Abstract
Otopetrins (Otop1–Otop3) comprise one of two known eukaryotic proton-selective channel families. Otop1 is required for otoconia formation and a candidate mammalian sour taste receptor. Here we report cryo-EM structures of zebrafish Otop1 and chicken Otop3 in lipid nanodiscs. The structures reveal a dimeric architecture, with each subunit forming twelve transmembrane helices divided into structurally similar amino (N) and carboxy (C) domains. Cholesterol-like molecules occupy various sites in Otop1 and Otop3 and occlude a central tunnel. In molecular dynamics simulations, hydrophilic vestibules formed by the N and C domains and in the intrasubunit interface between N and C domains form conduits for water entry into the membrane core, suggesting three potential proton conduction pathways. By mutagenesis, we test the roles of charged residues in each putative permeation pathway. Our results provide a structural basis for understanding selective proton permeation and gating of this conserved family of proton channels.
Proton channels mediate the passage of protons across cell membranes, thereby regulating the cellular and extracellular pH as well as membrane potential1. The diverse biological roles of proton channel activity include the triggering of bioluminescence in dinoflagellates2, regulation of pH in lung epithelia3, and the detection of sour taste4–6. Knowledge of molecular mechanisms of proton conduction and selectivity is largely derived from, and limited to, the M2 proton channel of influenza7 and the eukaryotic voltage-gated Hv1 proton channel8,9. Recently, Otopetrins were identified as a novel family of eukaryotic proton channels,10 with no structural similarity to M2, Hv1 or other ion channels. Mice have three related genes (Otop1, Otop2 and Otop3) that encode channels with distinct biophysical properties. For example, the current amplitudes of mouse Otop1 and mouse Otop3 increase linearly as a function of extracellular pH over a range of pH 6 – pH 4, while the current of mouse Otop2 saturates at ~pH 510. Otop1 has been demonstrated to be proton-selective, with a remarkable > 2 × 105-fold selectivity for protons over Na+ 10.
Because they were only recently characterized, the physiological roles of Otopetrins are just now beginning to be uncovered. Notably, Otop1 is required for proton currents in murine cells that detect sour taste and is a likely candidate for a sour taste receptor10. In addition, prior genetic studies identified Otop1 as the gene mutated in a spontaneously occurring vestibular disorder in mice and required in the vestibular system for normal otoconia development in mice11 and zebrafish12,13. Otop1 is also highly expressed in brown and white adipose tissue and plays a role in insulin resistance14. Otop2 and Otop3 have been detected in the digestive tract and elsewhere in the body10,15,16 suggesting yet unappreciated roles for proton conduction in various cell types. The recent characterization of Otopetrins as proton channels has opened avenues to decipher their physiological, biophysical, and biochemical characteristics.
Results
Otopetrin channel function, overall structure and domain organization
To provide a starting point for mechanistic studies on Otopetrins, we used fluorescence detection size exclusion chromatography17 (FSEC) to identify orthologs suitable for structure determination when expressed as N-terminal GFP fusions (see methods). We chose zebrafish Otop1 and chicken Otop3, which are 30% identical to each other by sequence and share 44% and 59% identity with human OTOP1 and OTOP3, respectively (Supplementary Fig. 1). When expressed in HEK-293 cells, both zebrafish Otop1 and chicken Otop3 conduct proton currents in response to lowering the extracellular pH like their mammalian counterparts10 (Fig. 1a, Supplementary Fig. 2). Hereafter we refer to these proteins as Otop1 and Otop3, respectively.
Figure 1. Proton channel function and structures of Otop1 and Otop3 homodimers in lipid nanodiscs.
a, currents measured by whole-cell patch clamp recording in HEK-293 cells expressing zfOtop1 (top) or chOtop3 (bottom) in response to acidic extracellular solutions with the indicated pH (pHi = 7.4, Vm = −80 mV). Inset of top panel shows inhibition of zfOtop1 currents by 1 mM Zn2+ (pink bar). b, orthogonal views of sharpened cryo-EM maps of zfOtop1. One subunit is colored dark and light shades of purple, while the other subunit is colored green and light green. Each subunit has a structurally homologous N domain (dark shade) and C domain (light shade). The center of the dimer contains a tunnel occupied by cholesterol-like densities. Putative cholesterol and lipid densities are colored light orange. c, orthogonal views of sharpened cryo-EM maps of chOtop3, colored as in b. d, side (left) and top (right) views of model of Otop1 dimer showing TM helices as cylinders. One subunit is colored gray while in the other subunit each of the twelve TM helices are colored differently and numbered. In the right panel, the central two-fold symmetry axis is depicted as a black oval, and pseudosymmetry axes relating the N domain and C domain within each subunit are depicted as empty red ovals. e, schematic of Otop1 structure, with TM helices colored corresponding to c. Loops not modeled due to poor density are depicted as dashed gray lines. f, structural alignment of the N domain (red) and C domain (blue).
We purified and reconstituted full-length proteins in lipid nanodiscs and determined C2-symmetric structures at overall resolutions of 3.0 Å for Otop1 and 3.3 Å for Otop3 (Fig. 1b,c, Supplementary Figs. 3, 4, Table 1). The quality of the cryo-EM maps was sufficient to define side chain orientation of almost all amino acids in the transmembrane helices (Supplementary Fig. 5). Notably, there are numerous lipid-like densities surrounding the channels or embedded in an internal tunnel at the dimer interface (Fig. 1b, c). These lipids were either carried over from cells or added during purification and nanodisc reconstitution. Of these, two densities adopted distinctive shapes and were modeled as cholesterol while an additional six densities corresponded to cholesteryl hemisuccinate (CHS) in the Otop1 map, whereas two CHS molecules were built into the Otop3 map (Supplementary Fig. 5c–f).
Cryo-EM data collection, refinement and validation statistics
| zfOtop1 in nanodiscs (EMDB 9360, PDB 6NF4) |
chOtop3 in nanodiscs (EMDB 9361, PDB 6NF6) |
|
|---|---|---|
| Data collection and processing | ||
| Magnification | 29,000 | 29,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e−/Å2) | 50 | 50 |
| Defocus range (μm) | 0.6–1.8 | 0.6–1.8 |
| Pixel size (Å) | 1.03 | 1.03 |
| Symmetry imposed | C2 | C2 |
| Initial particle images (no.) | 1,517,125 | 1,386,058 |
| Final particle images (no.) | 67,425 | 43,667 |
| Map resolution (Å) | 2.98 | 3.32 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | ||
| Refinement | ||
| Initial model used (PDB code) | N/A | 6NF4 |
| Model resolution (Å) | 3.06 | 3.35 |
| FSC threshold | 0.5 | 0.5 |
| Model resolution range (Å) | 2.9–8 | 3.3–8 |
| Map sharpening B factor (Å2) | −74 | −50 |
| Model composition | ||
| Nonhydrogen atoms | 6,970 | 6,600 |
| Protein residues | 842 | 822 |
| Ligands | 8 | 2 |
| B factors (Å2) | ||
| Protein | 24.8 | 53.8 |
| Ligand | 71.8 | 49.2 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.01 | 0.02 |
| Bond angles (°) | 1.00 | 0.95 |
| Validation | ||
| MolProbity score | 1.00 | 1.56 |
| Clashscore | 1.08 | 4.54 |
| Poor rotamers (%) | 0.27 | 0 |
| Ramachandran plot | ||
| Favored (%) | 96.8 | 95.2 |
| Allowed (%) | 3.2 | 4.5 |
| Disallowed (%) | 0 | 0.3 |
The structures show that Otop1 and Otop3 are cuboid-shaped homodimers with dimensions of roughly 70 Å x 50 Å x 50 Å, and almost all of their ordered mass resides within the membrane (Fig. 1b, c). A single Otopetrin subunit can be divided into two halves, the N (amino-terminal) and C (carboxy-terminal) domains. In the homodimeric arrangement, the N and C domains from two subunits occupy four quadrants surrounding a central axis, resulting in a pseudotetrameric organization (Fig. 1b, c). A cavernous tunnel containing lipids and/or cholesterols coincides with the central two-fold axis of Otop1 and Otop3. Intersubunit and intrasubunit interfaces between N and C domains appear to stabilize the overall assembly (Fig. 1b,c).
The topological organization of Otop1 (Fig. 1d) is shared by Otop3 (Supplementary Fig. 6a), and therefore likely to be adopted among all Otopetrins. Sequence conservation of Otopetrins mapped onto the structural model is shown in Supplementary Fig. 6b–d. Each Otop1 subunit contains twelve transmembrane helices (TM1–TM12) of which the first six (TM1–6) form the N domain while the remaining six (TM7–TM12) form the C domain. The N and C domains are structurally similar and are related by a pseudo two-fold symmetry axis at the intrasubunit interface (Fig. 1d–f). This domain organization is reminiscent of, but distinct from, the major facilitator superfamily (MFS) transporters18,19 and resistance-nodulation-cell division (RND) family of membrane proteins20 (Supplementary Fig. 6e–g). The intrasubunit interface of Otop1 is mediated mainly by TM6 and TM12, and is highly conserved, suggesting a critical role in structural and functional integrity (Supplementary Figs. 6b, 7a–d). The intersubunit interface, mediated by TM1 and TM9, is less conserved between Otop1 and Otop3 and across Otopetrins (Supplementary Figs. 6c, 7e–h). Two tryptophan residues (W394 and W398) in TM9 of zebrafish Otop1 are buried at the interface. FSEC analysis of the W394A and W398A mutants indicated that each express as monomeric species (Supplementary Fig. 7i). These monomeric mutants also resulted in loss of proton channel function, due to apparently impaired surface expression (Supplementary Fig. 7j–m). Therefore, bulky hydrophobic residues at the intersubunit interface stabilize dimer assembly, and surface trafficking likely requires dimerization.
Lipids including cholesterols in Otop1 and Otop3
Facilitated by nanodisc reconstitution, numerous annular and bound lipids were captured and resolved in the structures of Otop1 and Otop3, revealing how the molecules interact with the membrane (Fig. 2). Most remarkably, the central tunnel of Otop1 harbors six robust densities that were modeled as CHS molecules (Fig. 2a, b, Supplementary Fig. 5c, d). Otop3 has a distinct arrangement of lipids in the central tunnel, including two identifiable CHS molecules (Fig. 2a,d, Supplementary Fig. 5e, f). CHS, which was added during purification of Otop1 and Otop3, greatly enhances the monodispersity of detergent-solubilized proteins as shown by FSEC (Supplementary Figs. 3a, 4a). In native bilayers, cholesterol or cholesterol-like lipids likely bind in the central tunnel to stabilize the dimeric protein assembly21,22. To test this, we performed all-atom molecular dynamics (MD) simulations on both Otop1 and Otop3 with cholesterol molecules placed in the central tunnel (and at the intrasubunit interface for Otop1, see also below and Methods). The arrangement of cholesterol molecules in the tunnel is maintained throughout the sub-microsecond MD simulations, indicating that they represent stable binding poses for cholesterol-like moieties (Supplementary Fig. 8a–d). Indeed, in simulations of Otop1 from which cholesterol was omitted, we observed considerable conformational drift (Supplementary Fig. 8e–g) and unrestricted diffusion of water and ions through the wide central tunnel. These together demonstrate how bound cholesterol molecules may contribute to the stability of the dimeric assembly and exclude the passage of unwanted water and solutes.
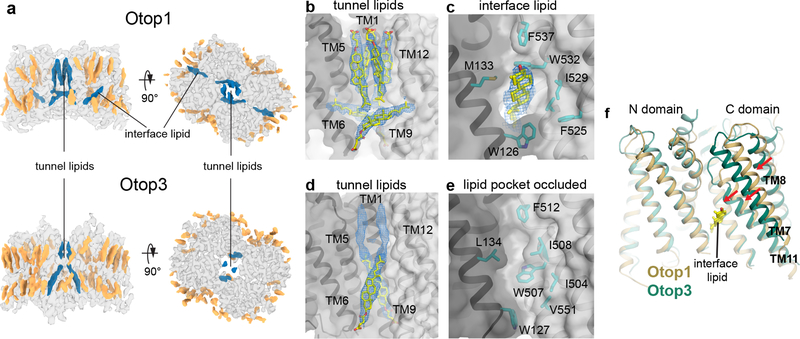
Figure 2. Lipid binding in Otop1 and Otop3.
a side (left) and top (right) views of cryo-EM map of Otop1 (top) and Otop3 (bottom). Protein density is transparent gray, annular lipid densities are colored gold, and lipid/cholesterol densities at the N domain-C domain intrasubunit interface and lipid tunnel are colored dark blue. b, densities (blue mesh, 3σ) corresponding to three CHS molecules (yellow and red sticks) in the lipid tunnel are apparent in Otop1. c, Density (blue mesh, 3σ) corresponding to a cholesterol molecule is also present at the intrasubunit interface. d, densities corresponding to a CHS molecule (blue mesh, 3σ) and an unassigned lipid (blue mesh, 3σ) are present in the Otop3 lipid tunnel. e, there is no lipid at the intrasubunit interface in Otop3, and the pocket is occluded. f, superimposition of Otop1 and Otop3 single subunits based on the N-domain shows that TM7, TM8, and TM11 of Otop3 is shifted relative to Otop1, likely due to absence of interface lipid.
(please relabel figure 2, so panel a, d are simply parts of panel a; otherwise, we’d have to rewrite the legend completely)
At the intrasubunit interface of Otop1, a well-resolved cholesterol-like density is sandwiched in a pocket between the N and C domains in the lower leaflet (Fig. 2c). In contrast to those in the central tunnel, the interfacial cholesterols show greater mobility and in most cases they reposition themselves during simulations (Supplementary Fig. 8a, c). This lipid is absent in the Otop3 density map, despite high sequence conservation in this region (Supplementary Fig. 6b). Instead, the pocket is occluded by a conserved tryptophan (W507 in Otop3), resulting in a shift of the C-domain relative to the N-domain (Fig. 2e,f). Occupancy of this interface lipid site therefore appears to influence the conformation of Otopetrins. Overall, these observations underscore the importance of lipids, in particular cholesterol-like molecules, in shaping Otopetrin structure, and suggest that lipids play a central role in dimer stability, function, and/or membrane localization. The differences in lipid-binding observed when comparing Otop1 and Otop3 suggest that lipid modulation is subtype-specific.
Putative pores in the N and C domains
Protons are believed to transfer across membrane proteins by ‘hopping’ along a hydrogen-bonded network consisting of water molecules and/or amino acid moieties1,23–27. The proton conduction pathway(s) in Otopetrins would thus likely be at least partially hydrated. Two structurally analogous vestibule-shaped openings in each Otop1/Otop3 subunit could represent loci for proton permeation, one housed by the N domain and the other by the C domain (Fig. 3). The N domain vestibule is lined by TM2–TM6 (Fig. 3a), while the C domain vestibule is lined by TM8–TM12 (Fig. 3b). Both of the vestibules contain numerous polar and charged residues, many of which are conserved (Fig. 3a, b). Below the bilayer midpoint lies a region of hydrophobic residues that constrict the N and C domain vestibules in both Otop1 and Otop3, potentially serving as hydrophobic ‘plugs’ that regulate water or ion accessibility. Notably, hydrophobic gates are exploited across many ion channel families, including Hv128,29.
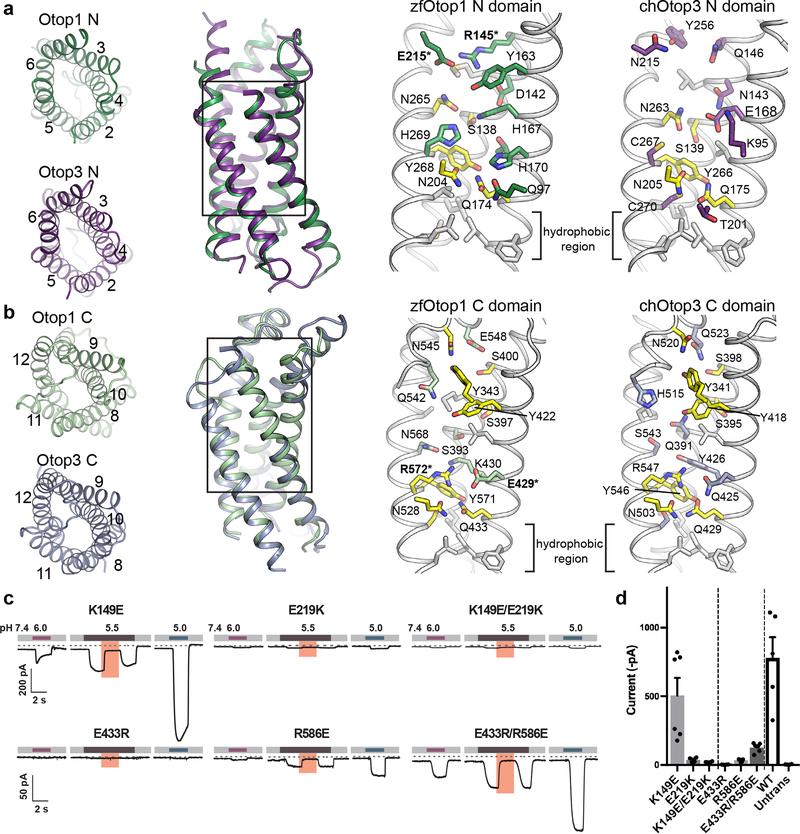
Figure 3. N and C domain vestibules Otop1 and Otop3.
a, various views of Otop1 N domain and Otop3 N domain. Top views in which TM1 is omitted (far left panels) are shown, as well as side view (middle left) of the aligned N domains of Otop1 (green) and Otop3 (purple). Middle right and far right panels show expanded views, with residues conserved between Otop1 and Otop3 colored yellow. b, various views of Otop1 and Otop3 C domains, with panels arranged as in a. TM7 is omitted from the top views. In both a and b, residue numbering refers to zebrafish Otop1 and chicken Otop3. c, currents measured by whole-cell patch clamp recording in HEK-293 cells expressing each mouse Otop1 mutant in response to acidic extracellular solutions with pH indicated (pHi = 7.4, Vm = −80 mV). The currents elicited in response to pH 5.5 were inhibited by 1 mM Zn2+ (pink bars). Numbering is according to the mouse Otop1 sequence. The residues in zfOtop1 that are equivalent to those mutated in mOtop1 are bolded and marked with an asterisk in panels a and b (K149 in mOtop1 corresponds to R145 in zfOtop1; E219 = E215; E433 = E429; R586 = R572). d, averaged current magnitude in response to pH 5.0 from experiments as in the left panel. Individual data points, means and s.e.m are shown, for K149E (n=6 cells), E219K (n=4) , K149E/E219K (n=4), E433R(n=4), R586E(n=4), E433R/R586E (n=6), WT (n=5), and untransfected (n=4). Current magnitudes for K149E/E219K were not significantly different compared to E219K (P=0.17, but they were significantly smaller compared with K149E (P=0.013, two-tailed Student’s T-test). Currents magnitudes for E433R/R586E were significantly larger as compared with E433R and R586E (P= 0.0003 and P=0.0006, respectively, by two-tailed Student’s T-test).
In Hv1, an arginine-aspartic acid salt bridge has been proposed to function as a ‘selectivity filter’ in the proton conduction pathway27,30. Each of the vestibules of Otop1 also contains an apparent salt bridge: R145-E215 (K149-E219 in mouse Otop1) in the N domain vestibule (Fig. 3a) and E429-R572 (E433-R586 in mOtop1) in the C domain vestibule (Fig. 3b). To test the functional significance of these putative salt bridges, we introduced charge reversal mutations in mOtop1 and tested their proton channel activity (Fig. 3c,d). The K149E mutant functioned similar to wildtype, while currents were greatly reduced in the E219K mutant, and not ‘rescued’ in the K149E/E219K double mutant. These results argue against the functional significance for a salt bridge in the N domain vestibule of Otop1, consistent with the observation that Q146 in Otop3, the equivalent residue to K149 in mOtop1, does not make interactions with neighboring side chains (Fig. 3b). In contrast, similar experiments provide strong support for a functional salt bridge in the C domain; the greatly diminished current amplitude observed from each of the single mutants (E433R, R586E) was partly rescued in the double mutant (E433R/R586E). Therefore, this electrostatic interaction in the C-domain of Otop1 apparently supports proper proton channel function and the exact positioning of the residues is not essential. Indeed, the equivalent residues in Otop3 (Q425, R547) interact with each other in a similar fashion via apparent hydrogen bonding (Fig. 3b), though the functional relevance of this interaction in Otop3 is unknown.
Another highly conserved feature of the putative permeation pathways within the N and C domains of Otop1 and Otop3 is a constriction composed of glutamine-asparagine-tyrosine, which we abbreviate as the QNY triad. (Q174/N204/Y268 and Q433/N528/Y571 in Otop1, Q175/N205/Y266 and Q429/N503/Y546 in Otop3) (Fig. 3a,b, Supplementary Fig. 1). Superposition of the domains shows that these triads occupy analogous positions (Supplementary Fig. 9a, b), and their side chains are close enough to interact directly or through intervening waters (Supplementary Fig. 9c–f). The function of this triad is uncertain, but a role in proton transfer seems possible considering it is conserved in both N and C domains, and across Otopetrin orthologues.
Water penetration of Otop1 in N and C domains
We conducted all-atom molecular dynamics (MD) simulations of Otop1 (Fig. 4) and Otop3 (Supplementary Fig. 10) in a mixed lipid bilayer (80:20 POPC:CHOL) to examine areas susceptible to water penetration and thus to explore potential proton permeation pathways. Though simulations of both subtypes yielded similar results, we focus our discussion on the simulations of Otop1 because of the higher resolution of the starting structure. The N and C domain vestibules both allowed water entry from the extracellular milieu into the membrane plane as well as the intrasubunit interface between N and C domains (Fig. 4a, b), while water permeation through the central tunnel is completely blocked by the cholesterol molecules (Fig. 4c). Close-up views of each of the water entry points are shown in Fig 4d. We observe the stochastic formation and breaking of a water wire in both the N and C domains (Supplementary Data Set 1). The interruption of the water wires occurs in the cytoplasmic half of the pathways, below the QNY triad, in part due to the presence of hydrophobic residues (Fig. 4d). Various hydrophilic side chains, including that of the QNY triad, contact water molecules and facilitate water penetration. The formation of a water wire during MD simulation suggests that proton conduction could occur through a water-hopping mechanism. The average (across three simulations of two subunits) distributions of water molecules along the putative pores (Fig. 4e) show a marked difference in the degree of wetting between the extracellular and cytoplasmic halves of the N and C domain pathways. An extended dryer region in the cytoplasmic half would not be expected to be conducive for proton conduction in the experimentally observed conformation, suggesting this might correspond to a hydrophobic gate region. However, we also note that proton conduction may not require a fully intact water wire31.
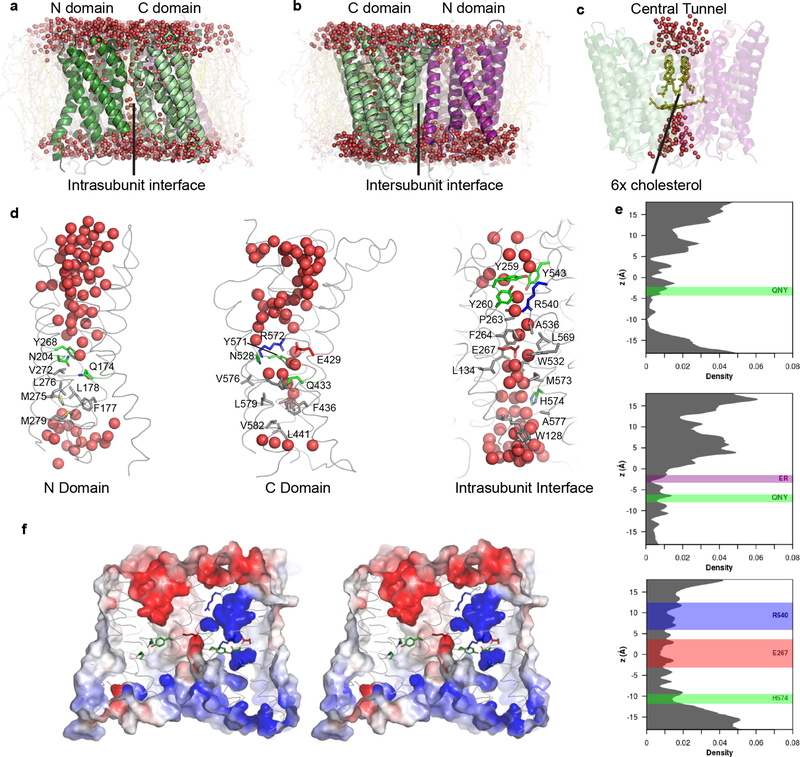
Figure 4. MD simulations reveal hydration of potential proton pathways in Otop1.
a–d, snapshots at the end of a 100-ns all-atom simulation (Supplementary Data Set 1, Run 3; d shows snapshots from subunit 1). A continuous presence of water is observed at the intrasubunit interface (a) but not at the intersubunit interface (b). c, cholesterol molecules in the central tunnel exclude water passage completely. e, the density distributions of water oxygen atoms along the three potential proton pathways, averaged across two subunits and three simulations. Positions of side chains noted in the text are drawn as rectangles, with the bounds representing μ ± 2σ. f, Electrostatic surface potential (contoured from −5 kT (red) to 5 kT (blue)) of the same snapshot in a–d and same view as a, in stereo representation. The surface is clipped ~15 Å from the front to highlight the potential along the putative pores. Highly negative potentials are found in the extracellular half of the N domain, and in the middle of the intrasubunit interface due to the conserved glutamic acid (E429). This is in contrast to the highly positive potential regions in the C domain.
Further, we calculated the electrostatic profile of an Otop1 subunit (Fig. 4f). The extracellular half of the N domain is highly negative throughout, while the C domain transits from negative in the extracellular side to a positive region in the membrane plane. That the N domain is overall more electronegative may suggest that it is more ideal for proton conduction than the C domain. Indeed, it has been proposed for aquaporins that an electropositive region of the permeation pathway is responsible for proton exclusion32. However, we are cautious in making conclusions from surface electrostatic potential because the exact role of electrostatics in in proton conduction remains unclear, and minor movements associated with gating could have a drastic impact on the surface charge distribution.
The Intrasubunit interface is a third putative pore
Surprisingly, the intrasubunit interface flanked by the N and C domains also permitted penetration of water from both sides of the bilayer, resulting in formation of a water wire (Fig. 4d). Most of the residues surrounding the water molecules at this interface are hydrophobic, with the notable exceptions of a glutamate (E267), an arginine (R540) and a histidine (H574) (Fig. 4d). E267 and H574 are highly conserved across Otopetrins (Supplementary Fig. 1), and the E267 side chain produces an electronegative region near the bilayer midplane point of the interface (Fig. 4f). The position of the cholesterol-like molecule at the intrasubunit interface does not appear to be deep enough to occlude the water wire. As noted above, the interactions of the cholesterol molecules in this region here are quite dynamic in our simulations, and the lipid molecules are able to diffuse away. On average the water distribution along the interface is more uniform than that of the N and C domains, without the clear break/hydrophobic gate seen in the latter.
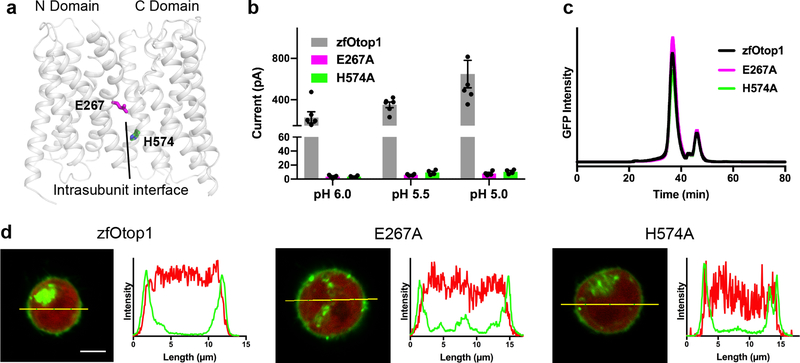
We tested the functional role of the two conserved charged residues (E267 and H574) that are at the intrasubunit interface (Fig. 5a) in zebrafish Otop1 by expressing alanine mutants in HEK cells. Strikingly, both mutants resulted in complete loss of proton channel activity, despite cell surface expression and biochemical behavior that resembled wild-type (Fig. 5b–d). Therefore, E267 and H574 are critical for proper proton channel function in zebrafish Otop1. Overall, these results suggest that the intrasubunit interface presents a third possible avenue for proton conduction in Otop1. That this interface is the most conserved region in Otopetrins (Supplementary Fig. 6b) further underscores its likely functional importance.
Figure 5. Mutation of charged residues at intrasubunit interface results in loss of function.
a, Cartoon model of a single subunit of zebrafish Otop1, with two charged residues (E267 and H574) at the intrasubunit interface shown as sticks. b, Current magnitude at various pH values in HEK-293 cells expressing N-terminally GFP fused zebrafish Otop1 (gray, n = 6), or its mutants E267A (magenta, n = 4) and H574A (green, n = 4), where n is the number of recorded cells. Individual data points, mean and s.e.m. are shown c, FSEC profiles of detergent-solubilized HEK-293 cells expressing the same constructs shown in b. d, confocal images (left panels) showing HEK-293 cells expressing tag-RFP and the Otop1 constructs shown in b, c. Each image is at the same scale, and scale bar (5 μm) is shown in the wild type image. Right panels show fluorescence intensity for GFP (green trace) and RFP (red trace) channels along the yellow line in the images.
Discussion
Despite their broad roles in biology, knowledge of proton channels has lagged behind that of other types of ion channels. The recent characterization of Otopetrins, previously implicated in vestibular function11,12, provides impetus for investigations of a novel proton channel family10. Here we have described cryo-EM structures of Otop1 and Otop3 in lipidic nanodiscs, elucidating the topology and dimeric organization of Otopetrins in a native-like bilayer environment. We also note that a recent study33 showed that detergent-solubilized Otop3 from Xenopus tropicalis adopts the same overall homodimeric architecture as the structures we have described here.
Our structures and MD simulations point to three possible routes for proton conduction per subunit: aqueous vestibules in the N and C domains, and the intrasubunit interface. At this point it remains uncertain which of these three pathways (or their combination) contributes to the proton currents recorded in electrophysiological experiments. We found mutations in each of the putative pores that result in loss of function. but it is unclear whether these phenotypes were caused by direct perturbation of a proton conduction pathway or through allosteric effects.
Despite these advances, clarifying the proton conduction pathway(s), as well as gating and selectivity mechanisms, will require further studies. Our results set the stage for such investigations on a family of proton channels involved in sour taste perception, vestibular function, and other as-yet identified roles.
Methods
Constructs
Zebrafish Otop1 (zfOtop1, Uniprot entry Q7ZWK8) was obtained by PCR from zebrafish whole animal cDNA and confirmed by sequencing. Chicken Otop3 (chOtop3, Uniprot entry R4GK65) was synthesized and codon-optimized for expression in human cell lines. Both cDNAs were cloned into a pcDNA3.1 vector with an N-terminal fusion tag consisting of an octahistidine tag followed by eGFP, a Gly-Thr-Gly-Thr linker, and 3C protease cleavage site (LEVLFQGP). We call these constructs GFP-zfOtop1 and GFP-chOtop3 below. We note that the Uniprot entry for chicken Otop3 used in our study is a 561 amino acid sequence that has been updated since we had it synthesized to a 555 amino acid sequence. The two sequences are identical excepting a short region starting at residue 176. In our chOtop3 the sequence is 176AFFLWHHSKDCIQVQHNLTR195. The corresponding sequence in the updated Uniprot entry is 176VMQIPSWQLTHSL189. In addition, alanine at position 438 in the Uniprot entry is lysine in our sequence of chicken Otop3. The sequence we used is more highly conserved and likely to be correct. Mutations were introduced into the mouse Otop1 cDNA and sequenced as described in Tu et al10.
FSEC and purification screening
Approximately one dozen Otopetrin orthologues fused to GFP at the N-terminus were screened by FSEC as previously described17. We identified GFP-zfOtop1 and GFP-chOtop3 as promising candidates for structure determination. To conduct FSEC, HEK293F cells suspended in Freestyle 293 expression medium were transfected with N-terminal GFP fusion constructs when they reached a cell density of ~1.5 × 106 / mL and incubated in an orbital shaker at 37° C supplemented with 8% CO2 for two days. 1 mL of cells was pelleted and resuspended in 200 μL of detergent-containing buffer and rotated at 4 °C for one hour, then centrifuged at greater than 50,000 x g in a tabletop ultracentrifuge, then injected onto a Superose 6 increase column (equilibrated to 20 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM n-dodecyl-β-D-maltoside, 1 mM EDTA) in line with a fluorimeter tracking GFP fluorescence.
Sample preparation
Similar procedures were used to prepare cryo-EM samples for zfOtop1 and chOtop3 in nanodiscs. HEK293F cells suspended in Freestyle 293 expression medium at a cell density of ~1.5 × 106 / mL were transfected with 1 mg of zfOTOP1 or chOTOP3 DNA and 3 mg polyethylenimines and incubated at 37°C for supplemented with 8% CO2 for 48 hours. Crude cell pellets were washed with cold PBS then resuspended in buffer containing 20 mM Tris pH 8.0, 150 mM NaCl, 1% n-dodecyl-β-D-maltoside, 0.15% cholesteryl hemisuccinate, 2 μg/μL leupeptin, 2 μg/μL aprotinin, 1 mM phenylmethylsulfonyl fluoride, 2 μM pepstatin A, 2 mM dithiothreitol (DTT) then stirred at 4 °C for 1 hour. The lysate was clarified by centrifugation in a JLA 16.25 rotor at 34,000 x g for 1 hour. Anti-GFP nanobody-coupled CNBr-Activated Sepharose 4B resin32 was added to the clarified lysate, and the mixture was rotated at 4°C for 1.5 hours. Resin was washed with more than 10 resin volumes of wash buffer (20 mM Tris pH 8.0, 150 mM NaCl, 0.07% n-dodecyl-β-D-maltoside, 0.01% cholesteryl hemisuccinate, 0.4 μg/μL aprotinin, 0.4 μM pepstatin A, 2 mM DTT). Washed resin was resuspended in wash buffer to make a ~40% slurry and 50 ug of prescission protease was added, then incubated at 4°C for five hours. Flowthrough containing cleaved zfOtop1 or chOtop3 was collected, concentrated in 100 kDa cutoff centrifugal filter, then injected into a Superose 6 increase column equilibrated with wash buffer. Peak fractions corresponding to zfOtop1 or chOtop3 were collected and concentrated to ~1 mg/mL, then mixed with MSP2N2 nanodisc scaffold protein33 and soybean polar lipid extract at a molar ratio of approximately 1:1.67:13.33 (monomer:MSP2N2:lipids) for zfOTOP1 and approximately 1:2.67:26.67 for chOTOP3. The total volume of the mixture was approximately 0.5 mL. The mixture was incubated on ice for one hour, then 100 mg of biobeads was added and nanodisc assembly was initiated by rotation overnight at 4° C. After removing biobeads by centrifugation, the nanodisc assembly mixture was injected into a Superose 6 increase column equilibrated with buffer containing 20 mM Tris pH 8.0, 150 mM NaCl, 0.4 μg/μL aprotinin, 0.4 μM pepstatin A, 2 mM DTT. Peak fractions were concentrated to 2.1 mg/mL (for zfOtop1) or 1.1 mg/mL (for chOtop3) using a 100 kDa cutoff centrifugation filter. 3.5 μL of purified protein was applied to previously plasma-cleaned UltrAuFoil 1.2/1.3 300 mesh grids and blotted once for 3.5 seconds with blot force 0 after a wait time of 10 seconds. Blotted grids were plunge frozen into nitrogen-cooled liquid ethane using a Vitrobot Mark IV operated at 10 °C and 100% humidity.
Cryo-EM data collection
Images were collected at 300 kV using a Titan Krios coupled with a K2 Summit direct electron detector (Gatan) at a nominal magnification of 29,000 x with a pixel size of 1.03 Å. 53 frames (for zfOtop1) or 45 frames (for chOTOP3) of 250 ms exposure time were collected per movie, resulting in a total accumulated dose of ~50 electrons per Å2. Automated micrograph collection was performed using Leginon software34 with a target defocus range of 0.6–1.8 μM.
Cryo-EM Data Processing
Micrographs were aligned and dose-weighted using MotionCor235. Contrast transfer function (CTF) parameters were obtained with Gctf36. Data processing for zfOtop1 and chOtop3 were carried out similarly. The following processing procedure was used for zfOtop1; the procedure for chOtop3 is depicted as a flow chart in Supplementary Fig. 4d. 1,517,125 particles were picked from 2,138 micrographs using AutoPick in RELION-2.137 applying a gaussian blob as a reference. Picked particles were extracted without binning using RELION-2.1 with box size of 200 pixels (206 Å) and exported to cryoSPARC v0.6.538. Contaminants and particles not containing features of zfOTOP1 were removed by two rounds of 2D classification, resulting in 792,311 particle images that were retained for further processing. These particles were then subjected to ab initio reconstruction without symmetry, requesting 3 classes and a maximum resolution of 7 Å. The most populated class contained 435,468 particles and had secondary structure features apparent. These particles were transferred to RELION-2.1 and refined with C2 symmetry applied and using the ab initio reconstruction low pass filtered to 12 Å resolution as the initial reference. This refinement resulted in a 3.26 Å resolution map after postprocessing that displayed anisotropic features. The particles were then recentered and reextracted on the basis of their refined coordinates, followed by 3D classification without alignment using k=6 and tau fudge value of 8. This classification run was repeated twice, and the particles from the highest resolution class from each run were combined and duplicates removed, resulting in 76,546 particles. Masked refinement of these particles resulted in a 3.16 Å resolution map after postprocessing that did not exhibit anisotropic features. The refined particles were then subjected to local CTF and beam tilt estimation using the ctfrefine jobtype in RELION-3.039. Subsequent masked refinement of these particles resulted in a 3.07 Å resolution map. The refinement parameters and reconstruction from this refinement run were used as inputs for Bayesian polishing in RELION-3.039. After Bayesian polishing, masked refinement yielded a 3.02 Å resolution reconstruction. The polished particles were then subjected to a final round of 3D classification without alignment (k=3, tau fudge=8). Masked refinement of 67,425 particles from the highest resolution class resulted in the final 2.98 Å resolution map. For all refinements and classification jobs, C2 symmetry was applied unless otherwise noted, and stated resolutions were calculated at FSC=0.143 by postprocessing in RELION using a soft mask around the zfOtop1 protein and nanodisc belt.
Model building, refinement, and visualization
Cryo-EM maps were flipped to the correct handedness using the ‘vop zflip’ command in UCSF chimera40 and sharpened with phenix.autosharpen41 prior to model building in coot42. Manual building was iterated with real space refinement using phenix43 and rosetta44. The zfOtop1 model was built first. Most side chains in the TM helices were clearly resolved, allowing de novo model building using bioinformatic prediction of the positions of TM segments45 as a guide. To build the chOtop3 model, a homology model of chOtop3 was produced from the zfOtop1 structure using SWISS-MODEL46 and docked into the Otop3 map, then manually adjusted and refined. Geometric restraints for cholesterol and CHS molecules were produced using the ELBOW program in phenix. The distal N-terminus and portions of linkers connecting TM2–TM3, TM5–TM6, TM6–TM7, TM10–TM11 were omitted from either model due to poor density. The model of zfOtop1 (586 residues in full-length) contains residues 46–113, 123–221, 248–283, 302–443, 510–585. The model of Otop3 (561 residues in full-length) contains residues 49–116, 124–227, 246–283, 304–366, 375–438, 487–560. Structural figures were made in PyMOL47, UCSF Chimera48, or UCSF ChimeraX49. Sequence conservation scores were calculated and mapped onto the structure of zfOtop1 using Consurf50,51, with ~100 Otopetrin sequences obtained from UniprotKB as input, aiming for even distribution between Otop1, Otop2, and Otop3 subtypes. Sequence alignment was calculated using Clustal Omega52 and represented using ESPript 3.053.
Cell culture and transfection for electrophysiology and confocal microscopy
HEK-293 cells (CRL-1573, ATCC) were cultured in DMEM containing 10% fetal calf serum and 50 μg/ml gentamycin. Cells were transfected in 35mm petri dishes, with approximately 600–750 ng DNA and 2 μl TransIT-LT1 transfection reagent (Mirus Bio Corporation) following manufacturer’s protocol. Constructs were expressed as N-terminal GFP fusion (for zfOtop1 and chOtop3, see above) or a N-terminal YFP fusion (mOtop1 mutants). For imaging, zfOtop1constructs were co-transfected with a plasmid encoding Tag-RFP at a ratio of 2:1. The cells were lifted using Trypsin-EDTA 24h after transfection and plated onto a coverslip for patch clamp recordings. GFP fluorescence was used to select transfected cells for recording or imaging.
Patch clamp electrophysiology
Whole-cell patch clamp recording was performed as previously described10. Briefly, recordings were made with an Axonpatch 200B or 700B amplifier, digitized with a Digidata 1322a 16-bit data acquisition system, acquired with pClamp 8.2 and analyzed with Clampfit 8.2 (Molecular devices, Palo Alto, CA). Records were sampled at 5 kHz and filtered at 1 kHz. Patch pipettes with resistance of 2 – 4 MΩ were fabricated from borosilicate glass and only recordings in which a gigaohm seal was achieved were used in the analysis. For most of the experiments, the membrane potential was held at −80 mV or ramped from −80 mV to +80 mV (1V/s) once per second and solutions were exchanged with a Warner fast step system.
Confocal microscopy
HEK293 cells co-expressing the cytosolic fluorescent protein, Tag-RFP, and either WT or mutant channels fused to GFP were imaged using an Olympus Fluoview 1000 cofocal microscope. Surface expression of the channels was evident as a ring of GFP fluorescence that surrounded the RFP fluorescence which filled the cell. Images shown are representative of n>3 cells for each channel mutant.
Patch clamp electrophysiology solutions
Standard pipette solution contained 120 mM Cs-aspartate, 15 mM CsCl, 2 mM Mg-ATP, 5 mM EGTA, 2.4 mM CaCl2 (100 nM free Ca2+), and 10 mM HEPES (pH 7.3 with CsOH; 290 mosm). Proton currents were evoked in Na+-free, NMDG+-based extracellular solutions that contained 160 mM NMDG-Cl, 2mM CaCl2, and either 10 mM HEPES (for pH 7.4), 10 mM MES (for pH 6–5.5), or 10 mM HomoPIPES (for pH 5–4.5), pH adjusted with HCl. ZnCl2 was added to these solutions, and pH adjusted at concentrations indicated. Prior to recording, cells were bathed in Tyrode’s solution contained 145 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 20mM D-Glucose, 10mM HEPES (pH 7.4 with NaOH). Statistical analyses were performed with Microsoft Excel and data were plotted with Graphpad Prism.
Building the atomistic MD simulation system
All molecular dynamics simulations were carried out using GROMACS 5.1.254 on the nanodisc-embedded zfOtop1 and chOtop3 structures. In each case, the cholesterol/CHS molecules associated with the structures were first removed, then converted into a coarse-grained (CG) representation (force field: MARTINI 2.2)55 using MemProtMD56. The protein was embedded in a band of 80%:20% POPC:cholesterol molecules that are randomly oriented. A control using 100% POPC was also performed for zfOtop1. The periodic simulation box (~125×120×100 Å) was completed with the addition of water and 0.15 M NaCl. A 100-ns CG simulation with protein backbone beads position restrained (force constant: 1000 kJ mol–1 nm–2) permitted lipid self-assembly. The final frame was then converted to atomistic (AT) detail using CG2AT-Align57, with the CHARMM36 force field58 and solvated again in TIP3P water59 and 0.15 M NaCl. Cholesterol/CHS molecules associated with the structures were added back to the sets of simulations in the mixed membrane, after the conversion to the AT system via manual alignment to the original models. Any water molecules, ions or lipid molecules (one POPC molecule usually ends up in the central tunnel in CG self-assembly) that resulted in steric clashes in the AT system (within 6 Å) were removed. Eight cholesterol molecules were added back to the zfOtop1 system, in accordance with the structure. For chOtop3 however, having two cholesterol molecules in the central tunnel was insufficient to prevent the flow of water via the central tunnel in our simulations. We therefore placed six cholesterols in the central tunnel of chOtop3 in the same starting position as zfOtop1 as an approximation. We however note that the distribution of cholesterols and other lipid-like densities in our EM maps of zfOtop1 and chOtop3 are different.
Atomistic MD simulations
The system with added the cholesterol molecules was energy minimised to maximum force 1000 kJ mol–1 nm–2, before running a 1-ns simulation (time-step: 1 fs) with the protein backbone atoms position restrained (force constant: 1000 kJ mol–1 nm–2). Three repeats of 100-ns production run (time-step: 2 fs) were then performed based on the final snapshot of the short simulation, without position restraints. Here, distance restraints were applied between endings of the missing loops (force constant: 1000 kJ mol–1 nm–2; lower and upper bound values r0 and r1 were ±1 Å of the starting distances). The AT simulations were performed as NPT ensembles held at 1 bar and 310 K. A semi-isotropic Parrinello-Rahman barostat60 (coupling constant: 1 ps; compressibility: 4.5×10–5 bar–1) and a velocity-rescaling thermostat61 (coupling constant: 0.1 ps) were used. All covalent bonds were constrained using the LINCS algorithm62. Electrostatics were modelled with a Particle Mesh Ewald (PME) model63, and van der Waals’ interactions were modelled using a cut-off scheme, both with cut-off distances at 10 Å.
MD simulations trajectory analysis
Protein-cholesterol distances were calculated with the distance tool in GROMACS based on their centres of geometry (Supplementary Fig. 8c–d). Root-mean-square deviations measurements were performed using the RMSD Trajectory Tool in VMD64. Alignments and calculations were based on the backbone atoms of transmembrane residues (Supplementary Fig. 8e–g). For the extraction of water trajectories (Supplementary Data Set 1), the trajectories were aligned either at the domain level (for the selection in the N and C domains) or at the subunit level (for the selection at the interface). Water molecules within the N domain pathway were selected based on their proximity (within 5 Å) to the domain excluding TM1 (starting from V81 for zfOtop1, A81 for chOtop3), while excluding those found within 6 Å of lipid molecules or within 8 Å of the other three domains. A similar method was used to select waters in the C domain pathway (within 5 Å of the entire domain) and with the same exclusions as the N domain. A cuboid selection measuring 10×12×42 Å3 was used to select water molecules along the intrasubunit interface, which covers the outer half of interface. MD-related figures were rendered using PyMOL47. Electrostatic profiles (Fig. 4f, Supplementary Fig. 10h) were generated using the APBS Electrostatics plugin in PyMOL. When estimating the water density along the putative pathways (Supplementary Data Set 1 and Fig. 4e), kernel density estimations (KDEs) were performed on the z-position of oxygen atoms of water molecules using the density function in R65. Gaussian kernels were used with the bandwidths chosen using the Sheather & Jones method.
Data availability
Cryo-EM maps and atomic coordinates have been deposited to the Electron Microscopy Data Bank and wwPDB, respectively, with accession codes EMD-9360 and PDB 6NF4 (zebrafish Otop1) and EMD-9361 and PDB 6NF6 (chicken Otop3). All other data are available upon reasonable request to the corresponding authors.
Supplementary Material
Acknowledgements
We thank W. Anderson for managing the electron microscopy facility at Scripps Research, H. Turner and J. Torres for help with data collection, C. Bowman for assistance with computation, D. Artiga for help with generating cDNA constructs and Vsevolod Katritch for providing expert advice on design of expression constructs. We acknowledge members of the Ward, Liman, Sansom, and Patapoutian labs for helpful advice. This work was supported by a Ray Thomas Edwards Foundation grant to A.B.W, funding from the NIH (NIDCD013741) to E.R.L, Wellcome (grant 208361/Z/17/Z), BBSRC (grants BB/N000145/1 and BB/R00126X/1), and EPSRC (grant EP/R004722/1) to M.S.P.S. K.S. is a postdoctoral fellow of the Jane Coffin Childs Memorial Fund for Medical Research. C.C.A.T. is supported by the Skaggs-Oxford Scholarship and the Croucher Foundation.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.DeCoursey TE The Voltage-Gated Proton Channel: A Riddle, Wrapped in a Mystery, inside an Enigma. Biochemistry-Us 54, 3250–3268, doi: 10.1021/acs.biochem.5b00353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fogel M & Hastings JW Bioluminescence: mechanism and mode of control of scintillon activity. Proc Natl Acad Sci U S A 69, 690–693 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer H Function of Proton Channels in Lung Epithelia. Wiley Interdiscip Rev Membr Transp Signal 1, 247–258, doi: 10.1002/wmts.17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang RB, Waters H & Liman ER A proton current drives action potentials in genetically identified sour taste cells. Proc Natl Acad Sci U S A 107, 22320–22325, doi: 10.1073/pnas.1013664107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richter TA, Caicedo A & Roper SD Sour taste stimuli evoke Ca2+ and pH responses in mouse taste cells. J Physiol 547, 475–483, doi: 10.1113/jphysiol.2002.033811 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyall V et al. Decrease in rat taste receptor cell intracellular pH is the proximate stimulus in sour taste transduction. Am J Physiol-Cell Ph 281, C1005–C1013 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Decoursey TE Voltage-gated proton channels. Compr Physiol 2, 1355–1385, doi: 10.1002/cphy.c100071 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramsey IS, Moran MM, Chong JA & Clapham DE A voltage-gated proton-selective channel lacking the pore domain. Nature 440, 1213–1216, doi: 10.1038/nature04700 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki M, Takagi M & Okamura Y A voltage sensor-domain protein is a voltage-gated proton channel. Science 312, 589–592, doi: 10.1126/science.1122352 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Tu YH et al. An evolutionarily conserved gene family encodes proton-selective ion channels. Science 359, 1047–1050, doi: 10.1126/science.aao3264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurle B et al. Non-syndromic vestibular disorder with otoconial agenesis in tilted/mergulhador mice caused by mutations in otopetrin 1. Hum Mol Genet 12, 777–789 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Hughes I et al. Otopetrin 1 is required for otolith formation in the zebrafish Danio rerio. Dev Biol 276, 391–402, doi: 10.1016/j.ydbio.2004.09.001 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sollner C, Schwarz H, Geisler R & Nicolson T Mutated otopetrin 1 affects the genesis of otoliths and the localization of Starmaker in zebrafish. Dev Genes Evol 214, 582–590, doi: 10.1007/s00427-004-0440-2 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Wang GX et al. Otopetrin 1 Protects Mice From Obesity-Associated Metabolic Dysfunction Through Attenuating Adipose Tissue Inflammation. Diabetes 63, 1340–1352, doi: 10.2337/db13-1139/-/DC1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10, R130, doi: 10.1186/gb-2009-10-11-r130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parikh K et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567, 49–55, doi: 10.1038/s41586-019-0992-y (2019). [DOI] [PubMed] [Google Scholar]
- 17.Kawate T & Gouaux E Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681, doi: 10.1016/j.str.2006.01.013 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Quistgaard EM, Low C, Guettou F & Nordlund P Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat Rev Mol Cell Biol 17, 123–132, doi: 10.1038/nrm.2015.25 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Abramson J et al. Structure and mechanism of the lactose permease of Escherichia coli. Science 301, 610–615, doi: 10.1126/science.1088196 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Murakami S, Nakashima R, Yamashita E & Yamaguchi A Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419, 587–593, doi: 10.1038/nature01050 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Gupta K et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature 541, 421–424, doi: 10.1038/nature20820 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu HX et al. Structure of a Class C GPCR Metabotropic Glutamate Receptor 1 Bound to an Allosteric Modulator. Science 344, 58–64, doi: 10.1126/science.1249489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagle JF & Morowitz HJ Molecular mechanisms for proton transport in membranes. Proc Natl Acad Sci U S A 75, 298–302 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong M & DeGrado WF Structural basis for proton conduction and inhibition by the influenza M2 protein. Protein Sci 21, 1620–1633, doi: 10.1002/pro.2158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinto LH et al. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci U S A 94, 11301–11306 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsey IS et al. An aqueous H+ permeation pathway in the voltage-gated proton channel Hv1. Nat Struct Mol Biol 17, 869–875, doi: 10.1038/nsmb.1826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudev T et al. Selectivity Mechanism of the Voltage-gated Proton Channel, HV1. Sci Rep 5, 10320, doi: 10.1038/srep10320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chamberlin A et al. Hydrophobic plug functions as a gate in voltage-gated proton channels. Proc Natl Acad Sci U S A 111, E273–282, doi: 10.1073/pnas.1318018111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aryal P, Sansom MS & Tucker SJ Hydrophobic gating in ion channels. J Mol Biol 427, 121–130, doi: 10.1016/j.jmb.2014.07.030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan D et al. Peregrination of the selectivity filter delineates the pore of the human voltage-gated proton channel hHV1. J Gen Physiol 142, 625–640, doi: 10.1085/jgp.201311045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeCoursey TE CrossTalk proposal: Proton permeation through HV 1 requires transient protonation of a conserved aspartate in the S1 transmembrane helix. J Physiol 595, 6793–6795, doi: 10.1113/JP274495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H et al. Charge delocalization in proton channels, I: the aquaporin channels and proton blockage. Biophys J 92, 46–60, doi: 10.1529/biophysj.106.091934 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Q, Zeng W, She J, Bai X & Jiang Y Structural and functional characterization of an otopetrin family proton channel. Elife 8, doi: 10.7554/eLife.46710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchhofer A et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 17, 133–U162, doi: 10.1038/nsmb.1727 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Grinkova YV, Denisov IG & Sligar SG Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers. Protein Eng Des Sel 23, 843–848, doi: 10.1093/protein/gzq060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suloway C et al. Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60, doi: 10.1016/j.jsb.2005.03.010 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12, doi: 10.1016/j.jsb.2015.11.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimanius D, Forsberg BO, Scheres SHW & Lindahl E Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5, doi: 10.7554/eLife.18722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296, doi: 10.1038/nmeth.4169 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, doi: 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Afonine PV et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr D Struct Biol 74, 814–840, doi: 10.1107/S2059798318009324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr D 60, 2126–2132, doi: 10.1107/S0907444904019158 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 66, 213–221, doi: 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang RYR et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife 5, doi: 10.7554/eLife.17219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes I et al. Identification of the Otopetrin Domain, a conserved domain in vertebrate otopetrins and invertebrate otopetrin-like family members. BMC Evol Biol 8, 41, doi: 10.1186/1471-2148-8-41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waterhouse A et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303, doi: 10.1093/nar/gky427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schrodinger L The PyMOL Molecular Graphics System, Version 2.0.
- 49.Pettersen EF et al. UCSF chimera - A visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Goddard TD et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25, doi: 10.1002/pro.3235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashkenazy H et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44, W344–350, doi: 10.1093/nar/gkw408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landau M et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 33, W299–302, doi: 10.1093/nar/gki370 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sievers F et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539, doi: 10.1038/msb.2011.75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robert X & Gouet P Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42, W320–324, doi: 10.1093/nar/gku316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25, doi: 10.1016/j.softx.2015.06.001 (2015). [DOI] [Google Scholar]
- 56.de Jong DH et al. Improved Parameters for the Martini Coarse-Grained Protein Force Field. J Chem Theory Comput 9, 687–697, doi: 10.1021/ct300646g (2013). [DOI] [PubMed] [Google Scholar]
- 57.Stansfeld PJ et al. MemProtMD: Automated Insertion of Membrane Protein Structures into Explicit Lipid Membranes. Structure 23, 1350–1361, doi: 10.1016/j.str.2015.05.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stansfeld PJ & Sansom MSP From Coarse Grained to Atomistic: A Serial Multiscale Approach to Membrane Protein Simulations. J Chem Theory Comput 7, 1157–1166, doi: 10.1021/ct100569y (2011). [DOI] [PubMed] [Google Scholar]
- 59.Best RB et al. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone phi, psi and Side-Chain chi(1) and chi(2) Dihedral Angles. J Chem Theory Comput 8, 3257–3273, doi: 10.1021/ct300400x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW & Klein ML Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys 79, 926–935, doi:Doi 10.1063/1.445869 (1983). [DOI] [Google Scholar]
- 61.Parrinello M & Rahman A Polymorphic Transitions in Single-Crystals - a New Molecular-Dynamics Method. J Appl Phys 52, 7182–7190, doi:Doi 10.1063/1.328693 (1981). [DOI] [Google Scholar]
- 62.Bussi G, Donadio D & Parrinello M Canonical sampling through velocity rescaling. J Chem Phys 126, doi: 10.1063/1.2408420 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Hess B P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J Chem Theory Comput 4, 116–122, doi: 10.1021/ct700200b (2008). [DOI] [PubMed] [Google Scholar]
- 64.Essmann U et al. A Smooth Particle Mesh Ewald Method. J Chem Phys 103, 8577–8593, doi:Doi 10.1063/1.470117 (1995). [DOI] [Google Scholar]
- 65.Humphrey W, Dalke A & Schulten K VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps and atomic coordinates have been deposited to the Electron Microscopy Data Bank and wwPDB, respectively, with accession codes EMD-9360 and PDB 6NF4 (zebrafish Otop1) and EMD-9361 and PDB 6NF6 (chicken Otop3). All other data are available upon reasonable request to the corresponding authors.