Abstract
Formins are a family of multidomain scaffold proteins involved in actin-dependent morphogenetic events. In Aspergillus nidulans, the formin SEPA participates in two actin-mediated processes, septum formation and polarized growth. In this study, we use a new null mutant to demonstrate that SEPA is required for the formation of actin rings at septation sites. In addition, we find that a functional SEPA::GFP fusion protein localizes simultaneously to septation sites and hyphal tips, and that SEPA colocalizes with actin at each site. Using live imaging, we show that SEPA localization at septation sites and hyphal tips is dynamic. Notably, at septation sites, SEPA forms a ring that constricts as the septum is deposited. Moreover, we demonstrate that actin filaments are required to maintain the proper localization pattern of SEPA, and that the amino-terminal half of SEPA is sufficient for localization at septation sites and hyphal tips. In contrast, only localization at septation sites is affected by loss of the sepH gene product. We propose that specific morphological cues activate common molecular pathways to direct SEPA localization to the appropriate morphogenetic site.
INTRODUCTION
The actin cytoskeleton functions in numerous cellular processes, including cell motility, organelle and vesicle transport, morphogenesis, and cytokinesis. To perform these multiple tasks, the actin cytoskeleton is controlled by numerous regulatory and accessory proteins that direct the polymerization of actin monomers into filaments and the cross-linking of filaments into a network (Ayscough, 1998; Chen et al., 2000). Assembly and organization of the actin cytoskeleton is linked to environmental and intracellular signals by multiple pathways (Schmidt and Hall, 1998). Key regulators of these pathways are members of the Rho family of low molecular weight GTPases. Rho GTPases are membrane-bound proteins that act as molecular switches to relay spatial and temporal information to effectors that reorganize the actin cytoskeleton (Tanaka and Takai, 1998). Among the effectors of Rho GTPases are proteins that contain multiple protein–protein interaction domains and appear to function as molecular scaffolds (Bishop and Hall, 2000). Scaffold proteins integrate incoming signals with actin cytoskeleton dynamics by interacting with both the signaling proteins and actin-binding proteins. Examples include the ezrin/radixin/moesin, enabled/vasodilator-stimulated phosphoprotein, Wiskott-Aldrich syndrome protein/Wiskott-Aldrich syndrome protein-interacting protein, and formin families of proteins (Frazier and Field, 1997; Beckerle, 1998; Wasserman, 1998; Bretscher, 1999; Ramesh et al., 1999; Zeller et al., 1999; Mullins, 2000).
Formins are conserved from fungi to humans and are characterized by the presence of two conserved carboxy-terminal regions, the FH1 and FH2 domains (Emmons et al., 1995). The proline-rich FH1 domain interacts with SH3 and WW domain-containing proteins, as well as with the actin monomer-binding protein profilin (Chan et al., 1996; Manseau et al., 1996; Uetz et al., 1996; Bedford et al., 1997; Chang et al., 1997; Evangelista et al., 1997; Imamura et al., 1997; Watanabe et al., 1997; Kamei et al., 1998). The FH2 domain interacts with the actin-binding proteins Bud6p and elongation factor 1α (Evangelista et al., 1997; Umikawa et al., 1998), as well as with Smy1p, a kinesin-related protein that may function in actin filament-based transport (Kikyo et al., 1999). Three other domains located in the amino-terminal half of formins may be conserved: the Rho GTPase-binding site (Kohno et al., 1996; Evangelista et al., 1997; Imamura et al., 1997; Watanabe et al., 1997), the FH3 domain (Petersen et al., 1995), and the Spa2p-binding domain (Fujiwara et al., 1998). The FH3 domain and Spa2p-binding domain are thought to regulate the localization of formins to sites of cell surface remodeling (Petersen et al., 1998; Ozaki-Kuroda et al., 2001).
Phenotypic characterization of formin mutants has provided evidence that formins are required for actin function. For example, during cytokinesis, actin rings fail to form in Schizosaccharomyces pombe cdc12 and Drosophila melanogaster dia mutants (Chang et al., 1997; Afshar et al., 2000). In contrast, actomyosin rings form at the mother/bud neck in Saccharomyces cerevisiae bni1 mutants, but they do not contract (Vallen et al., 2000). Furthermore, actin fails to localize to tips of mating projections during conjugation in bni1 and S. pombe fus1 mutants (Evangelista et al., 1997; Petersen et al., 1998). Consistent with their role in actin cytoskeletal organization, most formins colocalize with actin at sites of polarized growth or with the actin ring during cytokinesis (Chang et al., 1997; Evangelista et al., 1997; Fujiwara et al., 1998; Petersen et al., 1998; Swan et al., 1998; Afshar et al., 2000; Ozaki-Kuroda et al., 2001).
Conidiospores of the filamentous fungus Aspergillus nidulans undergo a series of morphogenetic events during germination (Harris, 1997). Initially, conidiospores undergo a period of isotropic swelling, which is followed by a switch to apical growth and the subsequent emergence of a germ tube. Apical (tip) growth is maintained throughout the life of a hypha, and, unlike what occurs in budding or fission yeast, it is not interrupted during septation. Septation occurs once cells have satisfied a size requirement and completed at least one round of mitosis (Wolkow et al., 1996). Experiments with cytochalasin A, an actin-depolymerizing drug, demonstrate that actin filaments are required for both apical growth and septation (Harris et al., 1994; Torralba et al., 1998). Consistent with its function, actin localizes to both hyphal tips and septation sites (Harris et al., 1994). Like actin, the sepA gene product functions in a number of morphogenetic processes; the temperature-sensitive (ts) sepA1 mutant fails to septate at restrictive temperature, displays a defective tip growth pattern, and has abnormally wide hyphae (Morris, 1976). Molecular characterization has revealed that sepA encodes a formin (Harris et al., 1997).
In this study, we characterize the role of SEPA during septum formation and tip growth by constructing a new null allele and by determining the localization pattern of a functional SEPA::GFP fusion protein. In addition, we provide initial insight into the mechanisms underlying the simultaneous targeting of SEPA to distinct structures within hyphal cells.
MATERIALS AND METHODS
Aspergillus Strains and Growth Methods
Strains used in this study are described in Table 1. All genetic manipulations were performed as described previously (Harris et al., 1994). The following media were used: MAG (2% dextrose, 2% malt extract, 0.2% peptone, trace elements, and vitamins; pH 6.5), YGV (2% dextrose, 0.5% yeast extract, and vitamins), and MN (1% dextrose, nitrate salts, trace elements, and biotin; pH 6.5). Arginine (1 mM), uridine (5 mM), and uracil (10 mM) were added as needed. Trace elements, nitrate salts, and vitamins were added as described in the appendix to Kafer (1977). For solid media, 1.5% agar was added.
Table 1.
A. nidulans strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| A28a | pabaA6 biA1 | FGSCa |
| AH13 | wA3; argB2; chaA1 | Lab stock |
| AJM68 | pyrG89; wA3; sepH1; pyroA4 | Bruno et al., 2001 |
| ALH1 | sepA4ΔBm | Harris et al., 1997 |
| AML9 | pyrG89 pabaA1; argB2 | Lab stock |
| AML13 | pabaA1 yA2; argB2; chaA1? | Lab stock |
| ASH35 | sepA1 yA2; argB2 | Harris et al., 1997 |
| ASH162 | pyrG89 pabaA1 yA2 | Lab stock |
| AKS6 | sepA6ΔFH∷argB; wA2; argB2; chaA1 | This study |
| AKS64 | sepA1 yA2; sepA∷gfp∷argB argB2 | This study |
| AKS65 | sepA1 yA2; sepA∷gfp∷argB argB2 | This study |
| AKS70 | sepA∷gfp∷pyr-4; pyrG89 pabaA1 yA2 | This study |
| AKS71 | sepA∷gfp∷pyr-4; pyrG89; wA3; sepH1; pyroA4 | This study |
| AKS76 | yA2; sepA∷gfp∷argB argB2; chaA1 | This study |
| AKS82 | sepA(aa1-838)∷gfp∷pyr-4; pyrG89 pabaA1 yA2 | This study |
| AKS84 | sepA(aa1-132)∷gfp∷pyr-4; pyrG89 pabaA1 yA2 | This study |
Fungal Genetics Stock Center, Department of Microbiology, University of Kansas Medical Center, Kansas City, KS 66160-7420.
DNA Techniques
Subcloning was performed using standard methods (Sambrook et al., 1989) except that the TOPO TA Cloning kit (Invitrogen, Carslbad, CA) was used to subclone polymerase chain reaction (PCR) fragments of sepA1 into pCR2.1-TOPO. PCRs were carried out using Vent or Taq polymerase (Invitrogen). GC-rich PCR was performed using the Advantage-GC Genomic PCR kit (CLONTECH, Palo Alto, CA) or the PCRX Enhancer System (Invitrogen). PCR primer sequences are available on request. Sequencing and oligonucleotide synthesis were performed by the Molecular Core Facility at the University of Connecticut Health Center. The sepA sequence has been updated in GenBank (accession number U83658).
Isolation of DNA from A. nidulans and transformations were performed using standard procedures (Timberlake, 1990; Oakley and Osmani, 1993). Southern blots were analyzed using digoxigenin-labeled probes and nonradioactive detection (Roche Molecular Biochemicals, Indianapolis, IN).
Plasmid Constructs
The plasmid containing sepA::gfp, pKES59, was constructed in multiple steps. A unique NotI site that replaced the sepA stop codon was constructed using PCR to amplify a 333-base pair fragment of sepA. This fragment was subcloned into pYESTrp (Invitrogen) by using an internal SphI site and the NotI site from the primer. Using the SphI site and a KpnI site 3′ to the NotI site, the fragment was ligated into pKES1, a plasmid containing most of sepA (HindIII to SphI) (Harris et al., 1997). The resulting plasmid, pKES30, contains the entire sepA gene (5422 base pairs without the stop codon) followed by unique NotI and KpnI sites, plus 964 base pairs of upstream sequence. PCR was used to incorporate NotI sites on both ends of gfp from pMCB32 (Fernandez-Abalos et al., 1998). pMCB32 contains a codon-modified version of green fluorescent protein (GFP), which is optimized for expression in mammals and plants and carries the S65T substitution. gfp was ligated in-frame with sepA in the NotI site of pKES30, resulting in the plasmid pKES46.
On the 5′ end, a KpnI site was incorporated just 3′ of the HindIII site by PCR of a 982-base pair fragment of sepA. This fragment was subcloned into pKES46 by using an internal SnaBI site and the HindIII site, resulting in a plasmid (pKES58) containing sepA::gfp flanked by KpnI sites. pKES59 was constructed by subcloning the sepA::gfp gene fusion by using the KpnI sites into the pyr-4–containing vector pRG3 (Waring et al., 1989).
Another plasmid containing sepA::gfp, pKES56, was also constructed from pKES46. pKES56 contains a truncated allele of argB that will target integration of sepA::gfp to the argB locus. The truncated allele of argB was made by PCR with pSDW194 (James et al., 1999). The truncated argB gene was then cloned into pCR2.1-TOPO resulting in pKES55. pKES56 was constructed by subcloning the truncated argB gene from pKES55 into pKES46 (sepA::gfp).
pKES20 was constructed by subcloning an ∼3.5-kb SacI fragment from pON48 (Harris et al., 1997) into pYESTrp (Invitrogen). This fragment contains the last 2779 base pairs of sepA and 0.7 kb of downstream sequence.
The sepA disruption plasmid pKES14 was constructed in three steps. First, a 5′ piece of sepA from pKES1 (a 1.3-kb HindIII-BamHI fragment) was ligated into pUC18, resulting in plasmid pKES12. Second, a BamHI fragment from pSalArgB (Miller et al., 1987) containing the argB gene was ligated into the BamHI site of pKES12, resulting in plasmid pKES13. Third, pKES14 was constructed by ligating a 3′ piece of sepA from pON48 (a 3-kb MfeI-EcoRI fragment) into the EcoRI site of pKES13.
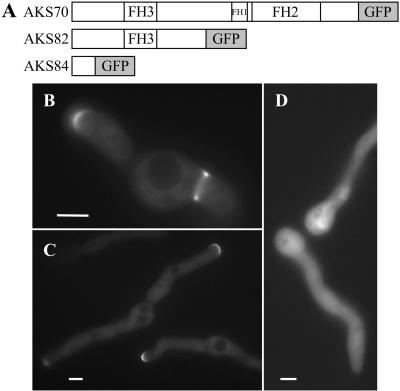
pKES63 and pKES64, two plasmids containing 5′ sepA fragments fused to gfp, were constructed from pKES58. A 1.3-kb KpnI-BamHI 5′ fragment of sepA was subcloned from pKES58 into pRG3, resulting in plasmid pKES61. PCR of pMCB32 was performed to incorporate a 5′ BamHI site and a 3′ SphI site onto the ends of gfp. The PCR fragment containing gfp was subcloned into pCR2.1-TOPO, resulting in plasmid pKES62. gfp from pKES62 was subsequently subcloned downstream of the sepA fragment in pKES61 by using the BamHI and SphI sites, resulting in plasmid pKES63. pKES63 contains 0.96 kb of upstream sequence plus 399 base pairs of sepA sequence fused in-frame to gfp, encoding a predicted fusion protein of 371 amino acids (aa) (132 aa of SEPA plus 239 aa of GFP). pKES64 was constructed by subcloning a 2115-base pair BamHI sepA fragment into the BamHI site of pKES63. pKES64 contains 0.96 kb of upstream sequence plus 2514 base pairs of sepA sequence fused in-frame to gfp, encoding a predicted fusion protein of 1077 aa (838 aa of SEPA plus 239 aa of GFP). The strains AKS84 and AKS82 (Figure 8A) were obtained by transforming strain ASH162 with pKES63 and pKES64, respectively.
Figure 8.
Amino terminal half of SEPA is sufficient for localization to both septation sites and hyphal tips. (A) Diagrams of three SEPA::GFP constructs (see MATERIALS AND METHODS). (B and C) Nearly normal localization (see text) of the amino terminal half of SEPA in strain AKS82. Bars, 4 μm. (D) Failure of a shorter amino-terminal fragment (in strain AKS84) to localize. Bar, 3 μm.
Immunofluorescence Microscopy and Live Imaging
Coverslips with adherent cells were processed for microscopy and stained with Calcofluor (American Cyanamid, Wayne, NJ) and Hoechst 33258 (Molecular Probes, Eugene, OR) to visualize septa and nuclei, respectively (Harris et al., 1994). Immunofluorescence microscopy for detection of the actin cytoskeleton was performed using standard protocols (Harris et al., 1999). Mouse C4 monoclonal anti-actin antibody (ICN Biomedicals, Aurora, OH) diluted at 1:400 was used as the primary antibody. Texas Red-conjugated goat antimouse antibodies diluted at 1:100 were used as secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA).
SEPA::GFP was observed by fluorescence microscopy with a standard fluorescein isothiocyanate filter, and images were captured using an Axioplan charge-coupled device camera. Slides were also viewed using an Olympus BX60 microscope. Images were processed and printed using Adobe Photoshop and Adobe Illustrator (Adobe Systems, Mountain View, CA) and Microsoft PowerPoint (Microsoft, Redmond, WA). Counts of septa, SEPA at tips and SEPA rings are based on the means of at least three separate experiments in which the number of cells counted (n) usually equaled 100. Cytochalasin A (Sigma, St. Louis, MO) was used at a final concentration of 2 μg/ml from a 1 mg/ml stock made in dimethyl sulfoxide (DMSO). We were unable to determine the localization of SEPA in two sep mutants, sepD5 and sepG1, because GFP is no longer stable at the lowest temperature that these mutants fail to form septa (i.e., 42°C).
Live imaging was performed using an Olympus IX70 inverted fluorescence microscope with a HiQ fluorescein filter set and 100-W Hg lamp. Cells were grown in YGV on coverslips sealed over a hole punched out of a Petri dish on a Bioptechs heated stage as described previously (Xiang et al., 2000). Images were obtained using a Princeton Instruments 5-MHz MicroMax cooled charge-coupled device camera system and IPLab software (Scanalytics, Fairfax, VA). Image sequences were then converted into Quicktime. Live imaging of SEPA::GFP at hyphal tips was acquired from 13-h and older hyphae (28°C).
RESULTS
The sepA1 Temperature-sensitive Mutation Lies in Highly Conserved Amino Acid in FH2 Domain
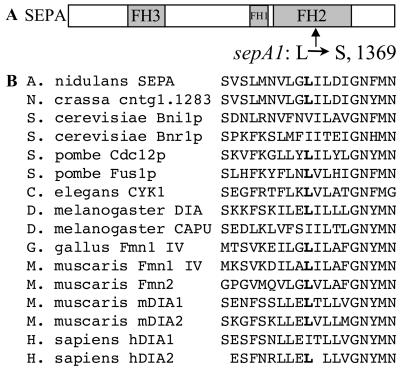
The sepA gene was originally identified by the ts sepA1 mutation (Morris, 1976). The nature and location of this mutation were determined by sequencing pooled PCR reactions of genomic DNA from a sepA1 strain, ASH35. To localize the sepA1 mutation, ASH35 was first transformed with fragments of sepA to determine which sequences can repair the mutation. Ts+ transformants were obtained using pKES20, a plasmid containing the last 2778 bases of sepA (our unpublished results). This region of sepA1 was therefore cloned and sequenced to identify the mutation. A single mutation was found, a T:A-to-C:G transition at base pair 4106, which changes amino acid 1369 from leucine (TTA; wild-type) to serine (TCA) (Figure 1A). Notably, this hydrophobic-to-hydrophilic substitution lies in a highly conserved residue within the FH2 domain (Figure 1B). The sepA1 mutation may disrupt protein–protein interactions of the FH2 domain, or the sepA1 protein product may be unfolded and degraded at high temperature.
Figure 1.
Location of the sepA1 mutation. (A) Schematic drawing of SEPA depicting the FH3, FH1, and FH2 domains. An arrow indicates the location within the FH2 domain of the L1369S mutation. (B) Alignment of formin family members highlighting the conservation of the hydrophobic residue (boldface) altered in sepA1. Sequences were aligned using the CLUSTALw program.
sepA Is Required for Septation
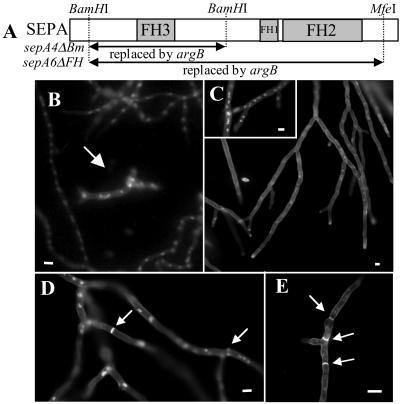
In previous work, a sepA disruption strain, ALH1 (sepA4ΔBm), was constructed and shown to have a ts growth defect (Harris et al., 1997). In addition, it was found that sepA4ΔBm strains produce septa after a long delay. Because the ts sepA1 allele never produces septa at the restrictive temperature, we questioned whether the sepA4ΔBm strain displays the true null phenotype. To test this, we constructed a new sepA disruption strain in which a larger piece of the gene, including the conserved FH1 and FH2 regions, was replaced. The new sepA6ΔFH strains are missing a 4668-base pair BamHI-MfeI fragment, whereas the sepA4ΔBm strain is missing a 2135-base pair BamHI fragment (Figure 2A).
Figure 2.
Comparison of two sepA deletion mutants, sepA4ΔBm and sepA6ΔFH. (A) Schematic drawing of sepA indicating the sequences replaced by argB in sepA4ΔBm and sepA6ΔFH. (B) sepA6ΔFH and SepA+ hyphae from a mixed spore population harvested from the AKS6/AML13 heterokaryon grown for 18 h in YGV medium and stained to visualize nuclei. An arrow indicates a sepA6ΔFH germling surrounded by wild-type hyphae. (C) sepA6ΔFH fails to produce septa and displays dichotomous branching. Strain AKS6 grown for 44 h in MN medium and stained to show nuclei and septa. Inset shows hyphae stained only to visualize nuclei. (D) sepA4ΔBm septates after a delay. Arrows indicate septa that have formed in ALH1 (sepA4ΔBm) grown for 17 h. (E) Multiple septa (arrows) in a wild-type strain (A28) after 24 h of growth. Bars, 12 μm.
The new sepA disruption strains were constructed by transforming strain AH13 with uncut pKES14. We screened 250 Arg+ transformants for restricted colonial growth because sepA1 strains exhibit this phenotype. Three transformants displayed restricted colonial growth and failed to produce conidiating colonies. These transformants (sepA6ΔFH) were propagated as heterokaryons with AML13 (argB). Growth of the two types of spores harvested from the heterokaryon can be controlled using different media; both wild-type and sepA6ΔFH hyphae grow in media supplemented with arginine such as YGV (Figure 2B), whereas in MN media, only sepA6ΔFH spores grow (Figure 2C, inset). Analysis of each transformant by Southern blotting of DNA obtained from strains grown in MN and YGV confirmed that sepA had been replaced with argB in sepA6ΔFH hyphae (our unpublished results). We thus conclude that sepA is not an essential gene, although it is required for conidiation.
Similar to sepA4ΔBm strains, the sepA6ΔFH disruptants display dichotomous branching (split tips) and contain hyphae that are 1.5–2.5 times wider than normal (Figure 2, B and C). However, unlike sepA4ΔBm colonies, sepA6ΔFH colonies display restricted colonial growth at all temperatures. Moreover, sepA6ΔFH strains do not septate, even after >40 h of growth (Figure 2C), whereas sepA4ΔBm strains (Figure 2D) and wild-type strains (Figure 2E) contain multiple septa after shorter periods of growth. From these observations, we conclude that SEPA is required for septum formation.
sepA Is Required for Actin Ring Formation
Septation in A. nidulans is preceded by the formation and constriction of an actin ring (Momany and Hamer, 1997). Because sepA is required for septation, we next asked whether it plays a role in actin ring formation. A mixed spore population from the sepA6ΔFH heterokaryon was inoculated into selective MN media, allowing only sepA6ΔFH spores to germinate, and actin was visualized by immunofluorescence after 20 h. In wild-type cells, actin localizes to hyphal tips, in rings at septation sites, and in cortical spots (Figure 3A). However, in sepA6ΔFH strains, actin was observed only at hyphal tips and in cortical spots; no actin rings were observed (n = 100; Figure 3, B and C). Thus, SEPA is required for actin ring formation, but not for actin localization at hyphal tips and cortical spots.
Figure 3.
sepA6ΔFH cells fail to make actin rings, but localize actin properly to hyphal tips and cortical spots. (A) A28 (wild-type) germlings stained with anti-actin antibodies. Arrows indicate rings at septation sites; the cortical spots are not visible in this image. (B and C) sepA6ΔFH spores from an AKS6/AML13 heterokaryon grown in MN medium and stained with anti-actin antibodies. The ungerminated spores in B are wild-type sepA spores, which do not grow in MN medium because they remain Arg−. Bars, 2 μm (A) or 4 μm (B and C).
SEPA Displays Dynamic Localization at Septation Sites and Hyphal Tips
Based on its role in septation and polarized growth, we predicted that SEPA would localize to sites of septation and to hyphal tips. To test these predictions, we constructed plasmids encoding SEPA::GFP fusion proteins (see MATERIALS AND METHODS). pKES56 also contains a truncated version of argB to target integration to the argB locus, and pKES59 contains the Neurospora crassa pyr-4 gene, which can complement the A. nidulans pyrG89 mutation. These constructs were transformed into strains ASH35 (sepA1) and ASH162 (wild-type), respectively. Analysis of Southern blots revealed that two ASH35 transformants, AKS64 and AKS65, contain a single copy of sepA::gfp integrated at the argB locus (our unpublished results). Because these transformants grow and septate like wild-type at the restrictive temperature (our unpublished results), we conclude that SEPA::GFP is functional.
The strains transformed with pKES59 displayed differences in brightness. These differences may be due to different numbers of plasmid integration events, resulting in variable levels of expression of SEPA::GFP. We isolated strains expressing SEPA::GFP at a level sufficient for good imaging. For example, one of these strains, AKS70, was shown by Southern blotting to contain more than five copies of the sepA::gfp construct (our unpublished results). To ensure that the pattern of SEPA::GFP localization in these strains was not an artifact of overexpression, we also examined strain AKS76, a daughter of a cross between AKS65 and AH13 (note that AKS76 was used because it possesses a wild-type allele at the sepA locus; Table 1). The SEPA::GFP localization pattern in AKS76 was indistinguishable from the pattern exhibited by the brighter strains (compare Figure 4A with B). In addition, we have observed seemingly aberrant localization of SEPA::GFP when it is overexpressed under the inducible alcA promoter (our unpublished results). Because we do not observe a similar aberrant localization pattern in strains containing multiple copies of sepA::gfp, we conclude that these strains display normal localization of SEPA, only brighter.
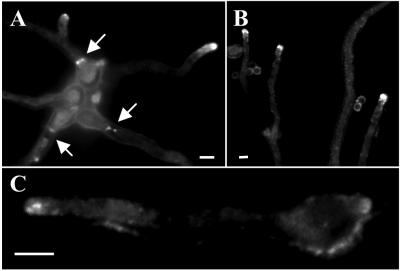
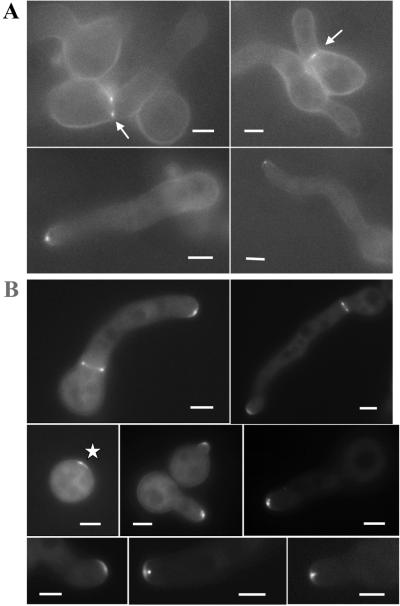
Figure 4.
SEPA::GFP localizes to sites of septation and to hyphal tips. (A) Localization of SEPA::GFP to septation sites and hyphal tips in a strain (AKS76) carrying one copy of sepA::gfp. Arrows indicate ring structures. (B) Localization of SEPA::GFP in a strain (AKS70) carrying multiple copies of sepA::gfp. The asterisk indicates SEPA::GFP located to a presumptive site of germ tube emergence. Bars, 3 μm.
In AKS70, SEPA::GFP localized faintly to the cytoplasm and brightly to the tips of hyphae and at septation sites (Figure 4B). Localization to both sites occurred simultaneously during septation. SEPA::GFP was found at hyphal tips in 100% of cells (n = 100), typically as a crescent (Figure 4B); 80% of the cells also showed a bright spot near the tip. SEPA::GFP was seen at sites of septation in 20% of cells (n = 100) after 13 h of growth at 28°C.
Using live imaging, we have observed the dynamics of SEPA::GFP localization at sites of septation. SEPA-GFP appears as a spot and then as a ring-like structure that constricts into a dot and eventually disappears (Figure 5A; supplemental movies 1, 2, and 8). Five such ring sequences were obtained and analyzed (Table 2). At 28°C, formation of the ring from a spot took from 1.5 to 9 min, the full-size ring was present from 3 to 11.5 min, and ring constriction and disappearance took from 21.5 to 35 min. The length of the entire process ranged from 29 to 43.5 min.
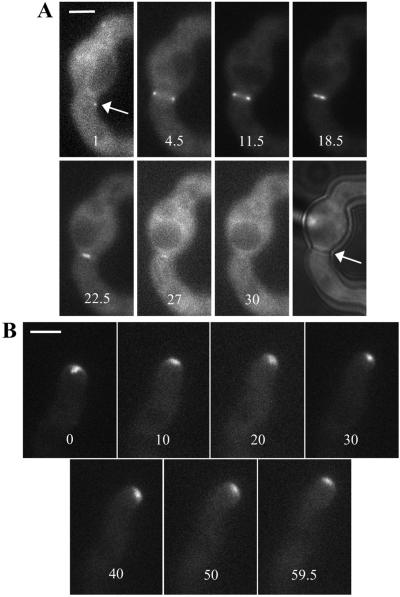
Figure 5.
SEPA::GFP ring dynamics. Times are indicated in minutes. (A) Stills from the live imaging sequence also presented as supplemental movie 1. The arrow indicates the spot of SEPA::GFP visualized before the ring (see DISCUSSION). The final still in the series shows a brightfield image of the same cell taken after the live imaging sequence. An arrow points to the septum that has formed. (B) Stills from the live imaging sequence also presented as supplemental movie 3. Bar, 3 μm.
Table 2.
Timing of SEPA ring dynamics (28°C)
| Ring dynamicsa | Ring
|
||||
|---|---|---|---|---|---|
| 1 | 2b | 3c | 4 | 5 | |
| • → ○ | 2 (min) | 8 | ∼5 | 1.5 | 9 |
| ○ (no change) | 3 | 11.5 | 4.5 | 7 | 4 |
| ○ → | 24 | ∼24 | 28 | 35 | 21.5 |
| Total time | 29 | ∼43.5 | ∼37.5 | 43.5 | 34.5 |
Three stages of ring dynamics are shown: formation of the ring from a single spot, maintenance of a full-size ring, and ring constriction (and disappearance).
Ring 2 goes out of focus, allowing only an approximation of the constriction time to be made.
Ring 3 is just starting to form at the start of the sequence, allowing only an approximation of the formation time to be made.
At hyphal tips, the SEPA::GFP spot and crescent were also dynamic. In the live image sequences, the location of the spot and crescent changed as the tip extended, usually moving toward the direction of growth (Figure 5B; supplemental movies 3 and 4). Notably, at sites of germ tube emergence (Figure 4B, asterisk) and at new branches (our unpublished results), SEPA appeared just before outgrowth. Thus, SEPA localization at both septation sites and hyphal tips is dynamic and may anticipate the location of growth and cell wall deposition.
SEPA and Actin Colocalize at Septation Sites and Hyphal Tips
Because SEPA localized to two cellular locations where actin is also present, and SEPA is required for actin ring formation, we asked whether SEPA and actin colocalize. In addition, we compared SEPA and actin localization to septum dynamics by localizing the major septum component, chitin. At sites of septation, SEPA::GFP colocalized with actin at all stages, including the full-size ring and the constricted rings, which appear as hourglass shapes (Figure 6, A-D). Hourglass shapes are not observed during live imaging and appear to be due to the separation of cell membranes from cell walls, which may occur during fixation. At tips of hyphae, actin appears in a tip-high gradient that extends subapically. SEPA colocalizes with actin at the very tip (Figure 6, E and F).
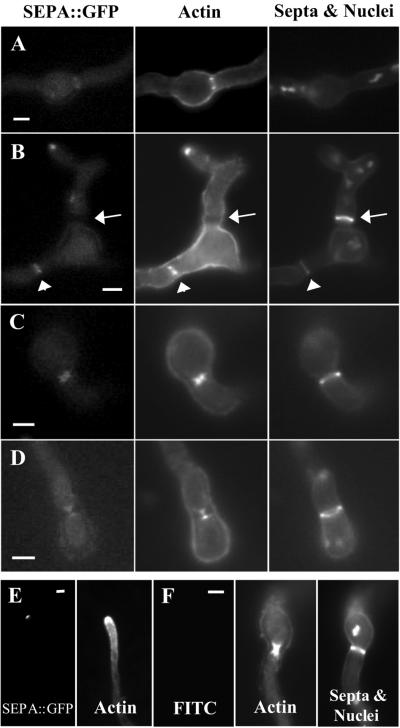
Figure 6.
SEPA::GFP and actin colocalize in ring structures and at the hyphal tip. (A-D) Strain AKS70 (sepA::gfp) was stained with anti-actin antibodies, Calcofluor, and Hoechst 33258 to visualize actin, septa, and nuclei. Images are ordered by their stage of septation from earliest to latest. (A) SEPA::GFP and actin rings are visible, but no septal material is present. (B) SEPA::GFP and actin rings have started to constrict and a faint septal ring is present (arrowheads). The arrow indicates a mature septum, which is brighter than the septal ring; SEPA::GFP and actin are absent at this stage. (C and D) SEPA::GFP and actin colocalize during constriction, appearing as hourglass shapes (see text). (E) SEPA::GFP and actin colocalize at the very tips of hyphae in strain AKS70. (F) Control showing that fluorescence due to actin staining is not visible using the fluorescein isothiocyanate filter used for GFP. Also shown are actin, septa, and nuclei in wild-type strain A28, which lacks SEPA::GFP. Bars, 3 μm.
Actin Filaments Are Required to Maintain Normal Patterns of SEPA Localization
Because actin and SEPA colocalize, actin filaments could conceivably regulate SEPA dynamics at septation sites, hyphal tips, or both. To test this notion, the location of SEPA::GFP was determined in strains treated with cytochalasin A, an inhibitor of actin polymerization (Torralba et al., 1998). At septation sites, cytochalasin A disrupted the constriction and disappearance of the SEPA ring. Instead, SEPA accumulated, forming “beads” at septation sites (Figure 7A; supplemental movie 5). In A. nidulans, cytochalasin A disrupts polarized growth at hyphal tips, causing them to swell (Torralba et al., 1998). Cytochalasin A also disrupted the pattern of SEPA localization at hyphal tips; SEPA accumulated and formed beads that dispersed around the swollen tips (Figure 7B; supplemental movie 6). Control treatment with DMSO did not affect either the dynamics of SEPA ring constriction or the dynamics of SEPA at hyphal tips (supplemental movies 7 and 8). In addition, benomyl, a microtubule-destabilizing drug, had no effect on SEPA ring constriction or localization at hyphal tips (our unpublished results). These observations suggest that actin filaments, but not microtubules, are required for the maintenance of dynamic SEPA structures at both hyphal tips and septation sites.
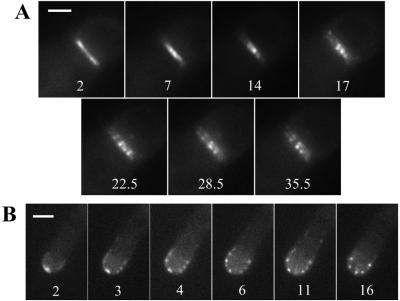
Figure 7.
Effect of cytochalasin A on SEPA::GFP ring dynamics. Strain AKS70 was grown for 13 h and treated with 2 mg/ml cytochalasin A. Times (min) since addition of the drug are indicated. (A) A septation site (see supplemental movie 5). Bar, 1.5 μm. (B) A hyphal tip (see supplemental movie 6). Bar, 3 μm.
sepH Is Required for SEPA Localization at Septation Sites
sepH1 was identified in a screen for ts mutants that fail to septate at restrictive temperatures of ≥37°C (Harris et al., 1994; Table 3). sepH encodes a ortholog of S. pombe Cdc7p, a protein kinase required for signaling during septation (Bruno et al., 2001). To determine whether the sepH gene product is required for SEPA localization, plasmid pKES59 (sepA::gfp) was transformed into the sepH1 strain, AJM68. Southern analysis showed that the resulting strain, AKS71, contains more than five copies of the sepA::gfp construct (our unpublished results). At 37°C, SEPA::GFP in AKS71 localizes to hyphal tips but not to septation sites (Table 3). Thus, SEPH is required for SEPA ring formation, but not for localization of SEPA at hyphal tips.
Table 3.
sepH is required for SEPA localization to septation sites
| Strain | Temperaturea (°C) | % of Cells with SEPA at tipsb | % of Cells with SEPA ringsb | % of Cells with septab |
|---|---|---|---|---|
| AKS70 (wt) | 28 | 100–0 | 20.3–4.4 | 35–13.0 |
| 37 | 99–0.6 | 17.3–5.7 | 82–2.6 | |
| AKS71 (sepH1) | 28 | 99–0.6 | 14–1.7 | 18.7–3.2 |
| 37 | 99–0.6 | 0.3–0.3 | 0–0 |
Cells were grown in YGV for 12 h at 28°C or 10 h at 37°C.
Counts are the mean − standard error of the mean of three independent experiments (n = 100).
The Amino-Terminal Half of SEPA Is Sufficient to Localize GFP to Septation Sites and Hyphal Tips
We hypothesized that different domains within SEPA might direct it to septation sites and the hyphal tip. To begin defining such domains, we made SEPA::GFP constructs that contained the first 132 or the first 838 amino acids of SEPA fused to GFP (see MATERIALS AND METHODS; Figure 8A). The longer protein (in strain AKS82) localized to both septation sites and hyphal tips, indicating that the amino-terminal half of SEPA (including the FH3 domain), can direct SEPA to each of these locations (Figure 8, B and C). In contrast, the shorter protein failed to localize to specific sites and remained cytoplasmic (Figure 8D).
Interestingly, the pattern of GFP localization at hyphal tips in AKS82 cells was different from that of full-length SEPA::GFP. Specifically, the crescents were broader, and no bright spots are observed in AKS82 cells (Figure 8C; cf. Figure 4B). Thus, the carboxy terminus of SEPA, which includes the FH1 and FH2 domains, is required for the formation of SEPA spots and for confining SEPA localization within the hyphal tip. In contrast, it appears to be dispensable for localization to septation sites.
Notably, although strain AKS82 still contains a wild-type sepA gene, its cells are also delayed in septation. After 12 h of growth in YGV, hyphae expressing full-length sepA::gfp (AKS70) and AKS82 hyphae are similar in size and have undergone a similar number of mitoses (our unpublished results). However, the majority (60 ± 11%) of AKS70 hyphae have undergone septation, whereas only a minority (8 ± 5%) of AKS82 hyphae have septated. After 16 h, all hyphae of both strains possess septa. The ASK82 hyphae are also wider (3.7 ± 0.1 μm; n = 20) than AKS70 and wild-type (A28) hyphae (3.1 ± 0.1 μm; n = 20). Presumably, the amino-terminal half of SEPA out-competes endogenous SEPA for binding to a protein required for SEPA function in both septation and maintenance of the appropriate hyphal width.
DISCUSSION
SEPA Is Required for Septation and Actin Ring Formation
In this study, we constructed a sepA disruption strain (sepA6ΔFH) that eliminates additional sequence compared with the previously reported deletion mutant (sepA4ΔBm; Harris et al., 1997). We have found that sepA6ΔFH mutants fail to septate and can only form tiny colonies that lack conidia. In contrast, sepA4ΔBm mutants undergo septation after a lengthy delay and fail to produce normal colonies only at higher temperatures (Harris et al., 1997). We therefore conclude that sepA is required for the formation of septa, but is not essential for vegetative growth in A. nidulans. This phenotype is similar to that described for the other late-acting sep mutants in A. nidulans (i.e., sepD, sepG, and sepH; Harris et al., 1994; Bruno et al., 2001). It is not known how sepA4ΔBm mutants are able to septate despite missing the 5′ half of the gene. It is possible that residual expression of the 3′ half of the gene is sufficient to allow septation.
By analyzing the phenotype of sepA6ΔFH, we have found that SEPA is required for the formation of actin rings at septation sites. This observation is consistent with the known roles of other formins in promoting cytokinesis (Wasserman, 1998). In contrast, actin localizes to the hyphal tip in the absence of SEPA, although the polarity defects observed in sepA mutants suggest that the tip-associated actin may not function in a normal manner. Accordingly, although other pathways may be available for recruiting actin to the hyphal tip, we propose that SEPA is required for the overall fidelity of actin function at this site.
Dynamic Localization of SEPA at Septation Sites and Hyphal Tips
In this study, we have found that SEPA localizes to septation sites and hyphal tips. Simultaneous localization to distinct subcellular sites is a unique feature of SEPA that is not shared by other fungal formins. The localization pattern mirrors that of actin, which is also located simultaneously at septation sites and hyphal tips in A. nidulans (Harris, 1997). In contrast, in yeast cells, actin relocalizes from cell tips to division sites concomitant with septation. Accordingly, yeast formins either localize to sites of cytokinesis or polarized growth, but not both at the same time (Chang et al., 1997; Petersen et al., 1998; Ozaki-Kuroda et al., 2001). The ability of SEPA to simultaneously organize distinct actin structures at different sites implies that its localization is subject to strict spatial and temporal control.
Using live imaging, we have determined the dynamics of SEPA at both septation sites and hyphal tips. At septation sites, the SEPA ring forms quickly after the initial appearance of an asymmetric SEPA spot. The presence of coiled-coil regions in SEPA (Harris et al., 1997) suggests that the spot may contain multimers that subsequently organize into a higher order ring structure. In S. pombe, Cdc12p also appears as a cortical spot before forming a medial ring (Chang, 1999). However, unlike SEPA, the Cdc12p spot is mobile and persists for a much longer period of time.
Like Cdc12p rings (Chang et al., 1997), SEPA rings constrict coincident with the deposition of septal wall material. In contrast, Bni1p rings do not constrict during septation in S. cerevisiae (Ozaki-Kuroda et al., 2001). Regardless of whether formin ring structures undergo constriction during septum formation, they are likely to guide the formation and subsequent contraction of actomyosin rings at septation sites (Vallen et al., 2000). The colocalization of SEPA and actin rings at all stages of constriction is consistent with this notion. The contraction of actomyosin rings is presumed to guide membrane insertion and wall deposition during septum formation (Vallen et al., 2000). However, it also remains possible that ring dynamics are driven by the force of centripetal cell wall deposition.
Although SEPA is found transiently in a ring structure during septation, it localizes continuously at sites of polarized growth. The dynamic localization of SEPA at hyphal tips generally correlates with the direction of tip extension. Similarly, in S. cerevisiae, the location of Bni1p in bud tips corresponds with the direction of bud growth (Ozaki-Kuroda et al., 2001). The pattern of localization at hyphal tips is a crescent usually subtended by a bright spot. The crescent of SEPA appears to be located at hyphal tips in a thin subcortical layer, whereas the bright spot is found just behind or within the crescent. The spot may colocalize with a dense collection of vesicles found at hyphal tips known as the Spitzenkörper, which is thought to be an organizing center for vesicles that are targeted to the growing tip (Bartnicki-Garcia et al., 1989). Notably, movement of the Spitzenkörper has also been shown to correlate with changes in the direction of hyphal extension (Riquelme et al., 1998).
Actin Filaments Function Interdependently with SEPA
We have found that actin filaments are required to maintain the proper pattern of SEPA localization at septation sites and hyphal tips. Specifically, treatment of hyphae with cytochalasin A causes preexisting SEPA structures to collapse into bead-like patches that remain at those sites. In contrast, maintenance of SEPA structures is not affected by the loss of microtubule integrity. Because SEPA is required for actin ring formation and is presumably involved in organizing actin structures at the hyphal tip, these observations suggest that actin filaments and SEPA function in an interdependent manner. For example, profilin, or another actin-associated protein capable of interacting with SEPA, may promote or stabilize interactions between SEPA and filamentous actin. Alternatively, the septins, a conserved family of GTP-binding proteins capable of forming filaments (Trimble, 1999; Momany et al., 2001), may coordinate the localization of SEPA and actin. The observation that yeast Bni1p displays genetic interactions with a septin supports this notion (Longtine et al., 1996).
SEPH Functions Upstream of SEPA and Actin Ring Formation at Septation Sites
In A. nidulans, signals activating cytokinesis are thought to emanate from mitotic nuclei, which somehow determine and/or activate the septation site (Wolkow et al., 1996). SEPH, by analogy to the role of Cdc7p in S. pombe, may be part of the signaling pathway that determines the timing and/or location of septum formation (McCollum and Gould, 2001). We have shown that SEPH function is required for SEPA localization at septation sites. Consistent with its role upstream of SEPA, sepH strains also do not form actin rings at restrictive temperature (our unpublished results; Bruno et al., 2001). Based on these observations, we suggest that SEPH may function in a regulatory pathway that coordinates actin ring formation with the events of mitosis. Notably, the role of SEPH is different than that of its orthologs, S. pombe Cdc7p and S. cerevisiae Cdc15p. Neither Cdc7p nor Cdc15p control actin ring formation; rather, both proteins are components of pathways that regulate actin ring constriction (McCollum and Gould, 2001).
The Amino-Terminal Half of SEPA Localizes to Septation Sites and as a Wider Crescent at Hyphal Tips
We have found that SEPA is targeted to septation sites and hyphal tips through sequences in its amino terminus. Because these sequences include the entire FH3 domain, we propose that it is primarily responsible for targeting GFP to these sites (Petersen et al., 1998). Moreover, our observation implies that the protein(s) that binds the FH3 domain is also located at septation sites and hyphal tips. Other domains of SEPA may modify the localization pattern. Consistent with this idea, we have found that the amino-terminal half of SEPA is not sufficient to direct GFP to a tight crescent at hyphal tips or to form the subtending bright spot. Presumably, interactions between the FH1 or FH2 domains and localized components of the actin cytoskeleton facilitate the recruitment of SEPA to specific sites. This may also explain the ability of sepA4ΔBm hyphae, which possess only the FH1 and FH2 domains, to septate after a lengthy delay. In addition, we have found that expression of the amino-terminal half of SEPA delays septation and causes hyphae to appear wider than normal. These dominant negative effects may occur because the amino-terminal half of SEPA interferes with the ability of endogenous SEPA to interact with an FH3-binding protein. Similarly, it has been found that overexpression of formin FH3 constructs in S. pombe and in human macrophages interferes with their normal function (Petersen et al., 1998; Yayoshi-Yamamoto et al., 2000).
Summary
Our findings support a model in which SEPA acts in a multiprotein complex to control the dynamic assembly and disassembly of functionally distinct actin structures. We suggest that the recruitment of SEPA to septation sites and hyphal tips is directed by specific morphological cues. Downstream of these cues, we propose that similar pathways involving Rho GTPases and septins ensure that SEPA forms the appropriate dynamic structure. Further analysis of the requirements for SEPA localization should reveal how fungal hyphae simultaneously direct growth at spatially distinct sites.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ron Morris and his laboratory members for the use of their microscope, charge-coupled device, and computer software to obtain the live imaging sequences. We also thank John Pringle and Peter Kraus for constructive comments that greatly improved the manuscript. This work was supported by a grant from the National Science Foundation (MCB-9723711).
Abbreviations used:
- aa
amino acid
- GFP
green fluorescent protein
- ts
temperature-sensitive
Footnotes
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–07-0356. Article and publication date are at www.molbiolcell.org/cgi/10.1091/mbc.01–07-0356.
REFERENCES
- Afshar K, Stuart B, Wasserman SA. Functional analysis of the Drosophila Diaphanous FH protein in early embryonic development. Development. 2000;127:1887–1897. doi: 10.1242/dev.127.9.1887. [DOI] [PubMed] [Google Scholar]
- Ayscough KR. In vivo functions of actin-binding proteins. Curr Opin Cell Biol. 1998;10:102–111. doi: 10.1016/s0955-0674(98)80092-6. [DOI] [PubMed] [Google Scholar]
- Bartnicki-Garcia S, Hergert F, Gierz G. Computer simulation of fungal morphogenesis and the mathematical basis for hyphal (tip) growth. Protoplasma. 1989;153:46–57. [Google Scholar]
- Beckerle MC. Spatial control of actin filament assembly: lessons from Listeria. Cell. 1998;95:741–748. doi: 10.1016/s0092-8674(00)81697-9. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Chan DC, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Regulation of cortical structure by the ezrin-radixin-moesin protein family. Curr Opin Cell Biol. 1999;11:109–116. doi: 10.1016/s0955-0674(99)80013-1. [DOI] [PubMed] [Google Scholar]
- Bruno KS, Morrell JL, Hamer JE, Staiger CS. SEPH, a Cdc7p ortholog from Aspergillus nidulans, functions upstream of actin ring formation during cytokinesis. Mol Microbiol. 2001;42:3–12. doi: 10.1046/j.1365-2958.2001.02605.x. [DOI] [PubMed] [Google Scholar]
- Chan DC, Bedford MT, Leder P. Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- Chang F. Movement of a cytokinesis factor Cdc12p to the site of cell division. Curr Biol. 1999;9:849–852. doi: 10.1016/s0960-9822(99)80372-8. [DOI] [PubMed] [Google Scholar]
- Chang F, D. Drubin D, Nurse P. Cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Bernstein BW, Bamburg JR. Regulating actin-filament dynamics in vivo. Trends Biochem Sci. 2000;25:19–23. doi: 10.1016/s0968-0004(99)01511-x. [DOI] [PubMed] [Google Scholar]
- Emmons S, Phan H, Calley J, Chen W, James B, Manseau L. cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 1995;9:2482–2494. doi: 10.1101/gad.9.20.2482. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Fernandez-Abalos JM, Fox H, Pitt C, Wells B, Doonan JH. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus Aspergillus nidulans. Mol Microbiol. 1998;27:121–130. doi: 10.1046/j.1365-2958.1998.00664.x. [DOI] [PubMed] [Google Scholar]
- Frazier JA, Field CM. Actin cytoskeleton: are FH proteins local organizers? Curr Biol. 1997;7:R414–R417. doi: 10.1016/s0960-9822(06)00205-3. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implications in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SD. The duplication cycle in Aspergillus nidulans. Fungal Genet Biol. 1997;22:1–13. doi: 10.1006/fgbi.1997.0990. [DOI] [PubMed] [Google Scholar]
- Harris SD, Hamer L, Sharpless KE, Hamer JE. The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J. 1997;16:3474–3483. doi: 10.1093/emboj/16.12.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SD, Hofmann AF, Tedford HW, Lee MP. Identification and characterization of genes required for hyphal morphogenesis in the filamentous fungus Aspergillus nidulans. Genetics. 1999;151:1015–1025. doi: 10.1093/genetics/151.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SD, Morrell JL, Hamer JE. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics. 1994;136:517–532. doi: 10.1093/genetics/136.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SW, et al. nimO, an Aspergillus gene related to budding yeast Dbf4, is required for DNA synthesis and mitotic checkpoint control. J Cell Sci. 1999;112:1313–1324. doi: 10.1242/jcs.112.9.1313. [DOI] [PubMed] [Google Scholar]
- Kafer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. J Biol Chem. 1998;273:28341–28345. doi: 10.1074/jbc.273.43.28341. [DOI] [PubMed] [Google Scholar]
- Kikyo M, Tanaka K, Kamei T, Ozaki K, Fujiwara T, Inoue E, Takita Y, Ohya Y, Takai Y. An FH domain-containing Bnr1p is a multifunctional protein interacting with a variety of cytoskeletal proteins in Saccharomyces cerevisiae. Oncogene. 1999;18:7046–7054. doi: 10.1038/sj.onc.1203184. [DOI] [PubMed] [Google Scholar]
- Kohno, et al. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Manseau L, Calley J, Phan H. Profilin is required for posterior patterning of the Drosophila oocyte. Development. 1996;122:2109–2116. doi: 10.1242/dev.122.7.2109. [DOI] [PubMed] [Google Scholar]
- McCollum D, Gould KL. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- Miller BL, Miller KY, Roberti KA, Timberlake WE. Position-dependent and -independent mechanisms regulate cell-specific expression of the SpoC1 gene cluster of Aspergillus nidulans. Mol Cell Biol. 1987;7:427–434. doi: 10.1128/mcb.7.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany M, Hamer JE. Relationship of actin, microtubules, and crosswall synthesis during septation in Aspergillus nidulans. Cell Motil Cytoskeleton. 1997;38:373–384. doi: 10.1002/(SICI)1097-0169(1997)38:4<373::AID-CM7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Momany M, Zhao J, Lindsey R, Westfall PJ. Characterization of the Aspergilus nidulans septin (asp) gene family. Genetics. 2001;157:969–977. doi: 10.1093/genetics/157.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris NR. Mitotic mutants of Aspergillus nidulans. Genet Res. 1976;26:237–254. doi: 10.1017/s0016672300016049. [DOI] [PubMed] [Google Scholar]
- Mullins RD. How WASP-family proteins, and the Arp2/3 complex convert intracellular signals into cytoskeletal structures. Curr Opin Cell Biol. 2000;12:91–96. doi: 10.1016/s0955-0674(99)00061-7. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Osmani SA. Cell cycle analysis using the filamentous fungus Aspergillus nidulans. In: Fantes P, Brooks R, editors. The Cell Cycle: A Practical Approach. New York, NY: IRL Press; 1993. pp. 127–142. [Google Scholar]
- Ozaki-Kuroda K, Yamamoto Y, Nohara H, Kinoshita M, Fujiwara T, Irie K, Takai Y. Dynamics localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:827–839. doi: 10.1128/MCB.21.3.827-839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Nielsen O, Egel R, Hagan IM. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J Cell Biol. 1998;141:1217–1228. doi: 10.1083/jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Weilguny D, Egel R, Nielsen O. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol Cell Biol. 1995;15:3697–3707. doi: 10.1128/mcb.15.7.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh N, Anton IM, Martinez-Quiles N, Geha RS. Waltzing with WASP. Trends Cell Biol. 1999;9:15–19. doi: 10.1016/s0962-8924(98)01411-1. [DOI] [PubMed] [Google Scholar]
- Riquelme M, Reynaga-Pena CG, Gierz G, Bartnicki-Garcia S. What determines growth direction in fungal hyphae? Fungal Genet Biol. 1998;24:101–109. doi: 10.1006/fgbi.1998.1074. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmidt A, Hall MN. Signaling to the actin cytoskeleton. Annu Rev Cell Dev Biol. 1998;14:305–338. doi: 10.1146/annurev.cellbio.14.1.305. [DOI] [PubMed] [Google Scholar]
- Swan KA, Severson AF, Carter JC, Martin PR, Schnabel H, Schnabel R, Bowerman B. cyk-1: a C. elegans FH gene required for a late step in embryonic cytokinesis. J Cell Sci. 1998;111:2017–2027. doi: 10.1242/jcs.111.14.2017. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Takai Y. Control of reorganization of the actin cytoskeleton by Rho family small GTP-binding proteins in yeast. Curr Opin Cell Biol. 1998;10:112–116. doi: 10.1016/s0955-0674(98)80093-8. [DOI] [PubMed] [Google Scholar]
- Timberlake WE. Molecular genetics of Aspergillus development. Annu Rev Genet. 1990;24:5–36. doi: 10.1146/annurev.ge.24.120190.000253. [DOI] [PubMed] [Google Scholar]
- Torralba S, Raudaskoski M, Pedregosa AM, Laborda F. Effect of cytochalasin A on apical growth, actin cytoskeleton organization and enzyme secretion in Aspergillus nidulans. Microbiology. 1998;144:45–53. doi: 10.1099/00221287-144-1-45. [DOI] [PubMed] [Google Scholar]
- Trimble WS. Septins: a highly conserved family of membrane-associated GTPases with functions in cell division and beyond. J Membr Biol. 1999;169:75–81. doi: 10.1007/s002329900519. [DOI] [PubMed] [Google Scholar]
- Uetz P, Fumagalli S, James D, Zeller R. Molecular interaction between limb deformity proteins (formins) and Src family kinases. J Biol Chem. 1996;271:33525–33530. doi: 10.1074/jbc.271.52.33525. [DOI] [PubMed] [Google Scholar]
- Umikawa M, Tanaka K, Kamei T, Shimizu K, Imamura H, Sasaki T, Takai Y. Interaction of Rho1p target Bni1p with F-actin-binding elongation factor 1α: implication in Rho1p-regulated reorganization of the actin cytoskeleton in Saccharomyces cerevisiae. Oncogene. 1998;16:2011–2016. doi: 10.1038/sj.onc.1201724. [DOI] [PubMed] [Google Scholar]
- Vallen EA, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring RB, May GS, Morris NR. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-encoding genes. Gene. 1989;79:119–130. doi: 10.1016/0378-1119(89)90097-8. [DOI] [PubMed] [Google Scholar]
- Wasserman S. FH proteins as cytoskeletal organizers. Trends Cell Biol. 1998;8:111–115. doi: 10.1016/s0962-8924(97)01217-8. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkow TD, Harris SD, Hamer JE. Cytokinesis in Aspergillus nidulans is controlled by cell size, nuclear positioning and mitosis. J Cell Sci. 1996;109:2179–2188. doi: 10.1242/jcs.109.8.2179. [DOI] [PubMed] [Google Scholar]
- Xiang X, Han G, Winkelmann DA, Zuo W, Morris NR. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr Biol. 2000;10:603–606. doi: 10.1016/s0960-9822(00)00488-7. [DOI] [PubMed] [Google Scholar]
- Yayoshi-Yamamoto S, Taniuchi I, Watanabe T. FRL, a novel formin-related protein, binds to Rac and regulates cell motility and survival of macrophages. Mol Cell Biol. 2000;20:6872–6881. doi: 10.1128/mcb.20.18.6872-6881.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R, Haramis AG, Zuniga A, McGuigan C, Dono R, Davidson G, Chabanis S, Gibson T. Formin defines a large family of morphoregulatory genes and their functions in establishment of the polarising region. Cell Tissue Res. 1999;296:85–93. doi: 10.1007/s004410051269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.