1046–1047. doi: 10.1038/nmeth.2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Akbani R, Zhao W, Lu Y, Weinstein JN, Mills GB, Liang H (2017) Explore, Visualize, and Analyze Functional Cancer Proteomic Data Using the Cancer Proteome Atlas. Cancer Res 77 (21):e51–e54. doi: 10.1158/0008-5472.CAN-17-0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cokelaer T, Chen E, Iorio F, Menden MP, Lightfoot H, Saez-Rodriguez J, Garnett MJ (2017) GDSCTools for Mining Pharmacogenomic Interactions in Cancer. Bioinformatics. doi: 10.1093/bioinformatics/btx744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hafner M, Niepel M, Chung M, Sorger PK (2016) Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods 13 (6):521–527. doi: 10.1038/nmeth.3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris LA, Frick PL, Garbett SP, Hardeman KN, Paudel BB, Lopez CF, Quaranta V, Tyson DR (2016) An unbiased metric of antiproliferative drug effect in vitro. Nat Methods 13 (6):497–500. doi: 10.1038/nmeth.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geeleher P, Cox NJ, Huang RS (2016) Cancer biomarker discovery is improved by accounting for variability in general levels of drug sensitivity in pre-clinical models. Genome Biol 17 (1):190. doi: 10.1186/s13059-016-1050-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikolova O, Moser R, Kemp C, Gonen M, Margolin AA (2017) Modeling gene-wise dependencies improves the identification of drug response biomarkers in cancer studies. Bioinformatics 33 (9):1362–1369. doi: 10.1093/bioinformatics/btw836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen B, Ma L, Paik H, Sirota M, Wei W, Chua MS, So S, Butte AJ (2017) Reversal of cancer gene expression correlates with drug efficacy and reveals therapeutic targets. Nat Commun 8:16022. doi: 10.1038/ncomms16022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Li X, Zhang L, Gao Q (2017) Improved anticancer drug response prediction in cell lines using matrix factorization with similarity regularization. BMC Cancer 17 (1):513. doi: 10.1186/s12885-017-3500-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJ, Quackenbush J (2013) Inconsistency in large pharmacogenomic studies. Nature 504 (7480):389–393. doi: 10.1038/nature12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Cell Line Encyclopedia C, Genomics of Drug Sensitivity in Cancer C (2015) Pharmacogenomic agreement between two cancer cell line data sets. Nature 528 (7580):84–87. doi: 10.1038/nature15736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Safikhani Z, El-Hachem N, Quevedo R, Smirnov P, Goldenberg A, Juul Birkbak N, Mason C, Hatzis C, Shi L, Aerts HJ, Quackenbush J, Haibe-Kains B (2016) Assessment of pharmacogenomic agreement. F1000Res 5:825. doi: 10.12688/f1000research.8705.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Safikhani Z, Smirnov P, Freeman M, El-Hachem N, She A, Rene Q, Goldenberg A, Birkbak NJ, Hatzis C, Shi L, Beck AH, Aerts H, Quackenbush J, Haibe-Kains B (2016) Revisiting inconsistency in large pharmacogenomic studies. F1000Res 5:2333. doi: 10.12688/f1000research.9611.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dancik V, Carrel H, Bodycombe NE, Seiler KP, Fomina-Yadlin D, Kubicek ST, Hartwell K, Shamji AF, Wagner BK, Clemons PA (2014) Connecting Small Molecules with Similar Assay Performance Profiles Leads to New Biological Hypotheses. J Biomol Screen 19 (5):771–781. doi: 10.1177/1087057113520226 [DOI] [PMC free article] [PubMed] [Google Scholar]