Abstract
Introduction
Long-term head-down bed rest (HDBR) results in musculoskeletal losses similar to those observed during long-term space flight. Agents such as testosterone, in addition to regular exercise, are effective countermeasures for reducing loss of skeletal muscle mass and function.
Objective
We investigated the skeletal muscle proteome of healthy men in response to long term HDBR alone (CON) and to HDBR with exercise (PEX) or exercise plus testosterone (TEX) countermeasures.
Method
Biopsies were performed on the vastus lateralis before (pre) HDBR and on HDBR days 32 (mid) and 64 (post). Extracted proteins from these skeletal muscle biopsies were subjected to 2-dimensional gel electrophoresis (2DE), stained for phosphoproteins (Pro-Q Diamond dye) and total proteins (Sypro Ruby dye). Proteins showing significant fold differences (t-test p ≤ 0.05) in abundance or phosphorylation state at mid or post were identified by mass spectroscopy (MS).
Results
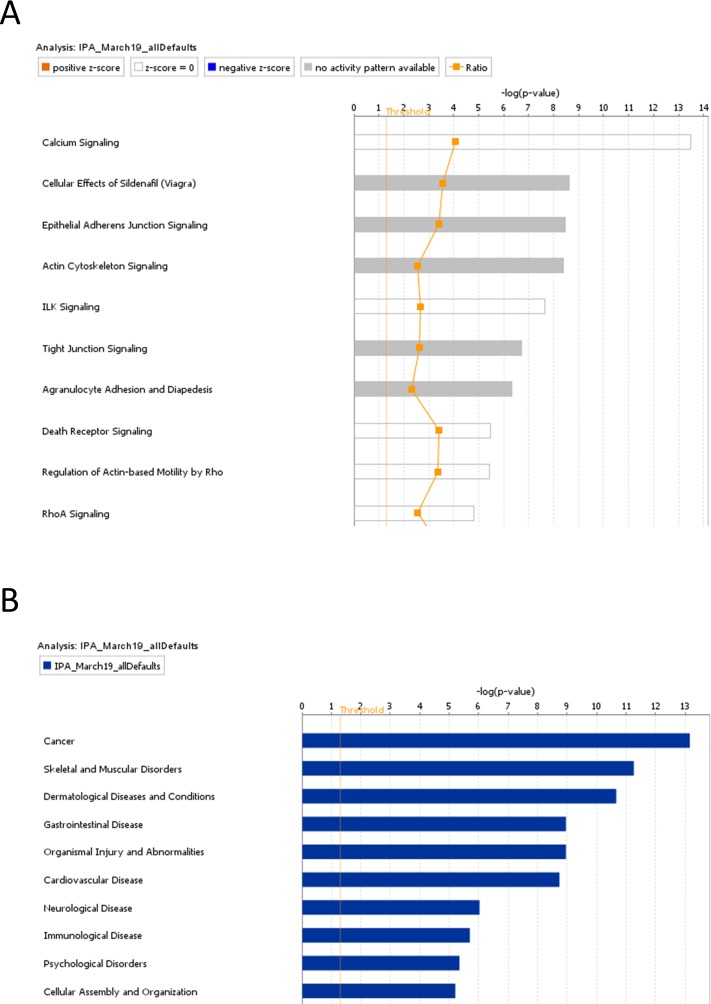
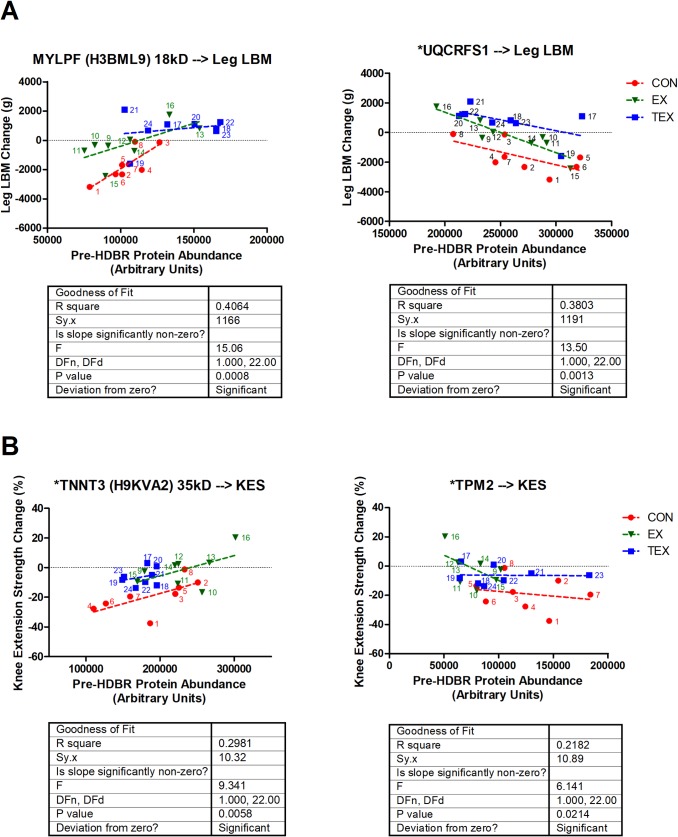
From a total of 932 protein spots, 130 spots were identified as potentially altered in terms of total protein or phosphoprotein levels due to HDBR and/or countermeasures, and 59 unique molecules emerged from MS analysis. Top canonical pathways identified through IPA included calcium signaling, actin cytoskeleton signaling, integrin linked kinase (ILK) signaling, and epithelial adherens junction signaling. Data from the pre-HDBR proteome supported the potential for predicting physiological post-HDBR responses such as the individual’s potential for loss vs. maintenance of muscle mass and strength.
Conclusions
HDBR resulted in alterations to skeletal muscle abundances and phosphorylation of several structural and metabolic proteins. Inclusion of exercise alone or in combination with testosterone treatment modulated the proteomic responses towards cellular reorganization and hypertrophy, respectively. Finally, the baseline proteome may aid in the development of personalized countermeasures to mitigate health risks in astronauts as related to loss of muscle mass and function.
Introduction
Skeletal muscle protein turnover is regulated through an intricate process involving biochemical and mechanical signals. Skeletal muscle size and composition is maintained when protein synthesis and breakdown is balanced while disruption of this balance can result in net gains or losses in muscle size and/or strength. Space flight related losses in muscle mass and strength are among the prime concerns for long duration space exploration missions and involve alterations in myofibrillar protein content and metabolism [1]. Skeletal muscle losses during space flight are largely attributed to absence of axial loading on weight bearing muscles, an environmental condition that can be mimicked adequately by bed rest studies on Earth. As such, the replacement of mechanical forces, via exercise, is among the primary operational countermeasures to mitigate muscle loss during space flight. While effective, exercise in space is time-consuming and only partially replaces the mechanical loading needed to completely prevent muscle atrophy and loss of function, particularly in highly susceptible muscle groups such as in the calf. Thus, additional interventions that complement inflight exercise countermeasures are sought. Testosterone has been considered as a potential countermeasure to be investigated due to its anabolic potential and known synergism with exercise. While exercise and testosterone are independently known to induce skeletal muscle protein synthesis, much is unknown regarding the differences and redundancies between the signals provided by the respective mechanical and biochemical stimuli.
The recent 70-day NASA-funded CFT70 bed rest campaign investigated the effects of strict, diet controlled, head down bed rest (HDBR) on lean body mass and muscle strength of healthy males, and the influence of a moderate to high-intensity exercise protocol (Sprint protocol), with or without testosterone supplementation, on mitigating these changes [2–4]. This NASA-led study was strictly monitored for all aspects known to affect skeletal muscle gain and loss (nutrition, exercise, axial loading, body movement etc.) and therefore provided a tremendously unique opportunity to investigate the effects of extended inactivity and unloading (with or without the inclusion of countermeasures) on changes in abundance and phosphorylation of skeletal muscle proteins in humans. We hypothesized that confinement to HDBR would alter the skeletal muscle proteome and that the inclusion of exercise alone or exercise with testosterone supplementation would each result in unique modifications of proteomic responses during HDBR. Furthermore, this unique opportunity afforded us the ability to perform post-hoc regression analyses to determine whether baseline proteomic data could be predictive of HDBR- or countermeasure-induced responses in muscle mass or strength.
Methods
Ethics
Subjects were recruited through the National Aeronautics and Space Administration (NASA) Human Research Program (HRP) testing facility at the Johnson Space Center (JSC) in Houston, TX. Screening, including the JSC Human Test Subject Facility physical examination and psychological evaluations were completed at NASA JSC. The study complied with the Declaration of Helsinki and was approved by The University of Texas Medical Branch (UTMB) Institutional Review Board (IRB) and by the NASA Committee for the Protection of Human Subjects (CPHS). Written informed consent was obtained from all subjects, and subjects were studied at the NASA Flight Analogs Research Unit (FARU) at UTMB. This research was conducted as part of a larger integrated NASA bed rest study campaign registered with ClinicalTrials.gov (NCT00891449).
Subjects
The bed rest study protocol and subject characteristics have been detailed in our previous report [3]. Study advertisement, recruitment, and randomization was conducted through the Human Test Subjects Facility at the NASA Johnson Space Center in Houston, TX. Healthy male volunteers (35 ± 8 years) were randomized (blocks of six) to one of 3 bed rest groups: placebo + non-exercise control (CON, n = 8), placebo + exercise (PEX, n = 8), and testosterone + exercise (TEX, n = 8) (Fig 1). Placebo vs. testosterone treatment assignments were blinded (CON) or double-blinded (PEX vs. TEX). The study was conducted at the Flight Analogs Research Unit (FARU) at UTMB in Galveston, TX. Testing was conducted at UTMB and NASA/JSC. PEX and TEX subjects followed a moderately intense exercise schedule throughout the HDBR phase [4]. Briefly, all PEX and TEX subjects followed an exercise protocol that included 6 days of high-intensity aerobic training, combined with 3 days of resistive strength exercise. Resistance exercise sessions were on the same day as the continuous aerobic exercise, separated by 4–6 h. Supine aerobic exercise was performed using the Standalone Zero Gravity Locomotion Simulator vertical treadmill and a supine cycle ergometer, and resistance exercise was performed on a horizontal squat device, a horizontal leg press (for leg press and calf raise exercise), and a prone leg curl machine. High-intensity interval aerobic exercise and continuous aerobic exercise were performed on alternating days. Starting one day before bed rest (BR-1), placebo (saline) or testosterone enanthate injections (100 mg, intramuscular) were administered in 2-week intervals (weekly testosterone enanthate for two weeks, followed by two weeks off, etc.) for the duration of the 70-day bed rest period. Thus, injections occurred immediately before bedrest (BR-1), and during bedrest (BR7, BR28, BR35, BR56, and BR63). Licensed nurses administered the IM injections in the gluteus maximus, alternating between sides of the body throughout the study. Clinical outcomes from this investigation were published previously and there were no adverse events in response or testosterone treatment [5].
Fig 1. Subject flow diagram.
This research was part of an integrated study registered with ClinicalTrials.gov (NCT00891449). Sample sizes were determined based on the primary outcomes from several independently funded investigations involved in the bed rest study campaign conducted between 2010 and 2014. A total of 24 subjects randomized to control (CON, n = 8), exercise plus placebo (PEX, n = 8) or exercise plus testosterone countermeasures (TEX, n = 8) completed this protocol during 70 days of head down bed rest (HDBR). Because of overlap in start-time between funded investigations, subject numbers may differ between reports that emanated from this bed rest campaign.
Muscle biopsy procedure
Muscle tissue was collected on BR-1 (pre), BR36 (mid), and BR64 (post). All biopsies were performed on the left leg of each subject. Each subject underwent the procedure three times during the study resulting in three biopsies forming a triangle between sites. The first site was approximately 10 cm proximal from the patella. The second site was approximately 4 cm proximal to the first site. The third site was between and approximately 3 cm lateral from the previous sites. Muscle biopsy procedures were performed as described elsewhere [6, 7]. Briefly, a site was marked on the vastus lateralis and cleaned with Betadine. Lidocaine (1%) was administered to the skin and deep muscle. An approximately 5 mm incision was made through the skin and fascia and a 5 mm Bergström needle was advanced into the muscle. While suction was applied, 100–200 mg of skeletal muscle tissue was collected by opening and closing the cutting window of the biopsy needle. The incision was sutured and covered with Bacitracin and steri-strips. Ice was applied to the site and ibuprofen was provided to the subject to alleviate soreness.
Skeletal muscle proteomics
Proteomic analyses were performed by the UTMB Biomolecular Resource Facility (BRF). Protein abundances were determined in fractionated muscle extracts using a Biofluids Analytical Platform (BAP) [8–10]. These analyses were completed in one continuous effort once all the muscle samples had been collected. The BAP fractionation component combines Superdex S-75 size-exclusion chromatography (SEC) of biofluids with electronically triggered fraction collection to create protein and peptide pools for subsequent separation and analysis. Fractionated samples were subjected to 2D gel electrophoresis (2DE) and stained for phosphoproteins (Pro-Q Diamond dye, ThermoFisher Scientific) or total proteins (Sypro Ruby dye, ThermoFisher Scientific). Pro-Q Diamond selectively stains phosphoproteins in gels and thus provides a convenient method for determining relative phosphorylation of proteins between samples–which is our purpose here—though not for pinpointing the sites (residues) of phosphorylation. The gels were imaged, and then analyzed using SameSpots software (TotalLab, Newcastle upon Tyne, UK), first aligning the images to a selected reference image, and quantitatively comparing log-transformed spot intensities between the groups (CON, PEX, TEX; pre, mid, post). Proteins showing significant p-value (≤ 0.05 in t-tests) and |fold differences| (≥1.50) between the groups (CON, PEX, TEX) or time points (pre, mid, post) were identified by matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF/TOF) mass spectroscopy (MS). This is a method used commonly for protein identification following 2DE analysis [11]. Lists of MS identified proteoforms were subjected to Ingenuity Pathway Analysis (IPA) to identify cell-signaling networks that responded to HDBR and/or countermeasures. Principal components analysis (PCA) was performed to characterize clustering in the three groups based on protein abundance and protein phosphorylation.
Data and statistical analyses
Data analyses included components of treatment (CON, PEX, TEX) and time (pre, post). The mid timepoint was included during the initial spot selection for MS identification and the data are presented in Tables 1–4. However, the discussion will focus on pre-post changes to facilitate interpretation of the findings.
Table 1. Changes in protein abundances.
Within-group changes of all identified spots. Ordering within the table is based on proteoform interpretation (i.e. intact, aggregate, fragment) and p-values (2-tailed, paired t-tests) of the pre to post changes in CON. P-values < 0.05 are shaded in yellow. Differences (%) within each comparison are shaded to indicate higher (red) or lower (blue) values relative to pre.
| CON: Mid vs. Pre |
CON: Post vs. Pre |
EX: Mid vs. Pre |
EX: Post vs. Pre |
TEX: Mid vs. Pre |
TEX: Post vs. Pre |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TABLE 1 | Protein Name | Symbol | Accession Number | pI | MW (kD) | UNIPROT MW (kd) | MS Protein Score | Proteoform Interpretation | P-Value | Diff. (%) | P-Value | Diff. (%) | P-Value | Diff. (%) | P-Value | Diff. (%) | P-Value | Diff. (%) | P-Value | Diff. (%) |
| metabolic | Adenylate kinase isoenzyme 1 | AK1 | P00568 | 9.61 | 24 | 22 | 178 | INTACT | 0.001 | 59.974 | 0.004 | 145.03 | 0.727 | 10.229 | 0.399 | 47.385 | 0.163 | -26.902 | 0.130 | 68.469 |
| metabolic | Heat shock protein beta-7 | HSPB7 | Q9UBY9 | 5.97 | 18 | 19 | 265 | INTACT | 0.020 | -7.022 | 0.004 | -21.175 | 0.571 | 4.724 | 0.047 | 9.858 | 0.995 | -0.416 | 0.036 | -20.085 |
| transport | Carbonic anhydrase 3 | CA3 | P07451 | 5.27 | 28 | 30 | 38 | INTACT | 0.093 | 17.191 | 0.006 | 64.159 | 0.253 | -14.058 | 0.238 | -11.829 | 0.497 | -11.237 | 0.482 | 8.895 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 10.2 | 18 | 17 | 36 | INTACT | 0.918 | 1.814 | 0.007 | 71.635 | 0.344 | -10.005 | 0.230 | -35.237 | 0.530 | 23.813 | 0.829 | -7.558 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.26 | 34 | 33 | 592 | INTACT | 0.277 | 13.529 | 0.010 | 55.081 | 0.963 | -0.473 | 0.131 | -24.622 | 0.322 | -13.284 | 0.800 | 4.170 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 9.67 | 195 | 223 | 451 | INTACT | 0.557 | -7.499 | 0.011 | -67.284 | 0.501 | 15.787 | 0.556 | 10.373 | 0.430 | -15.782 | 0.395 | -20.017 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.51 | 15 | 17 | 36 | INTACT | 0.063 | 34.890 | 0.011 | 62.836 | 0.784 | 12.622 | 0.843 | 1.338 | 0.882 | -16.726 | 0.439 | -37.644 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.57 | 37 | 40 | 325 | INTACT | 0.047 | -39.684 | 0.018 | -56.656 | 0.477 | 8.655 | 0.081 | 32.353 | 0.134 | 18.456 | 0.078 | 25.168 |
| transcription | Elongation factor 1-alpha 1 | EEF1A1 | P68104 | 9.56 | 53 | 50 | 58 | INTACT | 0.559 | 8.409 | 0.019 | 87.905 | 0.613 | 4.918 | 0.663 | 11.792 | 0.285 | -22.551 | 0.305 | 23.348 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.71 | 17 | 15 | 282 | INTACT | 0.343 | 24.089 | 0.019 | 184.10 | 0.838 | 0.765 | 0.566 | -14.978 | 0.952 | -7.445 | 0.012 | 123.57 |
| metabolic | Heat shock protein beta-1 | HSPB1 | P04792 | 5.34 | 26 | 23 | 270 | INTACT | 0.951 | 1.147 | 0.021 | 91.377 | 0.266 | -21.389 | 0.923 | 5.150 | 0.482 | -16.309 | 0.306 | 21.900 |
| degradation | Tripartite motif-containing protein 72 | TRIM72 | Q6ZMU5 | 6.15 | 49 | 53 | 343 | INTACT | 0.425 | -2.615 | 0.023 | -13.380 | 0.213 | 19.753 | 0.153 | 11.280 | 0.388 | 7.455 | 0.791 | -0.799 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.30 | 14 | 16 | 468 | INTACT | 0.667 | 10.124 | 0.031 | 112.43 | 0.749 | 6.978 | 0.716 | -0.824 | 0.114 | -40.203 | 0.298 | 41.607 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 9.59 | 200 | 223 | 417 | INTACT | 0.648 | -4.298 | 0.035 | -46.738 | 0.930 | 1.842 | 0.582 | -4.187 | 0.881 | -11.862 | 0.635 | -23.619 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 6.12 | 38 | 42 | 181 | INTACT | 0.589 | -5.966 | 0.036 | -22.725 | 0.213 | -13.162 | 0.349 | -7.231 | 0.181 | -11.152 | 0.841 | 2.550 |
| metabolic | Dihydrolipoyl dehydrogenase, mitochondrial | DLD | E9PEX6 | 6.82 | 54 | 52 | 271 | INTACT | 0.695 | -2.649 | 0.039 | -27.562 | 0.638 | 4.489 | 0.322 | -11.232 | 0.013 | 11.906 | 0.565 | 5.057 |
| structural | Actinin, alpha 2, isoform CRA_b | ACTN2 | B2RCS5 | 4.91 | 100 | 104 | 62 | INTACT | 0.632 | -14.040 | 0.042 | -94.054 | 0.872 | -2.195 | 0.745 | -8.746 | 0.114 | 34.274 | 0.792 | -0.894 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 4.95 | 42 | 42 | 642 | INTACT | 0.839 | -4.678 | 0.074 | -30.931 | 0.003 | 53.973 | 0.163 | 22.737 | 0.275 | 12.075 | 0.573 | -6.547 |
| transport | Hemoglobin subunit delta | HBD | P02042 | 8.76 | 14 | 16 | 297 | INTACT | 0.494 | -3.928 | 0.077 | 88.845 | 0.932 | -0.590 | 0.880 | -0.367 | 0.401 | -11.742 | 0.208 | 26.399 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.73 | 14 | 15 | 455 | INTACT | 0.316 | 13.294 | 0.108 | 54.162 | 0.544 | -17.414 | 0.737 | -1.063 | 0.542 | -6.699 | 0.097 | 20.322 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.46 | 37 | 40 | 288 | INTACT | 0.207 | -18.536 | 0.109 | -30.051 | 0.015 | 20.824 | 0.108 | 19.527 | 0.265 | -13.319 | 0.164 | 13.295 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 8.68 | 19 | 17 | 46 | INTACT | 0.579 | 20.153 | 0.142 | 56.028 | 0.028 | 62.469 | 0.528 | 11.082 | 0.044 | -59.236 | 0.997 | -7.796 |
| structural | Isoform 5 of Myosin-binding protein C, slow-type | MYBPC1 | Q00872-5 | 5.94 | 131 | 128 | 733 | INTACT | 0.186 | 17.231 | 0.146 | -25.507 | 0.843 | 0.791 | 0.329 | -13.199 | 0.042 | 30.194 | 0.323 | 18.923 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | Q5T8M7 | 5.51 | 44 | 38 | 416 | INTACT | 0.952 | 0.179 | 0.243 | -10.434 | 0.098 | -16.613 | 0.029 | -19.957 | 0.114 | -8.800 | 0.188 | -4.148 |
| structural | Isoform 5 of Radixin | RDX | P35241-5 | 6.16 | 74 | 69 | 65 | INTACT | 0.840 | -4.186 | 0.275 | 39.290 | 0.043 | 19.269 | 0.008 | 25.274 | 0.454 | 30.954 | 0.008 | 50.948 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.27 | 14 | 16 | 321 | INTACT | 0.535 | -3.584 | 0.283 | 43.307 | 0.712 | -14.745 | 0.534 | -18.281 | 0.243 | -19.456 | 0.275 | 37.422 |
| Ca | Protein S100-A13 | S100A13 | Q99584 | 5.55 | 13 | 11 | 72 | INTACT | 0.353 | 54.063 | 0.308 | 30.116 | 0.422 | 44.903 | 0.141 | 55.994 | 0.471 | -11.489 | 0.008 | 87.546 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.24 | 21 | 21 | 673 | INTACT | 0.809 | -1.707 | 0.319 | -13.318 | 0.305 | 7.972 | 0.409 | 4.897 | 0.341 | -5.513 | 0.106 | -9.445 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.50 | 37 | 40 | 143 | INTACT | 0.325 | -7.626 | 0.321 | -12.151 | 0.296 | 10.300 | 0.250 | 20.423 | 0.012 | 29.500 | 0.099 | 21.342 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.26 | 18 | 17 | 40 | INTACT | 0.767 | -5.559 | 0.340 | 27.256 | 0.099 | -16.789 | 0.368 | -14.984 | 0.631 | 1.243 | 0.090 | -34.953 |
| metabolic | Creatine kinase M-type | CKM | P06732 | 6.95 | 41 | 43 | 983 | INTACT | 0.882 | 0.440 | 0.361 | -7.476 | 0.241 | -9.951 | 0.077 | -9.531 | 0.700 | 1.895 | 0.885 | -1.366 |
| metabolic | Creatine kinase M-type | CKM | P06732 | 7.43 | 42 | 43 | 717 | INTACT | 0.644 | -3.791 | 0.387 | -11.375 | 0.670 | -2.217 | 0.325 | -5.465 | 0.746 | 5.274 | 0.937 | -0.089 |
| transport | Serum albumin | ALB | P02768 | 5.76 | 81 | 69 | 167 | INTACT | 0.918 | -1.211 | 0.400 | 35.737 | 0.003 | -20.865 | 0.102 | -13.992 | 0.624 | 8.361 | 0.052 | 24.045 |
| transport | Myoglobin | MB | P02144 | 5.51 | 17 | 17 | 57 | INTACT | 0.978 | 5.363 | 0.474 | -9.961 | 0.744 | -16.770 | 0.500 | -17.267 | 0.005 | -40.099 | 0.004 | -53.808 |
| contractile | Isoform MLC3 of Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976-2 | 4.92 | 22 | 21 | 98 | INTACT | 0.218 | 16.147 | 0.528 | 29.802 | 0.933 | -0.148 | 0.186 | -37.780 | 0.562 | -15.066 | 0.049 | -92.457 |
| contractile | Myosin-1 | MYH1 | P12882 | 9.48 | 200 | 223 | 551 | INTACT | 0.952 | -0.057 | 0.540 | -6.049 | 0.997 | 5.274 | 0.523 | -15.468 | 0.272 | -17.639 | 0.708 | -15.474 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.42 | 37 | 42 | 525 | INTACT | 0.522 | -7.899 | 0.548 | -6.458 | 0.918 | 0.813 | 0.208 | 20.450 | 0.074 | 30.566 | 0.372 | 10.649 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.20 | 35 | 33 | 120 | INTACT | 0.599 | 5.012 | 0.591 | -5.877 | 0.111 | -40.878 | 0.015 | -50.238 | 0.399 | -13.347 | 0.032 | -26.507 |
| Ca contractile | Troponin C type 2 (Fast), isoform CRA_a | TNNC2 | C9J7T9 | 4.59 | 18 | 16 | 328 | INTACT | 0.642 | 14.527 | 0.594 | 15.640 | 0.868 | -1.177 | 0.473 | -17.612 | 0.824 | 0.013 | 0.273 | -18.883 |
| metabolic | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | SDHA | D6RFM5 | 6.22 | 68 | 64 | 155 | INTACT | 0.785 | 5.171 | 0.600 | -9.833 | 0.783 | 1.307 | 0.345 | -12.345 | 0.567 | 18.575 | 0.349 | 9.239 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.67 | 18 | 17 | 36 | INTACT | 0.246 | -17.864 | 0.643 | 19.374 | 0.109 | -41.598 | 0.685 | -9.005 | 0.712 | -5.156 | 0.178 | -28.094 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | Q5T8M7 | 5.84 | 42 | 38 | 362 | INTACT | 0.939 | 1.110 | 0.661 | -1.566 | 0.029 | -29.561 | 0.361 | -8.812 | 0.326 | -12.461 | 0.861 | 0.764 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.93 | 23 | 21 | 360 | INTACT | 0.362 | -6.866 | 0.713 | 8.042 | 0.065 | -34.360 | 0.549 | -2.921 | 0.234 | -10.932 | 0.615 | -2.652 |
| contractile | Isoform MLC3 of Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976-2 | 5.17 | 20 | 21 | 199 | INTACT | 0.893 | 2.231 | 0.734 | -1.826 | 0.395 | 7.284 | 0.856 | 4.598 | 0.351 | -9.690 | 0.153 | -9.954 |
| structural | Alpha-actinin-2 | ACTN2 | P35609 | 10.2 | 104 | 104 | 162 | INTACT | 0.862 | 0.766 | 0.738 | 8.605 | 0.658 | -3.903 | 0.786 | 8.146 | 0.500 | 0.141 | 0.189 | -33.998 |
| Ca contractile | Isoform 4 of Tropomyosin alpha-1 chain | TPM1 | P09493-4 | 5.71 | 32 | 33 | 572 | INTACT | 0.685 | -4.241 | 0.758 | 22.837 | 0.353 | -18.897 | 0.841 | -2.851 | 0.462 | -14.688 | 0.395 | -22.715 |
| metabolic | Isoform 2 of Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADVL | P49748-2 | 8.74 | 68 | 70 | 96 | INTACT | 0.426 | 37.394 | 0.789 | 30.595 | 0.420 | 38.918 | 0.003 | 43.275 | 0.573 | -15.173 | 0.056 | -43.753 |
| glycolysis | Fructose-bisphosphate aldolase | ALDOA | H3BQN4 | 9.33 | 38 | 39 | 470 | INTACT | 0.056 | 16.485 | 0.798 | 2.631 | 0.407 | -5.423 | 0.100 | 9.765 | 0.708 | -1.715 | 0.536 | 6.644 |
| metabolic | Creatine kinase M-type | CKM | P06732 | 7.13 | 40 | 43 | 1010 | INTACT | 0.087 | 12.563 | 0.807 | -0.736 | 0.168 | -9.625 | 0.282 | -6.291 | 0.001 | 13.993 | 0.713 | 1.124 |
| metabolic | Fructose-bisphosphate aldolase A | ALDOA | P04075 | 7.44 | 38 | 39 | 380 | INTACT | 0.841 | -1.072 | 0.822 | 1.448 | 0.546 | 8.271 | 0.382 | -18.878 | 0.785 | 2.113 | 0.112 | -42.546 |
| structural | Desmin | DES | P17661 | 5.37 | 53 | 54 | 1170 | INTACT | 0.564 | 7.166 | 0.826 | 2.144 | 0.686 | -12.459 | 0.203 | 28.061 | 0.183 | 35.983 | 0.808 | -4.190 |
| structural | Desmin | DES | P17661 | 5.32 | 50 | 54 | 509 | INTACT | 0.869 | -10.445 | 0.842 | -0.409 | 0.880 | 6.754 | 0.035 | 56.153 | 0.229 | 20.552 | 0.241 | 18.764 |
| structural | Isoform 2 of Myosin-binding protein C, slow-type | MYBPC1 | Q00872-2 | 5.70 | 141 | 128 | 625 | INTACT | 0.854 | -0.973 | 0.868 | -7.483 | 0.259 | -16.945 | 0.079 | -17.626 | 0.547 | -46.173 | 0.462 | -66.339 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 6.00 | 41 | 42 | 454 | INTACT | 0.800 | 3.267 | 0.920 | 1.965 | 0.025 | -37.937 | 0.417 | -7.988 | 0.169 | -13.643 | 0.021 | 13.234 |
| contractile | Myosin-7 | MYH7 | P12883 | 7.09 | 205 | 223 | 381 | INTACT | 0.126 | 17.051 | 0.937 | 6.753 | 0.264 | 10.577 | 0.623 | 5.165 | 0.875 | 1.846 | 0.019 | -52.137 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.93 | 100 | 17 | 39 | AGG | 0.106 | -100.35 | 0.003 | -155.76 | 0.629 | -10.803 | 0.734 | -13.913 | 0.008 | 47.153 | 0.491 | -7.530 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.71 | 18 | 15 | 262 | AGG | 0.157 | 63.270 | 0.009 | 415.75 | 0.441 | 2.707 | 0.343 | -23.911 | 0.739 | -38.058 | 0.053 | 139.57 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 8.35 | 98 | 17 | 39 | AGG | 0.382 | 20.881 | 0.012 | 79.331 | 0.257 | 36.695 | 0.186 | 15.099 | 0.001 | -40.615 | 0.857 | -3.641 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.60 | 28 | 15 | 314 | AGG | 0.010 | 30.814 | 0.040 | 54.520 | 0.821 | -2.394 | 0.569 | -7.323 | 0.659 | -3.149 | 0.008 | 51.527 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.27 | 99 | 15 | 111 | AGG | 0.277 | 15.679 | 0.042 | 44.222 | 0.186 | 23.029 | 0.994 | 3.615 | 0.454 | -18.138 | 0.301 | 23.053 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.35 | 173 | 17 | 53 | AGG | 0.134 | -22.543 | 0.062 | -33.595 | 0.103 | -23.564 | 0.195 | -17.598 | 0.025 | 31.607 | 0.366 | 18.612 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.73 | 20 | 17 | 34 | AGG | 0.266 | 28.664 | 0.068 | 54.273 | 0.680 | 8.872 | 0.705 | -7.459 | 0.090 | 27.672 | 0.675 | 3.845 |
| degradation | E3 ubiquitin-protein ligase listerin | LTN1 | H7BYG8 | 5.14 | 126 | 91 | 44 | AGG | 0.833 | -0.946 | 0.077 | 67.094 | 0.267 | 5.934 | 0.770 | -0.629 | 0.841 | 6.157 | 0.074 | 22.611 |
| unkknown | Putative BCoR-like protein 2 | BCORP1 | Q8N888 | 8.96 | 98 | 16 | 38 | AGG | 0.935 | -2.249 | 0.086 | 26.192 | 0.085 | 29.614 | 0.782 | 9.203 | 0.290 | -21.309 | 0.412 | 4.834 |
| metabolic | Cytochrome b-c1 complex subunit Rieske, mitochondrial | UQCRFS1 | P47985 | 6.40 | 53 | 30 | 114 | AGG | 0.553 | -2.568 | 0.087 | -11.196 | 0.687 | 2.829 | 0.479 | 5.080 | 0.092 | 5.508 | 0.200 | 4.522 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.19 | 28 | 16 | 157 | AGG | 0.680 | -2.005 | 0.109 | 60.473 | 0.684 | -7.793 | 0.473 | -13.581 | 0.511 | -8.955 | 0.261 | 33.918 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.75 | 65 | 42 | 112 | AGG | 0.362 | 10.744 | 0.215 | 55.922 | 0.006 | -88.787 | 0.308 | -35.754 | 0.622 | 5.977 | 0.273 | -41.640 |
| transcription | Ataxin-3 | ATXN3 | G3V3T0 | 8.62 | 19 | 11 | 37 | AGG | 0.971 | 7.140 | 0.221 | 68.754 | 0.054 | 56.045 | 0.893 | -1.350 | 0.002 | -65.779 | 0.346 | -15.426 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.01 | 159 | 33 | 769 | AGG | 0.044 | -35.390 | 0.254 | -17.162 | 0.710 | 2.746 | 0.015 | 46.717 | 0.964 | 1.442 | 0.509 | -4.394 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.92 | 141 | 42 | 78 | AGG | 0.845 | -20.853 | 0.269 | -74.239 | 0.912 | -6.392 | 0.085 | 57.682 | 0.038 | 72.087 | 0.119 | 43.965 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 5.86 | 28 | 16 | 169 | AGG | 0.510 | -51.417 | 0.442 | 25.138 | 0.944 | -6.076 | 0.937 | -3.527 | 0.058 | 25.631 | 0.280 | ###### |
| Ca contractile | TNNT1 protein | TNNT1 | Q3B759 | 5.31 | 28 | 23 | 113 | AGG | 0.335 | -25.058 | 0.466 | 20.421 | 0.066 | -33.933 | 0.153 | -8.681 | 0.680 | -6.696 | 0.166 | 20.004 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.32 | 123 | 42 | 626 | AGG | 0.532 | -16.642 | 0.495 | -9.226 | 0.414 | -25.874 | 0.067 | -42.148 | 0.307 | 33.196 | 0.035 | 20.538 |
| Ca contractile | Troponin T, fast skeletal muscle | TNNT3 | H9KVA2 | 6.21 | 35 | 28 | 234 | AGG | 0.551 | 3.151 | 0.537 | -5.074 | 0.014 | -59.731 | 0.029 | -14.901 | 0.471 | -3.728 | 0.520 | -4.376 |
| metabolic | Isoform 2 of Glycogen phosphorylase, muscle form | PYGM | P11217-2 | 6.78 | 250 | 97 | 266 | AGG | 0.625 | 10.307 | 0.579 | 8.766 | 0.035 | 36.550 | 0.384 | 14.501 | 0.983 | 9.053 | 0.003 | -48.413 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.68 | 31 | 17 | 42 | AGG | 0.911 | 1.010 | 0.639 | 15.589 | 0.096 | -42.988 | 0.125 | -53.243 | 0.809 | -0.123 | 0.971 | -1.098 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.94 | 38 | 21 | 415 | AGG | 0.301 | 7.757 | 0.646 | -5.955 | 0.065 | -22.812 | 0.560 | -6.973 | 0.484 | 8.737 | 0.923 | -2.902 |
| transcription | Ataxin-3 | ATXN3 | G3V3T0 | 7.14 | 18 | 11 | 41 | AGG | 0.353 | -19.374 | 0.656 | -1.152 | 0.154 | 55.507 | 0.004 | 55.089 | 0.896 | 0.544 | 0.610 | -13.225 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.69 | 195 | 42 | 93 | AGG | 0.885 | -3.023 | 0.846 | 1.722 | 0.974 | -5.658 | 0.822 | -1.582 | 0.640 | 19.791 | 0.358 | 31.037 |
| Ca contractile | Isoform 4 of Tropomyosin alpha-1 chain | TPM1 | P09493-4 | 4.98 | 127 | 33 | 562 | AGG | 0.890 | -3.529 | 0.886 | 3.224 | 0.003 | 74.828 | 0.003 | 64.046 | 0.583 | 5.678 | 0.466 | -9.164 |
| metabolic | Mitochondrial inner membrane protein | IMMT | C9J406 | 5.75 | 87 | 73 | 148 | AGG | 0.353 | -6.863 | 0.896 | -0.931 | 0.792 | -2.951 | 0.429 | -4.939 | 0.023 | -12.604 | 0.853 | 1.353 |
| transport | Carbonic anhydrase 3 | CA3 | P07451 | 7.16 | 111 | 30 | 204 | AGG | 0.967 | -7.339 | 0.931 | -10.820 | 0.091 | 49.858 | 0.410 | 17.701 | 0.992 | -7.544 | 0.857 | -3.378 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.78 | 100 | 42 | 65 | AGG | 0.407 | 5.839 | 0.946 | -3.859 | 0.061 | -32.013 | 0.774 | -6.043 | 0.187 | 38.621 | 0.406 | 24.586 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.32 | 97 | 42 | 632 | AGG | 0.847 | -2.549 | 0.969 | 5.319 | 0.121 | -59.774 | 0.007 | -61.271 | 0.525 | 17.744 | 0.011 | 24.509 |
| structural | Keratin, type II cytoskeletal 2 epidermal | KRT2 | P35908 | 5.90 | 29 | 65 | 235 | FRAG | 0.105 | -33.779 | 0.010 | -68.189 | 0.068 | 25.534 | 0.401 | 61.275 | 0.014 | 72.284 | 0.601 | -11.301 |
| metabolic | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADS | P16219 | 6.30 | 38 | 44 | 95 | FRAG | 0.119 | -20.021 | 0.011 | -47.653 | 0.624 | -4.929 | 0.590 | 2.441 | 0.353 | 10.737 | 0.211 | 21.749 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 5.24 | 13 | 15 | 152 | FRAG | 0.836 | 4.416 | 0.011 | 40.645 | 0.243 | -23.019 | 0.236 | -22.989 | 0.315 | -13.330 | 0.481 | 10.203 |
| transport | Myosin-7 | MYH7 | P12883 | 9.27 | 99 | 223 | 119 | FRAG | 0.277 | 15.679 | 0.042 | 44.222 | 0.186 | 23.029 | 0.994 | 3.615 | 0.454 | -18.138 | 0.301 | 23.053 |
| metabolic | Calsequestrin-1 | CASQ1 | P31415 | 4.75 | 34 | 45 | 117 | FRAG | 0.134 | 18.064 | 0.047 | 57.142 | 0.430 | 19.514 | 0.930 | 6.253 | 0.057 | -30.206 | 0.293 | -15.611 |
| structural | Nebulin | NEB | F8WCL5 | 9.35 | 113 | 773 | 112 | FRAG | 0.285 | 29.891 | 0.049 | 37.168 | 0.268 | 38.281 | 0.869 | 4.306 | 0.593 | 13.530 | 0.159 | 32.437 |
| structural | Desmin | DES | P17661 | 5.13 | 37 | 54 | 254 | FRAG | 0.640 | 10.914 | 0.061 | 33.296 | 0.173 | -28.680 | 0.980 | 2.587 | 0.238 | -18.324 | 0.626 | 5.982 |
| glycolysis | Enolase (Fragment) | ENO3 | E5RGZ4 | 7.90 | 111 | 30 | 104 | FRAG | 0.887 | -6.345 | 0.076 | -37.105 | 0.112 | 31.686 | 0.670 | -14.817 | 0.330 | 26.891 | 0.329 | -31.928 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.28 | 13 | 16 | 374 | FRAG | 0.886 | 6.702 | 0.077 | 69.345 | 0.483 | -19.381 | 0.771 | 2.695 | 0.138 | -29.700 | 0.578 | 26.524 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.86 | 18 | 13 | 54 | FRAG | 0.968 | 5.957 | 0.084 | 23.218 | 0.766 | -4.935 | 0.635 | 7.572 | 0.311 | -17.507 | 0.103 | -27.369 |
| structural | Unconventional myosin-XIX (Fragment) | MYO19 | K7EMZ0 | 4.83 | 13 | 8 | 33 | FRAG | 0.453 | 11.383 | 0.087 | 46.201 | 0.488 | -8.860 | 0.666 | -2.953 | 0.983 | 6.363 | 0.508 | 14.896 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 4.13 | 13 | 16 | 423 | FRAG | 0.890 | -7.386 | 0.094 | 110.30 | 0.240 | -37.858 | 0.399 | -15.232 | 0.716 | 27.964 | 0.803 | 16.778 |
| contractile | Actin, alpha cardiac muscle 1 | ACTC1 | P68032 | 5.14 | 31 | 42 | 402 | FRAG | 0.894 | 19.072 | 0.107 | 66.675 | 0.419 | -11.198 | 0.712 | 20.182 | 0.004 | -78.646 | 0.717 | -3.449 |
| transcription | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 6.77 | 18 | 59 | 69 | FRAG | 0.466 | -5.725 | 0.109 | 45.959 | 0.544 | -15.433 | 0.597 | -5.846 | 0.653 | -9.658 | 0.098 | -15.187 |
| metabolic | Acyl-coenzyme A synthetase ACSM2B, mitochondrial (Fragment) | ACSM2B | H3BQ84 | 4.93 | 29 | 12 | 42 | FRAG | 0.044 | -24.131 | 0.131 | -31.352 | 0.965 | -3.970 | 0.159 | -37.024 | 0.185 | 30.108 | 0.773 | 8.325 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 4.16 | 13 | 16 | 261 | FRAG | 0.850 | -0.625 | 0.141 | 73.879 | 0.106 | -42.471 | 0.153 | -21.963 | 0.925 | 15.889 | 0.495 | 19.139 |
| contractile | Actin, alpha cardiac muscle 1 | ACTC1 | P68032 | 5.35 | 34 | 42 | 334 | FRAG | 0.104 | 32.048 | 0.182 | 34.174 | 0.593 | -15.202 | 0.147 | -27.973 | 0.545 | -4.691 | 0.002 | -32.503 |
| transport | Myoglobin (Fragment) | MB | B0QYF8 | 9.37 | 16 | 16 | 164 | FRAG | 0.011 | 123.22 | 0.183 | 19.268 | 0.529 | 9.365 | 0.832 | -8.550 | 0.629 | -18.888 | 0.248 | -31.431 |
| unknown | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Fragment) | MACF1 | H0Y390 | 9.41 | 137 | 506 | 40 | FRAG | 0.117 | 19.416 | 0.184 | 29.488 | 0.052 | 20.698 | 0.272 | 14.966 | 0.178 | -14.149 | 0.002 | 48.663 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 5.17 | 16 | 13 | 621 | FRAG | 0.758 | 2.304 | 0.191 | -14.022 | 0.364 | 9.847 | 0.880 | 0.257 | 0.597 | -0.315 | 0.852 | 0.309 |
| metabolic | Acyl-coenzyme A synthetase ACSM2B, mitochondrial (Fragment) | ACSM2B | H3BQ84 | 5.31 | 18 | 12 | 47 | FRAG | 0.811 | -14.188 | 0.205 | -20.103 | 0.366 | -11.135 | 0.000 | -39.328 | 0.313 | -6.814 | 0.262 | 10.793 |
| metabolic | Isoform 2 of Glycogen phosphorylase, muscle form | PYGM | P11217-2 | 5.98 | 28 | 97 | 155 | FRAG | 0.175 | 23.378 | 0.210 | 17.216 | 0.655 | 5.662 | 0.592 | 8.229 | 0.007 | -69.859 | 0.506 | -12.780 |
| contractile | Myosin-1 | MYH1 | P12882 | 9.92 | 191 | 223 | 358 | FRAG | 0.221 | -11.060 | 0.214 | -19.219 | 0.496 | 5.161 | 0.743 | -10.136 | 0.356 | 2.791 | 0.875 | -4.025 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BN54 | 5.20 | 16 | 15 | 797 | FRAG | 0.943 | -1.750 | 0.289 | -9.329 | 0.236 | 26.980 | 0.850 | -0.055 | 0.223 | 11.650 | 0.976 | -2.807 |
| structural | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 5.63 | 21 | 59 | 200 | FRAG | 0.886 | -4.688 | 0.300 | 26.584 | 0.668 | -17.821 | 0.172 | -33.551 | 0.509 | -6.357 | 0.671 | 6.536 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.33 | 18 | 13 | 129 | FRAG | 0.265 | 22.324 | 0.300 | 12.639 | 0.515 | -10.523 | 0.593 | 11.125 | 0.253 | -15.755 | 0.277 | -13.101 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.14 | 16 | 9 | 107 | FRAG | 0.507 | -7.698 | 0.301 | -15.272 | 0.866 | -2.988 | 0.688 | -8.587 | 0.189 | 33.805 | 0.886 | 1.382 |
| structural | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 5.22 | 14 | 59 | 250 | FRAG | 0.197 | -14.400 | 0.326 | -12.827 | 0.412 | -4.456 | 0.833 | 4.158 | 0.142 | -7.949 | 0.121 | -9.987 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 5.38 | 36 | 42 | 171 | FRAG | 0.594 | 9.301 | 0.327 | -12.032 | 0.217 | -14.127 | 0.068 | -27.710 | 0.279 | -16.151 | 0.014 | -32.629 |
| structural | Keratin, type II cytoskeletal 6A | KRT6A | P02538 | 6.13 | 33 | 60 | 435 | FRAG | 0.484 | -8.428 | 0.334 | -12.101 | 0.949 | 0.855 | 0.232 | 11.988 | 0.790 | -0.641 | 0.415 | -12.316 |
| metabolic | Pyruvate kinase (Fragment) | PKM | H3BTN5 | 8.16 | 57 | 53 | 528 | FRAG | 0.314 | 5.868 | 0.335 | -12.819 | 0.019 | 16.637 | 0.088 | 41.052 | 0.046 | 17.824 | 0.072 | -18.709 |
| structural | Cofilin-1 (Fragment) | CFL1 | E9PLJ3 | 6.04 | 17 | 9 | 66 | FRAG | 0.924 | 0.646 | 0.362 | -7.083 | 0.745 | 6.323 | 0.094 | 22.069 | 0.862 | 0.001 | 0.085 | -17.436 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BPK4 | 6.22 | 17 | 22 | 90 | FRAG | 0.536 | 6.965 | 0.389 | 13.416 | 0.561 | -10.970 | 0.483 | 7.869 | 0.181 | -24.561 | 0.678 | -5.782 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 5.60 | 143 | 223 | 384 | FRAG | 0.213 | 16.716 | 0.448 | 14.754 | 0.706 | -5.083 | 0.228 | -21.723 | 0.387 | 13.554 | 0.286 | 20.638 |
| structural | Keratin, type I cytoskeletal 9 | KRT9 | P35527 | 7.05 | 18 | 62 | 173 | FRAG | 0.270 | -25.589 | 0.459 | -30.517 | 0.249 | 38.229 | 0.005 | 62.927 | 0.346 | -5.986 | 0.726 | 16.306 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.22 | 31 | 9 | 74 | FRAG | 0.878 | 1.741 | 0.491 | 25.302 | 0.061 | -44.487 | 0.620 | -2.151 | 0.293 | 33.642 | 0.617 | 15.100 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.75 | 16 | 9 | 115 | FRAG | 0.633 | 3.271 | 0.504 | 0.139 | 0.073 | 51.451 | 0.507 | 12.151 | 0.061 | 27.606 | 0.829 | 2.775 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.36 | 18 | 13 | 280 | FRAG | 0.542 | -4.968 | 0.526 | 16.596 | 0.454 | -22.196 | 0.695 | 5.523 | 0.069 | -27.513 | 0.508 | -4.763 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BN54 | 5.17 | 37 | 15 | 662 | FRAG | 0.305 | 18.757 | 0.601 | -12.798 | 0.195 | 26.187 | 0.854 | 4.732 | 0.433 | -4.446 | 0.248 | 5.422 |
| unknown | Serine/threonine-protein phosphatase 4 regulatory subunit 4 | PPP4R4 | Q6NUP7 | 7.00 | 18 | 99 | 32 | FRAG | 0.422 | -12.066 | 0.603 | 31.295 | 0.731 | 19.620 | 0.281 | 38.654 | 0.210 | -59.963 | 0.295 | -44.620 |
| transcription | Histidine protein methyltransferase 1 homolog | METTL18 | O95568 | 4.89 | 20 | 42 | 38 | FRAG | 0.129 | -55.293 | 0.641 | -5.231 | 0.730 | -14.733 | 0.885 | -1.598 | 0.045 | 46.197 | 0.346 | 9.834 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.89 | 17 | 9 | 112 | FRAG | 0.935 | 7.103 | 0.641 | 0.917 | 0.009 | 36.433 | 0.951 | -3.017 | 0.148 | 12.042 | 0.999 | -8.156 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.10 | 16 | 13 | 64 | FRAG | 0.965 | 1.738 | 0.936 | 3.122 | 0.743 | -6.538 | 0.718 | 10.729 | 0.473 | -12.942 | 0.792 | -0.488 |
| transport | Fatty acid-binding protein, heart (Fragment) | FABP3 | S4R371 | 5.99 | 14 | 15 | 574 | FRAG | 0.198 | -22.190 | 0.953 | 3.971 | 0.174 | 8.128 | 0.420 | -11.975 | 0.078 | -13.499 | 0.465 | -7.175 |
| structural | Nebulin | NEB | F8WCL5 | 8.96 | 109 | 773 | 67 | FRAG | 0.791 | -5.050 | 0.985 | 8.001 | 0.326 | 58.027 | 0.743 | 15.439 | 0.167 | -19.911 | 0.895 | -2.046 |
| structural | Keratin, type II cytoskeletal 1 | KRT1 | P04264 | 4.87 | 31 | 66 | 41 | FRAG | 0.259 | -17.766 | 0.985 | 5.204 | 0.081 | 30.037 | 0.491 | 15.385 | 0.920 | -3.073 | 0.420 | -14.843 |
| unknown | #N/A | #N/A | #N/A | 10.2 | 19 | #N/A | #N/A | #N/A | 0.588 | -9.415 | 0.633 | 17.080 | 0.025 | -62.364 | 0.039 | -39.293 | 0.601 | 0.763 | 0.215 | -28.756 |
| CON | CON | EX | EX | TEX | TEX | |||||||||||||||
| Total | Mid vs. Pre | Post vs. Pre | Mid vs. Pre | Post vs. Pre | Mid vs. Pre | Post vs. Pre | ||||||||||||||
| INTACT | 55 | 3 | 17 | 7 | 6 | 6 | 9 | |||||||||||||
| AGG | 29 | 2 | 5 | 4 | 5 | 6 | 4 | |||||||||||||
| FRAG | 46 | 2 | 6 | 2 | 2 | 5 | 3 | |||||||||||||
| TOTAL | 130 | 7 | 28 | 13 | 13 | 17 | 16 | |||||||||||||
Table 4. Protein phosphorylation differences in PEX vs TEX.
Differences between PEX and TEX in all identified spots. Ordering within the table is based on proteoform interpretation (i.e. intact, aggregate, fragment) and p-values (2-tailed, unpaired t-tests) of the post comparison between PEX and TEX. P-values < 0.05 are shaded in yellow. Differences (%) within each comparison are shaded to indicate higher (red) or lower (blue) values in PEX relative to TEX.
| pre | mid | post | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEX compared to TEX | PEX compared to TEX | PEX compared to TEX | ||||||||||||
| TABLE 4 | Protein Name | Symbol | Accession Number | pI | MW (kD) | UNIPROT MW (kd) | MS Protein Score | Proteoform Interpretation | P-Value | Diff. (%) | P-Value | Diff. (%) | P-Value | Diff. (%) |
| metabolic | Creatine kinase M-type | CKM | P06732 | 7.13 | 40 | 43 | 1010 | INTACT | 0.236 | -20.578 | 0.256 | 13.685 | 0.001 | 28.608 |
| metabolic | Creatine kinase M-type | CKM | P06732 | 6.95 | 41 | 43 | 983 | INTACT | 0.390 | -7.588 | 0.273 | 10.779 | 0.001 | 32.480 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 9.59 | 200 | 223 | 417 | INTACT | 0.780 | -2.484 | 0.171 | 48.475 | 0.006 | 66.349 |
| contractile | Myosin-1 | MYH1 | P12882 | 9.48 | 200 | 223 | 551 | INTACT | 0.794 | -1.674 | 0.138 | 41.428 | 0.006 | 62.886 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 6.00 | 41 | 42 | 454 | INTACT | 0.714 | 4.888 | 0.275 | 13.461 | 0.008 | 32.904 |
| metabolic | Creatine kinase M-type | CKM | P06732 | 7.43 | 42 | 43 | 717 | INTACT | 0.439 | 7.649 | 0.855 | -4.380 | 0.012 | 40.507 |
| Ca contractile | Troponin C type 2 (Fast), isoform CRA_a | TNNC2 | C9J7T9 | 4.59 | 18 | 16 | 328 | INTACT | 0.085 | -26.115 | 0.702 | 5.751 | 0.021 | 41.717 |
| metabolic | Fructose-bisphosphate aldolase A | ALDOA | P04075 | 7.44 | 38 | 39 | 380 | INTACT | 0.706 | 3.300 | 0.805 | 4.543 | 0.025 | 40.745 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 9.67 | 195 | 223 | 451 | INTACT | 0.775 | -3.146 | 0.039 | 71.296 | 0.026 | 41.678 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | Q5T8M7 | 5.51 | 44 | 38 | 416 | INTACT | 0.310 | 16.739 | 0.203 | 23.749 | 0.038 | 41.029 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 8.68 | 19 | 17 | 46 | INTACT | 0.898 | -22.045 | 0.695 | 5.506 | 0.076 | -25.687 |
| Ca contractile | Isoform 4 of Tropomyosin alpha-1 chain | TPM1 | P09493-4 | 5.71 | 32 | 33 | 572 | INTACT | 0.903 | -13.820 | 0.912 | 3.034 | 0.080 | -42.144 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | Q5T8M7 | 5.84 | 42 | 38 | 362 | INTACT | 0.835 | 1.528 | 0.173 | 17.333 | 0.087 | 18.782 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.57 | 37 | 40 | 325 | INTACT | 0.955 | 1.949 | 0.479 | 9.030 | 0.107 | 24.714 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.50 | 37 | 40 | 143 | INTACT | 0.986 | 3.174 | 0.767 | 1.585 | 0.115 | 24.493 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.46 | 37 | 40 | 288 | INTACT | 0.925 | 0.014 | 0.389 | 9.426 | 0.115 | 23.931 |
| metabolic | Dihydrolipoyl dehydrogenase, mitochondrial | DLD | E9PEX6 | 6.82 | 54 | 52 | 271 | INTACT | 0.123 | 36.434 | 0.332 | -25.548 | 0.119 | -40.722 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.26 | 34 | 33 | 592 | INTACT | 0.333 | -14.320 | 0.900 | -1.025 | 0.120 | -41.172 |
| structural | Alpha-actinin-2 | ACTN2 | P35609 | 10.2 | 104 | 104 | 162 | INTACT | 0.389 | 14.501 | 0.721 | 8.119 | 0.130 | -45.507 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.93 | 23 | 21 | 360 | INTACT | 0.110 | 61.540 | 0.822 | 0.830 | 0.141 | 13.119 |
| structural | Desmin | DES | P17661 | 5.37 | 53 | 54 | 1170 | INTACT | 0.567 | 8.370 | 0.185 | 17.928 | 0.143 | 24.408 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.27 | 14 | 16 | 321 | INTACT | 0.344 | -12.978 | 0.519 | -14.885 | 0.148 | -49.523 |
| metabolic | Heat shock protein beta-1 | HSPB1 | P04792 | 5.34 | 26 | 23 | 270 | INTACT | 0.626 | 8.640 | 0.062 | -21.255 | 0.158 | -23.606 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.42 | 37 | 42 | 525 | INTACT | 0.748 | -3.572 | 0.723 | -8.574 | 0.160 | 24.234 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.30 | 14 | 16 | 468 | INTACT | 0.684 | -3.805 | 0.616 | 3.738 | 0.199 | -72.209 |
| transport | Myoglobin | MB | P02144 | 5.51 | 17 | 17 | 57 | INTACT | 0.195 | -26.622 | 0.515 | -17.249 | 0.210 | -17.022 |
| contractile | Isoform MLC3 of Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976-2 | 4.92 | 22 | 21 | 98 | INTACT | 0.944 | 1.879 | 0.896 | 1.562 | 0.239 | 16.453 |
| metabolic | Adenylate kinase isoenzyme 1 | AK1 | P00568 | 9.61 | 24 | 22 | 178 | INTACT | 0.748 | 12.248 | 0.716 | 1.923 | 0.299 | -13.293 |
| transcription | Elongation factor 1-alpha 1 | EEF1A1 | P68104 | 9.56 | 53 | 50 | 58 | INTACT | 0.394 | 13.572 | 0.548 | -21.987 | 0.304 | -33.996 |
| contractile | Isoform MLC3 of Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976-2 | 5.17 | 20 | 21 | 199 | INTACT | 0.082 | -27.611 | 0.128 | -23.383 | 0.338 | -14.496 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.26 | 18 | 17 | 40 | INTACT | 0.233 | -23.950 | 0.142 | -33.146 | 0.359 | -12.714 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.73 | 14 | 15 | 455 | INTACT | 0.752 | 1.851 | 0.764 | -5.764 | 0.386 | -12.319 |
| structural | Actinin, alpha 2, isoform CRA_b | ACTN2 | B2RCS5 | 4.91 | 100 | 104 | 62 | INTACT | 0.114 | 32.876 | 0.141 | 27.388 | 0.390 | -16.466 |
| glycolysis | Fructose-bisphosphate aldolase | ALDOA | H3BQN4 | 9.33 | 38 | 39 | 470 | INTACT | 0.053 | 41.254 | 0.869 | 4.377 | 0.396 | -20.963 |
| transport | Hemoglobin subunit delta | HBD | P02042 | 8.76 | 14 | 16 | 297 | INTACT | 0.115 | 27.180 | 0.701 | 5.088 | 0.404 | -28.722 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.24 | 21 | 21 | 673 | INTACT | 0.005 | -33.802 | 0.870 | -1.636 | 0.409 | -8.236 |
| Ca | Protein S100-A13 | S100A13 | Q99584 | 5.55 | 13 | 11 | 72 | INTACT | 0.336 | 24.532 | 0.663 | 14.737 | 0.422 | -11.854 |
| transport | Serum albumin | ALB | P02768 | 5.76 | 81 | 69 | 167 | INTACT | 0.634 | -5.665 | 0.520 | 6.823 | 0.483 | 6.108 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 10.2 | 18 | 17 | 36 | INTACT | 0.806 | 13.608 | 0.796 | -13.325 | 0.486 | -17.497 |
| degradation | Tripartite motif-containing protein 72 | TRIM72 | Q6ZMU5 | 6.15 | 49 | 53 | 343 | INTACT | 0.814 | 2.039 | 0.115 | 26.199 | 0.493 | 42.350 |
| transport | Carbonic anhydrase 3 | CA3 | P07451 | 5.27 | 28 | 30 | 38 | INTACT | 0.315 | 8.136 | 0.763 | 4.157 | 0.494 | -9.057 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 6.12 | 38 | 42 | 181 | INTACT | 0.972 | -0.010 | 0.208 | 12.809 | 0.499 | 12.872 |
| metabolic | Heat shock protein beta-7 | HSPB7 | Q9UBY9 | 5.97 | 18 | 19 | 265 | INTACT | 0.378 | -16.100 | 0.930 | -8.510 | 0.520 | 11.246 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.20 | 35 | 33 | 120 | INTACT | 0.745 | -10.235 | 0.152 | -33.648 | 0.558 | -4.471 |
| metabolic | Isoform 2 of Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADVL | P49748-2 | 8.74 | 68 | 70 | 96 | INTACT | 0.534 | -8.172 | 0.728 | -7.823 | 0.582 | 14.958 |
| metabolic | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | SDHA | D6RFM5 | 6.22 | 68 | 64 | 155 | INTACT | 0.989 | -4.287 | 0.690 | 8.013 | 0.587 | -11.022 |
| structural | Desmin | DES | P17661 | 5.32 | 50 | 54 | 509 | INTACT | 0.791 | 5.346 | 0.922 | -1.059 | 0.676 | 6.647 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.67 | 18 | 17 | 36 | INTACT | 0.579 | -1.988 | 0.278 | -26.803 | 0.677 | -2.112 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.71 | 17 | 15 | 282 | INTACT | 0.225 | 20.503 | 0.896 | -0.789 | 0.707 | 4.098 |
| structural | Isoform 5 of Myosin-binding protein C, slow-type | MYBPC1 | Q00872-5 | 5.94 | 131 | 128 | 733 | INTACT | 0.614 | -9.207 | 0.690 | -6.968 | 0.729 | -9.249 |
| structural | Isoform 5 of Radixin | RDX | P35241-5 | 6.16 | 74 | 69 | 65 | INTACT | 0.641 | -4.392 | 0.976 | 2.510 | 0.806 | -12.052 |
| structural | Isoform 2 of Myosin-binding protein C, slow-type | MYBPC1 | Q00872-2 | 5.70 | 141 | 128 | 625 | INTACT | 0.655 | 2.407 | 0.982 | 5.312 | 0.823 | -0.430 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 4.95 | 42 | 42 | 642 | INTACT | 0.131 | 18.485 | 0.451 | -7.035 | 0.959 | -0.796 |
| contractile | Myosin-7 | MYH7 | P12883 | 7.09 | 205 | 223 | 381 | INTACT | 0.122 | -31.226 | 0.902 | 3.242 | 0.976 | -3.985 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.51 | 15 | 17 | 36 | INTACT | 0.481 | 16.275 | 0.884 | 3.724 | 0.986 | -1.701 |
| transport | Carbonic anhydrase 3 | CA3 | P07451 | 7.16 | 111 | 30 | 204 | AGG | 0.998 | 3.046 | 0.125 | -29.160 | 0.005 | -72.200 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.27 | 99 | 15 | 111 | AGG | 0.836 | 9.037 | 0.575 | -11.737 | 0.018 | -69.453 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.35 | 173 | 17 | 53 | AGG | 0.560 | -5.339 | 0.540 | -21.994 | 0.021 | -52.825 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.19 | 28 | 16 | 157 | AGG | 0.867 | 0.853 | 0.321 | 26.987 | 0.073 | -34.374 |
| Ca contractile | TNNT1 protein | TNNT1 | Q3B759 | 5.31 | 28 | 23 | 113 | AGG | 0.134 | -29.878 | 0.058 | -36.719 | 0.117 | -19.609 |
| transcription | Ataxin-3 | ATXN3 | G3V3T0 | 7.14 | 18 | 11 | 41 | AGG | 0.500 | -10.578 | 0.524 | -12.336 | 0.169 | 20.236 |
| unkknown | Putative BCoR-like protein 2 | BCORP1 | Q8N888 | 8.96 | 98 | 16 | 38 | AGG | 0.504 | 9.082 | 0.456 | -19.685 | 0.247 | -20.274 |
| metabolic | Mitochondrial inner membrane protein | IMMT | C9J406 | 5.75 | 87 | 73 | 148 | AGG | 0.199 | -20.645 | 0.879 | -7.151 | 0.267 | 12.084 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.78 | 100 | 42 | 65 | AGG | 0.063 | 37.899 | 0.099 | -46.297 | 0.268 | 14.692 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.32 | 123 | 42 | 626 | AGG | 0.575 | -13.189 | 0.802 | -4.538 | 0.291 | -22.864 |
| Ca contractile | Troponin T, fast skeletal muscle | TNNT3 | H9KVA2 | 6.21 | 35 | 28 | 234 | AGG | 0.578 | 7.548 | 0.559 | -11.346 | 0.310 | 15.966 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.92 | 141 | 42 | 78 | AGG | 0.574 | 11.190 | 0.843 | 2.769 | 0.394 | 15.285 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.69 | 195 | 42 | 93 | AGG | 0.217 | 22.229 | 0.645 | 34.760 | 0.411 | -13.675 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.71 | 18 | 15 | 262 | AGG | 0.937 | -0.605 | 0.840 | 2.289 | 0.472 | 10.152 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.32 | 97 | 42 | 632 | AGG | 0.579 | 7.133 | 0.110 | -27.469 | 0.527 | -12.206 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 8.35 | 98 | 17 | 39 | AGG | 0.630 | 13.637 | 0.570 | 14.054 | 0.564 | -28.371 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.68 | 31 | 17 | 42 | AGG | 0.246 | 21.596 | 0.644 | -6.208 | 0.573 | -5.674 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.01 | 159 | 33 | 769 | AGG | 0.969 | 3.989 | 0.840 | 2.313 | 0.652 | -13.105 |
| degradation | E3 ubiquitin-protein ligase listerin | LTN1 | H7BYG8 | 5.14 | 126 | 91 | 44 | AGG | 0.046 | 13.411 | 0.934 | -0.232 | 0.654 | -6.187 |
| metabolic | Cytochrome b-c1 complex subunit Rieske, mitochondrial | UQCRFS1 | P47985 | 6.40 | 53 | 30 | 114 | AGG | 0.458 | 8.470 | 0.886 | 0.245 | 0.661 | -3.256 |
| metabolic | Isoform 2 of Glycogen phosphorylase, muscle form | PYGM | P11217-2 | 6.78 | 250 | 97 | 266 | AGG | 0.082 | -31.134 | 0.572 | 10.577 | 0.665 | -23.037 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.75 | 65 | 42 | 112 | AGG | 0.226 | 31.042 | 0.164 | -52.378 | 0.708 | 3.643 |
| transcription | Ataxin-3 | ATXN3 | G3V3T0 | 8.62 | 19 | 11 | 37 | AGG | 0.842 | -0.571 | 0.332 | 20.915 | 0.722 | 17.997 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.73 | 20 | 17 | 34 | AGG | 0.829 | 2.310 | 0.613 | -5.029 | 0.820 | -4.577 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.94 | 38 | 21 | 415 | AGG | 0.744 | -3.570 | 0.170 | 32.708 | 0.882 | -2.190 |
| Ca contractile | Isoform 4 of Tropomyosin alpha-1 chain | TPM1 | P09493-4 | 4.98 | 127 | 33 | 562 | AGG | 0.074 | -39.608 | 0.892 | 1.636 | 0.892 | 10.182 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.93 | 100 | 17 | 39 | AGG | 0.521 | 4.711 | 0.026 | 57.228 | 0.915 | 6.274 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 5.86 | 28 | 16 | 169 | AGG | 0.317 | -11.886 | 0.549 | 15.431 | 0.928 | -6.836 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.60 | 28 | 15 | 314 | AGG | 0.995 | 1.731 | 0.300 | -24.725 | 0.968 | 2.617 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.28 | 13 | 16 | 374 | FRAG | 0.417 | -10.748 | 0.702 | -6.219 | 0.016 | -86.174 |
| transport | Myosin-7 | MYH7 | P12883 | 9.27 | 99 | 223 | 119 | FRAG | 0.836 | 9.037 | 0.575 | -11.737 | 0.018 | -69.453 |
| unknown | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Fragment) | MACF1 | H0Y390 | 9.41 | 137 | 506 | 40 | FRAG | 0.044 | -28.026 | 0.903 | -0.069 | 0.031 | -42.117 |
| metabolic | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADS | P16219 | 6.30 | 38 | 44 | 95 | FRAG | 0.531 | 9.903 | 0.205 | 13.690 | 0.041 | 22.255 |
| metabolic | Calsequestrin-1 | CASQ1 | P31415 | 4.75 | 34 | 45 | 117 | FRAG | 0.200 | -12.681 | 0.657 | -3.224 | 0.050 | -32.049 |
| structural | Nebulin | NEB | F8WCL5 | 8.96 | 109 | 773 | 67 | FRAG | 0.458 | -11.145 | 0.665 | 2.703 | 0.054 | -25.416 |
| structural | Nebulin | NEB | F8WCL5 | 9.35 | 113 | 773 | 112 | FRAG | 0.560 | -12.816 | 0.637 | -3.076 | 0.067 | -62.524 |
| structural | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 5.63 | 21 | 59 | 200 | FRAG | 0.343 | 13.479 | 0.503 | -8.069 | 0.093 | -29.026 |
| metabolic | Pyruvate kinase (Fragment) | PKM | H3BTN5 | 8.16 | 57 | 53 | 528 | FRAG | 0.798 | -2.231 | 0.478 | -8.463 | 0.094 | -22.549 |
| glycolysis | Enolase (Fragment) | ENO3 | E5RGZ4 | 7.90 | 111 | 30 | 104 | FRAG | 0.332 | 24.513 | 0.007 | 60.388 | 0.097 | -36.006 |
| transport | Fatty acid-binding protein, heart (Fragment) | FABP3 | S4R371 | 5.99 | 14 | 15 | 574 | FRAG | 0.678 | -6.509 | 0.759 | 3.900 | 0.111 | -45.740 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.14 | 16 | 9 | 107 | FRAG | 0.971 | 7.119 | 0.184 | -38.576 | 0.130 | 42.751 |
| structural | Unconventional myosin-XIX (Fragment) | MYO19 | K7EMZ0 | 4.83 | 13 | 8 | 33 | FRAG | 0.549 | 20.606 | 0.832 | -3.361 | 0.131 | -22.350 |
| contractile | Myosin-1 | MYH1 | P12882 | 9.92 | 191 | 223 | 358 | FRAG | 0.666 | -7.265 | 0.030 | 33.180 | 0.179 | 25.737 |
| structural | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 5.22 | 14 | 59 | 250 | FRAG | 0.556 | -5.304 | 0.676 | 7.043 | 0.208 | 18.660 |
| metabolic | Isoform 2 of Glycogen phosphorylase, muscle form | PYGM | P11217-2 | 5.98 | 28 | 97 | 155 | FRAG | 0.118 | -43.708 | 0.518 | -9.793 | 0.256 | -34.741 |
| contractile | Actin, alpha cardiac muscle 1 | ACTC1 | P68032 | 5.35 | 34 | 42 | 334 | FRAG | 0.960 | -2.923 | 0.554 | 10.697 | 0.272 | 15.956 |
| structural | Keratin, type I cytoskeletal 9 | KRT9 | P35527 | 7.05 | 18 | 62 | 173 | FRAG | 0.093 | -43.326 | 0.516 | -13.255 | 0.354 | 12.637 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.10 | 16 | 13 | 64 | FRAG | 0.496 | -2.281 | 0.627 | -22.567 | 0.380 | 13.938 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BN54 | 5.17 | 37 | 15 | 662 | FRAG | 0.253 | -22.430 | 0.340 | -45.149 | 0.406 | -29.019 |
| metabolic | Acyl-coenzyme A synthetase ACSM2B, mitochondrial (Fragment) | ACSM2B | H3BQ84 | 5.31 | 18 | 12 | 47 | FRAG | 0.049 | -34.420 | 0.218 | -17.235 | 0.435 | -9.440 |
| contractile | Actin, alpha cardiac muscle 1 | ACTC1 | P68032 | 5.14 | 31 | 42 | 402 | FRAG | 0.940 | -1.558 | 0.735 | 3.549 | 0.458 | -13.188 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 4.16 | 13 | 16 | 261 | FRAG | 0.255 | 26.733 | 0.151 | -48.021 | 0.522 | -16.155 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BN54 | 5.20 | 16 | 15 | 797 | FRAG | 0.536 | -9.301 | 0.702 | -4.549 | 0.531 | 8.694 |
| metabolic | Acyl-coenzyme A synthetase ACSM2B, mitochondrial (Fragment) | ACSM2B | H3BQ84 | 4.93 | 29 | 12 | 42 | FRAG | 0.323 | 15.002 | 0.502 | 18.870 | 0.575 | 6.125 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.75 | 16 | 9 | 115 | FRAG | 0.940 | 7.459 | 0.992 | 9.985 | 0.628 | 17.635 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 5.17 | 16 | 13 | 621 | FRAG | 0.277 | -25.666 | 0.684 | 4.901 | 0.685 | 5.549 |
| structural | Cofilin-1 (Fragment) | CFL1 | E9PLJ3 | 6.04 | 17 | 9 | 66 | FRAG | 0.262 | -20.414 | 0.414 | -24.698 | 0.704 | 3.539 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.36 | 18 | 13 | 280 | FRAG | 0.379 | -12.307 | 0.458 | -23.247 | 0.710 | -0.061 |
| transcription | Histidine protein methyltransferase 1 homolog | METTL18 | O95568 | 4.89 | 20 | 42 | 38 | FRAG | 0.268 | 22.890 | 0.807 | -0.273 | 0.728 | -1.962 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 4.13 | 13 | 16 | 423 | FRAG | 0.128 | 34.368 | 0.004 | -75.666 | 0.778 | -6.020 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 5.24 | 13 | 15 | 152 | FRAG | 0.104 | 15.375 | 0.173 | 13.997 | 0.780 | 3.232 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.22 | 31 | 9 | 74 | FRAG | 0.368 | 16.711 | 0.020 | -92.540 | 0.784 | -8.679 |
| structural | Keratin, type II cytoskeletal 1 | KRT1 | P04264 | 4.87 | 31 | 66 | 41 | FRAG | 0.910 | 4.862 | 0.672 | 13.940 | 0.799 | 3.891 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.89 | 17 | 9 | 112 | FRAG | 0.731 | 9.086 | 0.902 | 1.484 | 0.833 | -4.530 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 5.38 | 36 | 42 | 171 | FRAG | 0.466 | -14.468 | 0.841 | 11.175 | 0.837 | 10.738 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.86 | 18 | 13 | 54 | FRAG | 0.445 | -8.197 | 0.986 | -2.011 | 0.850 | -8.166 |
| structural | Keratin, type II cytoskeletal 6A | KRT6A | P02538 | 6.13 | 33 | 60 | 435 | FRAG | 0.815 | -4.139 | 0.730 | -2.768 | 0.852 | 0.892 |
| transcription | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 6.77 | 18 | 59 | 69 | FRAG | 0.519 | -2.711 | 0.405 | -32.530 | 0.857 | -8.675 |
| structural | Desmin | DES | P17661 | 5.13 | 37 | 54 | 254 | FRAG | 0.195 | -32.632 | 0.463 | -25.495 | 0.870 | 1.254 |
| structural | Keratin, type II cytoskeletal 2 epidermal | KRT2 | P35908 | 5.90 | 29 | 65 | 235 | FRAG | 0.077 | 31.922 | 0.637 | 12.027 | 0.888 | -6.467 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.33 | 18 | 13 | 129 | FRAG | 0.297 | -20.016 | 0.390 | -23.974 | 0.918 | -10.137 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 5.60 | 143 | 223 | 384 | FRAG | 0.195 | 24.861 | 0.555 | 10.880 | 0.923 | -1.428 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BPK4 | 6.22 | 17 | 22 | 90 | FRAG | 0.510 | -15.747 | 0.346 | -26.993 | 0.941 | -8.377 |
| transport | Myoglobin (Fragment) | MB | B0QYF8 | 9.37 | 16 | 16 | 164 | FRAG | 0.007 | 46.073 | 0.318 | -18.533 | 0.965 | -2.515 |
| unknown | Serine/threonine-protein phosphatase 4 regulatory subunit 4 | PPP4R4 | Q6NUP7 | 7.00 | 18 | 99 | 32 | FRAG | 0.178 | -29.181 | 0.958 | -0.391 | 0.982 | 0.879 |
| unknown | #N/A | #N/A | #N/A | 10.2 | 19 | #N/A | #N/A | #N/A | 0.922 | -11.276 | 0.180 | -30.059 | 0.930 | 2.672 |
| Total | pre | mid | post | |||||||||||
| intact | 55 | 1 | 1 | 10 | ||||||||||
| aggregate | 29 | 1 | 1 | 3 | ||||||||||
| fragment | 46 | 3 | 4 | 4 | ||||||||||
| TOTAL | 130 | 5 | 6 | 17 | ||||||||||
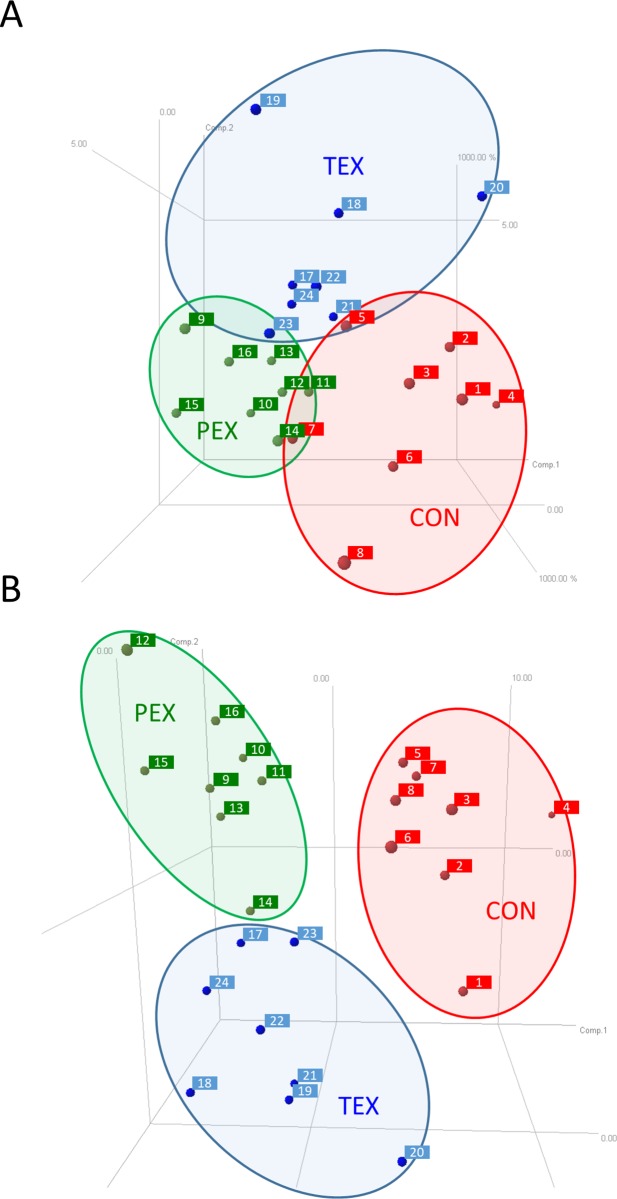
The t-tests described above and shown in Tables 1–4 were conducted according to standard gel spot picking procedures and compared data sets spot by spot (univariate analysis). In addition, we performed multivariate data analysis to reveal patterns, looking at our data more globally. For this purpose, we used Principal Components Analysis (PCA), a “dimension reduction” algorithm. PCA was performed using the analysis program TIBCO Spotfire. PCA transforms the original set of variables (spot intensities for selected samples) to a new, orthogonal, set of variables (the principal components) such that most of the variability in the original data is captured in the top few of the new components. In general, it is sufficient to examine the first 2 or 3 components to discover any existing clustering in the data. Our approach was to plot PCA1 vs. PCA2 vs. PCA3. We performed this calculation for the total abundance and phosphorylation data separately.
Results
From a total of 932 protein spots detected, 130 spots emerged as potentially altered in terms of phosphoprotein or total protein abundance due to HDBR (pre, mid, post) and/or countermeasures (CON, PEX, TEX), and were subjected to MS analysis for protein identification. Out of 130 spots, 129 were identified by MS while 1 remained unidentified. Among the identified spots, 55 proteoforms had molecular weights (MW) that were within 15% of predicted MW (UNIPROT) and were assumed to be full size proteins (INTACT). An additional 29 spots corresponded to MW that exceeded the predicted MW by at least 15% and were assumed to include protein modifications, dimers and aggregates (AGG). The remaining 45 spots were at least 15% below the predicted MW and were assumed to be protein fragments and peptides (FRAG).
Subjects
As previously reported in detail, LBM significantly decreased in CON, remained near/below baseline in PEX, and increased in TEX. Conversely, FM increased in both CON and PEX and remained near/below baseline in TEX [3]. HDBR resulted in decreased strength while exercise with or without testosterone countermeasures were protective against such losses in load bearing muscles [3].
Pre to post HDBR changes
Within-group abundance changes of all identified spots (pre-post, 2-tailed, paired t-tests) are shown in Table 1. Ordering in the table was based on Proteoform interpretation (i.e. intact, aggregate, fragment) and p-values of pre-post changes in CON. In CON, HDBR induced significant changes in the abundances of 17 intact proteoforms, 5 aggregates, and 4 fragments. The intact proteins included structural proteins (ANKRD2, ACTN2), Ca regulation and contractile proteins (TPM2, MYH2 (2 spots), BEST3 (2 spots), ACTG2), metabolic regulators (AK1, HSPB7, EEF1A1, HSPB1, TRIM72, DLD), and transport proteins (CA3, HBA1, HBB). The number of proteoforms that were significantly altered during HDBR were lower in PEX (6 intact, 5 aggregates, 3 fragments) and TEX (9 intact, 4, aggregates, 3 fragments) when compared to CON. Two of the proteins that underwent abundance changes in CON also changed in TEX. HSPB7 abundance went down in both CON and TEX, while it significantly increased in PEX. HBA1 increased in CON and TEX but underwent no significant change in PEX during HDBR. Testosterone has known erythropoietic properties, however, it remains unclear whether findings of increased HBA/HBB were physiologically relevant to skeletal muscle metabolism since circulating hematocrit did not change during the course of this study [3].
Spots that were differentially affected in response to countermeasures during HDBR included several structural and Ca regulation/contractile proteoforms as well as a few metabolic and transport proteins. Radixin (RDX) increased while TPM2 decreased respectively in PEX and TEX but not in CON. ACADVL and DES increased in PEX but not in TEX or CON. S100A13 increased in TEX but not in PEX or CON. MB, MYL1, and MYH7 each decreased in TEX but not in PEX or CON. Two spots representing ACTA1 were differentially affected in PEX and TEX respectively. ACTA1 (P68133, 41kD) increased in TEX but did not change in PEX or CON, while ACTA1 (Q5T8M7, 44kD) decreased in PEX but this change failed to reach significance in CON or TEX.
Changes in the phosphorylated proteoforms within each group (pre-post) are shown in Table 2. Significant HDBR-induced changes in phosphorylation status were observed in CON for 10 intact, 2 aggregate, and 11 fragment protein spots. The intact proteoforms included structural proteins (ANKRD2, MYBPC1), contractile proteins (ACTA1, TPM1), metabolic proteins (DLD, ACADVL), and transport proteins (ALB, CA3, HBA1, HBB). In TEX, 16 intact, 4 aggregate, and 5 fragment spots were significantly altered. Similar to changes during HDBR in CON, phosphorylated DLD and CA3 increased in TEX. The number of alterations in phosphorylated spots was lowest among the PEX group which included 7 intact, 3 aggregate, and 5 fragment proteoforms. Among the intact proteins, phosphorylated ALB increased in PEX, opposite to that observed in CON.
Table 2. Changes in protein phosphorylation.
Within-group changes of all identified spots. Ordering within the table is based on proteoform interpretation (i.e. intact, aggregate, fragment) and p-values (2-tailed, paired t-tests) of the pre to post changes in CON. P-values < 0.05 are shaded in yellow. Differences (%) within each comparison are shaded to indicate higher (red) or lower (blue) values relative to pre.
| CON: Mid vs. Pre |
CON: Post vs. Pre |
EX: Mid vs. Pre |
EX: Post vs. Pre |
TEX: Mid vs. Pre |
TEX: Post vs. Pre |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TABLE 2 | Protein Name | Symbol | Accession Number | pI | MW (kD) | UNIPROT MW (kd) | MS Protein Score | Proteoform Interpretation | P-Value | Diff. (%) | P-Value | Diff. (%) | P-Value | Diff. (%) | P-Value | Diff. (%) | P-Value | Diff. (%) | P-Value | Diff. (%) |
| transport | Serum albumin | ALB | P02768 | 5.76 | 81 | 69 | 167 | INTACT | 0.088 | -16.377 | 0.002 | -31.046 | 0.021 | 25.426 | 0.022 | 18.927 | 0.473 | 8.244 | 0.221 | 6.072 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.57 | 37 | 40 | 325 | INTACT | 0.172 | -18.960 | 0.003 | -36.411 | 0.445 | 9.137 | 0.191 | 21.639 | 0.702 | 6.374 | 0.674 | -0.568 |
| structural | Isoform 2 of Myosin-binding protein C, slow-type | MYBPC1 | Q00872-2 | 5.70 | 141 | 128 | 625 | INTACT | 0.232 | -21.211 | 0.004 | -65.499 | 0.778 | 2.954 | 0.399 | -17.345 | 0.885 | -0.849 | 0.795 | -14.097 |
| metabolic | Dihydrolipoyl dehydrogenase, mitochondrial | DLD | E9PEX6 | 6.82 | 54 | 52 | 271 | INTACT | 0.153 | 52.829 | 0.005 | 218.78 | 0.008 | -71.616 | 0.089 | -33.710 | 0.814 | 5.129 | 0.006 | 43.588 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | Q5T8M7 | 5.84 | 42 | 38 | 362 | INTACT | 0.674 | -1.895 | 0.008 | -29.860 | 0.007 | 24.080 | 0.218 | 13.959 | 0.385 | 6.230 | 0.896 | -2.664 |
| Ca contractile | Isoform 4 of Tropomyosin alpha-1 chain | TPM1 | P09493-4 | 5.71 | 32 | 33 | 572 | INTACT | 0.183 | 25.928 | 0.010 | 67.594 | 0.373 | -7.904 | 0.262 | -8.037 | 0.731 | -25.334 | 0.342 | 15.594 |
| transport | Carbonic anhydrase 3 | CA3 | P07451 | 5.27 | 28 | 30 | 38 | INTACT | 0.196 | 18.463 | 0.016 | 20.965 | 0.863 | 4.034 | 0.288 | 9.667 | 0.185 | 6.057 | 0.041 | 29.330 |
| metabolic | Isoform 2 of Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADVL | P49748-2 | 8.74 | 68 | 70 | 96 | INTACT | 0.353 | 21.067 | 0.021 | 73.591 | 0.813 | 7.247 | 0.250 | 33.994 | 0.723 | 10.603 | 0.905 | 7.753 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.73 | 14 | 15 | 455 | INTACT | 0.838 | 1.024 | 0.021 | 72.832 | 0.475 | -11.995 | 0.555 | 3.463 | 0.833 | 2.112 | 0.136 | 18.360 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.30 | 14 | 16 | 468 | INTACT | 0.109 | 21.358 | 0.028 | 123.45 | 0.484 | -13.910 | 0.252 | -17.658 | 0.102 | -19.114 | 0.596 | 41.000 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.50 | 37 | 40 | 143 | INTACT | 0.372 | -9.830 | 0.051 | -26.410 | 0.334 | 11.352 | 0.138 | 24.249 | 0.227 | 16.195 | 0.787 | 2.972 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 6.00 | 41 | 42 | 454 | INTACT | 0.604 | 3.814 | 0.055 | -21.468 | 0.265 | 10.738 | 0.226 | 13.879 | 0.858 | 0.702 | 0.188 | -11.268 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.93 | 23 | 21 | 360 | INTACT | 0.230 | 14.509 | 0.058 | 43.123 | 0.022 | -30.028 | 0.197 | -53.165 | 0.125 | -11.542 | 0.487 | -7.254 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.27 | 14 | 16 | 321 | INTACT | 0.717 | 8.427 | 0.059 | 75.777 | 0.909 | -6.750 | 0.320 | 8.646 | 0.490 | -7.064 | 0.229 | 43.790 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.42 | 37 | 42 | 525 | INTACT | 0.380 | -15.755 | 0.062 | -47.995 | 0.272 | 13.704 | 0.216 | 25.413 | 0.120 | 17.955 | 0.545 | -2.598 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.46 | 37 | 40 | 288 | INTACT | 0.479 | -9.245 | 0.063 | -40.259 | 0.097 | 15.277 | 0.118 | 27.472 | 0.352 | 8.164 | 0.925 | 2.871 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 9.67 | 195 | 223 | 451 | INTACT | 0.739 | 6.430 | 0.114 | -35.182 | 0.222 | 51.786 | 0.013 | 37.336 | 0.053 | -15.551 | 0.606 | -6.407 |
| metabolic | Adenylate kinase isoenzyme 1 | AK1 | P00568 | 9.61 | 24 | 22 | 178 | INTACT | 0.279 | 4.450 | 0.131 | 32.941 | 0.518 | 7.271 | 0.895 | 1.406 | 0.618 | 12.585 | 0.056 | 28.956 |
| structural | Isoform 5 of Radixin | RDX | P35241-5 | 6.16 | 74 | 69 | 65 | INTACT | 0.449 | 10.794 | 0.132 | 48.955 | 0.419 | 13.037 | 0.079 | 26.030 | 0.546 | 8.795 | 0.694 | 35.279 |
| metabolic | Heat shock protein beta-1 | HSPB1 | P04792 | 5.34 | 26 | 23 | 270 | INTACT | 0.344 | -12.279 | 0.134 | 37.997 | 0.048 | -33.508 | 0.837 | -0.741 | 0.796 | 1.528 | 0.032 | 33.298 |
| structural | Desmin | DES | P17661 | 5.32 | 50 | 54 | 509 | INTACT | 0.044 | 28.847 | 0.154 | 28.380 | 0.504 | 11.396 | 0.024 | 41.797 | 0.150 | 28.569 | 0.048 | 40.068 |
| degradation | Tripartite motif-containing protein 72 | TRIM72 | Q6ZMU5 | 6.15 | 49 | 53 | 343 | INTACT | 0.340 | -15.023 | 0.158 | 22.035 | 0.251 | 12.505 | 0.264 | 53.223 | 0.743 | -1.363 | 0.528 | 9.833 |
| transport | Hemoglobin subunit delta | HBD | P02042 | 8.76 | 14 | 16 | 297 | INTACT | 0.723 | 3.198 | 0.160 | 70.750 | 0.192 | -17.710 | 0.023 | -30.196 | 0.980 | -5.420 | 0.104 | 25.740 |
| metabolic | Creatine kinase M-type | CKM | P06732 | 7.13 | 40 | 43 | 1010 | INTACT | 0.104 | 10.247 | 0.164 | -10.460 | 0.093 | 17.080 | 0.896 | -1.438 | 0.125 | -18.079 | 0.011 | -57.303 |
| structural | Desmin | DES | P17661 | 5.37 | 53 | 54 | 1170 | INTACT | 0.319 | 3.512 | 0.184 | -19.519 | 0.420 | 8.405 | 0.740 | 2.925 | 0.954 | 3.150 | 0.190 | -11.537 |
| metabolic | Creatine kinase M-type | CKM | P06732 | 6.95 | 41 | 43 | 983 | INTACT | 0.191 | -6.565 | 0.205 | -9.055 | 0.805 | 2.641 | 0.081 | -9.345 | 0.063 | -19.369 | 0.006 | -55.851 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 9.59 | 200 | 223 | 417 | INTACT | 0.631 | 2.829 | 0.230 | -23.686 | 0.458 | 35.924 | 0.036 | 39.686 | 0.059 | -12.888 | 0.171 | -22.045 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | Q5T8M7 | 5.51 | 44 | 38 | 416 | INTACT | 0.615 | 5.951 | 0.236 | -23.175 | 0.228 | 18.554 | 0.815 | 1.339 | 0.480 | 4.657 | 0.391 | -19.210 |
| contractile | Isoform MLC3 of Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976-2 | 4.92 | 22 | 21 | 98 | INTACT | 0.342 | -9.033 | 0.236 | -13.189 | 0.497 | 10.302 | 0.335 | 15.418 | 0.977 | -3.022 | 0.932 | 0.973 |
| contractile | Isoform MLC3 of Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976-2 | 5.17 | 20 | 21 | 199 | INTACT | 0.082 | 23.429 | 0.238 | 7.781 | 0.978 | -4.331 | 0.771 | -0.439 | 0.370 | -6.626 | 0.276 | -11.943 |
| contractile | Myosin-1 | MYH1 | P12882 | 9.48 | 200 | 223 | 551 | INTACT | 0.690 | -4.372 | 0.247 | -15.043 | 0.360 | 31.637 | 0.075 | 44.326 | 0.192 | -11.514 | 0.147 | -14.749 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.67 | 18 | 17 | 36 | INTACT | 0.421 | 16.006 | 0.272 | 40.554 | 0.056 | -87.545 | 0.293 | -40.258 | 0.030 | -42.169 | 0.161 | -40.087 |
| structural | Actinin, alpha 2, isoform CRA_b | ACTN2 | B2RCS5 | 4.91 | 100 | 104 | 62 | INTACT | 0.544 | -7.832 | 0.273 | 70.093 | 0.521 | -12.846 | 0.123 | -30.903 | 0.231 | -10.466 | 0.323 | 18.221 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.26 | 34 | 33 | 592 | INTACT | 0.225 | -11.486 | 0.280 | 35.618 | 0.557 | -5.287 | 0.631 | -3.109 | 0.091 | -15.055 | 0.499 | 19.765 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 4.95 | 42 | 42 | 642 | INTACT | 0.655 | -5.770 | 0.289 | -21.530 | 0.666 | -5.218 | 0.910 | 3.015 | 0.018 | 21.377 | 0.254 | 23.030 |
| glycolysis | Fructose-bisphosphate aldolase | ALDOA | H3BQN4 | 9.33 | 38 | 39 | 470 | INTACT | 0.753 | 0.156 | 0.294 | 14.020 | 0.554 | -6.965 | 0.416 | -10.814 | 0.065 | 20.680 | 0.017 | 54.191 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.71 | 17 | 15 | 282 | INTACT | 0.533 | 8.613 | 0.348 | 20.792 | 0.551 | -11.979 | 0.538 | -7.798 | 0.853 | 1.860 | 0.570 | 7.385 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 6.12 | 38 | 42 | 181 | INTACT | 0.418 | -11.161 | 0.384 | -15.655 | 0.086 | 17.568 | 0.010 | 48.739 | 0.261 | 8.075 | 0.010 | 31.764 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.24 | 21 | 21 | 673 | INTACT | 0.266 | -11.106 | 0.405 | -4.221 | 0.014 | 18.968 | 0.078 | 20.512 | 0.175 | -13.107 | 0.698 | -2.580 |
| transcription | Elongation factor 1-alpha 1 | EEF1A1 | P68104 | 9.56 | 53 | 50 | 58 | INTACT | 0.749 | 3.233 | 0.420 | 43.295 | 0.625 | -12.507 | 0.328 | -18.533 | 0.613 | 19.422 | 0.226 | 28.387 |
| metabolic | Creatine kinase M-type | CKM | P06732 | 7.43 | 42 | 43 | 717 | INTACT | 0.682 | -7.129 | 0.483 | -12.718 | 0.112 | -17.283 | 0.047 | -20.348 | 0.732 | -4.286 | 0.008 | -57.083 |
| structural | Alpha-actinin-2 | ACTN2 | P35609 | 10.2 | 104 | 104 | 162 | INTACT | 0.785 | 2.241 | 0.484 | -7.086 | 0.487 | 13.078 | 0.684 | 6.734 | 0.507 | 21.551 | 0.021 | 77.826 |
| contractile | Myosin-7 | MYH7 | P12883 | 7.09 | 205 | 223 | 381 | INTACT | 0.085 | 16.181 | 0.493 | 14.459 | 0.051 | 34.110 | 0.215 | 33.495 | 0.766 | 2.589 | 0.945 | 5.783 |
| structural | Isoform 5 of Myosin-binding protein C, slow-type | MYBPC1 | Q00872-5 | 5.94 | 131 | 128 | 733 | INTACT | 0.658 | 6.238 | 0.513 | -11.562 | 0.555 | -5.429 | 0.358 | 7.851 | 0.195 | -9.493 | 0.725 | 7.892 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.20 | 35 | 33 | 120 | INTACT | 0.149 | -40.841 | 0.559 | -18.088 | 0.838 | 7.614 | 0.183 | -28.029 | 0.196 | 22.866 | 0.068 | -35.092 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.26 | 18 | 17 | 40 | INTACT | 0.057 | -66.450 | 0.569 | 4.364 | 0.057 | -46.859 | 0.248 | -18.825 | 0.229 | -31.220 | 0.115 | -30.671 |
| Ca | Protein S100-A13 | S100A13 | Q99584 | 5.55 | 13 | 11 | 72 | INTACT | 0.057 | -87.694 | 0.577 | -19.353 | 0.838 | 10.193 | 0.968 | -2.678 | 0.518 | 17.630 | 0.031 | 35.661 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 10.2 | 18 | 17 | 36 | INTACT | 0.907 | 7.117 | 0.600 | 20.084 | 0.527 | 3.366 | 0.811 | -15.321 | 0.338 | 24.510 | 0.452 | 15.752 |
| Ca contractile | Troponin C type 2 (Fast), isoform CRA_a | TNNC2 | C9J7T9 | 4.59 | 18 | 16 | 328 | INTACT | 0.397 | 19.862 | 0.639 | 3.340 | 0.296 | 18.049 | 0.083 | 19.016 | 0.297 | -17.195 | 0.001 | -50.171 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.51 | 15 | 17 | 36 | INTACT | 0.619 | 0.778 | 0.673 | 1.644 | 0.624 | 3.669 | 0.620 | 3.489 | 0.146 | 7.461 | 0.017 | 22.379 |
| transport | Myoglobin | MB | P02144 | 5.51 | 17 | 17 | 57 | INTACT | 0.932 | 1.814 | 0.687 | 11.851 | 0.519 | -19.525 | 0.867 | -17.269 | 0.188 | -18.921 | 0.227 | -26.890 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 8.68 | 19 | 17 | 46 | INTACT | 0.955 | -4.365 | 0.707 | 20.252 | 0.883 | -5.957 | 0.230 | -37.166 | 0.726 | -30.286 | 0.803 | -33.192 |
| metabolic | Heat shock protein beta-7 | HSPB7 | Q9UBY9 | 5.97 | 18 | 19 | 265 | INTACT | 0.230 | 19.379 | 0.741 | 10.231 | 0.195 | -33.663 | 0.702 | -19.585 | 0.081 | -31.832 | 0.019 | -54.451 |
| metabolic | Fructose-bisphosphate aldolase A | ALDOA | P04075 | 7.44 | 38 | 39 | 380 | INTACT | 0.282 | -6.333 | 0.854 | -4.964 | 0.650 | -3.678 | 0.050 | -23.399 | 0.745 | -3.947 | 0.016 | -68.130 |
| metabolic | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | SDHA | D6RFM5 | 6.22 | 68 | 64 | 155 | INTACT | 0.157 | -56.986 | 0.866 | -2.947 | 0.245 | 32.462 | 0.072 | 63.241 | 0.050 | 22.634 | 0.017 | 73.782 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.94 | 38 | 21 | 415 | AGG | 0.091 | -25.492 | 0.014 | -41.151 | 0.172 | 31.383 | 0.229 | 13.347 | 0.545 | -3.836 | 0.331 | 11.836 |
| Ca contractile | Isoform 4 of Tropomyosin alpha-1 chain | TPM1 | P09493-4 | 4.98 | 127 | 33 | 562 | AGG | 0.639 | 12.663 | 0.038 | 38.043 | 0.089 | 46.624 | 0.031 | 69.072 | 0.548 | -6.232 | 0.605 | 9.914 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.27 | 99 | 15 | 111 | AGG | 0.457 | 34.506 | 0.075 | 92.095 | 0.727 | -14.350 | 0.420 | -29.088 | 0.597 | 12.895 | 0.058 | 43.132 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 8.35 | 98 | 17 | 39 | AGG | 0.339 | 16.525 | 0.078 | 45.482 | 0.527 | -15.922 | 0.295 | -30.296 | 0.293 | -13.917 | 0.835 | 11.958 |
| transport | Carbonic anhydrase 3 | CA3 | P07451 | 7.16 | 111 | 30 | 204 | AGG | 0.693 | 21.650 | 0.092 | 72.191 | 0.446 | -17.725 | 0.456 | -19.421 | 0.246 | 19.841 | 0.022 | 48.588 |
| metabolic | Cytochrome b-c1 complex subunit Rieske, mitochondrial | UQCRFS1 | P47985 | 6.40 | 53 | 30 | 114 | AGG | 0.138 | 19.466 | 0.117 | 27.956 | 0.508 | 3.635 | 0.360 | 5.698 | 0.053 | 16.997 | 0.017 | 18.384 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.69 | 195 | 42 | 93 | AGG | 0.130 | -56.075 | 0.138 | 34.406 | 0.337 | 45.939 | 0.359 | 26.428 | 0.119 | 36.950 | 0.017 | 75.663 |
| Ca contractile | Troponin T, fast skeletal muscle | TNNT3 | H9KVA2 | 6.21 | 35 | 28 | 234 | AGG | 0.879 | 0.463 | 0.142 | -21.683 | 0.681 | -3.007 | 0.864 | 1.513 | 0.503 | 9.799 | 0.471 | -6.220 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.73 | 20 | 17 | 34 | AGG | 0.900 | -2.481 | 0.174 | 28.277 | 0.350 | 17.663 | 0.716 | 4.050 | 0.262 | 15.046 | 0.449 | 11.325 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.71 | 18 | 15 | 262 | AGG | 0.558 | 8.003 | 0.183 | 18.206 | 0.834 | -5.019 | 0.066 | 23.395 | 0.657 | -8.004 | 0.465 | 11.350 |
| Ca contractile | TNNT1 protein | TNNT1 | Q3B759 | 5.31 | 28 | 23 | 113 | AGG | 0.374 | 6.084 | 0.197 | -14.342 | 0.706 | 7.487 | 0.048 | 20.841 | 0.285 | 7.194 | 0.121 | 11.286 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.93 | 100 | 17 | 39 | AGG | 0.035 | -83.244 | 0.222 | -42.503 | 0.248 | 15.770 | 0.971 | 7.077 | 0.204 | -31.210 | 0.436 | 5.503 |
| transcription | Ataxin-3 | ATXN3 | G3V3T0 | 8.62 | 19 | 11 | 37 | AGG | 0.501 | 16.650 | 0.289 | 34.968 | 0.686 | -25.806 | 0.689 | -11.199 | 0.204 | -48.190 | 0.281 | -31.961 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.19 | 28 | 16 | 157 | AGG | 0.348 | 21.979 | 0.336 | 20.300 | 0.565 | 17.868 | 0.054 | -22.384 | 0.323 | -8.531 | 0.212 | 10.734 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 5.86 | 28 | 16 | 169 | AGG | 0.034 | -35.599 | 0.369 | -1.744 | 0.327 | 27.851 | 0.226 | 23.574 | 0.840 | -0.431 | 0.532 | 17.996 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.01 | 159 | 33 | 769 | AGG | 0.904 | -5.087 | 0.476 | 2.766 | 0.698 | -7.272 | 0.502 | -8.195 | 0.343 | -10.860 | 0.993 | 8.709 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.32 | 123 | 42 | 626 | AGG | 0.810 | -0.928 | 0.518 | -13.951 | 0.477 | -10.261 | 0.999 | 3.879 | 0.336 | -19.552 | 0.257 | 12.758 |
| transcription | Ataxin-3 | ATXN3 | G3V3T0 | 7.14 | 18 | 11 | 41 | AGG | 0.329 | -21.376 | 0.522 | 3.707 | 0.384 | -23.430 | 0.809 | -7.606 | 0.208 | -17.320 | 0.068 | -43.067 |
| metabolic | Mitochondrial inner membrane protein | IMMT | C9J406 | 5.75 | 87 | 73 | 148 | AGG | 0.785 | -7.492 | 0.621 | -28.581 | 0.309 | 19.853 | 0.024 | 26.239 | 0.988 | 4.047 | 0.264 | -7.117 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.78 | 100 | 42 | 65 | AGG | 0.251 | 22.075 | 0.636 | 19.697 | 0.008 | -38.309 | 0.331 | 10.892 | 0.022 | 52.418 | 0.020 | 33.330 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.60 | 28 | 15 | 314 | AGG | 0.996 | 0.397 | 0.641 | 5.530 | 0.351 | -11.447 | 0.624 | -4.382 | 0.672 | 11.715 | 0.487 | -5.292 |
| unkknown | Putative BCoR-like protein 2 | BCORP1 | Q8N888 | 8.96 | 98 | 16 | 38 | AGG | 0.984 | -2.929 | 0.659 | 15.608 | 0.149 | -32.953 | 0.475 | -5.274 | 0.665 | 0.041 | 0.079 | 24.625 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.75 | 65 | 42 | 112 | AGG | 0.989 | -4.139 | 0.792 | 14.342 | 0.286 | -25.358 | 0.495 | 10.230 | 0.142 | 59.503 | 0.050 | 39.371 |
| degradation | E3 ubiquitin-protein ligase listerin | LTN1 | H7BYG8 | 5.14 | 126 | 91 | 44 | AGG | 0.237 | -17.638 | 0.805 | -2.634 | 0.561 | 3.976 | 0.637 | -3.253 | 0.044 | 19.805 | 0.154 | 16.633 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.35 | 173 | 17 | 53 | AGG | 0.293 | 26.579 | 0.835 | 9.887 | 0.367 | -24.147 | 0.375 | -20.227 | 0.587 | -2.237 | 0.079 | 20.672 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.92 | 141 | 42 | 78 | AGG | 0.159 | -55.460 | 0.845 | -17.202 | 0.937 | 5.064 | 0.168 | 28.258 | 0.740 | 3.499 | 0.377 | 23.702 |
| metabolic | Isoform 2 of Glycogen phosphorylase, muscle form | PYGM | P11217-2 | 6.78 | 250 | 97 | 266 | AGG | 0.958 | -10.494 | 0.882 | -8.007 | 0.114 | 22.257 | 0.779 | 1.048 | 0.080 | -19.479 | 0.505 | -5.476 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.68 | 31 | 17 | 42 | AGG | 0.419 | -24.676 | 0.884 | -10.046 | 0.180 | -34.873 | 0.056 | -31.471 | 0.706 | 0.260 | 0.728 | -2.315 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.32 | 97 | 42 | 632 | AGG | 0.136 | 36.669 | 0.894 | -6.617 | 0.139 | -21.983 | 0.628 | -5.594 | 0.370 | 16.156 | 0.517 | 13.842 |
| structural | Nebulin | NEB | F8WCL5 | 9.35 | 113 | 773 | 112 | FRAG | 0.545 | 24.170 | 0.007 | 97.004 | 0.525 | 12.149 | 0.339 | -16.293 | 0.829 | 1.738 | 0.327 | 23.878 |
| transport | Fatty acid-binding protein, heart (Fragment) | FABP3 | S4R371 | 5.99 | 14 | 15 | 574 | FRAG | 0.427 | 10.456 | 0.013 | 66.293 | 0.888 | -1.004 | 0.683 | -2.839 | 0.610 | -5.919 | 0.225 | 33.056 |
| contractile | Actin, alpha cardiac muscle 1 | ACTC1 | P68032 | 5.14 | 31 | 42 | 402 | FRAG | 0.616 | -8.374 | 0.013 | 66.647 | 0.416 | -12.373 | 0.898 | 9.155 | 0.219 | -18.341 | 0.244 | 21.654 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BN54 | 5.20 | 16 | 15 | 797 | FRAG | 0.580 | -15.458 | 0.021 | -63.103 | 0.404 | 16.359 | 0.600 | -13.293 | 0.486 | 9.229 | 0.024 | -34.595 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 5.60 | 143 | 223 | 384 | FRAG | 0.759 | -5.655 | 0.022 | -53.665 | 0.875 | 0.954 | 0.008 | -20.826 | 0.393 | 16.657 | 0.345 | 4.816 |
| unknown | Serine/threonine-protein phosphatase 4 regulatory subunit 4 | PPP4R4 | Q6NUP7 | 7.00 | 18 | 99 | 32 | FRAG | 0.337 | 21.480 | 0.022 | 49.030 | 0.402 | -42.126 | 0.490 | -36.055 | 0.014 | -73.364 | 0.007 | -77.302 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 5.24 | 13 | 15 | 152 | FRAG | 0.050 | 20.765 | 0.026 | 31.505 | 0.451 | -4.047 | 0.132 | -8.209 | 0.920 | -0.373 | 0.868 | 3.284 |
| structural | Keratin, type I cytoskeletal 9 | KRT9 | P35527 | 7.05 | 18 | 62 | 173 | FRAG | 0.373 | 12.484 | 0.026 | 37.369 | 0.334 | -43.696 | 0.848 | -17.835 | 0.003 | -69.265 | 0.015 | -90.232 |
| glycolysis | Enolase (Fragment) | ENO3 | E5RGZ4 | 7.90 | 111 | 30 | 104 | FRAG | 0.493 | -12.362 | 0.027 | 47.906 | 0.300 | 28.137 | 0.600 | -18.914 | 0.551 | 2.809 | 0.150 | 42.410 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 5.17 | 16 | 13 | 621 | FRAG | 0.800 | 4.657 | 0.046 | -25.100 | 0.029 | 38.986 | 0.738 | 4.163 | 0.857 | 3.329 | 0.076 | -27.338 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.86 | 18 | 13 | 54 | FRAG | 0.632 | 11.869 | 0.049 | 64.910 | 0.357 | -66.069 | 0.521 | -51.517 | 0.037 | -62.721 | 0.184 | -51.561 |
| transcription | Histidine protein methyltransferase 1 homolog | METTL18 | O95568 | 4.89 | 20 | 42 | 38 | FRAG | 0.696 | 23.242 | 0.069 | 98.219 | 0.467 | 7.996 | 0.860 | -2.406 | 0.152 | 27.597 | 0.043 | 22.357 |
| transport | Myosin-7 | MYH7 | P12883 | 9.27 | 99 | 223 | 119 | FRAG | 0.457 | 34.506 | 0.075 | 92.095 | 0.727 | -14.350 | 0.420 | -29.088 | 0.597 | 12.895 | 0.058 | 43.132 |
| transcription | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 6.77 | 18 | 59 | 69 | FRAG | 0.328 | 18.298 | 0.095 | 60.439 | 0.091 | ###### | 0.450 | -62.068 | 0.122 | -50.316 | 0.154 | -53.174 |
| metabolic | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADS | P16219 | 6.30 | 38 | 44 | 95 | FRAG | 0.417 | -11.555 | 0.097 | -22.539 | 0.323 | 9.613 | 0.631 | 2.673 | 0.602 | 7.540 | 0.620 | -8.344 |
| structural | Keratin, type II cytoskeletal 6A | KRT6A | P02538 | 6.13 | 33 | 60 | 435 | FRAG | 0.020 | -25.855 | 0.101 | -32.417 | 0.197 | 6.726 | 0.143 | 8.603 | 0.227 | 6.486 | 0.670 | 3.364 |
| structural | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 5.22 | 14 | 59 | 250 | FRAG | 0.934 | -1.998 | 0.116 | -20.881 | 0.210 | 34.289 | 0.009 | 42.113 | 0.138 | 24.580 | 0.445 | 13.732 |
| metabolic | Isoform 2 of Glycogen phosphorylase, muscle form | PYGM | P11217-2 | 5.98 | 28 | 97 | 155 | FRAG | 0.324 | -31.697 | 0.125 | -40.609 | 0.548 | 3.120 | 0.189 | 16.016 | 0.125 | -27.728 | 0.670 | 8.776 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.28 | 13 | 16 | 374 | FRAG | 0.849 | -1.401 | 0.125 | 70.949 | 0.264 | -15.643 | 0.341 | -15.074 | 0.154 | -21.239 | 0.149 | 46.085 |
| contractile | Myosin-1 | MYH1 | P12882 | 9.92 | 191 | 223 | 358 | FRAG | 0.609 | -6.091 | 0.126 | -31.534 | 0.054 | 43.161 | 0.105 | 39.252 | 0.845 | 2.181 | 0.885 | 3.248 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 4.13 | 13 | 16 | 423 | FRAG | 0.382 | 14.695 | 0.128 | 22.979 | 0.130 | -32.324 | 0.256 | -19.558 | 0.001 | 58.875 | 0.299 | 19.154 |
| structural | Keratin, type II cytoskeletal 1 | KRT1 | P04264 | 4.87 | 31 | 66 | 41 | FRAG | 0.349 | 16.586 | 0.140 | 100.12 | 0.197 | -33.032 | 0.472 | -16.206 | 0.038 | -41.007 | 0.317 | -15.130 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.36 | 18 | 13 | 280 | FRAG | 0.145 | 20.827 | 0.184 | 28.320 | 0.225 | -50.349 | 0.988 | -15.087 | 0.121 | -28.392 | 0.253 | -29.172 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 4.16 | 13 | 16 | 261 | FRAG | 0.423 | 10.065 | 0.193 | 38.468 | 0.029 | -36.403 | 0.319 | -21.181 | 0.364 | 21.832 | 0.176 | 21.477 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.89 | 17 | 9 | 112 | FRAG | 0.032 | 48.747 | 0.201 | 36.641 | 0.491 | -20.197 | 0.224 | -28.763 | 0.819 | -8.808 | 0.529 | -12.923 |
| contractile | Actin, alpha cardiac muscle 1 | ACTC1 | P68032 | 5.35 | 34 | 42 | 334 | FRAG | 0.300 | -12.549 | 0.236 | 15.391 | 0.709 | 9.240 | 0.490 | -5.697 | 0.384 | -5.801 | 0.090 | -26.145 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.10 | 16 | 13 | 64 | FRAG | 0.233 | 22.680 | 0.258 | 27.140 | 0.418 | -39.179 | 0.654 | -6.463 | 0.418 | -8.783 | 0.316 | -24.070 |
| metabolic | Acyl-coenzyme A synthetase ACSM2B, mitochondrial (Fragment) | ACSM2B | H3BQ84 | 5.31 | 18 | 12 | 47 | FRAG | 0.720 | -15.501 | 0.271 | -36.934 | 0.343 | 13.249 | 0.430 | 7.399 | 0.646 | -6.673 | 0.154 | -14.364 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.75 | 16 | 9 | 115 | FRAG | 0.170 | 37.563 | 0.330 | 37.392 | 0.442 | -19.641 | 0.039 | -35.987 | 0.394 | -22.248 | 0.064 | -48.865 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.14 | 16 | 9 | 107 | FRAG | 0.550 | 6.080 | 0.437 | 15.893 | 0.378 | -29.285 | 0.905 | 3.866 | 0.798 | 12.402 | 0.052 | -28.304 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BN54 | 5.17 | 37 | 15 | 662 | FRAG | 0.905 | -7.540 | 0.457 | -0.369 | 0.119 | 10.644 | 0.435 | 11.671 | 0.790 | 7.402 | 0.420 | 17.681 |
| metabolic | Calsequestrin-1 | CASQ1 | P31415 | 4.75 | 34 | 45 | 117 | FRAG | 0.818 | 3.327 | 0.515 | 12.230 | 0.086 | 16.615 | 0.922 | 0.317 | 0.682 | 3.102 | 0.141 | 17.560 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.33 | 18 | 13 | 129 | FRAG | 0.015 | 32.227 | 0.527 | 16.578 | 0.355 | -27.585 | 0.763 | -15.196 | 0.187 | -16.661 | 0.304 | -25.529 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BPK4 | 6.22 | 17 | 22 | 90 | FRAG | 0.703 | 5.638 | 0.548 | 0.771 | 0.240 | -48.094 | 0.798 | -10.036 | 0.308 | -23.879 | 0.663 | -17.519 |
| structural | Cofilin-1 (Fragment) | CFL1 | E9PLJ3 | 6.04 | 17 | 9 | 66 | FRAG | 0.351 | 14.784 | 0.576 | 9.863 | 0.090 | -48.829 | 0.877 | -13.256 | 0.106 | -33.426 | 0.052 | -41.203 |
| metabolic | Acyl-coenzyme A synthetase ACSM2B, mitochondrial (Fragment) | ACSM2B | H3BQ84 | 4.93 | 29 | 12 | 42 | FRAG | 0.667 | 3.979 | 0.582 | 11.406 | 0.897 | 2.847 | 0.546 | -9.938 | 0.370 | -4.131 | 0.863 | -1.452 |
| transport | Myoglobin (Fragment) | MB | B0QYF8 | 9.37 | 16 | 16 | 164 | FRAG | 0.936 | 1.008 | 0.604 | 16.059 | 0.046 | -45.512 | 0.032 | -22.197 | 0.334 | 15.562 | 0.197 | 22.545 |
| unknown | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Fragment) | MACF1 | H0Y390 | 9.41 | 137 | 506 | 40 | FRAG | 0.620 | -4.088 | 0.628 | 5.432 | 0.346 | 17.411 | 0.601 | 14.243 | 0.357 | -11.605 | 0.026 | 26.817 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.22 | 31 | 9 | 74 | FRAG | 0.995 | -6.011 | 0.668 | 14.068 | 0.027 | -59.994 | 0.225 | -18.566 | 0.445 | 28.736 | 0.625 | 6.979 |
| structural | Desmin | DES | P17661 | 5.13 | 37 | 54 | 254 | FRAG | 0.495 | 9.080 | 0.710 | -8.268 | 0.748 | 6.110 | 0.923 | -2.209 | 0.809 | -0.115 | 0.110 | -37.263 |
| structural | Unconventional myosin-XIX (Fragment) | MYO19 | K7EMZ0 | 4.83 | 13 | 8 | 33 | FRAG | 0.997 | -1.975 | 0.712 | 2.683 | 0.395 | -26.914 | 0.105 | -39.817 | 0.937 | -0.938 | 0.661 | 5.539 |
| structural | Nebulin | NEB | F8WCL5 | 8.96 | 109 | 773 | 67 | FRAG | 0.251 | -16.458 | 0.725 | 0.249 | 0.798 | -6.351 | 0.930 | -0.511 | 0.042 | -24.959 | 0.152 | 12.266 |
| structural | Keratin, type II cytoskeletal 2 epidermal | KRT2 | P35908 | 5.90 | 29 | 65 | 235 | FRAG | 0.030 | -39.062 | 0.756 | 1.992 | 0.047 | -24.031 | 0.016 | -38.113 | 0.985 | -2.752 | 0.996 | 1.694 |
| metabolic | Pyruvate kinase (Fragment) | PKM | H3BTN5 | 8.16 | 57 | 53 | 528 | FRAG | 0.127 | -13.560 | 0.760 | -1.189 | 0.554 | -6.602 | 0.078 | -11.098 | 0.800 | 2.887 | 0.528 | 7.900 |
| structural | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 5.63 | 21 | 59 | 200 | FRAG | 0.136 | -23.353 | 0.958 | 2.664 | 0.539 | -12.350 | 0.367 | -14.524 | 0.500 | 8.158 | 0.159 | 27.849 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 5.38 | 36 | 42 | 171 | FRAG | 0.470 | -4.668 | 0.992 | -0.574 | 0.973 | 13.280 | 0.171 | -18.861 | 0.050 | -19.235 | 0.055 | -50.668 |
| unknown | #N/A | #N/A | #N/A | 10.2 | 19 | #N/A | #N/A | #N/A | 0.910 | -4.952 | 0.893 | 2.706 | 0.169 | -28.707 | 0.863 | -3.407 | 0.691 | -16.698 | 0.841 | -18.142 |
| CON | CON | EX | EX | TEX | TEX | |||||||||||||||
| Total | Mid vs. Pre | Post vs. Pre | Mid vs. Pre | Post vs. Pre | Mid vs. Pre | Post vs. Pre | ||||||||||||||
| INTACT | 55 | 1 | 10 | 6 | 7 | 2 | 16 | |||||||||||||
| AGG | 29 | 2 | 2 | 1 | 3 | 2 | 4 | |||||||||||||
| FRAG | 46 | 5 | 11 | 5 | 5 | 7 | 5 | |||||||||||||
| TOTAL | 130 | 8 | 23 | 12 | 15 | 11 | 25 | |||||||||||||
Differences in PEX vs TEX
The pre-post effects of the two countermeasure groups (PEX vs TEX) were compared to provide insight on the influence of testosterone treatment vs. the underlying effects of exercise on proteomic responses during HDBR. Comparisons of the protein abundances between PEX and TEX subjects before (pre) and after (post) HDBR are shown in Table 3. Ordering in the table was based on proteoform interpretation (i.e. intact, aggregate, fragment) and p-values of post differences between PEX compared to TEX (positive values reflect higher expression in PEX compared to TEX). The abundances of 7 intact proteoforms, 5 aggregates, and 4 fragments were found to differ between PEX and TEX following HDBR. The intact proteins included structural proteins DES (2 spots) and RDX, oxygen transport proteins HBB (2 spots), HBD, and ALDOA, a glycolytic enzyme integral to the function and structure of the sarcoplasmic reticulum [12, 13]. In contrast, at baseline there were 3 intact, 6 aggregates, and 6 fragment spots found to be different between PEX and TEX. The intact proteins included ALB (transport protein), MYBPC1 (structural protein), and MYL1 (contractile protein), none of which were found to be different between the two exercise groups following HDBR.
Table 3. Protein abundance differences in PEX vs TEX.
Differences between PEX and TEX in all identified spots. Ordering within the table is based on proteoform interpretation (i.e. intact, aggregate, fragment) and p-values (2-tailed, unpaired t-tests) of the post comparison between PEX and TEX. P-values < 0.05 are shaded in yellow. Differences (%) within each comparison are shaded to indicate higher (red) or lower (blue) values in PEX relative to TEX.
| pre | mid | post | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEX compared to TEX | PEX compared to TEX | PEX compared to TEX | ||||||||||||
| TABLE 3 | Protein Name | Symbol | Accession Number | pI | MW (kD) | UNIPROT MW (kd) | MS Protein Score | Proteoform Interpretation | P-Value | Diff. (%) | P-Value | Diff. (%) | P-Value | Diff. (%) |
| structural | Desmin | DES | P17661 | 5.32 | 50 | 54 | 509 | INTACT | 0.080 | -48.410 | 0.137 | 44.153 | 0.006 | 95.133 |
| structural | Desmin | DES | P17661 | 5.37 | 53 | 54 | 1170 | INTACT | 0.241 | -26.728 | 0.525 | -20.752 | 0.013 | 69.090 |
| structural | Isoform 5 of Radixin | RDX | P35241-5 | 6.16 | 74 | 69 | 65 | INTACT | 0.225 | 11.022 | 0.581 | -24.488 | 0.016 | -33.774 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.27 | 14 | 16 | 321 | INTACT | 0.340 | 15.504 | 0.813 | -7.165 | 0.022 | -87.744 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.30 | 14 | 16 | 468 | INTACT | 0.092 | 51.528 | 0.744 | 5.507 | 0.023 | -116.34 |
| transport | Hemoglobin subunit delta | HBD | P02042 | 8.76 | 14 | 16 | 297 | INTACT | 0.156 | 28.821 | 0.721 | -9.102 | 0.048 | -63.425 |
| glycolysis | Fructose-bisphosphate aldolase | ALDOA | H3BQN4 | 9.33 | 38 | 39 | 470 | INTACT | 0.078 | -16.432 | 0.464 | 13.864 | 0.048 | 19.839 |
| contractile | Myosin-7 | MYH7 | P12883 | 7.09 | 205 | 223 | 381 | INTACT | 0.384 | 6.656 | 0.687 | 5.378 | 0.051 | 50.011 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.20 | 35 | 33 | 120 | INTACT | 0.776 | 5.255 | 0.136 | -26.258 | 0.052 | -25.000 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.71 | 17 | 15 | 282 | INTACT | 0.684 | -9.920 | 0.554 | 26.266 | 0.069 | -133.86 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.46 | 37 | 40 | 288 | INTACT | 0.062 | 31.041 | 0.686 | 4.410 | 0.077 | -24.209 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.26 | 34 | 33 | 592 | INTACT | 0.778 | 3.525 | 0.543 | 10.321 | 0.094 | -34.394 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.73 | 14 | 15 | 455 | INTACT | 0.555 | 4.742 | 0.646 | -9.176 | 0.103 | -27.367 |
| metabolic | Heat shock protein beta-7 | HSPB7 | Q9UBY9 | 5.97 | 18 | 19 | 265 | INTACT | 0.231 | 12.992 | 0.481 | -7.272 | 0.117 | 16.755 |
| transport | Serum albumin | ALB | P02768 | 5.76 | 81 | 69 | 167 | INTACT | 0.027 | -21.759 | 0.753 | -5.935 | 0.135 | -16.132 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 8.68 | 19 | 17 | 46 | INTACT | 0.121 | 63.119 | 0.165 | 35.844 | 0.142 | -36.226 |
| degradation | Tripartite motif-containing protein 72 | TRIM72 | Q6ZMU5 | 6.15 | 49 | 53 | 343 | INTACT | 0.808 | -5.893 | 0.311 | 18.660 | 0.150 | 18.779 |
| structural | Actinin, alpha 2, isoform CRA_b | ACTN2 | B2RCS5 | 4.91 | 100 | 104 | 62 | INTACT | 0.166 | -29.194 | 0.686 | -9.578 | 0.182 | 19.866 |
| metabolic | Creatine kinase M-type | CKM | P06732 | 7.13 | 40 | 43 | 1010 | INTACT | 0.066 | -27.345 | 0.908 | 2.031 | 0.184 | 18.477 |
| metabolic | Heat shock protein beta-1 | HSPB1 | P04792 | 5.34 | 26 | 23 | 270 | INTACT | 0.402 | 20.749 | 0.255 | -20.710 | 0.195 | -39.983 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.67 | 18 | 17 | 36 | INTACT | 0.906 | -4.629 | 0.194 | -31.746 | 0.197 | 22.953 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.42 | 37 | 42 | 525 | INTACT | 0.727 | -11.529 | 0.506 | -8.823 | 0.213 | 21.408 |
| transport | Carbonic anhydrase 3 | CA3 | P07451 | 5.27 | 28 | 30 | 38 | INTACT | 0.857 | 0.233 | 0.975 | -1.180 | 0.221 | -22.061 |
| metabolic | Creatine kinase M-type | CKM | P06732 | 6.95 | 41 | 43 | 983 | INTACT | 0.088 | -26.852 | 0.206 | 13.941 | 0.227 | 17.396 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.51 | 15 | 17 | 36 | INTACT | 0.953 | 15.526 | 0.924 | 4.923 | 0.238 | 20.740 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 9.67 | 195 | 223 | 451 | INTACT | 0.858 | 6.756 | 0.167 | 27.252 | 0.245 | 24.083 |
| Ca | Protein S100-A13 | S100A13 | Q99584 | 5.55 | 13 | 11 | 72 | INTACT | 0.476 | 17.335 | 0.364 | 43.234 | 0.256 | -41.068 |
| metabolic | Fructose-bisphosphate aldolase A | ALDOA | P04075 | 7.44 | 38 | 39 | 380 | INTACT | 0.987 | 0.173 | 0.739 | 2.399 | 0.304 | 19.704 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.24 | 21 | 21 | 673 | INTACT | 0.356 | 8.794 | 0.550 | 3.973 | 0.325 | 5.525 |
| structural | Alpha-actinin-2 | ACTN2 | P35609 | 10.2 | 104 | 104 | 162 | INTACT | 0.645 | 18.989 | 0.214 | -19.991 | 0.347 | 21.788 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 9.59 | 200 | 223 | 417 | INTACT | 0.607 | -3.027 | 0.555 | 16.704 | 0.348 | 22.243 |
| Ca contractile | Isoform 4 of Tropomyosin alpha-1 chain | TPM1 | P09493-4 | 5.71 | 32 | 33 | 572 | INTACT | 0.892 | 4.122 | 0.909 | -2.763 | 0.349 | 14.589 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.57 | 37 | 40 | 325 | INTACT | 0.869 | -5.352 | 0.924 | -0.371 | 0.363 | 11.399 |
| transcription | Elongation factor 1-alpha 1 | EEF1A1 | P68104 | 9.56 | 53 | 50 | 58 | INTACT | 0.621 | 15.888 | 0.525 | 12.458 | 0.404 | -27.866 |
| metabolic | Adenylate kinase isoenzyme 1 | AK1 | P00568 | 9.61 | 24 | 22 | 178 | INTACT | 0.458 | 12.384 | 0.411 | 28.689 | 0.419 | -28.461 |
| structural | Isoform 5 of Myosin-binding protein C, slow-type | MYBPC1 | Q00872-5 | 5.94 | 131 | 128 | 733 | INTACT | 0.010 | -48.654 | 0.752 | 10.405 | 0.432 | 10.425 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.26 | 18 | 17 | 40 | INTACT | 0.113 | 34.200 | 0.222 | -65.228 | 0.473 | -14.341 |
| contractile | Isoform MLC3 of Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976-2 | 5.17 | 20 | 21 | 199 | INTACT | 0.015 | 21.329 | 0.972 | -0.861 | 0.484 | -5.494 |
| metabolic | Isoform 2 of Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADVL | P49748-2 | 8.74 | 68 | 70 | 96 | INTACT | 0.079 | 102.84 | 0.733 | -25.798 | 0.511 | 1.538 |
| Ca contractile | Troponin C type 2 (Fast), isoform CRA_a | TNNC2 | C9J7T9 | 4.59 | 18 | 16 | 328 | INTACT | 0.779 | -10.323 | 0.548 | -6.770 | 0.558 | 11.516 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 4.95 | 42 | 42 | 642 | INTACT | 0.258 | 19.077 | 0.870 | -0.377 | 0.578 | 9.823 |
| structural | Ankyrin repeat domain-containing protein 2 | ANKRD2 | Q9GZV1 | 5.50 | 37 | 40 | 143 | INTACT | 0.717 | -6.417 | 0.383 | -6.892 | 0.624 | 5.611 |
| metabolic | Creatine kinase M-type | CKM | P06732 | 7.43 | 42 | 43 | 717 | INTACT | 0.309 | -10.434 | 0.733 | 0.937 | 0.717 | 4.805 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 6.12 | 38 | 42 | 181 | INTACT | 0.329 | -7.469 | 0.515 | 8.362 | 0.735 | -2.324 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 10.2 | 18 | 17 | 36 | INTACT | 0.357 | -19.358 | 0.560 | -10.515 | 0.743 | -5.342 |
| contractile | Myosin-1 | MYH1 | P12882 | 9.48 | 200 | 223 | 551 | INTACT | 0.681 | -6.553 | 0.377 | 30.166 | 0.761 | 6.559 |
| structural | Isoform 2 of Myosin-binding protein C, slow-type | MYBPC1 | Q00872-2 | 5.70 | 141 | 128 | 625 | INTACT | 0.545 | 55.274 | 0.163 | -38.197 | 0.774 | -9.801 |
| metabolic | Dihydrolipoyl dehydrogenase, mitochondrial | DLD | E9PEX6 | 6.82 | 54 | 52 | 271 | INTACT | 0.055 | -25.170 | 0.253 | 13.173 | 0.782 | 7.113 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | Q5T8M7 | 5.84 | 42 | 38 | 362 | INTACT | 0.302 | -12.461 | 0.854 | -0.356 | 0.787 | 2.569 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 6.00 | 41 | 42 | 454 | INTACT | 0.237 | -19.817 | 0.883 | 1.759 | 0.806 | -2.055 |
| transport | Myoglobin | MB | P02144 | 5.51 | 17 | 17 | 57 | INTACT | 0.175 | 24.261 | 0.970 | -3.611 | 0.808 | 5.552 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.93 | 23 | 21 | 360 | INTACT | 0.756 | -7.520 | 0.692 | -9.072 | 0.816 | 7.240 |
| contractile | Isoform MLC3 of Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976-2 | 4.92 | 22 | 21 | 98 | INTACT | 0.106 | 55.122 | 0.233 | -49.330 | 0.829 | -11.052 |
| metabolic | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | SDHA | D6RFM5 | 6.22 | 68 | 64 | 155 | INTACT | 0.223 | -28.107 | 0.548 | 14.639 | 0.918 | 4.386 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | Q5T8M7 | 5.51 | 44 | 38 | 416 | INTACT | 0.125 | -14.161 | 0.530 | 5.399 | 0.997 | -0.892 |
| unkknown | Putative BCoR-like protein 2 | BCORP1 | Q8N888 | 8.96 | 98 | 16 | 38 | AGG | 0.029 | 58.872 | 0.736 | -3.683 | 0.016 | -52.516 |
| transcription | Ataxin-3 | ATXN3 | G3V3T0 | 8.62 | 19 | 11 | 37 | AGG | 0.017 | 80.334 | 0.020 | 51.427 | 0.026 | -58.344 |
| Ca contractile | TNNT1 protein | TNNT1 | Q3B759 | 5.31 | 28 | 23 | 113 | AGG | 0.855 | 2.993 | 0.052 | -26.801 | 0.034 | -34.325 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.68 | 31 | 17 | 42 | AGG | 0.805 | 4.236 | 0.018 | -44.190 | 0.037 | -57.999 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.92 | 141 | 42 | 78 | AGG | 0.210 | -38.124 | 0.301 | -47.025 | 0.046 | 51.284 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.19 | 28 | 16 | 157 | AGG | 0.372 | 13.750 | 0.685 | -6.913 | 0.057 | -73.020 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.93 | 100 | 17 | 39 | AGG | 0.166 | -44.406 | 0.213 | -27.005 | 0.058 | 36.314 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.60 | 28 | 15 | 314 | AGG | 0.534 | -11.151 | 0.432 | 14.437 | 0.068 | -46.309 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.27 | 99 | 15 | 111 | AGG | 0.108 | 50.416 | 0.987 | 0.514 | 0.069 | -78.634 |
| metabolic | Isoform 2 of Glycogen phosphorylase, muscle form | PYGM | P11217-2 | 6.78 | 250 | 97 | 266 | AGG | 0.086 | 25.362 | 0.690 | -2.100 | 0.079 | 35.555 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 8.35 | 98 | 17 | 39 | AGG | 0.029 | 78.392 | 0.511 | 12.470 | 0.090 | -49.545 |
| Ca contractile | Troponin T, fast skeletal muscle | TNNT3 | H9KVA2 | 6.21 | 35 | 28 | 234 | AGG | 0.006 | -30.747 | 0.447 | -17.370 | 0.120 | 18.771 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.32 | 97 | 42 | 632 | AGG | 0.104 | -52.053 | 0.742 | -18.619 | 0.129 | -32.058 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 9.71 | 18 | 15 | 262 | AGG | 0.907 | 6.309 | 0.770 | 41.961 | 0.157 | -215.59 |
| degradation | E3 ubiquitin-protein ligase listerin | LTN1 | H7BYG8 | 5.14 | 126 | 91 | 44 | AGG | 0.705 | -2.566 | 0.984 | -1.767 | 0.218 | -20.295 |
| Ca contractile | Isoform 4 of Tropomyosin alpha-1 chain | TPM1 | P09493-4 | 4.98 | 127 | 33 | 562 | AGG | 0.111 | 39.660 | 0.704 | 1.264 | 0.237 | 28.225 |
| contractile | Actin, alpha skeletal muscle | ACTA1 | P68133 | 5.32 | 123 | 42 | 626 | AGG | 0.098 | -42.120 | 0.873 | -16.347 | 0.241 | -20.562 |
| transcription | Ataxin-3 | ATXN3 | G3V3T0 | 7.14 | 18 | 11 | 41 | AGG | 0.065 | 54.508 | 0.701 | 7.562 | 0.256 | 13.650 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.69 | 195 | 42 | 93 | AGG | 0.811 | -0.350 | 0.653 | -23.508 | 0.257 | -32.646 |
| Ca contractile | Tropomyosin beta chain | TPM2 | P07951 | 5.01 | 159 | 33 | 769 | AGG | 0.112 | 33.282 | 0.243 | -28.410 | 0.282 | 14.917 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.75 | 65 | 42 | 112 | AGG | 0.289 | -25.580 | 0.088 | -59.102 | 0.316 | 31.025 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 5.86 | 28 | 16 | 169 | AGG | 0.810 | 6.106 | 0.154 | -44.961 | 0.329 | -245.69 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 6.35 | 173 | 17 | 53 | AGG | 0.040 | -20.977 | 0.146 | -30.519 | 0.438 | -15.299 |
| Ca | Bestrophin-3 | BEST3 | F8VVX2 | 4.73 | 20 | 17 | 34 | AGG | 0.731 | -3.489 | 0.254 | -14.732 | 0.491 | -7.829 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 4.78 | 100 | 42 | 65 | AGG | 0.025 | -39.528 | 0.190 | -32.966 | 0.498 | 5.611 |
| transport | Carbonic anhydrase 3 | CA3 | P07451 | 7.16 | 111 | 30 | 204 | AGG | 0.423 | 30.272 | 0.373 | 29.812 | 0.688 | -7.064 |
| metabolic | Mitochondrial inner membrane protein | IMMT | C9J406 | 5.75 | 87 | 73 | 148 | AGG | 0.392 | -9.729 | 0.084 | 23.126 | 0.715 | 3.168 |
| contractile | Myosin light chain 1/3, skeletal muscle isoform | MYL1 | P05976 | 5.94 | 38 | 21 | 415 | AGG | 0.372 | -14.587 | 0.223 | -19.102 | 0.793 | 10.226 |
| metabolic | Cytochrome b-c1 complex subunit Rieske, mitochondrial | UQCRFS1 | P47985 | 6.40 | 53 | 30 | 114 | AGG | 0.885 | -1.203 | 0.962 | -0.515 | 0.807 | 1.743 |
| metabolic | Acyl-coenzyme A synthetase ACSM2B, mitochondrial (Fragment) | ACSM2B | H3BQ84 | 4.93 | 29 | 12 | 42 | FRAG | 0.344 | 16.126 | 0.017 | -66.970 | 0.007 | -72.367 |
| metabolic | Acyl-coenzyme A synthetase ACSM2B, mitochondrial (Fragment) | ACSM2B | H3BQ84 | 5.31 | 18 | 12 | 47 | FRAG | 0.960 | -1.433 | 0.666 | -13.467 | 0.008 | -52.183 |
| metabolic | Pyruvate kinase (Fragment) | PKM | H3BTN5 | 8.16 | 57 | 53 | 528 | FRAG | 0.782 | 0.569 | 0.982 | 0.676 | 0.013 | 66.495 |
| transport | Hemoglobin subunit alpha | HBA1 | P69905 | 5.24 | 13 | 15 | 152 | FRAG | 0.716 | 1.770 | 0.581 | -5.989 | 0.034 | -37.938 |
| structural | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 5.63 | 21 | 59 | 200 | FRAG | 0.962 | -6.831 | 0.884 | -4.235 | 0.057 | -33.182 |
| unknown | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Fragment) | MACF1 | H0Y390 | 9.41 | 137 | 506 | 40 | FRAG | 0.375 | 8.112 | 0.230 | 24.899 | 0.062 | -39.800 |
| transport | Myosin-7 | MYH7 | P12883 | 9.27 | 99 | 223 | 119 | FRAG | 0.108 | 50.416 | 0.987 | 0.514 | 0.069 | -78.634 |
| metabolic | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADS | P16219 | 6.30 | 38 | 44 | 95 | FRAG | 0.029 | -49.840 | 0.113 | 27.213 | 0.072 | 26.078 |
| structural | Unconventional myosin-XIX (Fragment) | MYO19 | K7EMZ0 | 4.83 | 13 | 8 | 33 | FRAG | 0.389 | 18.656 | 0.313 | -38.380 | 0.077 | -40.357 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 4.16 | 13 | 16 | 261 | FRAG | 0.589 | 18.059 | 0.101 | -95.360 | 0.081 | -71.546 |
| structural | Nebulin | NEB | F8WCL5 | 8.96 | 109 | 773 | 67 | FRAG | 0.011 | 50.238 | 0.696 | 23.149 | 0.131 | -27.535 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 6.28 | 13 | 16 | 374 | FRAG | 0.185 | 27.182 | 0.480 | -13.390 | 0.134 | -56.692 |
| transport | Fatty acid-binding protein, heart (Fragment) | FABP3 | S4R371 | 5.99 | 14 | 15 | 574 | FRAG | 0.051 | 19.332 | 0.817 | 3.139 | 0.155 | -24.675 |
| contractile | Myosin-2 | MYH2 | Q9UKX2 | 5.60 | 143 | 223 | 384 | FRAG | 0.281 | -19.685 | 0.820 | -7.798 | 0.159 | -22.691 |
| structural | Nebulin | NEB | F8WCL5 | 9.35 | 113 | 773 | 112 | FRAG | 0.620 | 5.200 | 0.687 | 19.867 | 0.177 | -33.573 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.10 | 16 | 13 | 64 | FRAG | 0.282 | -17.765 | 0.258 | 19.772 | 0.193 | 31.037 |
| contractile | Actin, alpha cardiac muscle 1 | ACTC1 | P68032 | 5.14 | 31 | 42 | 402 | FRAG | 0.055 | 76.671 | 0.877 | 0.631 | 0.249 | -42.101 |
| structural | Cofilin-1 (Fragment) | CFL1 | E9PLJ3 | 6.04 | 17 | 9 | 66 | FRAG | 0.130 | 19.294 | 0.247 | -21.046 | 0.260 | 20.168 |
| transport | Hemoglobin subunit beta | HBB | P68871 | 4.13 | 13 | 16 | 423 | FRAG | 0.695 | 23.772 | 0.130 | ###### | 0.267 | -66.555 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.89 | 17 | 9 | 112 | FRAG | 0.520 | 23.269 | 0.678 | -10.150 | 0.291 | -17.412 |
| unknown | Serine/threonine-protein phosphatase 4 regulatory subunit 4 | PPP4R4 | Q6NUP7 | 7.00 | 18 | 99 | 32 | FRAG | 0.257 | 48.855 | 0.358 | 31.896 | 0.292 | 34.710 |
| metabolic | Calsequestrin-1 | CASQ1 | P31415 | 4.75 | 34 | 45 | 117 | FRAG | 0.003 | 46.920 | 0.892 | 7.351 | 0.298 | -19.602 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 5.17 | 16 | 13 | 621 | FRAG | 0.361 | 11.836 | 0.877 | -5.452 | 0.309 | -11.894 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BN54 | 5.17 | 37 | 15 | 662 | FRAG | 0.441 | 16.372 | 0.668 | 8.082 | 0.326 | -17.139 |
| transcription | Histidine protein methyltransferase 1 homolog | METTL18 | O95568 | 4.89 | 20 | 42 | 38 | FRAG | 0.261 | -29.554 | 0.120 | -32.688 | 0.381 | 16.099 |
| structural | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 5.22 | 14 | 59 | 250 | FRAG | 0.007 | 22.120 | 0.219 | -18.091 | 0.449 | -6.599 |
| structural | Keratin, type II cytoskeletal 1 | KRT1 | P04264 | 4.87 | 31 | 66 | 41 | FRAG | 0.026 | 59.615 | 0.240 | -24.818 | 0.464 | -20.453 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BPK4 | 6.22 | 17 | 22 | 90 | FRAG | 0.100 | 27.375 | 0.748 | -9.993 | 0.471 | -11.629 |
| structural | Keratin, type II cytoskeletal 6A | KRT6A | P02538 | 6.13 | 33 | 60 | 435 | FRAG | 0.530 | 12.109 | 0.448 | -9.065 | 0.517 | 12.195 |
| glycolysis | Enolase (Fragment) | ENO3 | E5RGZ4 | 7.90 | 111 | 30 | 104 | FRAG | 0.833 | 3.854 | 0.755 | 1.807 | 0.584 | 10.639 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.86 | 18 | 13 | 54 | FRAG | 0.135 | 25.455 | 0.450 | -13.981 | 0.650 | 9.213 |
| structural | Keratin, type II cytoskeletal 2 epidermal | KRT2 | P35908 | 5.90 | 29 | 65 | 235 | FRAG | 0.328 | 30.399 | 0.085 | -73.101 | 0.650 | 37.656 |
| contractile | Isoform 2 of Actin, gamma-enteric smooth muscle | ACTG2 | P63267-2 | 5.38 | 36 | 42 | 171 | FRAG | 0.528 | -6.446 | 0.866 | 3.103 | 0.653 | 10.546 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.22 | 31 | 9 | 74 | FRAG | 0.656 | -6.354 | 0.018 | -80.282 | 0.656 | -10.552 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.33 | 18 | 13 | 129 | FRAG | 0.024 | 31.687 | 0.447 | -25.945 | 0.723 | -4.776 |
| contractile | Myosin-1 | MYH1 | P12882 | 9.92 | 191 | 223 | 358 | FRAG | 0.515 | -13.379 | 0.152 | 18.255 | 0.729 | 7.087 |
| transcription | Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 6.77 | 18 | 59 | 69 | FRAG | 0.801 | -6.667 | 0.832 | -3.348 | 0.786 | 16.080 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.14 | 16 | 9 | 107 | FRAG | 0.675 | -10.865 | 0.327 | -24.146 | 0.790 | 0.706 |
| contractile | Actin, alpha cardiac muscle 1 | ACTC1 | P68032 | 5.35 | 34 | 42 | 334 | FRAG | 0.697 | 2.570 | 0.397 | -14.601 | 0.808 | 0.945 |
| transport | Myoglobin (Fragment) | MB | F2Z337 | 4.75 | 16 | 9 | 115 | FRAG | 0.622 | 25.746 | 0.552 | -14.003 | 0.817 | -15.234 |
| structural | Keratin, type I cytoskeletal 9 | KRT9 | P35527 | 7.05 | 18 | 62 | 173 | FRAG | 0.098 | 35.935 | 0.430 | 11.375 | 0.873 | 3.053 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BN54 | 5.20 | 16 | 15 | 797 | FRAG | 0.935 | -4.170 | 0.635 | 6.043 | 0.881 | 7.035 |
| contractile | Myosin regulatory light chain 2, skeletal muscle isoform (Fragment) | MYLPF | H3BML9 | 6.36 | 18 | 13 | 280 | FRAG | 0.263 | 14.359 | 0.801 | -7.922 | 0.938 | -3.447 |
| transport | Myoglobin (Fragment) | MB | B0QYF8 | 9.37 | 16 | 16 | 164 | FRAG | 0.509 | 16.853 | 0.325 | 16.052 | 0.946 | 3.617 |
| structural | Desmin | DES | P17661 | 5.13 | 37 | 54 | 254 | FRAG | 0.707 | -5.214 | 0.847 | 3.286 | 0.958 | 1.844 |
| metabolic | Isoform 2 of Glycogen phosphorylase, muscle form | PYGM | P11217-2 | 5.98 | 28 | 97 | 155 | FRAG | 0.374 | 21.548 | 0.026 | 58.431 | 0.967 | 0.421 |
| unknown | #N/A | #N/A | #N/A | 10.2 | 19 | #N/A | #N/A | #N/A | 0.266 | -17.562 | 0.331 | -38.427 | 0.414 | 8.669 |
| Total | pre | mid | post | |||||||||||
| intact | 55 | 3 | 0 | 7 | ||||||||||
| aggregate | 29 | 6 | 2 | 5 | ||||||||||
| fragment | 46 | 6 | 3 | 4 | ||||||||||
| TOTAL | 130 | 15 | 5 | 16 | ||||||||||
Comparisons of protein phosphorylation status between PEX and TEX subjects before (PRE) and after (post) HDBR are shown in Table 4. While there were only a few pre-HDBR differences in phosphorylated proteoforms between PEX and TEX (1 intact, 1 aggregate, and 3 fragment), this difference increased to 10 intact, 3 aggregate, and 4 fragment proteoforms post-HDBR. The intact proteins that were significantly different (all higher in PEX following HDBR when compared to TEX) included contractile proteins (MYH1, MYH2 (2 spots), ACTA1 (2 spots), TNNC2) and metabolic proteins (CKM (3 spots) and ALDOA).
Principal Components Analyses (PCA)
Results from the PCA performed on post-HDBR data are shown in Fig 2. The purpose of this analysis was to reveal patterns of protein expression/phosphorylation in the post data that may distinguish the CON, PEX, and TEX sample groups from one another. For abundance PCA, the spot intensities of the 71 gel spots that showed significant expression difference among these three sample groups were used as input. Fig 2A is the resulting PCA1-PCA2-PCA3 plot. The 24 dots in the figure represent the eight CON (red), eight PEX (green), and eight TEX (blue) samples. Notably, there is distinct clustering among the groups, with the three sample groups occupying separable, non-overlapping, regions in the PCA plot. The results of a similar PCA, based on the 81 gel spots exhibiting significant phosphorylation differences is depicted in Fig 2B. The color coding of the 24 spots is the same as for the abundance PCA and the clustering of the three sample groups is even more evident for the phosphorylation PCA. The variability in data captured by PCA along PC1 and PC2 were 39% and 10% respectively in Fig 2A, and were 30% and 12% respectively in Fig 2B. While these numbers are moderate, the clear separation among CON, TEX, and PEX observed in the plots (which is especially noticeable in Fig 2B, the phosphorylation based-plot) suggests that, by the end of the study, there were clear differences in the patterns of protein expression/phosphorylation among the participant groups that underwent just bed rest (CON), exercise (PEX), and exercise + testosterone (TEX) treatment.
Fig 2. Principal Components Analyses (PCA).
Principal Components Analysis demonstrating post-HDBR differences in the proteomes of CON (red, subjects 1–8), PEX (green, subjects 9–16), and TEX (blue, subjects 17–24). Spots are numbered for consistent comparison of data from individual subjects between figures throughout the manuscript. (A) PCA based on post-HDBR differences in protein abundance. (B) PCA based on post-HDBR differences in protein phosphorylation.
Pathway analyses
HDBR resulted in proteomic changes within skeletal muscle and both countermeasures caused differential changes during long term HDBR. Major canonical signaling networks were identified through Ingenuity Pathway Analysis (IPA) as playing a role in the differential regulation of skeletal muscle in response to HDBR in the presence or absence of applied exercise and placebo or exercise and testosterone countermeasures. Several pathways such as Calcium Signaling, Cellular Effects of Sildenafil (Viagra), Epithelial Adherens Junction Signaling, Actin Cytoskeleton Signaling, and ILK Signaling can be associated with those signaling networks (Fig 3). Similar analyses revealed that the biological functions most likely impacted included Cellular Assembly and Organization, Cellular Function and Maintenance, Cell Death and Survival, Carbohydrate Metabolism, Molecular Transport, Organ Morphology, Tissue Development, Behavior, Cardiovascular System Development and Function, and Skeletal and Muscular System Development and Function. Finally, IPA associated a number of known pathologies with the observed changes, including musculoskeletal, dermatological, gastrointestinal, cardiovascular, neurological, immunological, and psychological disorders.
Fig 3. Ingenuity Pathway Analysis (IPA).
(A) Top pathways identified by preliminary IPA based on differential changes in protein abundances and phosphorylation in response to ~70 days of HDBR with or without countermeasures. (B) Top associated pathologies identified by IPA based on differential changes in protein abundances and phosphorylation in response to ~70 days of HDBR with or without countermeasures.
Accession numbers of proteoforms that showed significant pre-post HDBR changes were also submitted for gene ontology enrichment analyses of individual groups (http://www.geneontology.org) (see S1 Table, gene ontology). For instance, confinement to HDBR (CON) altered abundances of proteins related to biological process of muscle contraction (MYH2, ACTG2, TPM2, TRIM72, ANKRD2), consistent with the previously published losses in muscle mass and strength in this group [3]. Exercise countermeasures (PEX) uniquely affected cellular organization in skeletal muscle (ACTA1, TPM2, TPM1, KRT9, DES) while the addition of testosterone (TEX) affected mesenchymal migration proteins (ACTA1, ACTG2, ACTC1), which would be consistent with increased myogenesis [14] and previously published increases in lean body mass in the TEX group [3]. Thus, the countermeasures resulted in considerable modulation of the networks and pathways identified in our proteomic analyses and show overlap with known physiological responses that occur on Earth and during space flight.
Prediction models
Linear regression analyses using the discovery proteomics data identified several potentially predictive biomarkers for the individual subject susceptibility to HDBR and the effectiveness of countermeasures to prevent losses in lean leg mass and knee extension strength. There were significant (P < 0.05) relationships found between baseline (i.e., pre-HDBR) abundance levels of Myosin regulatory light chain 2, skeletal muscle isoform (fragment) (MYLPF H3BML9 (18kD); regulator of muscle contraction) (Fig 4A), Ubiquinol-cytochrome c reductase complex (UQCRFS1 P47985 (53kD); mitochondrial respiratory chain protein involved in energy metabolism), Adenylate kinase isoenzyme (AK1 P00568 (24kD); cytosolic protein involved in skeletal muscle ATP metabolism and energy homeostasis), and Desmin (DES P17661 (50kD); an intermediate filament protein) vs. subsequent changes in lean leg mass during HDBR. Baseline expression of Troponin T Type3, fast skeletal muscle type (TNNT3 H9KVA2 (35kD); anchoring protein necessary for skeletal muscle contraction) (Fig 4B), Ubiquinol-cytochrome c reductase complex (UQCRFS1 P47985 (53kD); energy metabolism), Actin A (ACTA1 P68133 (41kD) & ACTA1 Q5T8M7 (42kD); a muscle specific structural microfilament), and a 159 kD proteoform identified as an aggregate of Tropomyosin beta chain (TPM2 P07951 (159kD); a calcium induced contractile regulator expected at 32 kD) in the vastus lateralis were predictive of changes in strength performance of the relevant large muscle group (quadriceps femoris) during HDBR.
Fig 4. Prediction model.
(A) Baseline (Pre-HDBR) protein abundance levels of Myosin regulatory light chain 2 (MYLPF) in the vastus lateralis plotted against PRE-post changes in Leg LBM. There were no relationships between baseline Leg LBM and protein abundances (not shown). (B) Baseline (Pre-HDBR) protein abundance levels of Troponin T Type3 (TNNT3) in the vastus lateralis plotted against PRE-post changes in knee extension strength (KES). There were no relationships between baseline strength and protein abundances (not shown).
Discussion
The present study demonstrated notable alterations in the skeletal muscle proteome of healthy men in response to skeletal muscle unloading under extremely well-controlled long-term HDBR conditions. In addition, these changes were compared to the alterations that occur when exercise countermeasures with or without testosterone supplementation were included as part of the HDBR condition. We previously reported on the efficacy of these countermeasures in protecting against muscle atrophy [3]. Exercise was largely successful in protecting against HDBR-induced strength loss while the addition of testosterone to exercising subjects resulted in an accumulation of lean body mass. The addition of this proteomic investigation supplemented the existing data by identifying several proteins with known structural, contractile, or metabolic functions to be altered in response to bed rest, exercise, and/or testosterone. Overall, our results corroborate findings by others [15–18] that long duration bed rest results in changes in the abundances and post-translational phosphorylation of proteins with structural, contractile, and metabolic functions.
Structural changes
Confinement to bed rest is known to induce a downregulation of structural proteins [15, 16]. Likewise in this study, long duration HDBR resulted in decreased abundances and/or phosphorylation of several structural proteins (ANKRD2, ACTN2, MYBPC1) in CON, which was not observed in the countermeasure groups PEX and TEX. Conversely, high intensity and/or eccentric exercise protocols are known to promote synthesis of structural proteins such as DES as well as ERM (Ezrin, RDX and Moesin) proteins [19, 20]. The ERM proteins play an important role in signal transduction between the extracellular matrix and the cytoskeletal proteins that maintain cell structure [21] and are susceptible to gravitational stresses in vitro [22, 23]. In the current study, DES significantly increased in PEX which resulted in higher post-HDBR abundances compared to TEX while increases in RDX abundances in TEX subjects outpaced the increase observed in PEX subjects, resulting in higher post-HDBR expression among TEX subjects. Overall, these findings confirm the published data demonstrating benefits of exercise and testosterone countermeasures on protecting against losses of muscle mass in these subjects during long duration HDBR [3, 4].
Contractile changes
Bed rest induced alteration in abundances of contractile proteins were consistent with those reported elsewhere. For instance, increased tropomyosin (TPM2) and decreases in smooth muscle actin ACTG2 were previously reported in response to 60 days of bed rest [17]. Decreases in MYH2 abundances in the present report, along with unaltered abundances in MYH1, are also consistent with findings described during other bed rest studies [15, 17, 24, 25]. However, this direction of change is in contrast to the expected modest slow-to-fast transition from MYH1 towards MYH2a fibers in vastus lateralis reported by our colleagues [26], This disparity may be due to methodological differences between comparing ratios of fibers identified by the predominant protein type vs. measuring protein abundances in total sample homogenates. Thus, the decrease in myosin protein abundance may be more reflective of overall losses in skeletal muscle protein than of shifts in skeletal muscle fiber type.
Changes in abundances and phosphorylation of contractile protein abundances were also observed in the PEX and TEX exercise groups. Despite comparable responses in muscle strength between the exercise groups [3], phosphorylation of several contractile proteins were higher in PEX compared to TEX after HDBR. Specifically, phosphorylation of MYH1 and MYH2 tended to increase in PEX while (statistically nonsignificant) declines in TEX were observed. Similarly, opposite pre-post HDBR changes were observed in the phosphorylation of thin filament proteins ACTA1 and TNNC2 between PEX and TEX. post-HDBR phosphorylation among PEX was consistently higher for these proteins than that among TEX (Table 4). Phosphorylation of the contractile protein troponin 2 (TNNC2) decreased in TEX and tended to increase in PEX while no consistent changes were observed in spots related to myosin regulatory light chain (MYLPF).
Regulation of contractile proteins through phosphorylation is complex and it is difficult to relate the changes observed in the skeletal muscle proteome to the functional changes observed in the subjects. For instance, phosphorylation of myosin 1 heavy chain is important during cellular organization but does not affect the strength of its interaction with actin [27]. In contrast, phosphorylation of contractile proteins such as myosin regulatory light chain and troponin plays a role in maintaining Ca2+ sensitivity and improve force production, especially under suboptimal Ca concentrations [28, 29]. The mechanisms behind the differences in phosphorylation of the mentioned contractile proteins among exercising groups during HDBR remain unclear but could be related to a number of factors. Among the possibilities is that the need for cellular reorganization and functional optimization in response to exercise alone (PEX) was partially offset by a drive towards hypertrophy and generation of new muscle tissue in the subjects receiving testosterone (TEX). Abundance and phosphorylation of S100A13 increased in TEX. This protein family has been shown to increase in response to estrogens as well as androgens in various tissues and are regarded as early response genes involved in regulation of tissue growth, angiogenesis and inflammatory responses [30, 31]. The consistently higher level of phosphorylation among contractile proteins in PEX may be indicative of increased cellular restructuring and protein turnover in response to exercise induced mechanical stresses [32]. In contrast, administration of testosterone may have blunted this exercise-induced protein turnover and catabolism [33] contributing to the net increases in lean mass and protection of muscle strength in TEX.
Metabolic changes
Phosphorylation of ALDOA, a triad-associated protein involved in Ca+2 regulation and integral to excitation-contraction coupling of skeletal muscle [13], tended to decline in PEX and TEX. post-HDBR abundance as well as phosphorylation of this glycolytic enzyme were lower among TEX compared to PEX. Androgen induced downregulation of ALDOA has been reported in adipose tissue and this corresponds to a repression of pyruvate synthesis and decreased lipogenesis [34].
Abundance of ACADVL, a mitochondrial enzyme which catalyzes the first step in the beta oxidation pathway, increased in response to exercise (PEX) but not in testosterone treated subjects (TEX). Interestingly, HDBR alone (CON) resulted in increases in phosphorylated ACADVL. The mechanism behind this shift in the CON subjects is unclear as increased abundance of ACADVL has previously been associated with responses to exercise training [16, 20, 35]. Increased abundance (PEX) and/or activation by phosphorylation of ACADVL (CON) during HDBR may have been responses to offset shifts towards increased buildup of intracellular lipids [36, 37] and could be consistent with the increases in fat mass observed in these subjects [3]. Furthermore, HDBR has been reported to result in decreases in mitochondrial enzymes such as DLD [16], which is consistent with our results in CON. Although prevented by resistive vibration exercise (RVE) countermeasures [16], the exercise and testosterone countermeasures in the present study did not inhibit the decline in DLD. Similarly, there were no overt changes in the abundance of the energy transduction protein CKM, although phosphorylation of CKM declined in PEX as well as TEX during HDBR (Table 3). This decline was more profound in the TEX group resulting in significant post-HDBR differences between PEX and TEX (Table 4). Interestingly, the decline in phospho-CKM in TEX was somewhat offset by an increase in phosphorylation of the alternate ATP producing kinase, AK1 (P = 0.056, Table 3).
Prediction models
In addition to demonstrating that HDBR results in alterations in the skeletal muscle proteome that may be modulated by exercise and testosterone countermeasures, we identified a subset of proteins that appeared predictive of HDBR-induced changes in muscle mass and strength. Discovery of sensitive proteomic biomarkers may in the future allow for personalized medicine approaches by aiding in the development of more directed countermeasures based upon baseline proteome profiles. Our data identified several proteins at baseline that showed strong correlation with subsequent changes in muscle mass and strength during HDBR. These proteomic biomarkers offer a good potential for prediction of HDBR induced changes in body composition or strength (Fig 4). While countermeasures improved lean body mass and strength during HDBR compared to control, baseline abundances of these skeletal muscle proteins were predictive of the outcomes. Visually, the linear regression analyses lead to interesting interpretations when the pre-post responses within each countermeasure group are compared between the groups. For instance, in Fig 4A, low abundance of the 18kD proteoform for MYLPF in skeletal muscle tissue collected before bed rest was a strong predictor for the quantity of LBM lost during HDBR, especially in absence of countermeasures. Furthermore, these data suggest that exercise alone may be an excellent countermeasure for individuals with relatively mid to high baseline levels of 18kD MYLPF, and that exercise + testosterone may be effective in improving LBM independent of baseline levels of these proteins. Baseline levels of TNNT3 in vastus lateralis were good predictors of changes in knee extension strength during HDBR while exercise with or without testosterone countermeasures provided an upward shift in protection against loss of strength (Fig 4B). Subjects with best cases in terms of knee extension strength changes during HDBR were among those with highest pre-HDBR abundances in TNNT3.
Identifying individuals as either responders or non-responders to the effects of HDBR and/or countermeasures is complex and depends on a myriad of factors including, but not limited to, the individual’s susceptibility to HDBR induced muscle atrophy as well as the predicted effect of the countermeasures. As expected, no individual protein we identified was fully predictive for every subject or physiological function measured, and the development of accurate predictive models will likely involve algorithms that include panels of several proteins and factors. However, our simple approach illustrates the potential use of this predictive model for identifying the responses to either HDBR and/or countermeasures in individual subjects (Fig 4). For example, subject # 1 (CON) was among the individuals with the most severe muscle atrophy in terms of both mass as well as strength. Conversely, the pre-bed rest abundance levels of 3 out of the 4 proteins depicted in Fig 4 were good predictors as this subject was consistently towards the low end of the regression-line in the absence of countermeasures. Subject # 11 (PEX) was a good responder to the exercise countermeasure. Pre-HDBR data would have predicted losses in muscle mass and strength akin to those observed in subject #1 (CON) but subject #11 repeatedly performed better, consistent with the upward shifted lines in Fig 3A–3D. Subjects # 17 and #21 (TEX) were good responders to the testosterone + exercise countermeasure and these subjects consistently performed better than predicted by pre-HDBR proteomics data. Interestingly, subject 16 (PEX) was among the greatest gainers of muscle mass and strength. This subject was among the exercisers, although his data were consistently towards the upper end of the regression-line suggesting that even in the absence of countermeasures this subject might have been among those with the lowest severity of muscle atrophy.
While this study was not originally designed or powered for this type of investigation, the prediction modeling exercise was undertaken as proof-of-concept to probe for possible factors associated with individual variations in responses to countermeasures among the test subjects. We propose that our data may serve as a prelude to methods that utilize the baseline proteome in a personalized medicine approach to aid in the prediction of health and performance risks in response to the absence or presence of countermeasures. Further development of such methods could have clinical applications and would specifically help NASA and other agencies shape personalized prescriptions of countermeasures for crewmembers to follow during long duration space flight missions.
A potential clinical limitation of the current study is that it did not include a non-exercising group that received testosterone during HDBR. The addition of such a group was included in the early design of this study. However, a control + testosterone group was ultimately downselected during discussions with the NASA human research program (HRP) and the other investigative teams included in this bed rest campaign as it was deemed that such a group was of less operational interest to NASA given that exercise countermeasure protocols will continue to be high priority for all astronauts during flight. Inclusion of a non-exercise (CON) group remained a high priority for the complement of selected investigators and stakeholders that were participating in this bed rest campaign. Thus, the selected countermeasures for evaluation during this investigation included the Sprint exercise protocol and a combined Sprint + testosterone protocol vs. standard HDBR control conditions.
Conclusion
In summary, long-duration HDBR results in numerous proteomic alterations spanning a range of biological functions that are blunted or reversed by the addition of exercise countermeasures. During HDBR, exercise appeared to drive cellular reorganization in skeletal muscle while the addition of testosterone blunted catabolism and induced overall skeletal muscle hypertrophy. This investigation demonstrated that the inclusion of a low dose testosterone countermeasure partially modulates the effects of exercise providing a unique insight into the differential mechanical and biochemical regulators of muscle proteins during HDBR. Furthermore, the baseline proteomic data offered important insight and its potentially applicability as a powerful tool to predict changes in muscle mass and strength, and the effectiveness of exercise and hormonal countermeasures. Knowledge of the individual physiological susceptibility to functional declines during unloading may help tailor effective countermeasure strategies to the individual astronaut prior to embarking on a space flight mission.
Supporting information
Proteoforms that showed significant pre-post abundance changes in Table 1 were submitted for Enrichment Analyses (Annotation Version and Release Date: GO Ontology database Released 2018-08-09, http://www.geneontology.org/,). Table include only results with False Discovery Rate < 0.05. A. CON, B. PEX, C. TEX.
(PDF)
Raw protein abundance and phosphorylation data from MS Analyses. These data were submitted to the NASA Life Science Data Archive (https://lsda.jsc.nasa.gov/).
(XLSX)
The study complied with the Declaration of Helsinki and was approved by The University of Texas Medical Branch (UTMB) Institutional Review Board (IRB) and by the NASA Committee for the Protection of Human Subjects (CPHS).
(PDF)
This study adheres to CONSORT guidelines.
(DOC)
Acknowledgments
We acknowledge all the subjects, UTMB NASA FARU nurses, NASA monitors and colleagues that assisted with this project. This type of project takes a considerable amount of effort and we thank all who took part in this rewarding process.
Data Availability
Relevant data are within the manuscript and its Supporting Information files. Full data sets can be obtained through the NASA Life Science Data Archive (https://lsda.jsc.nasa.gov/).
Funding Statement
This research was part of an integrated study registered with ClinicalTrials.gov (NCT00891449) and was funded by the National Aeronautics and Space Administration (#NNX10AP86G, RJU/MSM). The study was conducted with the support of the Institute for Translational Sciences at the University of Texas Medical Branch through funding by a Clinical and Translational Science Award (UL1TR000071) from the National Center for Advancing Translational Sciences, National Institutes of Health and (RO1CA127971, MSM) from the National Cancer Institute.
References
- 1.Edgerton VR, Zhou MY, Ohira Y, Klitgaard H, Jiang B, Bell G, et al. Human fiber size and enzymatic properties after 5 and 11 days of spaceflight. J Appl Physiol (1985). 1995;78(5):1733–9. 10.1152/jappl.1995.78.5.1733 . [DOI] [PubMed] [Google Scholar]
- 2.Cromwell RL, Scott JM, Downs M, Yarbough PO, Zanello SB, Ploutz-Snyder L. Overview of the NASA 70-day Bed Rest Study. Med Sci Sports Exerc. 2018. Epub 2018/03/22. 10.1249/MSS.0000000000001617 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dillon EL, Sheffield-Moore M, Durham WJ, Ploutz-Snyder LL, Ryder JW, Danesi CP, et al. Efficacy of Testosterone plus NASA Exercise Countermeasures during Head-Down Bed Rest. Med Sci Sports Exerc. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ploutz-Snyder LL, Downs M, Goetchius E, Crowell B, English KL, Ploutz-Snyder R, et al. Exercise Training Mitigates Multi-System Deconditioning During Bed Rest Med Sci Sports Exerc. 2018. [DOI] [PMC free article] [PubMed]
- 5.Dillon EL, Sheffield-Moore M, Durham WJ, Ploutz-Snyder LL, Ryder JW, Danesi CP, et al. Efficacy of Testosterone plus NASA Exercise Countermeasures during Head-Down Bed Rest. Med Sci Sports Exerc. 2018;50(9):1929–39. 10.1249/MSS.0000000000001616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergstrom J. Percutaneous needle biopsy of skeletal muscle in physiological and clinical research. Scand J Clin Lab Invest. 1975;35(7):609–16. . [PubMed] [Google Scholar]
- 7.Sheffield-Moore M, Dillon EL, Casperson SL, Gilkison CR, Paddon-Jones D, Durham WJ, et al. A randomized pilot study of monthly cycled testosterone replacement or continuous testosterone replacement versus placebo in older men. J Clin Endocrinol Metab. 2011;96(11):E1831–7. 10.1210/jc.2011-1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiktorowicz JE, Stafford S, Rea H, Urvil P, Soman K, Kurosky A, et al. Quantification of cysteinyl S-nitrosylation by fluorescence in unbiased proteomic studies. Biochemistry. 2011;50(25):5601–14. Epub 2011/05/28. 10.1021/bi200008b . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savidge TC, Urvil P, Oezguen N, Ali K, Choudhury A, Acharya V, et al. Host S-nitrosylation inhibits clostridial small molecule-activated glucosylating toxins. Nat Med. 2011;17(9):1136–41. Epub 2011/08/23. 10.1038/nm.2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheffield-Moore M, Wiktorowicz JE, Soman KV, Danesi CP, Kinsky MP, Dillon EL, et al. Sildenafil increases muscle protein synthesis and reduces muscle fatigue. Clin Transl Sci. 2013;6(6):463–8. 10.1111/cts.12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soman KV, Stafford SJ, Pazdrak K, Wu Z, Luo X, White WI, et al. Activation of Human Peripheral Blood Eosinophils by Cytokines in a Comparative Time-Course Proteomic/Phosphoproteomic Study. J Proteome Res. 2017;16(8):2663–79. Epub 2017/07/21. 10.1021/acs.jproteome.6b00367 . [DOI] [PubMed] [Google Scholar]
- 12.Kramerova I, Kudryashova E, Wu B, Ottenheijm C, Granzier H, Spencer MJ. Novel role of calpain-3 in the triad-associated protein complex regulating calcium release in skeletal muscle. Hum Mol Genet. 2008;17(21):3271–80. 10.1093/hmg/ddn223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu KY, Zweier JL, Becker LC. Functional coupling between glycolysis and sarcoplasmic reticulum Ca2+ transport. Circ Res. 1995;77(1):88–97. . [DOI] [PubMed] [Google Scholar]
- 14.Bhasin S, Taylor WE, Singh R, Artaza J, Sinha-Hikim I, Jasuja R, et al. The mechanisms of androgen effects on body composition: mesenchymal pluripotent cell as the target of androgen action. J Gerontol A Biol Sci Med Sci. 2003;58(12):M1103–10. 10.1093/gerona/58.12.m1103 . [DOI] [PubMed] [Google Scholar]
- 15.Brocca L, Cannavino J, Coletto L, Biolo G, Sandri M, Bottinelli R, et al. The time course of the adaptations of human muscle proteome to bed rest and the underlying mechanisms. J Physiol. 2012;590(20):5211–30. Epub 2012/07/30. 10.1113/jphysiol.2012.240267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriggi M, Vasso M, Fania C, Capitanio D, Bonifacio G, Salanova M, et al. Long term bed rest with and without vibration exercise countermeasures: effects on human muscle protein dysregulation. Proteomics. 2010;10(21):3756–74. 10.1002/pmic.200900817 . [DOI] [PubMed] [Google Scholar]
- 17.Salanova M, Gelfi C, Moriggi M, Vasso M, Viganò A, Minafra L, et al. Disuse deterioration of human skeletal muscle challenged by resistive exercise superimposed with vibration: evidence from structural and proteomic analysis. FASEB J. 2014;28(11):4748–63. Epub 2014/08/13. 10.1096/fj.14-252825 . [DOI] [PubMed] [Google Scholar]
- 18.Cho Y, Ross RS. A mini review: Proteomics approaches to understand disused vs. exercised human skeletal muscle. Physiol Genomics. 2018;50(9):746–57. Epub 2018/06/29. 10.1152/physiolgenomics.00043.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hody S, Leprince P, Sergeant K, Renaut J, Croisier JL, Wang F, et al. Human muscle proteome modifications after acute or repeated eccentric exercises. Med Sci Sports Exerc. 2011;43(12):2281–96. 10.1249/MSS.0b013e318222edf3 . [DOI] [PubMed] [Google Scholar]
- 20.Holloway KV, O'Gorman M, Woods P, Morton JP, Evans L, Cable NT, et al. Proteomic investigation of changes in human vastus lateralis muscle in response to interval-exercise training. Proteomics. 2009;9(22):5155–74. 10.1002/pmic.200900068 . [DOI] [PubMed] [Google Scholar]
- 21.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11(4):276–87. 10.1038/nrm2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wehland M, Ma X, Braun M, Hauslage J, Hemmersbach R, Bauer J, et al. The impact of altered gravity and vibration on endothelial cells during a parabolic flight. Cell Physiol Biochem. 2013;31(2–3):432–51. 10.1159/000343380 . [DOI] [PubMed] [Google Scholar]
- 23.Ma X, Wehland M, Aleshcheva G, Hauslage J, Wasser K, Hemmersbach R, et al. Interleukin-6 expression under gravitational stress due to vibration and hypergravity in follicular thyroid cancer cells. PLoS One. 2013;8(7):e68140 10.1371/journal.pone.0068140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trappe S, Trappe T, Gallagher P, Harber M, Alkner B, Tesch P. Human single muscle fibre function with 84 day bed-rest and resistance exercise. J Physiol. 2004;557(Pt 2):501–13. 10.1113/jphysiol.2004.062166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arentson-Lantz EJ, English KL, Paddon-Jones D, Fry CS. Fourteen days of bed rest induces a decline in satellite cell content and robust atrophy of skeletal muscle fibers in middle-aged adults. J Appl Physiol (1985). 2016;120(8):965–75. 10.1152/japplphysiol.00799.2015 PubMed Central PMCID: PMCPMC4835912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murach KA, Minchev K, Grosicki GJ, Lavin K, Perkins RK, Ryder JW, et al. Myocellular Responses to Concurrent Flywheel Training during 70 Days of Bed Rest. Med Sci Sports Exerc. 2018. Epub 2018/03/22. 10.1249/MSS.0000000000001620 . [DOI] [PubMed] [Google Scholar]
- 27.Novak KD, Titus MA. The myosin I SH3 domain and TEDS rule phosphorylation site are required for in vivo function. Mol Biol Cell. 1998;9(1):75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li MX, Hwang PM. Structure and function of cardiac troponin C (TNNC1): Implications for heart failure, cardiomyopathies, and troponin modulating drugs. Gene. 2015;571(2):153–66. 10.1016/j.gene.2015.07.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryder JW, Lau KS, Kamm KE, Stull JT. Enhanced skeletal muscle contraction with myosin light chain phosphorylation by a calmodulin-sensing kinase. J Biol Chem. 2007;282(28):20447–54. 10.1074/jbc.M702927200 . [DOI] [PubMed] [Google Scholar]
- 30.Rani SG, Sepuru KM, Yu C. Interaction of S100A13 with C2 domain of receptor for advanced glycation end products (RAGE). Biochim Biophys Acta. 2014;1844(9):1718–28. 10.1016/j.bbapap.2014.06.017 . [DOI] [PubMed] [Google Scholar]
- 31.Hamzeh M, Robaire B. Identification of early response genes and pathway activated by androgens in the initial segment and caput regions of the regressed rat epididymis. Endocrinology. 2010;151(9):4504–14. 10.1210/en.2010-0023 . [DOI] [PubMed] [Google Scholar]
- 32.Boyle ST, Kular J, Nobis M, Ruszkiewicz A, Timpson P, Samuel MS. Acute compressive stress activates RHO/ROCK-mediated cellular processes. Small GTPases. 2018:1–17. 10.1080/21541248.2017.1413496 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossetti ML, Steiner JL, Gordon BS. Increased mitochondrial turnover in the skeletal muscle of fasted, castrated mice is related to the magnitude of autophagy activation and muscle atrophy. Mol Cell Endocrinol. 2018. 10.1016/j.mce.2018.01.017 . [DOI] [PubMed] [Google Scholar]
- 34.Bolduc C, Yoshioka M, St-Amand J. Transcriptomic characterization of the long-term dihydrotestosterone effects in adipose tissue. Obesity (Silver Spring). 2007;15(5):1107–32. 10.1038/oby.2007.623 . [DOI] [PubMed] [Google Scholar]
- 35.Schild M, Ruhs A, Beiter T, Zugel M, Hudemann J, Reimer A, et al. Basal and exercise induced label-free quantitative protein profiling of m. vastus lateralis in trained and untrained individuals. J Proteomics. 2015;122:119–32. 10.1016/j.jprot.2015.03.028 . [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Calvo R, Chanda D, Oligschlaeger Y, Miglianico M, Coumans WA, Barroso E, et al. Small heterodimer partner (SHP) contributes to insulin resistance in cardiomyocytes. Biochim Biophys Acta. 2017;1862(5):541–51. 10.1016/j.bbalip.2017.02.006 . [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Calvo R, Vazquez-Carrera M, Masana L, Neumann D. AICAR Protects against High Palmitate/High Insulin-Induced Intramyocellular Lipid Accumulation and Insulin Resistance in HL-1 Cardiac Cells by Inducing PPAR-Target Gene Expression. PPAR Res. 2015;2015:785783 10.1155/2015/785783 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteoforms that showed significant pre-post abundance changes in Table 1 were submitted for Enrichment Analyses (Annotation Version and Release Date: GO Ontology database Released 2018-08-09, http://www.geneontology.org/,). Table include only results with False Discovery Rate < 0.05. A. CON, B. PEX, C. TEX.
(PDF)
Raw protein abundance and phosphorylation data from MS Analyses. These data were submitted to the NASA Life Science Data Archive (https://lsda.jsc.nasa.gov/).
(XLSX)
The study complied with the Declaration of Helsinki and was approved by The University of Texas Medical Branch (UTMB) Institutional Review Board (IRB) and by the NASA Committee for the Protection of Human Subjects (CPHS).
(PDF)
This study adheres to CONSORT guidelines.
(DOC)
Data Availability Statement
Relevant data are within the manuscript and its Supporting Information files. Full data sets can be obtained through the NASA Life Science Data Archive (https://lsda.jsc.nasa.gov/).