Abstract
Injectable hydrogels present several advantages over prefabricated scaffolds including ease of delivery, shear-thinning property, and broad applicability in the fields of drug delivery and tissue engineering. Here, we report an approach to develop injectable hydrogels with sustained drug release properties, exploiting the chemical nature of the DNA backbone and silicate nanodisks. A two-step gelation method is implemented for generating a combination of noncovalent network points, leading to a physically cross-linked hydrogel. The first step initiates the development of an interconnected structure by utilizing DNA denaturation and rehybridization mechanism to form hydrogen bonds between complementary base pairs of neighboring DNA strands. The anisotropic charge distribution of two-dimensional silicate nanodisks (nSi) makes them an active center in the second step of the gelation process. Silicate nanodisks create additional network points via attractive electrostatic interactions with the DNA backbone, thereby enhancing the mechanical resilience of the formulated hydrogel. The thermally stable hydrogels displayed an increase in elasticity and yield stress as a function of nSi concentration. They were able to form self-supporting structures post injection due to their rapid recovery after removal of cyclic stress. Moreover, the presence of nanosilicate was shown to modulate the release of a model osteogenic drug dexamethasone (Dex). The bioactivity of released Dex was confirmed from in vitro osteogenic differentiation of human adipose stem cells and in vivo bone formation in a rat cranial bone defect model. Overall, our DNA-based nanocomposite hydrogel obtained from a combination of noncovalent network points can serve as an injectable material for bone regeneration and carrier for sustained release of therapeutics.
Keywords: nanocomposites, DNA, two-dimensional nanosilicates, physical cross-linking, injectable hydrogels, controlled release
Graphical Abstract

Physically cross-linked injectable hydrogels are three-dimensional networks where the polymeric chains are held together through noncovalent interactions such as electrostatic forces, hydrogen bonds, and hydrophobic interactions.1-3 They have been extensively investigated for biomedical and pharmaceutical applications due to their noninvasive nature and advantages with respect to chemically conjugated networks. The primary advantage of physical hydrogels is their ability to form an interconnected structure without the need of any toxic cross-linking agents which can hinder the biological activity of encapsulated biomolecules or pose biocompatibility issues. Shear-thinning physical hydrogels can easily be injected using a fine gauge needle, and gelation can occur almost instantly following injection owing to their ability to reassemble after the removal of the shear force. Hence, these materials have become increasingly important for bone regenerative applications, particularly for cranial repair.4,5 Currently, available treatment methods largely depend on invasive techniques, where the biomaterials are surgically placed in the bone defect sites.4 On the contrary, injectable materials offer the advantage of direct delivery into the defect site along with entrapped therapeutics. From a clinical perspective, minimally invasive procedures lower the risk of infection, reduce the extensive blood loss, and avoid surgical scar formation. Furthermore, injectable hydrogels can fill up any desired shape and hence can be used to treat irregular defects in contrast to prefabricated scaffolds.
Natural polysaccharides and proteins such as alginate, hyaluronic acid, chitosan, gelatin, or collagen have been extensively investigated for the preparation of injectable hydrogels due to their higher biocompatibility when compared to that of synthetic polymers.1,6,7 However, in most of the cases, there is a need for an extra functionalization step or the addition of a second polymer to introduce a reactive building block for physical cross-linking.1,8-10 A more advanced strategy to produce injectable hydrogel is by selection of polymers that can self-assemble into defined structures due to the presence of specific molecular recognition motifs. DNA is a natural biopolymer, and there is an immense potential for treating DNA as a chemical entity instead of a genetic material. DNA possesses many desirable properties such as high molecular weight and presence of available chemical functionalities on the backbone for chemical and physical bonding to other bioactive substances.11-13 Additionally, the base-pairing interactions are highly specific, leading to the well-known double-helix structure. Given these features, DNA can be employed for the fabrication of injectable hydrogels for biomedical applications. Several mechanisms have been explored to design DNA-based hydrogels, including ligase-mediated reactions, combination with cationic materials or ionic liquids, covalent bond formation, and complementary base pairing.12,14-18 However, only a few reports have investigated the possibility to form injectable hydrogels by exploring the inherent noncovalent interactions of DNA.19-21
Recent studies have shown the potential of nanosilicates to form a physically cross-linked network via noncovalent interactions with various polymers.22,23 Nanosilicates (Laponite XLS) are two-dimensional (2D) disk-shaped synthetic silicates with a diameter of 20–30 nm and thickness of ~1 nm.24-26 They have anisotropic charge distribution on their surface that can be leveraged for their interaction with phosphate groups of the DNA backbone. Specifically, the edges of the silicate nanodisks (nSi) have positive charge, whereas the top and bottom surfaces are negatively charged.27 Thus, they can form reversible electrostatic bonds with charged polymers which can dynamically form and break. This phenomenon makes nSi an ideal candidate to be used in the design of shear-thinning injectable hydrogels. The reversible nature of the electrostatic bonds allows for immediate recovery after injection, thereby restoring the prestrained moduli upon removal of shear. Hence, nSi can be used as a component in the DNA-based hydrogel to assist the formation of a three-dimensional (3D) network via electrostatic interactions. No previous study has focused on harnessing the anisotropic charge distribution of nanosilicates for the fabrication of DNA-based injectable hydrogels. Additionally, due to the aforementioned properties, silicate nanodisks can modulate the release of bioactive molecules including drugs, proteins, and vaccines.28 For instance, the high surface area and the presence of anisotropic charges enable the high loading efficiency and the sustained release of bioactive molecules including antigens.
Herein, we report the formation of an injectable hydrogel based on DNA and silicate nanodisks that can be prepared by a facile technique of heating and mixing. The proposed strategy presents the advantage of developing physically cross-linked networks without the need of chemical modification of the DNA backbone or design of specific DNA base sequences. The formulated hydrogel is created by the combination of two different types of network points: type A, interconnections between neighboring DNA strands induced by the heating step; type B, linkages due to the interaction between nSi and the DNA backbone. We examined the effect of nSi on shear-thinning and injectability properties of the hydrogel by investigating the rheological behavior of different formulations. Additionally, the specific electrostatic interactions between nSi and DNA, responsible for the network formation, were thoroughly investigated using X-ray photoelectron spectroscopy (XPS) analysis. Finally, as a proof of concept, the ability of the formulated nanocomposite hydrogels to modulate the release of the model drug dexamethasone (Dex) was monitored in vitro. We envision that the developed DNA-based nanocomposite hydrogel has the potential to be used in a broad range of biomedical applications, such as injectable hydrogels for tissue regeneration, drug-eluting functional coatings, sustained drug delivery vehicles, and bioinks for tissue engineering.
RESULTS AND DISCUSSION
Shear-Thinning Injectable DNA–nSi Nanocomposite Hydrogels with a Porous Structure.
In this study, DNA is used as the base polymer to form physical hydrogels. A fundamental structural characteristic of DNA is the specific base-pairing interactions. These interactions were exploited in the first step of thermal gelation, by promoting the base pairing among adjacent strands of DNA (Figure 1). As the first step, a 4% (w/v) DNA solution in water was heated at 90 °C for different time intervals ranging from 30 s to 30 min followed by a cooling step at 37 °C. DNA concentrations lower than 4% (w/v) were not studied as the sample analysis led to inconsistent results. A temperature of 90 °C is required to denature the double helix of DNA, by breaking hydrogen bonds between complementary base pairs.29 The subsequent cooling phase is necessary to allow the rehybridization of the DNA strands in a random conformation through interactions with base pairs of neighboring strands. The whole process leads to the formation of an interconnected structure (type A network) that exhibits different properties when compared to the original DNA solution that did not undergo the heating—cooling treatment.
Figure 1.
Schematic representation of the design strategy for the development of multifunctional nanocomposite hydrogels. DNA—nSi injectable hydrogels are formed via a two-step gelation method. The first step consists of an intermediate weak gel (pregel) formation by heating and subsequent cooling of double-stranded DNA. The denaturation of double-stranded DNA followed by rehybridization in a random fashion facilitates the development of interconnections between adjacent DNA strands (type A network points) via complementary base pairing. Introduction of nSi in the second step of the gelation process increases the number of network points (type B) via electrostatic interaction with the DNA backbone, resulting in a shear-thinning injectable hydrogel.
We monitored the physical changes in the different systems by performing rheological measurements on the samples to detect the most suitable heating time. The goal of this part of the study was to identify the optimum heating interval required to obtain an intermediate weak DNA-based network (pregel) that could be mixed with nSi to form the nanocomposite hydrogel (Figure 1). An increase in the duration of the heating step induced a change of the DNA solutions from a sol (G″ > G′) to a gel state (G′ > G″) (Figure S1a). Oscillatory shear rheology confirmed the formation of a solid-like material (G′ > G″) for heating times longer than 30 s. Precisely, the analysis of the tan δ profiles, which is defined as the ratio of loss modulus (G′′) to storage modulus (G′), displayed a constant decrease of this parameter with prolonged heating times (Figure S1b). At short heating times corresponding to 30 s, the supplied heat energy was not sufficient enough to unwind a substantial section of the DNA strand. Hence, this formulation displayed tan δ values greater than 1, indicative of a liquid-like material (G″ > G′) with similar properties to the DNA solution that did not receive any thermal treatment. On the contrary, tan δ values below 1 indicate the formation of a cross-linked network, and the lower is this parameter, the more interconnected is the corresponding hydrogel. Additionally, frequency sweeps carried out in the frequency range of 0.01 to 10 Hz showed an increase in the G′ values from 99 to 460 Pa at 1 Hz corresponding to the samples heated for 30 s and 30 min, respectively. However, the G′ profile for the 30 s sample was different from the other systems since this formulation was in a sol state, and thus the G′ values were strongly affected by the frequency applied (Figure S1c). These data demonstrate a direct relationship between the degree of network cross-linking and the duration of the heating step. A heating time of 45 s was sufficient enough to form a weak DNA-based gel (G′ > G″), and this parameter was kept constant in the following optimization studies. Longer heating intervals were not selected as the corresponding gels were too cross-linked to be mixed with nSi.
To further confirm the formation of an interconnected network with a heating process of only 45 s, we investigated the difference in viscosity following the thermal treatment. An increase in viscosity was observed in the whole range of investigated shear rates, which supports the hypothesis of structural changes occurring in the DNA solution during the process of pregel formation (Figure S2a). Furthermore, absorbance measurements of the DNA solution before and after 45 s of heating confirmed the separation of the DNA strands during the heating phase (Figure S2b,c). Both groups exhibited a maximum absorbance at ~260 nm because of the electronic structure of purine and pyrimidine bases present in DNA.30 However, the absorbance of the pregel at 260 nm was slightly higher than that of the starting DNA solution. This change is likely due to the DNA strand separation leading to the partial unwinding of the DNA double helix during the heating cycle. After the rehybridization step occurring during the cooling phase, some of the base pairs end up remaining free, which in turn are responsible for the increase in absorbance detected at 260 nm.29 These combined results indicate the successful formation of a pregel structure.
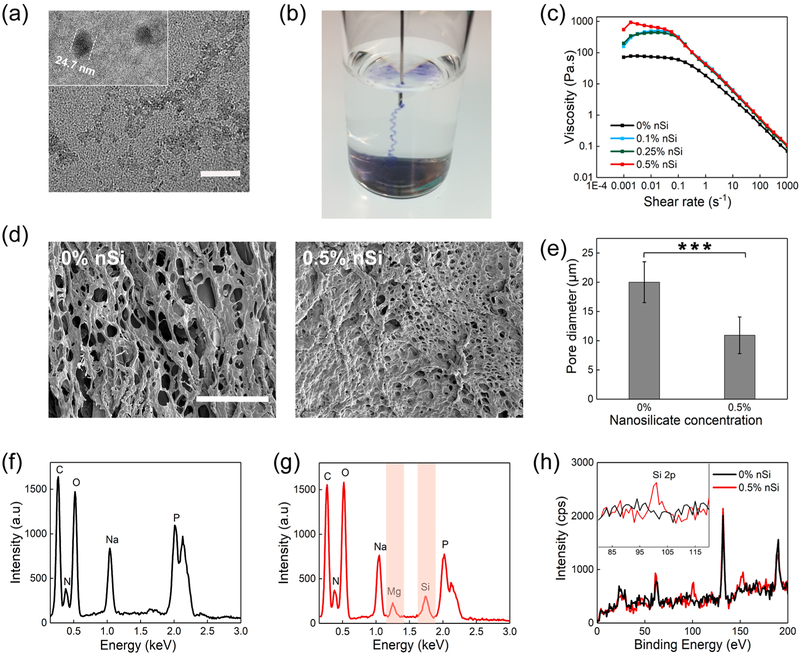
In the second step, different concentrations of nSi dispersions were mixed with the optimized pregel formulation to form the nanocomposite DNA-based hydrogel (Figure 1b). Silicate nanodisks are 20–25 nm in diameter, as confirmed using transmission electron microscopy (TEM) (Figure 2a). The anisotropically charged two-dimensional, ultrathin structure of nSi (1–2 nm) with high surface-to-volume ratio facilitates the physical interaction of the nanodisks with the DNA matrix to form physically cross-linked hydrogels via the development of type B network points.31 nSi was dispersed in Milli-Q water and vortexed to form an exfoliated dispersion with a high surface area, before inclusion into the DNA pregel. Thereafter, the nSi dispersion was added to the pregel at room temperature followed by a vigorous vortexing treatment to prevent the aggregation of the nanosilicate disks. Nano-composite hydrogels were developed consisting of three different nSi concentrations ranging from 0.1 to 0.5% (w/v) labeled as 0.1% nSi, 0.25% nSi, and 0.5% nSi. All of the formulated DNA—nSi physically cross-linked hydrogels could be easily injected using a 22 gauge needle (Figure 2b and Movie S1) due to their shear-thinning behavior (Figure 2c). This property is an essential requisite for injectable hydrogels as they should be able to flow with increasing shear rates.32
Figure 2.
Morphology, composition, and shear-thinning characteristics of the nanocomposite hydrogels. (a) TEM image displaying the nSi dispersion in water (scale bar = 100 nm). (b) Image showing the injection of the blue colored hydrogel through a 22 gauge surgical needle. (c) Viscosity vs shear rate plots illustrate an increase in the viscosity due to the presence of nSi. All the formulations display the typical shear-thinning behavior with a reduction in the viscosity as the shear rate increases. (d) Scanning electron microscopy images of the DNA-based hydrogel showing a highly porous structure. The inclusion of nSi reduces the pore size of the hydrogel network (scale bar =100 μm). (e) Measurement of pore diameters by ImageJ, displaying a significant decrease in pore size of the hydrogel network upon addition of nSi. Results are shown as mean ± standard deviation (n = 50) (*p < 0.05, **p < 0.01, ***p < 0.001). (f) Energy-dispersive X-ray spectra of 0% nSi (i.e., DNA gel without nSi) and (g) 0.5% nSi (i.e., DNA gel with 0.5% nSi) indicate the presence of Si and Mg in the nanocomposite hydrogel. (h) XPS of 0.5% nSi hydrogels confirming the presence of nSi in the nanocomposite hydrogel. The Si 2p peak was visible at around 100 eV.
The interaction between the DNA backbone and nSi was detected by monitoring the viscosity of the hydrogel formulations as a function of shear rate starting from 0.001 to 1000 s−1. As the concentration of nSi was increased, a corresponding change in viscosity was monitored (Figure 2c). As expected, an increase in the concentration of nSi resulted in an increment in viscosity, which is commonly observed for nSi-based nanocomposite systems. Additionally, we investigated the porosity of the hydrogels as this parameter can affect the drug release capabilities of the formulated injectable hydrogels. The DNA system without any silicate possesses a highly porous interconnected structure, as observed by scanning electron microscopy (SEM) analysis. The inclusion of nSi caused a reduction in the porosity of the hydrogel, which is indicative of a more compact network with a higher degree of physical cross-linking (Figure 2d). Specifically, the incorporation of nSi at 0.5% (w/v) caused a significant reduction in the average pore diameter from 20.22 to 10.81 μm (Figure 2e). These findings are consistent with previous reports focused on the characterization of nSi-based nanocomposite hydrogels.33 Further analysis of the internal porosity was obtained by performing a microcomputed tomography (microCT) analysis, which can better display the interconnected three-dimensional porous structure of the nanocomposite hydrogel compared to SEM analysis (Movie S2).
The presence of nSi in the hydrogel was detected via energy-dispersive X-ray spectroscopy (EDX). The EDX spectra of 0% nSi (Figure 2f) showed the peaks corresponding to carbon, nitrogen, oxygen, and phosphorus that are the constitutional elements found in the DNA. The spectra also displayed an additional peak relative to sodium because the DNA used in this study presented phosphate groups coupled with sodium counterions. The EDX spectra of the 0.5% nSi system showed the presence of Mg and Si signals, which are the major components of silicate nanodisks (Figure 2g). The presence of nSi in the cross-linked hydrogel network was further confirmed by XPS (Figure 2h). The peak at ~100 eV corresponded to Si 2p, which was absent in the system with 0% nSi.34 Overall, these results indicate the successful formation of an injectable shear-thinning nanocomposite hydrogel which displays a porous structure.
Mechanical and Structural Characterization of Shear-Thinning Hydrogels.
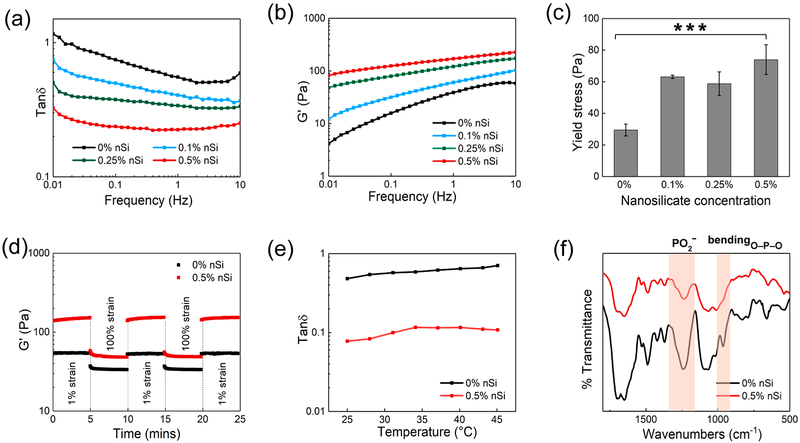
Oscillatory shear rheology was performed to investigate the effect of nSi on the mechanical properties of the nanocomposite DNA-based hydrogel. Preliminary strain sweep measurements were carried out to detect the linear viscoelastic (LVE) region of each sample. Subsequently, frequency sweep analysis was performed in the LVE region at frequencies ranging from 0.01 up to 10 Hz. The pregel formulation (0% nSi) exhibited the highest values of tan δ, and a corresponding decrease in this parameter was observed as the concentration of nSi was increased (Figure 3a). These results can be attributed to the ability of the nanosilicate to form a physically cross-linked network when combined with DNA. Silicate nanodisks interact electrostatically with the negatively charged phosphate groups of the DNA backbone due to the presence of positive charges on its edges.27,35 These interactions modulate the elasticity of the nanocomposite hydrogel, and the formation of a large number of these network points can be responsible for the observed increase in the storage moduli values (Figure 3b).
Figure 3.
Mechanical and structural characterization of nanocomposite hydrogels as injectable materials. (a) Tan δ (G″/G′) profiles for the nanocomposite hydrogels over a range of frequency from 0.01 to 10 Hz. (b) Frequency sweep experiments performed in the range of 0.01 to 10 Hz indicate an increase in storage modulus as the concentration of nSi was increased. (c) Plot of yield stress as a function of nSi concentration. Results are reported as mean ± standard deviation (n = 3) (***p < 0.001). (d) Recovery data obtained by monitoring the storage modulus of the nanocomposite hydrogels while subjecting them to alternating high (100%) and low (1%) strain conditions. Both 0% nSi (i.e., DNA gel without nSi) and 0.5% nSi (i.e., DNA gel with 0.5% nSi) exhibited more than 95% recovery. (e) Temperature sweeps carried out from 25 to 45 °C. Tan δ values displayed no significant changes in the range of experimental temperatures. (f) Fourier transform infrared spectra of the DNA hydrogel with and without nSi. The highlighted bands (asymmetric stretching and bending of phosphate group) indicate the electrostatic interactions between the nanosilicate edges and DNA.
An injectable material should form self-supporting structures following injection, which allows the hydrogel to remain in the defect area without flowing away into adjacent sites. Hence, we investigated the effect of nSi in modulating the yield stress of the DNA-based hydrogels. Yield stress was defined as the crossover point between G′ and G″ in the oscillatory shear stress sweeps.36 An increase in the yield stress from 29.45 ± 3.75 to 73.98 ± 9.42 Pa was observed with an increase in the nSi concentration from 0 to 0.5%, respectively (Figure 3c). The observed increment is likely due to the reinforcement of the DNA network induced by the dispersed nanosilicate. Upon injection, the nanocomposite hydrogel was capable of retaining its shape when compared to the starting DNA solution (Figure S3a). Such an effect has been reported in similar studies investigating the influence of nSi on the mechanical properties of polymeric nanocomposite hydrogels.37 No significant change in the values of yield stress was detected when the temperature was varied from 25 to 37 °C for all the formulations (Figure S3b). This result indicates that each one of the designed DNA-based nanocomposite hydrogels possesses thermal stability and can be injected without losing its structural integrity as the temperature increases. However, the system containing 0.5% w/v nSi exhibited the highest yield strength and elasticity, and this formulation was chosen as the optimal group for further testing. Hydrogels with nSi concentrations greater than 0.5% w/v were not studied as the starting nSi dispersions were too viscous to attain a uniform distribution of the nanodisks in the pregel matrix.
Another essential aspect to consider while designing injectable nanocomposite hydrogels is to verify whether the material can sustain cyclic strain and regain its original structure once the shear strain is removed. Oscillatory strain amplitude studies were carried out by applying two consecutive cycles of high (100%) and low (1%) strain. The variation of the storage modulus (G′) was monitored, and complete recovery of G′, within seconds, was observed for both the formulation with and without the nanosilicate (Figures 3d and S3c). However, a higher value of G′ was detected for the nanocomposite system. Overall, the formulated physically cross-linked network exhibited rapid recovery and restoration of the storage modulus following the removal of stress. Additionally, the hydrogel containing 0.5% nSi maintained its elastic properties as the temperature was increased from 25 to 45 °C (Figure 3e). These data confirmed the stability of the formulated nanocomposite hydrogel over a wide range of temperatures that the sample might experience during storage and post injection in vivo. All the advantages mentioned so far were possible using a relatively low concentration of silicate nanodisks (0.5% w/v) as compared to other nanocomposite systems containing nSi.38,39 Using a lower concentration of nanosilicate can also be beneficial to reduce the possible cytotoxic effects.40
Finally, we compared the rheological properties of the nanocomposite hydrogels with and without the preliminary heating step to evaluate the effect of thermal energy on the gel strength, elasticity, and self-healing properties. The unheated sample formed by mixing both DNA and nSi is composed of only type B network points (Figure S4a). The storage modulus (G′) of the unheated sample was higher than the one that did not undergo a preliminary heating step (Figure S4b). Despite the higher G′ values, the relatively similar tan δ for both the groups indicated a higher loss modulus for the unheated sample compared to that of the heated one (Figure S4c). This can be attributed to the absence of the base network of interconnected DNA strands (type A network points) in the unheated sample. The heating step guarantees the formation of a more stable and flexible network that displayed higher yield stress (Figure S4d) and a faster recovery of its original storage moduli values (Figure S4e). Additionally, the absence of a preheating step caused a reduction in the percentage of recovery which was only equal to 60% of the original G′ values for the unheated formulation (Figure S4f). Hence, it was confirmed that the initial heating step imparts a profound beneficial effect on the gel mechanical properties. The interconnections with neighboring DNA chains in the pregel are not sufficient to form a stable gel, but these preliminary linkages (type A network points) are necessary to maximize the strength and the elasticity of the hydrogel. Overall, the difference in rheological responses of the nanocomposite hydrogels provides conclusive evidence that both the heating step and addition of nSi assisted in the formation of a mechanically resilient elastic hydrogel, resulting from a combination of type A and type B network points.
To identify the electrostatic interactions between nSi and the DNA backbone, Fourier transform infrared spectroscopy (FTIR) was carried out on the samples with and without the nanosilicate. Pure DNA hydrogels exhibited asymmetric stretching vibrations of a phosphate group (PO2−) at 1241 cm−1 and O─P─O bending vibration at 963 cm−1 (Figure 3f). Both bands are characteristic spectral signatures of a double-stranded DNA structure.41-43 However, a red shift of the PO2- asymmetric stretching to 1236 cm−1 as well as the decrease in the peak intensity was observed upon addition of nSi. Moreover, the disappearance of the sharp peak at 963 cm−1 corresponding to the bending of phosphate anion suggested a possible change in the molecular environment. The observed results were consistent with previous studies reporting the interaction of DNA with various cationic elements.43 These findings indicate the binding of the positive edges of nSi with the phosphate group in the DNA backbone, which was further confirmed via XPS.
Electrostatic Interactions of Silicate Nanodisks with the DNA Backbone for Hydrogel Formation.
XPS studies were performed to validate the type of interactions between DNA and nSi and interpret the function of nSi in reinforcing the DNA-based hydrogel network. XPS spectra of oxygen (O 1s), phosphorus (P 2p), and nitrogen (N 1s) were recorded to evaluate the differences between the two formulations with and without nSi. Differences in peak shapes and peak width were found by comparing the O 1s spectrum of the DNA network (0% nSi) and the nanocomposite hydrogel (0.5% nSi) (Figure 4a). Similarly, the P 2p peak appeared at ~132.8 eV for the 0% nSi sample, whereas a shift of the same peak to higher binding energy at ~133.32 eV was observed for the sample containing nSi (Figure 4b). These results indicate a change in the electronic environment in the vicinity of oxygen and phosphorus. On the contrary, no significant difference was detected in the N 1s peak position and shape for the two systems (Figure 4c).
Figure 4.
XPS analysis of the nanocomposite DNA-based hydrogel. Comparison of high-resolution XPS spectra of 0% nSi (i.e., DNA gel without nSi) and 0.5% nSi (i.e., DNA gel with 0.5% nSi) for (a) oxygen (O 1s), (b) phosphorus (P 2p), and (c) nitrogen (N 1s). Comparison of the deconvoluted peaks of (d) O 1s, (e) P 2p, and (f) N 1s for both 0% nSi and 0.5% nSi systems. The experimental data points are shown as solid black lines. The rearrangement of the O 1s peak components and the shift of P 2p peak confirmed the presence of attractive electrostatic interactions between the oxygen atom of phosphate anion (PO2−) and silicate nanodisks. (g) Proposed mechanism for physical cross-linking between the DNA backbone and silicate nanodisks.
For a better insight, a more detailed analysis was performed by deconvoluting each core level spectra into their constituents. A summary of the peak positions for each subcomponent and their respective assignment to specific groups of DNA is presented in Table S1. The O 1s spectrum was deconvoluted into two components, O1 and O2 (Figure 4d). O1 at 530.86 eV was assigned to the C═O, P═O, and P─O− groups, whereas the O2 peak at 533.07 eV corresponded to the C─O─C and C─O─P groups of DNA. The results are in agreement with previous XPS reports investigating DNA.44 The O 1s spectrum of the nanocomposite hydrogel (0.5% nSi) exhibits an apparent redistribution between the O2 and O2 components (Figure 4d). The area under the peak of O1 diminishes with the simultaneous increase in the area under the peak for O2. Such an observation can be explained as a shift in a fraction of the O1 component to higher binding energy, thereby increasing the area under the peak O2. The shift can be attributed to a decrease in electron density in the vicinity of O1 sites due to the attractive electrostatic interactions with positively charged edges of silicate nanodisks.
The presence of attractive electrostatic forces with oxygen was also confirmed by analyzing the XPS spectrum of P 2p. The core level P 2p spectrum was split into two spin doubletss, P1 and P2, with an area ratio of 2:1, respectively. For 0% nSi sample, P1 at 131.95 eV corresponds to P 2p3/2, whereas P2 was assigned to P 2p1/2 at 132.8 eV (Figure 4e). These data are consistent with the reported literature.45 The nanocomposite hydrogel (0.5% nSi) exhibited the same peaks P1 and P2, but a shift toward higher binding energies was observed. Specifically, an increase in binding energy by 0.5 and 0.52 eV was detected for P1 and P2 peaks, respectively (Figure 4e). The electrostatic attraction between the electropositive region of nSi with the negatively charged oxygen of the phosphate group destabilizes the electronic environment of the nearest phosphorus atom. This leads to a decrease in electron density around phosphorus, resulting in the increment of binding energies.46 Similar effects are described for complexes of DNA-like molecules with cations such as Ce4+.47
To rule out the possibility of strong interaction of nSi with the nitrogenous bases of DNA, we compared the deconvoluted N 1s spectra for both systems. The core level N 1s was deconvoluted into two peaks, N1 and N2, originating due to the different chemical environment of the nitrogen atoms. The N1 peak was assigned to the nitrogen atoms of C═N─C and C─NH2 groups corresponding to a binding energy of 398.05 eV (Figure 4f). Additionally, the peak of N2 at 399.33 eV was generated from the nitrogen atoms present in N─C─O and N─C═O functionalities.44 There was no substantial change in the peak distributions of N1 and N2 upon introduction of nSi, indicating minimum interactions of the nanosilicate with the nitrogen atom of the DNA bases (Figure 4f). Overall, the XPS results confirmed the presence of attractive electrostatic interactions between nSi and the oxygen atom of the phosphate anion (PO2−) in the DNA backbone. The physical cross-linking mechanism derived from the XPS results depicts interconnection of neighboring DNA strands via silicate nanodisks (Figure 4g). These additional network points are the principal rationale for the observed increase in gel strength, elasticity, and yield stress compared to the DNA-based gel without nSi.
Several reports have focused on studying the effect of physical interactions between the specific binding sites of DNA and metal ions.48,49 Similarly, binding with positively charged ions has shown to alter the structure and conformation of DNA.49 To our knowledge, no previous study has leveraged the potential of DNA to form three-dimensional network structures with nSi.20,50-52 Our study moves a step forward in utilizing the noncovalent interactions of DNA to create shear-thinning nanocomposite hydrogels with rapid self-healing properties.
Multifunctionality of the Biocompatible Hydrogel.
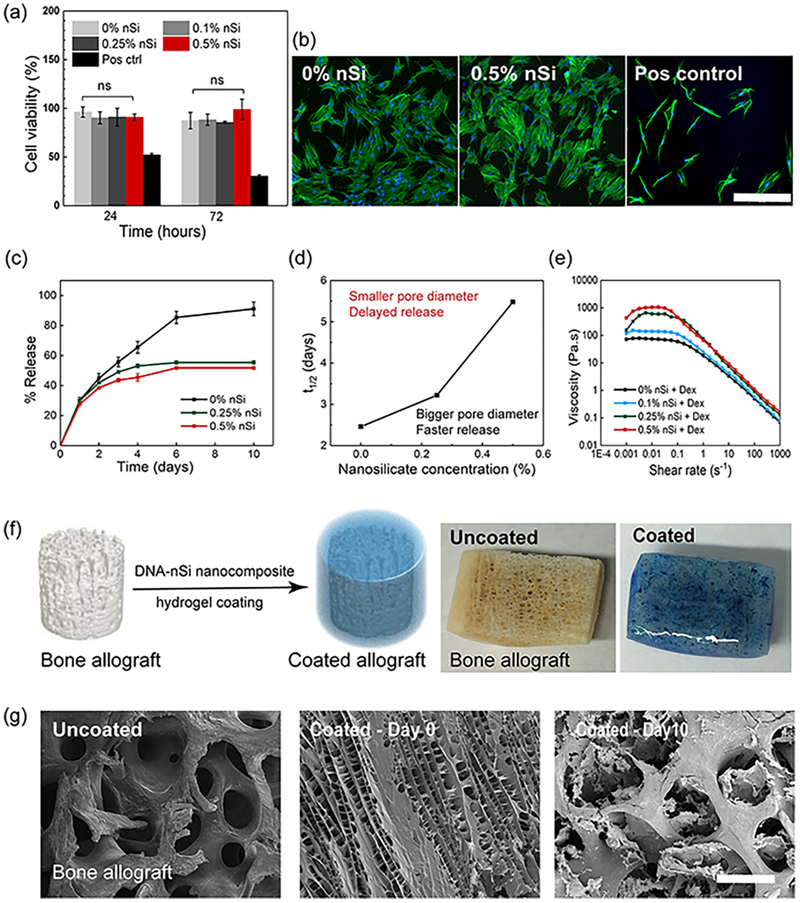
Biocompatibility is a prime requirement for a material to be used in biomedical applications. The biocompatibility of nanocomposite hydrogels was assessed by performing an MTS assay using human adipose stem cells (hASCs). hASCs were chosen for the in vitro studies for their easy accessibility and rapid rate of proliferation.53,54 hASCs were cultured in contact with the hydrogels containing different concentrations of nSi. The positive control was represented by cells grown under serum starvation. All the groups treated with the formulated hydrogel showed more than 85% cell viability even after 72 h (Figure 5a). No significant difference was observed with the incorporation of nSi over time. Live–dead assay performed after 24 h of gel treatment confirmed the MTS results, thus supporting the biocompatibility of the nanocomposite hydrogels (Figure S5a). Our results are consistent with previous studies, which have shown that nSi induces minimum cytotoxic effects.37
Figure 5.
In vitro biocompatibility and controlled release properties of the physically cross-linked hydrogels. (a) MTS assay at 24 and 72 h of hASCs in contact with the nanocomposite hydrogels revealed no significant difference in cell viability irrespective of the concentration of nSi used. Cells grown under serum-starved conditions served as a positive control. (b) Representative fluorescence images of actin stained with Alexa Fluor 488 Phalloidin (green) and nuclei stained with DAPI (blue), of hASCs after 72 h of contact with the hydrogels. Cells grown under serum-starved conditions served as a positive control (scale bar = 400 μm). (c) Comparison of release profiles of Dex from DNA hydrogels without nSi and the nanocomposite systems over a period of 10 days. Results are shown as the mean ± standard deviation (n = 3). (d) Half-release time (t1/2) of Dex as a function of nSi concentration confirmed the sustained release behavior of the nanocomposite hydrogels. (e) Viscosity vs shear rate plots of hydrogels loaded with the drug confirming the retention of shear-thinning behavior after Dex loading. (f) Schematic and corresponding images displaying a bone allograft before and after coating with the nanocomposite DNA-based hydrogel. (g) Time-lapse SEM micrographs of the allograft showing the erosion of the coating from the graft surface after 10 days of study (scale bar = 500 μm).
Furthermore, the influence of nSi on cell morphology and spreading was monitored over a period of 72 h. The morphology of hASCs was similar in all the groups after 24 and 72 h post-treatment (Figures 5b and S5b,c). In summary, the in vitro studies demonstrated that the nanocomposite hydrogels induce minimum cytotoxic effects and do not hinder cell spreading. These data strengthen their potential use as multifunctional hydrogels in regenerative medicine.
We anticipated that the formulated hydrogels could also function as sustained drug delivery vehicles and bioactive coatings in addition to their application as injectable hydrogels in minimally invasive therapies. To validate this concept, we encapsulated an osteogenic model drug Dex within the hydrogel and monitored its release over a period of 10 days.55
The release rate from the DNA-based hydrogel was observed to decrease with a higher concentration of nSi (Figure 5c). The half-life time of the release (t1/2) is defined as the time at which 50% of the drug is released from the hydrogel. An increase in t1/2 from 2.5 to 5.5 days with the increment in nSi concentration from 0 to 0.5% implies enhanced retention of Dex in the nSi containing hydrogels (Figure 5d). Extended-release times for the nanocomposite hydrogels can be attributed to their compact structure with reduced pore diameter compared to the pristine DNA-based hydrogels (0% nSi). As a comparison, we investigated the release profile of Dex from a gelatin—nSi hydrogel containing nSi at the concentration of 0.5% w/v. The amount of gelatin was optimized to obtain a gel with a viscosity comparable to that of the best performing DNA-based nanocomposite hydrogel (0.5% nSi) (Figure S6a). The gelatin-based hydrogel released approximately 80% of the loaded drug within 2 days, resulting in a much lower half-life time of release (t1/2 = 1.3 days) as compared to that of the DNA-based system (t1/2 = 5.5 days) (Figure S6b,c).
Finally, the DNA-based hydrogel formulations maintained their shear-thinning properties even after the drug loading, confirming their potential as injectable drug delivery systems (Figure 5e).
Aside from being injectable and demonstrating controlled drug release properties, the designed nanocomposite system can be successfully used as a bioactive coating. Specifically, as a proof of concept, the nanocomposite hydrogel was used to coat a human bone allograft isolated from the hip region (Figure 5f). Recent studies have demonstrated that the process of osteointegration of a bone allograft into the host tissue can be accelerated by the application of bioactive hydrogel coatings.56 These coatings can help promote the release of drugs or growth factors, as well as modulate the recruitment and osteogenic differentiation of host stem cells.57,58 Ideally, the formulated hydrogel can then act as a drug-eluting bioactive layer to enhance the therapeutic efficacy of bone allografts. SEM analysis of the uncoated and coated allografts revealed the uniform coverage of the microporous graft surface by the nanocomposite DNA-based hydrogel. The hydrogel coating started to erode away after being soaked in a phosphate buffer solution at pH 7.4, as evidenced by SEM analysis, thereby restoring the innate porous framework of the allograft (Figure 5g). Furthermore, the formulated nanocomposite hydrogel could be successfully bioprinted into defined geometries (Figure S7). However, careful selection in the concentration of DNA and the silicate nanodisks are the main key parameters that can influence the reproducibility of bioprinting. Future work in this direction would involve the enhancement of mechanical integrity of the bioink in order to inhibit pore collapse while printing three-dimensional complex structures with high resolution.39 Additionally, the high costs of the DNA extraction process and the limited amount of DNA that can be extracted are major issues that can limit the scalability of the bioprinting process.
To conclude, these promising data indicate that the designed injectable nanocomposite hydrogel could be potentially used as a multifunctional platform to modulate the release of osteogenic drugs or act as bioprinting ink as well as a coating for bone allografts.
Preserved Bioactivity of the Entrapped Drug: Modulation of in Vitro and in Vivo Osteogenic Differentiation.
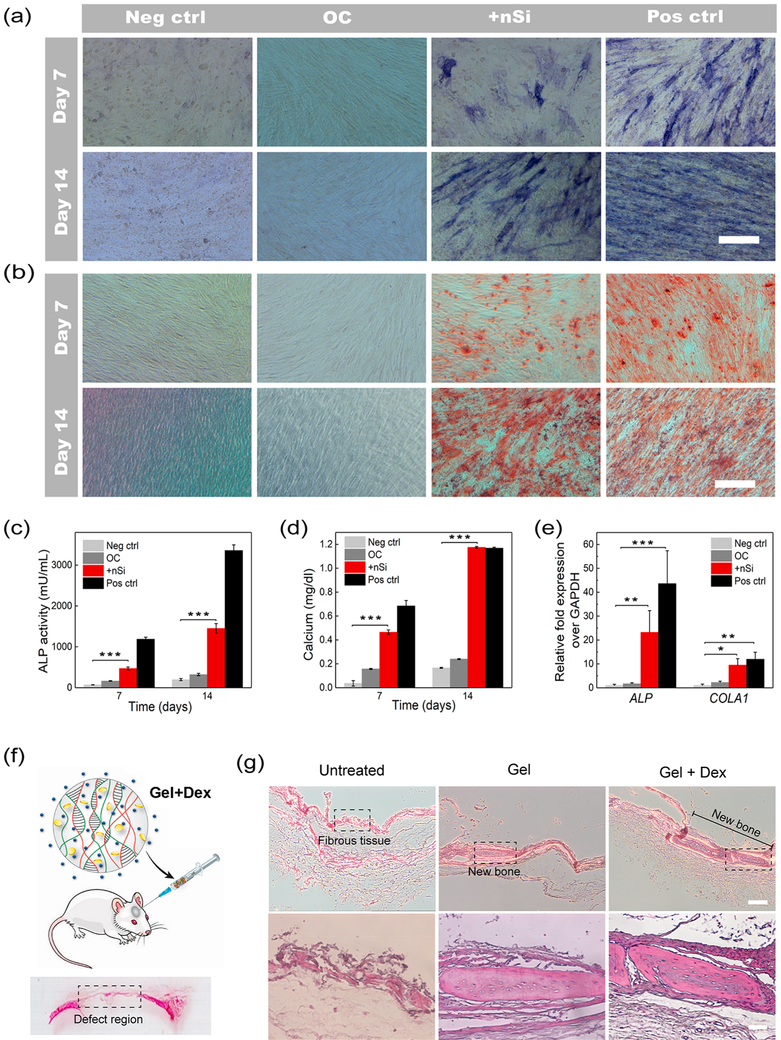
Aside from modulating the release of drugs, an efficient delivery vehicle should not induce any significant change in the bioactivity of the encapsulated cargo.59-62 To prove the efficacy of the released drug, we examined whether the Dex released from the nanocomposite hydrogel was still capable of promoting osteogenic differentiation of hASCs. Dex is a well-known osteogenic drug that stimulates stem cell osteogenic differentiation.55,63 hASCs were cultured for 14 days in osteoconductive media (α-MEM, 10% FBS, 1% penicillin/streptomycin, 50 μM ascorbic acid-2-phosphate, 10 mM β-glycerophosphate) supplemented with Dex obtained from the release study. Osteogenic differentiation of hASCs was evaluated by monitoring in vitro alkaline phosphatase (ALP) expression and calcium deposition after 7 and 14 days of drug treatment. As controls, hASCs were cultured in osteoinductive media (positive control), basal media (negative control), and osteoconductive media without the Dex (OC). The basal media contains α-MEM, 10% FBS, and 1% penicillin/streptomycin, whereas the osteoinductive medium is composed of α-MEM, 10% FBS, 1% penicillin/streptomycin, 50 μM ascorbic acid 2-phosphate, 10 mM β-glycerophosphate, and 10 nM Dex. Both qualitative and quantitative analyses revealed significant up-regulation of ALP in the Dex-treated group similar to the positive control group (Figure 6a,c). The observed trend was further validated by alizarin red colorimetric staining and quantification of the calcified matrix. At both 7 and 14 days, the Dex-treated group along with the positive control displayed a greater extent of calcium deposition. This resulted in intense alizarin red staining, which supports the osteogenic differentiation of stem cells (Figure 6b).
Figure 6.
Evaluation of in vitro and in vivo efficacy of the drug-loaded hydrogel for osteogenic differentiation. (a) Images of ALP staining of hASCs after 7 and 14 days of Dex treatment. Both the Dex-treated group (+nSi) and cells grown in osteoinductive media (pos ctrl) show high levels of ALP, representing osteogenic differentiation of hASCs (scale bar = 400 μM). (b) Alizarin red staining of hASCS after 7 and 14 days to detect the calcium deposition (scale bar = 400 μM). Intense red staining signifies osteogenic differentiation of hASCs. (c) ALP quantification demonstrated levels of ALP expression in the Dex-treated group significantly higher than those of the cells grown in basal media (neg ctrl) and osteoconductive media (OC). (d) Quantification of the deposited calcium confirmed significantly higher amounts of calcium in the Dex-treated group as compared to that in neg ctrl and OC. (e) qPCR analysis of different osteogenic markers after 14 days of Dex treatment. The fold expression increase of ALP and COLA1 was assessed following the ΔΔCt relative method. Results are shown as mean ± standard deviation (n = 3) (*p < 0.05, **p < 0.01, ***p < 0.001). Basal media, α-MEM, 10% FBS, 1% penicillin/streptomycin; osteoconductive media, α-MEM, 10% FBS, 1% penicillin/streptomycin, 50 μM ascorbic acid-2-phosphate, 10 mM β-glycerophosphate; osteoinductive media, α-MEM, 10% FBS, 1% penicillin/streptomycin, 50 μM ascorbic acid-2-phosphate, 10 mM β-glycerophosphate, and 10 nM Dex. (f) Schematic representation of the injection of the hydrogel (0.5% nSi + Dex) into a rat cranial defect and a representative image of the hematoxylin and eosin (H&E)-stained rat calvarial defect region. (g) H&E staining of tissue sections obtained from the rat calvarial defects 4 weeks post-treatment. Images were taken at two different magnifications (5× top row and 20× bottom row). Scale bars are 200 μm for the 5× images and 50 μm for the 20× images. The new bone formation in the central region of the cranial defects are marked in the images. The sections enclosed within the dotted lines are magnified in the bottom row images (n = 6).
Calcium quantification results provided confirmatory evidence in support of the staining images (Figure 6d). Finally, qPCR analysis after 14 days of treatment showed an increased upregulation of osteoblast markers in the Dex-treated group, thereby confirming the efficacy of the released Dex. Specifically, the fold increase expression of ALP and COLA1 were significantly higher in the Dex-treated group (23.23 ± 9.07 and 9.51 ± 2.68, respectively) and positive control (43.58 ± 13.78 and 11.93 ± 2.97, respectively) as compared to the other two groups (Figure 6e).64,65 However, the difference in gene expression of BGLAP and BSP was not statistically significant among the groups as these two markers are expressed in a later stage of osteogenic differentiation (Figure S8).66-69 Overall, these findings confirm the retention of the osteogenic property of Dex after being released from the nanocomposite hydrogel.
Finally, we assessed the in vivo osteogenic potential of the drug-loaded hydrogel in a rat cranial defect model (Figure 6f). Three different groups (n = 6) were tested including untreated rats (untreated), rats receiving the gel without any Dex (gel), and animals treated with the drug-loaded gel formulation (gel + Dex). Hematoxylin and eosin (H&E) staining was used to evaluate the formation of new bone within the defect after 4 weeks postsurgery. The untreated group showed the presence of soft tissue in the defect region without the formation of new bone (Figure 6g). On the contrary, small amounts of new bone were observed in the central defect region for both gel and gel + Dex groups (Figure 6g). The formation of bone in the gel group could be attributed to the presence of nanosilicate, which can promote in vitro osteogenic differentiation of stem cells.40 However, the limited bone regeneration capability shown by the Dex-loaded hydrogels could be ascribed to several factors including the short time frame of the study or the lack of additional osteogenic and angiogenic growth factors co-delivered in the defect area. To enhance the regenerative potential of the designed DNA-based hydrogel for cranial defect repair, further optimization regarding the type and dosage of possible growth factors, such as BMP-2, BMP-7, and VEGF, should be investigated.
CONCLUSION
We have successfully designed a shear-thinning DNA—nSi nanocomposite hydrogel with rapid self-healing properties. Silicate nanodisks electrostatically interact with the negatively charged phosphate backbone of DNA, thereby enhancing the physical and mechanical properties of the hydrogel. The physical crosslinking introduced by nSi resulted in an injectable hydrogel with increased structural integrity, elasticity, and thermal stability. The formulated nanocomposite hydrogels were also biocompatible and exhibited sustained release of the model drug Dex likely due to the reduced internal porosity induced by the inclusion of nSi. The drug maintained its bioactivity after release as confirmed by in vitro osteogenic differentiation studies of hASCs. Furthermore, the hydrogel was tested as a coating to prove its suitability as a drug-eluting degradable layer to modulate the surface properties of bone allografts, without sacrificing the long-term graft architecture. Preliminary studies were carried out in vivo by injecting the fabricated hydrogels in a rat cranial defect model showing minimal bone regeneration, which was observed mainly in the group containing Dex. Overall, the presented findings demonstrate an encouraging future for the formulated mechanically resilient DNA-based nanocomposite hydrogels as a class of injectable materials for diverse biomedical applications such as controlled drug delivery, tissue regeneration, and 3D bioprinting.
To further advance the applicability of the formulated hydrogel as a drug delivery system for bone regeneration, it is important to validate the suitability of this platform for the release of larger molecules including growth factors and proteins involved in the process of bone repair. Additionally, the possible in vivo inflammatory response, which plays a major role in the process of bone healing, needs to be thoroughly investigated for successful implementation of the material for future applications in a clinical setting.
EXPERIMENTAL SECTION
Preparation of the Pregel Formulation.
DNA at the concentration of 4% w/v (deoxyribonucleic acid sodium salt from salmon testes, Sigma-Aldrich, molecular weight ~1.3 × 106 g/mol, corresponding to ~2000 base pairs) was first dissolved in Milli-Q water at 35 °C for 2 days.12,13 The solution (300 μL) was then heated at 90 °C for 45 s. The pregel was formed by cooling the heated DNA solutions at 37 °C for 2 h.
Formation of the Nanocomposite Hydrogel.
To prepare nanocomposite hydrogels, the nanosilicates (Laponite XLG, Southern Clay Products Inc., USA) were dispersed in Milli-Q water with vigorous agitation (vortexing). Then, the nSi dispersions were added to the DNA-based pregel, and the mixture was vortexed for 5 min for further dispersion of the nanosilicates in the hydrogel matrix. The mixtures were then kept at 37 °C overnight for complete gelation. Three different concentrations of nSi (0.1, 0.25, and 0.5% w/v) were tested. The nanocomposite hydrogels were stored at 4 °C for further studies. The Dex-loaded hydrogels were formed by adding the required amount of the drug to the pregel formulation along with nSi dispersion to achieve a concentration of 875 ¼g/mL in the final hydrogel. The concentration of Dex was kept constant in all the hydrogels prepared for release studies. Equal volumes of a 14% (w/v) gelatin solution and 1% (w/v) nanosilicate dispersion were mixed together in order to form the gelatin-based hydrogels for a comparison of Dex release profiles. The Dex-loaded gelatin-based hydrogels were prepared by adding the drug to the gelatin solution along with the nSi dispersion to obtain a final concentration of Dex similar to the DNA-based hydrogel (0.5% nSi + Dex).
Physical and Chemical Characterization.
High-resolution TEM images of nSi dispersions were obtained using an FEI, Tecnai F20XT, USA system. The samples were prepared by placing a drop of nSi dispersion on top of carbon-coated 200-mesh copper grids, followed by drying the grids in vacuum for a few hours. The surface morphology of the nanocomposite hydrogels was evaluated using SEM with an in-lens detector, at an acceleration voltage ranging from 1 to 10 kV. The freeze-dried nanocomposite hydrogels were sputter coated with gold before being placed on the sample holder for imaging. The pore diameters for each group were analyzed using ImageJ software from five different images.
Elemental surface chemistry was characterized by EDX using the same samples analyzed by SEM. A Bruker Vector-22 FTIR spectrophotometer (PIKE Technologies, USA) was used for recording the FTIR spectra of the samples. Freeze-dried samples were mixed with KBr (Fischer, USA) and compressed to form a tablet for recording the spectra in the range of 400 up to 4000 cm−1 (resolution of 1 cm−1). MicroCT analysis of the freeze-dried hydrogel samples was performed using a Xradia MicroXCT tomographic X-ray microscope. An X-ray source of 50 kV was used to obtain a video of three-dimensional porosity of the hydrogel at a magnification of 20×.
Rheological Analysis.
All rheological analyses were performed in an AR2000 rheometer (TA Instruments, New Castle, DE). Nano-composite hydrogels were tested using a 25 mm steel cone and plate geometry, and a water trap was placed on the geometry to avoid excessive evaporation during the test. Viscosity measurements were performed at 25 °C, at shear rates ranging from 0.001 to 1000 s−1. Frequency sweeps in the range from 0.01 up to 10 Hz were recorded for all samples at 25 °C in the linear viscoelastic region at 1% of strain. Preliminary strain sweep tests were carried out in the range of 0.1 up to 100% of strain to define the range of viscoelastic region. Yield stress was evaluated by conducting oscillatory stress sweep measurements at 1 Hz from 0.1 to 200 Pa. The yield stress is identified as the crossover point where G″ exceeds the value of G′. Recovery experiments were performed by monitoring the G′ and G′′ at 1 Hz by applying 100% strain for 5 min followed by 1% strain for 5 min. Finally, temperature sweeps were conducted at a frequency of 1 Hz in the range of 25 to 45 °C with a temperature increase of 1 °C/min. A steel plate and plate geometry of 25 mm diameter with a solvent trap were used for the temperature sweep studies. All rheological experiments were carried out in triplicate.
X-ray Photoelectron Spectroscopy.
The presence of nSi in the hydrogel, as well as the formation of a physically cross-linked network between nSi and DNA backbone, was analyzed using XPS. A PHI 5000 VersaProbe II ultrahigh vacuum (1 × 10−9 bar) apparatus with an Al Kα X-ray source and a monochromator was used to obtain the XPS spectra. Freeze-dried samples were used for all the measurements. An X-ray beam size of 100 μm was used for all the tests. The survey spectra were recorded with a pass energy (PE) of 117 eV, a step size of 1 eV, and a dwell time of 20 ms. In contrast, a PE of 23.5 eV, a step size of 0.05 eV, and a dwell time of 20 ms were used to record the high-energy resolution spectra of O 1s, N 1s, and P 2p for the 0% nSi and 0.5% nSi hydrogel. Analyzer’s focal point was located by performing Auto-z (i.e., automated height adjustment to the highest intensity) before each measurement. The optimal signal-to-noise ratio was obtained by adjusting the number of average sweeps for each element (average number of sweeps was 1–20). All the raw data were processed and deconvoluted by Multipak peak fitting software version 2.3.15.
Coating of a Bone Allograft.
A human bone allograft cut from the hip bone (Curasan Inc., USA) was dip coated with the 0.5% nSi hydrogel. Briefly, the bone graft was immersed in the reaction mixture before gelation. The graft was removed from the solutions after coating, and the gel was allowed to form at room temperature. To study the integrity of the coating over time, the coated allograft was soaked in PBS with 1% penicillin/streptomycin (Thermo-Fischer Scientific, USA) and maintained at 37 °C for 10 days. The PBS solution was changed every alternate day. After 10 days, the graft was removed from the PBS solution and freeze-dried before SEM imaging. A bone allograft without any coating was imaged as well as a reference.
Bioprinting of the Nanocomposite Hydrogel.
The 0.5% nSi nanocomposite hydrogel was bioprinted to well-defined structures using an extrusion-based printing method. The structures were designed utilizing a repetier—host software and loaded via computer-aided manufacturing software. An Inkredible benchtop 3D bioprinter (Cellink, Sweden) was used to print the constructs. Printing was done at room temperature, using a 25 G needle (inner diameter = 0.25 mm). An XY-plane speed of 39 mm/s and an extrusion speed of 10 mm/s were employed to achieve a layer thickness of 0.2 mm. Phase contrast images of the printed construct were obtained by using a bright-field microscope EVOS cell imaging system (Thermo-Fischer Scientific, USA).
In Vitro Dexamethasone Release Studies.
Release studies were performed using slide-a-lyzer mini dialysis units with a molecular weight cut off of 2 kDa (Thermo Fisher Scientific Inc., Waltham, MA, USA).70 The dialysis cup was filled with 200 μL of each respective hydrogel. Experiments were conducted in a 48-well plate filled with 400 μL of PBS placed in a shaker incubator at 37 °C and a rotating speed of 60 rpm. The entire volume was withdrawn every 24 hours up to 10 days until a plateau was reached. The absorbance of Dex was detected using a Cytation 5 plate reader (BioTek, USA) at 240 nm. A standard calibration curve in the range of 0.01 to 0.4 mg/mL was used to quantify the amount of Dex released. The percentage of Dex was calculated as the mean ± standard deviation (n = 3). Hydrogels without the drug were also tested at 260 nm to detect any possible degradation and to assess if DNA chains were leaching out.
In Vitro Biocompatibility Studies.
Human adipose-derived stem cells (RoosterBio, USA) were grown in Invitrogen α-minimum essential medium (α-MEM) with 10% fetal bovine serum (FBS) (Thermo-Fischer Scientific, USA) and 1% penicillin/streptomycin (basal media). The cells were maintained at 37 °C and 5% CO2. Passages 3–5 were used for all the experiments. To evaluate the hydrogel biocompatibility, hASCs were cultured up to confluency in a 48-well plate and then exposed to the different nanocomposite hydrogel formulations. The gels were delivered to the cells after being dispersed in a PBS solution (20 μL of gel/200 μL of basal media). Subsequently, an MTS assay (Promega, USA) was performed after 24 and 72 h to test the biocompatibility of the hydrogels. As a positive control, the cells were grown under serum starvation conditions to disrupt cell growth. Furthermore, live/dead cell staining assay was performed to visualize cell viability after 24 h of contact with the nanocomposite systems. The hydrogel concentrations used for live/dead staining were similar to those used in the MTS studies. Live cells were stained with calcein AM as green, whereas the dead cells were stained with ethidium homodimer-1 as the red color. The effect of the formulated hydrogels on cell morphology and spreading was investigated by staining the actin filaments as well as the cell nuclei. The hASCs were seeded into chamber slides (4 × 104/mL) and allowed to grow in basal media for 24 h. Subsequently, the cells were treated with the different nanocomposite hydrogels (20 μL of gel/200 μL of basal media). After 24 and 72 h of exposure, the cells were fixed in 4% paraformaldehyde for 5 min at 37 °C followed by permeabilization with 0.1% Triton-X100 for 10 min at room temperature. Immunofluorescence images were obtained after counterstaining the F-actin with phalloidin-AlexaFluor488 (Invitrogen, USA) and nuclei with diamidino-2-phenylindole dilactate (DAPI, Invitrogen, USA).
In Vitro Differentiation.
The osteogenic differentiation of hASCs was investigated to evaluate the retained efficacy of Dex after being released. The hASCs (4 × 104/mL) were seeded into 24-well plates and allowed to grow in basal media for 24 h. The culture medium was changed after 24 h as per the respective groups. Four different groups used for the study were labeled as (1) negative control (Neg ctrl), hASCs cultured in basal media containing α-MEM with 10% FBS and 1% penicillin/streptomycin; (2) OC, hASCs cultured in osteoconductive media (α-MEM supplemented with 10% FBS, 50 μM ascorbic acid-2-phosphate, and 10 mM β-glycerophosphate); (3) +nSi, hASCs cultured in osteconductive media and supplemented with the Dex released from 0.5% nSi hydrogel; and (4) positive control (Pos ctrl), hASCs cultured in osteoinductive media (α-MEM, supplemented with 10% of FBS and 1% of penicillin/streptomycin, 50 μM ascorbic acid-2-phosphate, 10 mM β-glycerophosphate, and 10 nM of Dex). The final concentration of Dex in the +nSi group was maintained at 10 nM. The culture media of all the groups were changed every alternate day. The presence of ALP was visualized by staining the cells at respective time points following the standard protocol provided by the manufacturer (alkaline phosphatase kit, Sigma-Aldrich, USA). The enzyme activity was further quantified by an alkaline phosphatase fluorometric assay kit (Abcam, ab83371, USA), by measuring the fluorescence intensity at ex/em = 360/440 nm. Alizarin red S staining was performed following previously described protocols.71 Briefly, cells were fixed in 4% paraformaldehyde after being washed with PBS. Then they were incubated with 2% Alizarin red S (ScienCell Research Laboratories, USA) for 5 min. Excess stain was removed by washing with deionized water before taking the images of the stained samples. The calcium concentration was quantified by a colorimetric calcium assay kit (Cayman Chemical, USA) as per the manufacturer’s protocol.
Finally, qPCR analysis was performed to estimate the gene expression of several osteoblast markers including ALP, COLA1, BGLAP, and BSP using GAPDH as the control gene. Groups tested were the same as described above. mRNA from the respective samples were extracted at day 14. Three different samples were tested for each group (n = 3). The ΔΔCt relative method was used to calculate the fold increase expression. The experimental results were normalized based on the gene expression levels obtained for the group of hASCs cultured in basal media.
Animals and Surgical Procedures for Implantation of Nanocomposite Hydrogels.
The animal surgeries for in vivo experiments were performed at the University of Kansas Medical Center, Kansas City, Kansas. All experimental procedures with animals were approved by the Animal Care and Use Committee. A rat calvarial defect model with 8–9 weeks old female Sprague—Dawley rats were used in this study. All surgical procedures were performed under sterile conditions. The periosteum of the rats was removed after making a longitudinal skin incision over the midsagittal suture of the skull and a precise dissection. Then a dental burr was used to create an 8 mm diameter full thickness defect in the parietal bone. During the process, the wound area was constantly rinsed with normal saline to prevent overheating and remove any bone debris from the defect region. This was a very important step in this procedure for evaluating the formation of new bone within the cranial bone defect. The bone defects were filled with 0.5% nSi hydrogel (gel), 0.5%nSi hydrogel + dexamethasone (gel + Dex), or left untreated (negative control). An amount of 0.3 mg of Dex was used per animal, and six animals were used per group. The surgical wound was thereafter closed with a 5-0 nylon suture as previously described, and the day of surgery was designated as day “0”.72-74 Animals were sacrificed at 28 days after implantation to harvest the operated cranial bones for histological analysis. A time frame of 28 days was chosen as an end-point for this study based on our our previous studies which had demonstrated bone healing in 8 mm rat cranial defects 28–30 days after implantation of effective bone repair biomaterials.72-74
Histology Analysis.
The cranial samples (containing the bone defect and the surrounding host bone) were fixed in 10% buffered formalin (American MasterTech, Lodi, CA), decalcified in 25% formic acid (Sigma, St. Louis, MO), and embedded in paraffin (Leica Biosystems, Richmond, IL), sectioned at 5 μm, and stained with H&E to observe the cellular and tissue response to various treatments in the cranial bone defects. Six animals per group were examined.
Statistical Analysis.
Statistical analysis was carried out to determine whether a significant difference existed between the experimental groups. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests was performed. GraphPad Prism 5 was used for all the statistical analyses. A p value less than 0.05 was used to identify statistical significance (*p < 0.05, **p < 0.01, ***p < 0.001).
Supplementary Material
Movie S1: Video displaying the injection of 0.5% nSi through a 22 gauge surgical needle; the hydrogel is colored in blue for better visualization (AVI)
Movie S2: Video displaying the internal porosity of 0.5% nSi (AVI)
ACKNOWLEDGMENTS
The authors acknowledge the University of Kansas for funding and assistance which made possible the realization of this research project. A.P. acknowledges an investigator grant provided by the Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (NIGMS) of the NIH Award Number P20GM103638, University of Kansas New Faculty General Research Fund. J.W. would like to acknowledge Mary A. and Paul R. Harrington Distinguished Professorship Endowment and the funding support provided by the U.S. National Institutes of Health (NIH) under Award Number R01 DE018713. The authors acknowledge Prof. Stevin Gehrke for providing assistance and access to all his laboratory instruments. Finally, the authors are thankful to Dr. Prem Thapa from the University of Kansas Microscopy and Analytical Imaging Laboratory for his assistance with SEM, TEM imaging, XPS and EDX analysis.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.8b02434.
Rheological, structural and biological characterization of the nanocomposite hydrogels (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Park H; Woo EK; Lee KY Ionically Cross-linkable Hyaluronate-based Hydrogels for Injectable Cell Delivery. J. Controlled Release 2014, 196, 146–153. [DOI] [PubMed] [Google Scholar]
- (2).Tan H; Xiao C; Sun J; Xiong D; Hu X Biological Self-assembly of Injectable Hydrogel as Cell Scaffold via Specific Nucleobase Pairing. Chem. Commun. 2012, 48, 10289–10291. [DOI] [PubMed] [Google Scholar]
- (3).Glassman MJ; Chan J; Olsen BD Reinforcement of Shear Thinning Protein Hydrogels by Responsive Block Copolymer Self-Assembly. Adv. Funct. Mater. 2013, 23, 1182–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kretlow JD; Young S; Klouda L; Wong M; Mikos AG Injectable Biomaterials for Regenerating Complex Craniofacial Tissues. Adv. Mater. (Weinheim, Ger.) 2009, 21, 3368–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Gaihre B; Uswatta S; Jayasuriya A Reconstruction of Craniomaxillofacial Bone Defects Using Tissue-Engineering Strategies with Injectable and Non-Injectable Scaffolds. J. Funct. Biomater. 2017, 8, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Paul A; Hasan A; Kindi HA; Gaharwar AK; Rao VTS; Nikkhah M; Shin SR; Krafft D; Dokmeci MR; Shum-Tim D; Khademhosseini A Injectable Graphene Oxide/Hydrogel-Based Angiogenic Gene Delivery System for Vasculogenesis and Cardiac Repair. ACS Nano 2014, 8, 8050–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Pacelli S; Acosta F; Chakravarti AR; Samanta SG; Whitlow J; Modaresi S; Ahmed RPH; Rajasingh J; Paul A Nanodiamond-based Injectable Hydrogel for Sustained Growth Factor Release: Preparation, Characterization and In vitro Analysis. Acta Biomater. 2017, 58, 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lee KY; Mooney DJ Alginate: Properties and Biomedical Applications. Prog. Polym. Sci. 2012, 37, 106–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Khunmanee S; Jeong Y; Park H Crosslinking Method of Hyaluronic-based Hydrogel for Biomedical Applications. J. Tissue Eng. 2017, 8, 204173141772646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ahmadi F; Oveisi Z; Samani SM; Amoozgar Z Chitosan Based Hydrogels: Characteristics and Pharmaceutical Applications. Res. Pharm. Sci. 2015, 10, 1–16. [PMC free article] [PubMed] [Google Scholar]
- (11).Xu Y; Wu Q; Sun Y; Bai H; Shi G Three-Dimensional Self-Assembly of Graphene Oxide and DNA into Multifunctional Hydrogels. ACS Nano 2010, 4, 7358–7362. [DOI] [PubMed] [Google Scholar]
- (12).Topuz F; Okay O Formation of Hydrogels by Simultaneous Denaturation and Cross-Linking of DNA. Biomacromolecules 2009, 10, 2652–2661. [DOI] [PubMed] [Google Scholar]
- (13).Shin M; Ryu JH; Park JP; Kim K; Yang JW; Lee H DNA/Tannic Acid Hybrid Gel Exhibiting Biodegradability, Extensibility, Tissue Adhesiveness, and Hemostatic Ability. Adv. Funct. Mater. 2015, 25, 1270–1278. [Google Scholar]
- (14).Um SH; Lee JB; Park N; Kwon SY; Umbach CC; Luo D Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 2006, 5, 797–801. [DOI] [PubMed] [Google Scholar]
- (15).Guo W; Qi X-J; Orbach R; Lu C-H; Freage L; Mironi-Harpaz I; Seliktar D; Yang H-H; Willner I Reversible Ag+-crosslinked DNA hydrogels. Chem. Commun. 2014, 50, 4065–4068. [DOI] [PubMed] [Google Scholar]
- (16).Lee CK; Shin SR; Lee SH; Jeon J-H; So I; Kang TM; Kim SI; Mun JY; Han S-S; Spinks GM; Wallace GG; Kim SJ DNA Hydrogel Fiber with Self-Entanglement Prepared by Using an Ionic Liquid. Angew. Chem., Int. Ed. 2008, 47, 2470–2474. [DOI] [PubMed] [Google Scholar]
- (17).Pandey PK; Rawat K; Aswal VK; Kohlbrecher J; Bohidar HB DNA Ionogel: Structure and Self-assembly. Phys. Chem. Chem. Phys. 2017, 19, 804–812. [DOI] [PubMed] [Google Scholar]
- (18).Shao Y; Jia H; Cao T; Liu D Supramolecular Hydrogels Based on DNA Self-Assembly. Acc. Chem. Res. 2017, 50, 659–668. [DOI] [PubMed] [Google Scholar]
- (19).Li C; Rowland MJ; Shao Y; Cao T; Chen C; Jia H; Zhou X; Yang Z; Scherman OA; Liu D Responsive Double Network Hydrogels of Interpenetrating DNA and CB[8] Host—Guest Supramolecular Systems. Adv. Mater. 2015, 27, 3298–3304. [DOI] [PubMed] [Google Scholar]
- (20).Wang D; Hu Y; Liu P; Luo D Bioresponsive DNA Hydrogels: Beyond the Conventional Stimuli Responsiveness. Acc. Chem. Res. 2017, 50, 733–739. [DOI] [PubMed] [Google Scholar]
- (21).Nishikawa M; Ogawa K; Umeki Y; Mohri K; Kawasaki Y; Watanabe H; Takahashi N; Kusuki E; Takahashi R; Takahashi Y; Takakura Y Injectable, Self-Gelling, Biodegradable, and Immunomodulatory DNA Hydrogel for Antigen Delivery. J. Controlled Release 2014, 1S0, 25–32. [DOI] [PubMed] [Google Scholar]
- (22).Wu CJ; Gaharwar AK; Chan BK; Schmidt G Mechanically Tough Pluronic F127/Laponite Nanocomposite Hydrogels from Covalently and Physically Cross-Linked Networks. Macromolecules 2011, 44, 8215–8224. [Google Scholar]
- (23).Strandman S; Zhu XX Self-Healing Supramolecular Hydrogels Based on Reversible Physical Interactions. Gels 2016, 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Peak CW; Stein J; Gold KA; Gaharwar AK Nanoengineered Colloidal Inks for 3D Bioprinting. Langmuir 2018, 34, 917–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Waters R; Pacelli S; Maloney R; Medhi I; Ahmed RPH; Paul A Stem cell secretome-rich nanoclay hydrogel: a dual action therapy for cardiovascular regeneration. Nanoscale 2016, 8, 7371–7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Paul A; Manoharan V; Krafft D; Assmann A; Uquillas JA; Shin SR; Hasan A; Hussain MA; Memic A; Gaharwar AK; Khademhosseini A Nanoengineered Biomimetic Hydrogels for Guiding Human Stem Cell Osteogenesis in Three Dimensional Microenvironments. J. Mater. Chem. B 2016, 4, 3544–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Jatav S; Joshi YM Chemical Stability of Laponite in Aqueous Media. Appl. Clay Sci 2014, 97–98, 72–77. [Google Scholar]
- (28).Zhao L; Seth A; Wibowo N; Zhao C-X; Mitter N; Yu C; Middelberg AP Vaccine 2014, 32, 327–337. [DOI] [PubMed] [Google Scholar]
- (29).Wang X; Lim HJ; Son A Characterization of Denaturation and Renaturation of DNA for DNA Hybridization. Environ. Health Toxicol. 2014, 29, e2014007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Schmid FX Biological Macromolecules: UV-visible Spectrophotometry Encyclopedia of Life Sciences; John Wiley & Sons Ltd., 2001; pp 1–4. [Google Scholar]
- (31).Carrow JK; Cross LM; Reese RW; Jaiswal MK; Gregory CA; Kaunas R; Singh I; Gaharwar AK Widespread Changes in Transcriptome Profile of Human Mesenchymal Stem Cells Induced by Two-dimensional Nanosilicates. Proc. Natl. Acad. Sci. U. S. A. 2018, H5, E3905–E3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Guvendiren M; Lu HD; Burdick JA Shear-thinning Hydrogels for Biomedical Applications. Soft Matter 2012, 8, 260–272. [Google Scholar]
- (33).Thakur A; Jaiswal MK; Peak CW; Carrow JK; Gentry J; Dolatshahi-Pirouz A; Gaharwar AK Injectable Shear-Thinning Nanoengineered Hydrogels for Stem Cell Delivery. Nanoscale 2016, 8, 12362–12372. [DOI] [PubMed] [Google Scholar]
- (34).Cerofolini GF; Galati C; Renna L Si 2p XPS spectrum of the Hydrogen-Terminated (100) Surface of Device-Quality Silicon. Surf. Interface Anal. 2003, 35, 968–973. [Google Scholar]
- (35).Joshi YM; Reddy GRK; Kulkarni AL; Kumar N; Chhabra RP Rheological Behaviour of Aqueous Suspensions of Laponite: New Insights into the Ageing Phenomena. Proc. R. Soc. London, Ser. A 2008, 464, 469–489. [Google Scholar]
- (36).Wilson SA; Cross LM; Peak CW; Gaharwar AK Shear-Thinning and Thermo-Reversible Nanoengineered Inks for 3D Bioprinting. ACS Appl. Mater. Interfaces 2017, 9, 43449–43458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Gaharwar AK; Avery RK; Assmann A; Paul A; McKinley GH; Khademhosseini A; Olsen BD Shear-Thinning Nano-composite Hydrogels for the Treatment of Hemorrhage. ACS Nano 2014, 8, 9833–9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Schexnailder P; Loizou E; Porcar L; Butler P; Schmidt G Heterogeneity in Nanocomposite Hydrogels from Poly(ethylene oxide) Cross-linked with Silicate Nanoparticles. Phys. Chem. Chem. Phys. 2009, 11, 2760–2766. [DOI] [PubMed] [Google Scholar]
- (39).Jin Y; Liu C; Chai W; Compaan A; Huang Y Self-Supporting Nanoclay as Internal Scaffold Material for Direct Printing of Soft Hydrogel Composite Structures in Air. ACS Appl. Mater. Interfaces 2017, 9, 17456–17465. [DOI] [PubMed] [Google Scholar]
- (40).Gaharwar AK; Mihaila SM; Swami A; Patel A; Sant S; Reis RL; Marques AP; Gomes ME; Khademhosseini A Bioactive Silicate Nanoplatelets for Osteogenic Differentiation of Human Mesenchymal Stem Cells. Adv. Mater. 2013, 25, 3329–3336. [DOI] [PubMed] [Google Scholar]
- (41).Serec K; Babic SD; Podgornik R; Tomic S Effect of Magnesium Ions on the Structure of DNA Thin Films: An Infrared Spectroscopy Study. Nucleic Acids Res. 2016, 44, 8456–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ahmad R; Arakawa H; Tajmir-Riahi HA A Comparative Study of DNA Complexation with Mg(II) and Ca(II) in Aqueous Solution: Major and Minor Grooves Bindings. Biophys. J. 2003, 84, 2460–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Mello MLS; Vidal BC Changes in the Infrared Microspectroscopic Characteristics of DNA Caused by Cationic Elements, Different Base Richness and Single-Stranded Form. PLoS One 2012, 7, e43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Volkov IL; Smirnova A; Makarova AA; Reveguk ZV; Ramazanov RR; Usachov DY; Adamchuk VK; Kononov AI DNA with Ionic, Atomic, and Clustered Silver: An XPS Study. J. Phys. Chem. B 2017, 121, 2400–2406. [DOI] [PubMed] [Google Scholar]
- (45).Kulkarni SK; Ethiraj AS; Kharrazi S; Deobagkar DN; Deobagkar DD Synthesis and Spectral Properties of DNA Capped CdS Nanoparticles in Aqueous and Non-Aqueous Media. Biosens. Bioelectron. 2005, 21, 95–102. [DOI] [PubMed] [Google Scholar]
- (46).Korin E; Froumin N; Cohen S Surface Analysis of Nanocomplexes by X-ray Photoelectron Spectroscopy (XPS). ACS Biomater. Sci. Eng. 2017, 3, 882–889. [DOI] [PubMed] [Google Scholar]
- (47).Shigekawa H; Ikawa H; Yoshizaki R; Iijima Y; Sumaoka J; Komiyama M Core Level Photoelectron Spectroscopy on the Lanthanide-Induced Hydrolysis of DNA. Appl. Phys. Lett. 1996, 68, 1433–1435. [Google Scholar]
- (48).Shamsi M; Kraatz H Interactions of Metal Ions with DNA and Some Applications. J. Inorg. Organomet. Polym. Mater. 2013, 23, 4–23. [Google Scholar]
- (49).Turel I; Kljun J Interactions of Metal Ions with DNA, its Constituents and Derivatives, Which May be Relevant for Anticancer Research. Curr. Top. Med. Chem. 2011, 11, 2661–87. [DOI] [PubMed] [Google Scholar]
- (50).Gao Y; Qi H DNA-Based Bulk Hydrogel Materials and Biomedical Application. J. Nanotechnol Eng. Med. 2015, 6, 040802-040802-6. [Google Scholar]
- (51).Xiong X; Wu C; Zhou C; Zhu G; Chen Z; Tan W Responsive DNA-Based Hydrogels and their Applications. Macromol Rapid Commun. 2013, 34, 1271–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Kahn JS; Hu Y; Willner I Stimuli-Responsive DNA-Based Hydrogels: From Basic Principles to Applications. Acc. Chem. Res. 2017, 50, 680–690. [DOI] [PubMed] [Google Scholar]
- (53).Han DS; Chang HK; Kim KR; Woo SM Consideration of Bone Regeneration Effect of Stem Cells: Comparison of Bone Regeneration Between Bone Marrow Stem Cells and Adipose-Derived Stem Cells. J. Craniofac. Surg. 2014, 25, 196–201. [DOI] [PubMed] [Google Scholar]
- (54).Pacelli S; Basu S; Berkland C; Wang J; Paul A Design of a Cytocompatible Hydrogel Coating to Modulate Properties of Ceramic-Based Scaffolds for Bone Repair. Cell. Mol. Bioeng. 2018, 11, 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Moretti R. d. C.; Duailibi MT; Martins PO; dos Santos JA; Duailibi SE Osteoinductive Effects of Preoperative Dexamethasone in Human Dental Pulp Stem Cells Primary Culture. Future Sci. OA 2017, 3, FSO184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Hoffman MD; Xie C; Zhang X; Benoit DSW The Effect of Mesenchymal Stem Cells Deliveredvia Hydrogel-Based Tissue Engineered Periosteum on Bone Allograft Healing. Biomaterials 2013, 34, 8887–8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Huang C; Das A; Barker D; Tholpady S; Wang T; Cui Q; Ogle R; Botchwey E Local Delivery of FTY720 Accelerates Cranial Allograft Incorporation and Bone formation. Cell Tissue Res. 2012, 347, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Petrie Aronin CE; Shin SJ; Naden KB; Rios PD; Sefcik LS; Zawodny SR; Bagayoko ND; Cui Q; Khan Y; Botchwey EA The Enhancement of Bone Allograft Incorporation by the Local Delivery of the Sphingosine 1-phosphate Receptor Targeted Drug FTY720. Biomaterials 2010, 31, 6417–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Ekenseair AK; Kasper FK; Mikos AG Perspectives on the Interface of Drug Delivery and Tissue Engineering. Adv. Drug Delivery Rev. 2013, 65, 89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Caramella C; Conti B; Modena T; Ferrari F; Bonferoni MC; Genta I; Rossi S; Torre ML; Sandri G; Sorrenti M; Catenacci L; Dorati R; Tripodo G Controlled Delivery Systems for Tissue Repair and Regeneration. J. Drug Delivery Sci. Technol. 2016, 32, 206–228. [Google Scholar]
- (61).Vo TN; Kasper FK; Mikos AG Strategies for Controlled Delivery of Growth Factors and Cells for Bone Regeneration. Adv. Drug Delivery Rev. 2012, 64, 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Patel A; Cholkar K; Mitra AK Recent Developments in Protein and Peptide Parenteral Delivery Approaches. Ther. Delivery 2014, 5, 337–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Yuasa M; Yamada T; Taniyama T; Masaoka T; Xuetao W; Yoshii T; Horie M; Yasuda H; Uemura T; Okawa A; Sotome S Dexamethasone Enhances Osteogenic Differentiation of Bone Marrow- and Muscle-Derived Stromal Cells and Augments Ectopic Bone Formation Induced by Bone Morphogenetic Protein-2. PLoS One 2015, 10, e0116462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Glynn ERA; Londono AS; Zinn SA; Hoagland TA; Govoni KE Culture Conditions for Equine Bone Marrow Mesenchymal Stem Cells and Expression of Key Transcription Factors During their Differentiation into Osteoblasts. J. Anim. Sci. Biotechnol. 2013, 4, 40–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Jikko A; Harris SE; Chen D; Mendrick DL; Damsky CH Collagen Integrin Receptors Regulate Early Osteoblast Differentiation Induced by BMP-2. J. Bone Miner. Res. 1999, 14, 1075–83. [DOI] [PubMed] [Google Scholar]
- (66).Wang F; Okawa H; Kamano Y; Niibe K; Kayashima H; Osathanon T; Pavasant P; Saeki M; Yatani H; Egusa H Controlled Osteogenic Differentiation of Mouse Mesenchymal Stem Cells by Tetracycline-Controlled Transcriptional Activation of Amelogenin. PLoS One 2015, 10, e0145677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Ganss B; Kim RH; Sodek J Bone Sialoprotein. Crit. Rev. Oral Biol. Med. 1999, 10, 79–98. [DOI] [PubMed] [Google Scholar]
- (68).Kulterer B; Friedl G; Jandrositz A; Sanchez-Cabo F; Prokesch A; Paar C; Scheideler M; Windhager R; Preisegger K-H; Trajanoski Z Gene Expression Profiling of Human Mesenchymal Stem Cells Derived from Bone Marrow During Expansion and Osteoblast Differentiation. BMC Genomics 2007, 8, 70–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Aubin JE Bone Stem Cells. J. Cell. Biochem. 1998, 72, 73–82. [DOI] [PubMed] [Google Scholar]
- (70).Seib FP; Jones GT; Rnjak-Kovacina J; Lin Y; Kaplan DL pH-Dependent Anticancer Drug Release from Silk Nanoparticles. Adv. Healthcare Mater. 2013, 2, 1606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Pacelli S; Maloney R; Chakravarti AR; Whitlow J; Basu S; Modaresi S; Gehrke S; Paul A Controlling Adult Stem Cell Behavior Using Nanodiamond-Reinforced Hydrogel: Implication in Bone Regeneration Therapy. Sci. Rep. 2017, 7, 6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Wang J; Zhou HY; Salih E; Xu L; Wunderlich L; Gu X; Hofstaetter JG; Torres M; Glimcher MJ Site-Specific In vivo Calcification and Osteogenesis Stimulated by Bone Sialoprotein. Calcif. Tissue Int. 2006, 79, 179–89. [DOI] [PubMed] [Google Scholar]
- (73).Xu L; Anderson AL; Lu Q; Wang J Role of Fibrillar Structure of Collagenous Carrier in Bone Sialoprotein-Mediated Matrix Mineralization and Osteoblast Differentiation. Biomaterials 2007, 28, 750–761. [DOI] [PubMed] [Google Scholar]
- (74).Wang J; Yang R; Gerstenfeld LC; Glimcher MJ Characterization of Demineralized Bone Matrix-Induced Osteogenesis in Rat Calvarial Bone Defects: III. Gene and Protein expression. Calcif. Tissue Int. 2000, 67, 314–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1: Video displaying the injection of 0.5% nSi through a 22 gauge surgical needle; the hydrogel is colored in blue for better visualization (AVI)
Movie S2: Video displaying the internal porosity of 0.5% nSi (AVI)