Abstract
Phosphoinositides (PI) are synthesized and turned over by specific kinases, phosphatases, and lipases that ensure the proper localization of discrete PI isoforms at distinct membranes. We analyzed the role of the yeast synaptojanin-like proteins using a strain that expressed only a temperature-conditional allele of SJL2. Our analysis demonstrated that inactivation of the yeast synaptojanins leads to increased cellular levels of phosphatidylinositol (3,5)-bisphosphate and phosphatidylinositol (4,5)-bisphosphate (PtdIns(4,5)P2), accompanied by defects in actin organization, endocytosis, and clathrin-mediated sorting between the Golgi and endosomes. The phenotypes observed in synaptojanin-deficient cells correlated with accumulation of PtdIns(4,5)P2, because these effects were rescued by mutations in MSS4 or a mutant form of Sjl2p that harbors only PI 5-phosphatase activity. We utilized green fluorescent protein-pleckstrin homology domain chimeras (termed FLAREs for fluorescent lipid-associated reporters) with distinct PI-binding specificities to visualize pools of PtdIns(4,5)P2 and phosphatidylinositol 4-phosphate in yeast. PtdIns(4,5)P2 localized to the plasma membrane in a manner dependent on Mss4p activity. On inactivation of the yeast synaptojanins, PtdIns(4,5)P2 accumulated in intracellular compartments, as well as the cell surface. In contrast, phosphatidylinositol 4-phosphate generated by Pik1p localized in intracellular compartments. Taken together, our results demonstrate that the yeast synaptojanins control the localization of PtdIns(4,5)P2 in vivo and provide further evidence for the compartmentalization of different PI species.
INTRODUCTION
Phosphoinositides (PIs) control several cellular processes including cell signaling, cell growth, vesicular trafficking, transcription, and actin cytoskeletal arrangement (Fruman et al., 1998; Martin, 2001; Simonsen et al., 2001). Phosphatidylinositol (PtdIns) can be differentially phosphorylated on its inositol head group to form seven different PI isoforms that act as second messengers in various cellular pathways. Accordingly, PI synthesis is regulated by specific kinases localized to distinct membrane sites. In this manner, individual PI isoforms can recruit isoform-specific PI-binding proteins to distinct membranes. Moreover, the reversible phosphorylation of these lipids make them ideally suited to act as temporal and spatial regulators of numerous cellular processes. Correspondingly, a set of phosphatases and lipases regulate PI turnover, thereby controlling the duration and distribution of signaling events mediated by PI second messengers.
The spatial and temporal regulation of PI signaling is illustrated by several previous studies utilizing protein domains with distinct PI-binding specificities (Balla et al., 2000; Czech, 2000; Hurley and Meyer, 2001). For example, pleckstrin homology (PH) domains present in numerous cell-signaling and cytoskeletal proteins display a wide range of PI-binding specificities. The PH domain of phospholipase C δ1 (PLCδ) binds phosphatidylinositol (4,5)-bisphosphate (PtdIns(4,5)-P2) with high specificity and is efficiently recruited to the plasma membrane (Kavran et al., 1998; Stauffer et al., 1998; Honda et al., 1999; Botelho et al., 2000). Other PH domains present in proteins such as Akt/PKB and Grp1 have specificity for phosphatidylinositol (3,4,5)-triphosphate and play a role in recruiting these proteins to the cell surface in response to external stimuli (Kavran et al., 1998; Venkateswarlu et al., 1998; Meili et al., 1999; Servant et al., 2000). In contrast, the FYVE (for Fab1, YGL023, Vps27, and EEA1) domain present in a number of membrane-trafficking proteins binds phosphatidylinositol 3-phosphate (PtdIns(3)P) with high specificity and is sufficient for endosomal targeting (Burd and Emr, 1998; Gaullier et al., 1998; Gillooly et al., 2000).
In particular, PtdIns(4,5)P2 controls several cellular processes, including actin cytoskeletal organization and membrane trafficking, through interactions with PtdIns(4,5)P2-binding proteins (Janmey, 1994; Martin, 2001). PtdIns(4,5)P2-mediated signaling events are initiated by specific PI kinases and are terminated in part by a family of PI 5-phosphatases (5-Pases). PI 5-Pases regulate cellular levels of PtdIns(4,5)P2 by removing the phosphate at the D5 position of the inositol head group (Majerus et al., 1999). Several mammalian 5-Pases have been identified thus far with various substrate specificities for PtdIns(4,5)P2, PtdIns (3,4,5)-triphosphate, and soluble inositol phosphates (Majerus et al., 1999). Recently, the crystal structure of a PI 5-Pase domain was determined, suggesting a novel conserved catalytic mechanism for this family of phosphatases (Tsujishita et al., 2001).
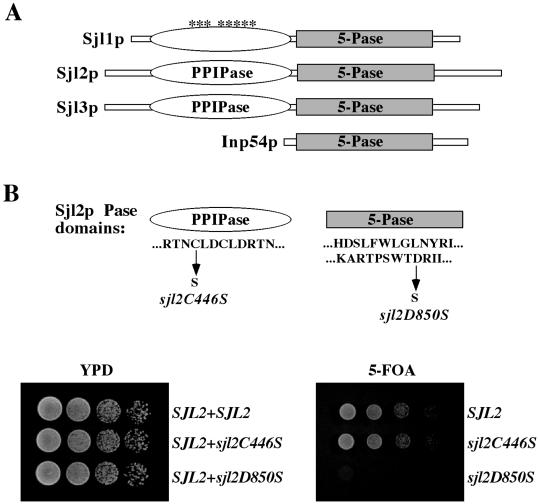
Four PI 5-Pases are present in the yeast Saccharomyces cerevisiae (Figure 1A). Three enzymes, Sjl1p, Sjl2p, and Sjl3p (also named Inp51p, Inp52p, and Inp53p), contain a Sac1-like domain and a 5-Pase domain, similar to mammalian synaptojanin. The Sac1-like domains of mammalian synaptojanin, Sjl2p, and Sjl3p, but not Sjl1p, possess polyphosphoinositide phosphatase (PPIPase) activity that dephosphorylates PtdIns(3)P, phosphatidylinositol 4-phosphate (PtdIns(4)P), and phosphatidylinositol 3,5-bisphosphate (PtdIns(3,5)P2) to PtdIns in vitro (Guo et al., 1999). The fourth yeast PI 5-Pase, Inp54p, possesses only 5-Pase activity and localizes to the endoplasmic reticulum (Wiradjaja et al., 2001). Although the yeast 5-Pase domains hydrolyze PtdIns(4,5)P2 to form PtdIns(4)P, they do not act on soluble inositol phosphates in vitro (Stolz et al., 1998b; Guo et al., 1999; Ooms et al., 2000; Raucher et al., 2000; Wiradjaja et al., 2001). Consistent with these in vitro activities, deletion of SJL1 leads to an increase in cellular PtdIns(4,5)P2 levels (Stolz et al., 1998b), whereas double deletion of SJL2 and SJL3 causes an increase in PtdIns(3,5)P2 levels in vivo (Guo et al., 1999).
Figure 1.
Growth of yeast synaptojanin mutant cells lacking either SacI-like PPIPase or 5-Pase activities. (A) Schematic representations of the domain structures of yeast PI 5-Pases. The SacI domains in Sjl1p, Sjl2p, and Sjl3p are shown by open ovals. The asterisks above the SacI-like domain of Sjl1p are to indicate that Sjl1p does not possess intrinsic PPIPase activity. The 5-Pase domains in Sjl1p, Sjl2p, Sjl3p, and Inp54p are indicated by gray bars. (B) The residues present in the conserved catalytic motif (CX5RTN) of the Sjl2p SacI domain are shown. The arrow indicates the substitution of the conserved cysteine residue that inactivates the PPIPase activity of Sjl2p in vivo. Conserved residues present in separate regions of the Sjl2p 5-Pase domain are shown. The arrow indicates the substitution of the conserved aspartate residue that inactivates the 5-Pase activity of Sjl2p in vivo. sjl1Δ sjl2Δ sjl3Δ cells harboring a URA3 CEN SJL2 plasmid (YCS156) were transformed with LEU2-marked CEN plasmids carrying the SJL2, sjl2C446S, or sjl2D850S allele. Tenfold serial dilutions of cells were spotted onto either YPD (left) or media containing 5-FOA (right) and incubated for 2 d at 30°C.
Deletion of all three yeast synaptojanin-like (SJL) genes is lethal (Srinivasan et al., 1997; Stolz et al., 1998a), suggesting that the yeast synaptojanins have essential but overlapping functions. Accordingly, single sjl1, sjl2, sjl3, and inp54 null mutants are viable and display no obvious phenotypes. However, sjl1Δ sjl2Δ and sjl2Δ sjl3Δ double mutants demonstrate several phenotypes, including impaired cell growth, defects in actin cytoskeletal organization, and aberrant cell surface and vacuole morphologies (Srinivasan et al., 1997; Stolz et al., 1998a). In contrast, sjl1Δ sjl3Δ double mutants display relatively few phenotypes (Srinivasan et al., 1997; Stolz et al., 1998a), suggesting that Sjl2p may provide overlapping functions in these cellular processes. Consistent with this, sjl1Δ sjl2Δ and sjl2Δ sjl3Δ cells display endocytic defects but not sjl1Δ sjl3Δ cells (Singer-Kruger et al., 1998). Moreover, although S. cerevisiae encodes multiple PI 5-Pases, genetic evidence suggests that Sjl1p and Sjl3p may have primary functions. Specific genetic interactions exist between sjl1 mutations and mutations in genes that encode actin-regulatory proteins, such as PAN1 and SAC6 (Singer-Kruger et al., 1998; Wendland and Emr, 1998). In contrast, Sjl3p is specifically implicated in clathrin-mediated protein sorting at the trans-Golgi network (TGN; Bensen et al., 2000).
Because previous work has suggested that Sjl2p provides overlapping functions, we generated a strain that expressed only a temperature-sensitive allele of SJL2. We found that inactivation of the yeast synaptojanins resulted in pleiotropic phenotypes, including effects on actin cytoskeletal organization, endocytic transport, cell surface morphology, and clathrin-dependent transport between the TGN and endosomes. Consequently, we examined whether regulation of PtdIns(4,5)P2 levels by the 5-Pase domain or control of other PI isoforms by the SacI-like domain of Sjl2p contributed to these phenotypes. Furthermore, we created two FLAREs to visualize pools of PtdIns(4,5)P2 and PtdIns(4)P in yeast by fusing GFP to PH domains with distinct PI-binding specificities. Accordingly, we examined the role of the yeast synaptojanins in regulating the steady-state distribution of these PIs in vivo. We found that PtdIns(4,5)P2 inappropriately accumulated in intracellular compartments on inactivation of the yeast synaptojanins, providing the first demonstration that these PI phosphatases were necessary to restrict the steady-state distribution of PtdIns(4,5)P2 to the plasma membrane. We propose that this spatial control of PtdIns(4,5)P2 within cells is critical for normal cell morphology and membrane trafficking.
MATERIALS AND METHODS
Reagents and Media
Enzymes used for recombinant DNA techniques were purchased from commercial sources and used as recommended by the suppliers. Standard recombinant DNA techniques were performed as previously described (Sambrook et al., 1989). Sources of growth media for yeast and bacterial strains have been described elsewhere (Gaynor et al., 1998), and standard yeast genetic methods were used throughout (Sherman et al., 1979). S. cerevisiae strains used in this study are listed in Table 1 and their constructions are described below. Primers used in this study are available upon request.
Table 1.
Strains used in this study
| Strains | Genotypes | Reference or source |
|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Robinson et al., 1988 |
| SEY6210.1 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Robinson et al., 1988 |
| SEY6210α/a | MATα/MATa homozygous diploid strain from a SEY6210/SEY6210.1 cross | Robinson et al., 1988 |
| 6210 arf1Δ | Same as SEY6210 except arflΔ::HIS3 | Gaynor et al., 1998 |
| JGY131 | Same as SEY6210 except sjl3Δ::TRP1 | Foti et al., 2001 |
| JGY132 | Same as SEY6210 except sjl2Δ::HIS3 sjl3Δ::TRP1 | Foti et al., 2001 |
| YCS62 | Same as SEY6210.1 except sjl1Δ::HIS3 | This study |
| YCS66 | Same as SEY6210.1 except sjl1Δ::HIS3 sjl2Δ::HIS3 | This study |
| YCS156 | Same as JGY132 except sjl1Δ::hisG and harboring pRS415SJL2 (LEU2 CEN6 SJL2) | This study |
| YCS157 | Same as YCS156 instead harboring pRS416SJL2 (URA3 CEN6 SJL2) | This study |
| YCS176 | Same as YCS156 except harboring pRS415sjl2ts-8 (LEU2 CEN6 sjl2ts-8) | This study |
| AAY102 | Same as SEY6210 except stt4Δ::HIS3 and harboring pRS415stt4-4 (LEU2 CEN6 stt4-4) | Audhya et al., 2000 |
| AAY104 | Same as SEY6210 except pik1Δ::HIS3 and harboring pRS314pik1-83 (TRP1 CEN6 pik1-83) | Audhya et al., 2000 |
| AAY106 | Same as SEY6210α/a except heterozygotic for mss4Δ::HIS3MX6/MSS4 | Audhya et al., 2000 |
| AAY143 | Same as JGY132 except sac1Δ::TRP1 and harboring pRS416sac1-23 (URA3 CEN6 sac1-23) | Foti et al., 2001 |
| AAY201 | Same as SEY6210 except mss4Δ::HIS3MX6 and harboring pRS416MSS4 (URA3 CEN6 MSS4) | This study |
| AAY202 | Same as AAY201 except harboring YCplac111mss4ts-102 (LEU2 CEN6 mss4ts-102) | This study |
| AAY219 | Same as AAY202 except sjl1Δ::HIS3 sjl2Δ::HIS3 | This study |
| AAY219.2 | Same as AAY219 except harboring pRS416MSS4 (URA3 CEN6 MSS4) | This study |
Plasmids, Strains, and Mutagenesis
A 5.6-kb SalI-SpeI fragment containing SJL2 was subcloned from a YEp24INP52 plasmid (generously supplied by J. York) into either pRS415 or pRS416 (Sikorski and Hieter, 1989), which had been cleaved with SpeI and SalI to create pRS415SJL2 and pRS416SJL2, respectively. A 3.8-kb SacI-PstI fragment containing SJL1 was subcloned from pRS316INP51 (Stolz et al., 1998b) into pRS415, which had been cleaved with SacI and PstI to create pRS415SJL1. To create an S. cerevisiae strain that expressed synaptojanin activity from SJL2 alone, a plasmid was first created to generate a chromosomal deletion of the SJL1 gene. This plasmid, pRS415sjl1Δ, was constructed by inserting a BamHI/BglII fragment containing a hisG-URA3-hisG cassette (Alani et al., 1987) into pRS415SJL1, which had been cleaved with BglII. The resulting construct was then digested with EcoRI and transformed into an sjl2::HIS3 sjl3::TRP1 strain (JGY132) carrying pRS415SJL2 to eliminate SJL1-coding sequences. PCR was used to confirm the SJL1 deletion. This strain was grown on media containing 5-fluoroorotic acid (5-FOA) to create an sjl1::hisG sjl2::HIS3 sjl3::TRP1 strain carrying pRS415SJL2 (YCS156). YCS156 was transformed with pRS416SJL2, and plasmid shuffle experiments were performed to isolate an sjl1Δ sjl2Δ sjl3Δ strain carrying pRS416SJL2 alone (YCS157).
To inactivate the Sac1-like PPIPase and 5-Pase activities of Sjl2p individually, we created two substitutions in Sjl2p, C446S and D850S, respectively. Mutations encoding the C446S and D850S substitutions were separately created in pRS415SJL2 by site-directed mutagenesis. The HpaI fragment containing the mutation encoding the C446S substitution was sequenced and subcloned into pRS416SJL2, which had been cleaved with HpaI to ensure that no other mutations were present in the pRS416sjl2C446S plasmid used in these studies. Accordingly, the NarI/SpeI fragment containing the mutation encoding the D850S substitution was sequenced and subcloned into pRS416SJL2, which had been cleaved with NarI and SpeI to create pRS416sjl2D850S. Finally, the 5.6-kb SalI/SpeI fragments containing only mutations encoding either the C446S or D850S substitutions were subcloned into pRS415, which had been cleaved with SalI and SpeI to create pRS415sjl2C446S and pRS415sjl2D850S, respectively.
A strain expressing only a temperature-conditional allele of SJL2 was generated in the following manner. SJL2-coding sequences were amplified by error-prone PCR (Muhlrad et al., 1992) and cotransformed with BglII-gapped pRS415SJL2 into YCS157. Ura+ Leu+ prototrophic transformants were selected and screened for growth on 5-FOA at 26 and 38°C. Cells now lacking pRS416SJL2 that grew at 26°C but not at 38°C on 5-FOA–containing media were selected for further study. From >12,000 transformants, four putative pRS415sjl2ts plasmids were isolated in Esherichia coli, retransformed into YCS157, retested for growth at 26°C but not 38°C on media containing 5-FOA, and tested for PI phosphatase activities at the nonpermissive temperature. A strain harboring the sjl2ts-8 allele (YCS176) displayed strong temperature-sensitive phenotypes and was selected for further characterization.
A strain expressing a temperature-conditional allele of MSS4 was generated in the following manner. MSS4-coding sequences were amplified by error-prone PCR and cotransformed with NdeI/StuI-gapped YCplac111MSS4 into AAY201. Transformants were selected and screened for growth on 5-FOA at 26 and 37°C. Transformants, now lacking pRS416MSS4, that grew at 26°C but not at 37°C on 5-FOA–containing media were selected for further study. From >13,000 transformants, six putative YCplac111mss4ts plasmids were isolated in E. coli, retransformed into AAY201, and retested for growth at 26°C but not 37°C on media containing 5-FOA and PtdIns(4)P 5-kinase activities at the nonpermissive temperature. One particular strain, AAY202, harboring the mss4-102 allele displayed the strongest temperature-sensitive phenotypes and was selected for further characterization.
To create YCS62, pMS1 (Wendland and Emr, 1998) was digested with FspI and AseI and transformed into SEY6210.1 to eliminate SJL1-coding sequences. PCR was used to confirm the SJL1 deletion. To construct an sjl1Δ sjl2Δ double mutant strain (YCS66), sjl1Δ cells (YCS62) and sjl2Δ sjl3Δ cells (JGY132) were crossed and sporulated, and tetrads were dissected. PCR was used to confirm disruptions in spores harboring the appropriate markers. To construct an sjl1Δ sjl2Δ mss4ts mutant strain (AAY219), mss4ts cells (AAY202) and sjl1Δ sjl2Δ cells (YCS66) were crossed and sporulated, and tetrads were dissected. With two different sets of primers for each gene, PCR was used to confirm disruptions in spores harboring the appropriate markers.
To visualize PtdIns(4)P and PtdIns(4,5)P2 in vivo, we constructed fusions between GFP and PH domains with distinct PI-binding specificities. To create a PtdIns(4,5)P2-specific FLARE, we fused two tandem copies of the PLCδ1 PH domain (Kavran et al., 1998) to GFP. PCR was used to generate sequences encoding two PLCδ1 PH domains flanked by either BamHI/EcoRI or EcoRI/SalI restriction sites. These PCR products were digested with BamHI/EcoRI or EcoRI/SalI and cloned in-frame into pGO35 (Burd and Emr, 1998), which had been cut with BglII and SalI to create a plasmid harboring a GFP-2×PH(PLCδ) fusion, pRS426GFP-2×PH(PLCδ). The PtdIns(4)P-binding PH domain of FAPPI (Dowler et al., 2000) was also fused to the C terminus of GFP to create a PtdIns(4)P-specific FLARE. A BamHI fragment containing the PH domain of FAPP1 was fused in-frame to GFP-coding sequences in the yeast GFP expression plasmid pGO35, which had been cleaved with BglII to create pRS426GFP-PH(FAPP1).
In Vivo PI Analysis
Analysis of PI levels was done essentially as described previously (Audhya et al., 2000; Foti et al., 2001). Briefly, before labeling, cells were grown in synthetic medium with the appropriate amino acids. Five OD600 units of cells from a log-phase culture were harvested, washed, and resuspended in inositol-free synthetic medium. Cells were next shifted to the appropriate temperature for 10 min, followed by the addition of 50 μCi of myo-[2-3H]inositol (Nycomed Amersham, Buckinghamshire, UK), and incubated for an additional 50 min at the appropriate temperature. Cells were lysed by mechanical agitation with glass beads in 4.5% perchloric acid to generate extracts. Further processing of extracts was as described previously (Stack et al., 1993). Analysis of 3H-labeled glycero-phosphoinositols was performed by separation on a Beckman (Fullerton, CA) System Gold HPLC and quantitated by liquid scintillation counting after either collecting fractions eluting from the HPLC column (Whatman, Clifton, NJ) every 0.66 min or by an on-line radiomatic detector (Packard Instrument, Meriden, CT).
Fluorescence and Electron Microscopy
For localization of actin, cells were grown to early log phase, shifted to the appropriate temperature for 2 h, fixed in 3.7% formaldehyde, and stained with rhodamine-phalloidin (Molecular Probes, Eugene, OR) as described previously (Audhya et al., 2000).
Labeling with FM4-64 (N-[3-triethylammoniumpropyl]-4-[p-diethylaminophenylhexatrienyl] pyridinium dibromide; Molecular Probes) was done essentially as described by Vida and Emr (1995). Briefly, cells were grown to early log phase in YPD and shifted to the appropriate temperature for 90 min. Cells (2 OD600 units) were harvested by centrifugation and labeled with 16 nM FM4-64 and 100 nM CMAC (Molecular Probes, Eugene, OR) in YPD prewarmed to the appropriate temperature, followed by a chase in YPD without the vital dyes at the appropriate temperature for either 0 or 30 min. Cells were concentrated and visualized by fluorescence microscopy.
To visualize PtdIns(4)P and PtdIns(4,5)P2 in vivo, cells expressing GFP-2×PH(PLCδ) or GFP-PH(FAPPI) were grown to early log at the permissive temperature. After shift to the appropriate temperature, cells were concentrated and visualized by fluorescence microscopy. For colabeling studies, cells expressing GFP-2×PH(PLCδ), which had been incubated at the appropriate temperature for 30 min, were killed with NaN3 plus NaF, stained with FM4-64 on ice, and observed by fluorescence microscopy.
All fluorescent images were observed using a Axiovert S1002TV inverted fluorescent microscope (Carl Zeiss, Thornwood, NY) and acquired and subsequently processed using a Delta Vision deconvolution system (Applied Precision, Seattle, WA). Observations were based on the examination of at least 100 cells.
For ultrastructural analysis, 50 OD600 units of log-phase cells incubated at the appropriate temperature were harvested from YPD medium and fixed in 3% glutaraldehyde, 0.1 M Na cacodylate (pH 7.4), 5 mM CaCl2, 5 mM MgCl2, and 2.5% sucrose for 1 h. Cells were further processed for electron microscopy as described previously (Rieder et al., 1996). Observations were based on the examination of at least 50 cells.
Metabolic Labeling and Immunoprecipitation
Cell labeling and immunoprecipitations were performed as described previously (Gaynor et al., 1998) with noted variations. Log-phase cultures were concentrated to 1–2 OD600/ml and labeled with 2 μl/OD600 Tran35S label (DuPont New England Nuclear, Boston, MA) for 10 min in YNB containing 100 μg/ml BSA and 20 μg/ml 2-macroglobulin. Cells were then chased with 5 mM methionine, 2 mM cysteine, and 0.2% yeast extract for the indicated times, and proteins were precipitated with 9% trichloroacetic acid. Temperature preshifts were conducted for 60 min at 38°C when appropriate. Extracts were immunoprecipitated with antisera against the mating pheromone α-factor, carboxypeptidase Y (CPY), alkaline phosphatase (ALP), or Hsp150p, which have been characterized previously (Gaynor et al., 1998; Audhya et al., 2000). Immunoprecipitated proteins were resuspended in sample buffer for resolution by SDS-PAGE and subjected to autofluorography.
RESULTS
The PI 5-Pase Activity of the Yeast Synaptojanins Is Essential for Cell Growth
Yeast encode four proteins that possess a PI 5-Pase domain (Figure 1A). Simultaneous deletion of SJL1, SJL2, and SJL3 is lethal, but double deletion of SJL2 and SJL3 is not (Srinivasan et al., 1997; Stolz et al., 1998a), suggesting that the 5-Pase activity is essential because Sjl1p lacks Sac-like PPIPase activity (Guo et al., 1999). To test this, we constructed a yeast strain that lacked the chromosomal copies of SJL1, SJL2, and SJL3 but carried SJL2 on a URA3-marked centromeric (CEN) plasmid. We chose to add back SJL2 on a CEN plasmid rather than SJL1 or SJL3 because sjl1Δ sjl3Δ cells display fewer effects on cell viability, morphology, and vesicular transport compared with sjl1Δ sjl2Δ and sjl2Δ sjl3Δ cells. Next, mutant forms of Sjl2p that bore substitutions in highly conserved residues of either the SacI-like domain (C446S) or 5-Pase domain (D850S) were generated (Figure 1B). Each substitution was sufficient to impair either the Sac1-like PPIPase activity or the 5-Pase activity of Sjl2p in vivo, respectively, as assessed below. However, neither of these substitutions affected the steady-state expression level of Sjl2p (Stefan, Audhya, and Emr, unpublished results).
We performed two tests to demonstrate that the C446S substitution inactivated the Sac1-like PPIPase activity of Sjl2p in vivo. Deletion of SAC1 and SJL3 in combination has been shown to be lethal, suggesting that the Sac1 domains of these proteins were indispensable for viability (Foti et al., 2001). Likewise, sac1tsf sjl2Δ sjl3Δ cells have been shown to be viable at 26°C but were unable to grow at 38°C (Foti et al., 2001). Similarly, sac1tsf sjl2Δ sjl3Δ cells expressing sjl2C446S from a LEU2 CEN plasmid were viable at 26°C but were unable to grow at 38°C (Stefan, Audhya, and Emr, unpublished results). In contrast, the growth defect of sac1tsf sjl2Δ sjl3Δ cells was complemented by coexpression of the SJL2 or sjl2D850S alleles. Moreover, sac1tsf sjl2Δ sjl3Δ cells have been shown to accumulate very high levels of PtsIns(4)P and PtdIns(3,5)P2 at the nonpermissive temperature (Foti et al., 2001). At the nonpermissive temperature, sac1tsf sjl2C446S sjl3Δ cells accumulated PtsIns(4)P and PtdIns(3,5)P2 at levels similar to those in sac1tsf sjl2Δ sjl3Δ cells (Table 2). In contrast, the increases in PtdIns(4)P and PtdIns(3,5)P2 observed in sac1tsf sjl2Δ sjl3Δ cells were partly complemented by coexpression of SJL2 (Table 2), similar to levels previously described in sac1tsf sjl3Δ cells (Foti et al., 2001). Taken together, these results indicated that the sjl2C446S allele behaved as a null allele with regard to the Sac1-like activity of Sjl2p. Confirmation that the D850S substitution impaired the 5-Pase activity of Sjl2p in vivo is described below.
Table 2.
Phosphoinositide levels in wild-type and various PI phosphatase and kinase mutant cells
| Strain | PtdIns levels (% of total 3H-labeled PI)
|
|||
|---|---|---|---|---|
| PtdIns(3)P | PtdIns(4)P | PtdIns(3,5)P2 | PtdIns(4,5)P2 | |
| sac1tsf sjl2Δ sjl3Δ 38°C | 0.75 ± 0.09 | 27.4 ± 3.9 | 0.80 ± 0.13 | 0.97 ± 0.17 |
| sac1tsf sjl2Δ sjl3Δ (CEN sjl2C446S)a 38°C | 0.88 ± 0.12 | 28.1 ± 4.4 | 0.75 ± 0.16 | 1.04 ± 0.11 |
| sac1tsf sjl2Δ sjl3Δ (CEN SJL2)a 38°C | 0.68 ± 0.15 | 12.0 ± 1.7 | 0.27 ± 0.09 | 1.18 ± 0.24 |
| sjl1Δ sjl2ts sjl3Δ 38°C | 0.90 ± 0.10 | 1.21 ± 0.12 | 0.20 ± 0.04 | 2.93 ± 0.30 |
| sjl1Δ sjl2ts sjl3Δ (CEN sjl2D850S)b 38°C | 0.87 ± 0.13 | 1.10 ± 0.11 | 0.10 ± 0.03 | 2.78 ± 0.16 |
| sjl1Δ sjl2ts sjl3Δ (CEN SJL2)b 38°C | 0.94 ± 0.11 | 0.80 ± 0.10 | 0.10 ± 0.02 | 1.30 ± 0.10 |
| Wild type 26°C | 0.88 ± 0.03 | 0.99 ± 0.08 | 0.05 ± 0.01 | 0.89 ± 0.05 |
| Wild type 38°C | 0.85 ± 0.04 | 1.14 ± 0.10 | 0.08 ± 0.02 | 1.28 ± 0.14 |
| mss4ts 26°C | 0.86 ± 0.08 | 0.88 ± 0.06 | 0.02 ± 0.01 | 0.51 ± 0.06 |
| mss4ts 38°C | 0.79 ± 0.05 | 1.05 ± 0.08 | 0.09 ± 0.01 | 0.17 ± 0.05 |
| sjl1Δ sjl2Δ 26°C | 0.69 ± 0.12 | 0.91 ± 0.11 | 0.04 ± 0.02 | 1.90 ± 0.26 |
| sjl1Δ sjl2Δ mss4ts 26°C | 0.74 ± 0.07 | 0.86 ± 0.09 | 0.06 ± 0.02 | 0.94 ± 0.13 |
Yeast strains incubated at the indicated temperatures were labeled with myo-[3H]inositol. Lipids were extracted and deacylated for analysis by HPLC as described in Materials and Methods. The mean peak area (cpm) of each PtdIns species is reported as a percentage of total 3H-labeled PIs. The values reported are the means (± SD) of at least two or three independent experiments. The appropriate PI species in each subset of experiments have been indicated in bold text to highlight relevant comparisons.
Derived from AAY143 (Foti et al., 2001) by transformation with pRS415SJL2 or pRS415sjl2C446S.
Derived from YCS176 by transformation with pRS415SJL2 or pRS415sjl2D850S.
Next, we performed plasmid shuffle experiments to determine which mutant form of Sjl2p was sufficient for viability. LEU2-marked CEN plasmids carrying the SJL2, sjl2D850S, or sjl2C446S alleles were tranformed into sjl1Δ sjl2Δ sjl3Δ triple mutant cells also harboring a URA3 CEN SJL2 plasmid. As shown in Figure 1B, sjl1Δ sjl2Δ sjl3Δ cells expressing wild-type Sjl2p from the LEU2-marked CEN plasmid were able to grow on 5-FOA plates after loss of the URA3 CEN plasmid carrying SJL2. Likewise, sjl1Δ sjl2Δ sjl3Δ cells expressing Sjl2C446S from a LEU2-marked CEN plasmid were able to form colonies on 5-FOA plates after the loss of the URA3 CEN SJL2 plasmid. However, sjl1Δ sjl2Δ sjl3Δ cells carrying sjl2D850S on a LEU2-marked CEN plasmid were unable to grow on media containing 5-FOA and thus were unable to lose the URA3 CEN SJL2 plasmid (Figure 1B). These results indicated that the 5-Pase activity was in fact the essential function conferred by SJL1, SJL2, and SJL3.
The Yeast Synaptojanins Regulate Cellular PtdIns(3,5)P2 and PtdIns(4,5)P2 Levels
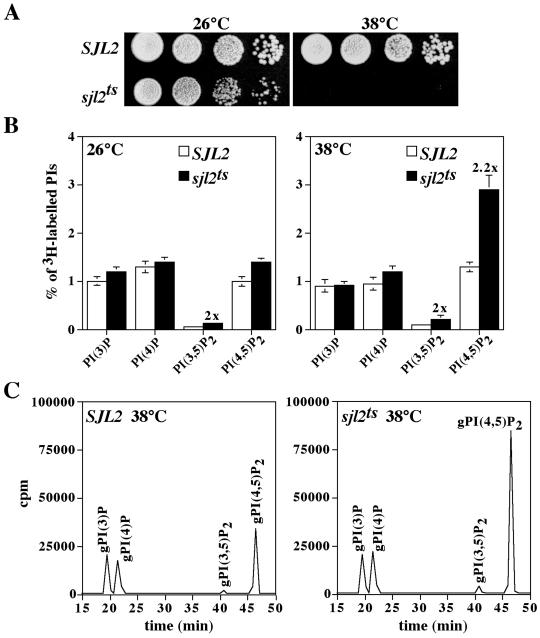
Our initial results suggested that proper homeostasis of cellular PtdIns(4,5)P2 levels was necessary for viability. To examine the immediate effects of inactivating the yeast synaptojanins, we generated temperature-conditional alleles of SJL2. The sjl1Δ sjl2Δ sjl3Δ strain harboring SJL2 on a URA3-marked CEN plasmid (YCS157) was transformed with pools of PCR-mutagenized SJL2 carried on LEU2-marked CEN plasmids. We then performed plasmid shuffle experiments to isolate mutants that were able to form colonies on 5-FOA plates at 26°C but not at 38°C. Approximately 12,000 transformants were screened for temperature-sensitive growth on media containing 5-FOA. Plasmids from transformants that were positive in this assay were recovered in E. coli and rescreened in YCS157 for the ability to confer temperature-sensitive growth on 5-FOA. One strain harboring an sjl2ts allele that conferred a strong temperature-sensitive growth phenotype (Figure 2A) was chosen for further characterization.
Figure 2.
sjl1Δ sjl2ts sjl3Δ cells display growth defects and generate increased levels of PtdIns(4,5)P2 at the restrictive temperature. (A) sjl1Δ sjl2Δ sjl3Δ cells expressing either SJL2 (YCS157) or sjl2ts (YCS176) from LEU2 CEN plasmids were grown in YPD at the permissive temperature. Tenfold serial dilutions of cells were spotted onto YPD and grown for 2 d at 26°C or 38°C. (B and C) Cells were preincubated at either 26°C or 38°C for 10 min and labeled with myo-[2-3H]inositol for 50 min at the permissive or nonpermissive temperatures. Lipids were then deacylated from cellular membranes, and glycero-phosphoinositols were extracted and analyzed by HPLC. (B) Quantitative comparisons of glycero-phosphoinositols generated by sjl1Δ SJL2 sjl3Δ cells (YCS157; open columns) and sjl1Δ sjl2ts sjl3Δ cells (YCS176; black columns) at 26°C (left) and 38°C (right) are shown. These data represent the means ± SD of three independent experiments. (C) Typical HPLC elution profiles are shown for sjl1Δ SJL2 sjl3Δ cells (YCS157; left) and sjl1Δ sjl2ts sjl3Δ cells (YCS176; right) at 38°C. Peaks corresponding to glycero-Ins(3)P, glycero-Ins(4)P, glycero-Ins(3,5)P2, and glycero-Ins(4,5)P2 are indicated.
Next, we analyzed changes in PI synthesis/turnover rates in sjl1Δ sjl2Δ sjl3Δ cells expressing either the SJL2 or sjl2ts alleles (hereafter referred to as SJL2 and sjl2ts cells, respectively). SJL2 and sjl2ts cells were pulse-labeled with myo-[2-3H]inositol at 26 and 38°C. Subsequent analysis of labeled lipids by HPLC enabled us to monitor the levels of glycero-phosphoinositols derived from PtdIns(3)P, PtdIns(4)P, PtdIns(3,5)P2, and PtdIns(4,5)P2. At the permissive temperature, PI levels in sjl2ts cells were nearly identical to those in SJL2 cells, except for an increase (twofold) in PtdIns(3,5)P2 levels (Figure 2B). Moreover, a shift to the restrictive temperature resulted in a specific increase in PtdIns(4,5)P2 levels (2.2-fold) in sjl2ts cells compared with SJL2 cells (Figure 2, B and C), consistent with our initial results demonstrating a requirement for the 5-Pase activity of Sjl2p for viability.
On sequence analysis of the sjl2ts allele used in these studies, we found several mutations throughout the SJL2-coding region. Subsequent mapping experiments indicated that substitutions in the Sac1 domain of Sjl2p were responsible for both the temperature-independent and temperature-sensitive phenotypes conferred by this sjl2ts allele (Stefan, Audhya, and Emr, unpublished results). The temperature-independent increases in PtdIns(3,5)P2 levels observed in sjl2ts cells suggested that these substitutions impaired the Sac1-like activity of Sjl2p in vivo, consistent with the increase in PtdIns(3,5)P2 levels previously observed in sjl2Δ sjl3Δ cells (Guo et al., 1999). More importantly, these substitutions caused an increase in PtdIns(4,5)P2 levels only at the restrictive temperature, consistent with the temperature-sensitive growth phenotype observed for these cells.
We used the sjl2ts cells to examine the effect of the Sjl2D850S substitution on the Sac1-like and 5-Pase activities of Sjl2p. At the nonpermissive temperature, sjl1Δ sjl2Δ sjl3Δ cells expressing both the sjl2ts and sjl2D850S alleles did not exhibit an increase in PtdIns(3,5)P2 levels as compared with SJL2 cells, but still displayed a twofold increase in PtdIns(4,5)P2 levels (Table 2). In contrast, coexpression of SJL2 complemented the effects on PtdIns(3,5)P2 and PtdIns(4,5)P2 levels observed in sjl2ts cells at the nonpermissive temperature (Table 2). These results indicated that the Sjl2D850S substitution impaired the 5-Pase activity of Sjl2p without affecting the Sac1-like PPIPase activity in vivo.
By measuring total cellular PI levels, we found that the yeast synaptojanin-like proteins regulated both PtdIns(3,5)P2 and PtdIns(4,5)P2 levels in vivo. Next, we assessed the physiological consequences of inactivating the yeast synaptojanins. Moreover, we addressed whether the SacI-like PPIPase or 5-Pase activities were specifically involved in the control of these various cellular processes.
The Yeast Synaptojanins Control Organization of the Actin Cytoskeleton by Regulating Cellular PtdIns(4,5)P2 Levels
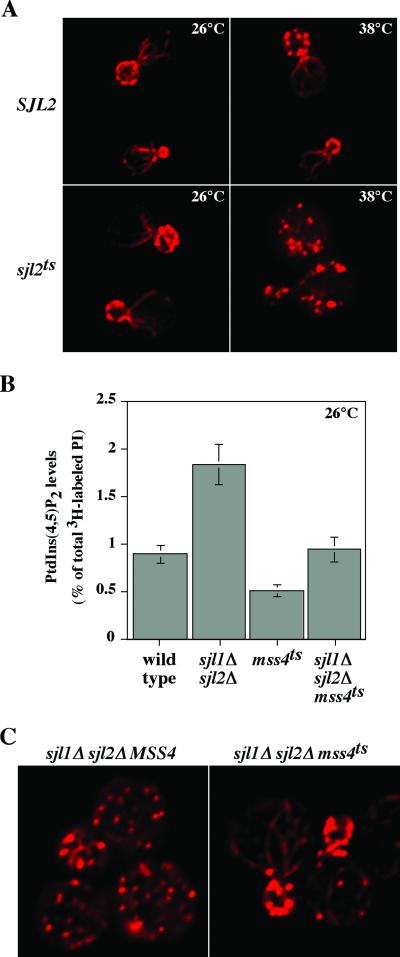
PIs, particularly PtdIns(4,5)P2, have been implicated in the organization of the actin cytoskeleton (Janmey, 1994). Because temperature shift of sjl2ts cells resulted in a significant increase in PtdIns(4,5)P2 levels, we investigated whether this resulted in effects on the actin cytoskeleton. At both the permissive and restrictive temperatures, SJL2 and sjl2ts cells were fixed, stained with rhodamine-phalloidin, and observed by fluorescence microscopy. At the permissive temperature, SJL2 and sjl2ts cells displayed actin cables in the mother cell aligned toward cortical actin patches concentrated in the bud, similar to previously published patterns of actin filaments in wild-type cells (Figure 3A; Karpova et al., 1998). However, after a shift to restrictive temperature, >90% of sjl2ts cells displayed actin patches randomly distributed throughout both mother and daughter cells, indicating that cells lacking synaptojanin activity fail to properly repolarize their actin cytoskeletons after exposure to high temperature (Figure 3A). After an identical temperature shift, >80% of SJL2 cells exhibited normal patterns of actin cables and cortical patches (Figure 3A). These results demonstrated that the yeast synaptojanins control actin organization and that the defects in actin cytoskeletal organization observed in sjl2ts cells correlated with increases in PtdIns(4,5)P2 levels.
Figure 3.
Cells deficient in synaptojanin-encoded PI 5-Pase activity fail to properly organize their actin cytoskeletons. (A) Actin cytoskeletal organization in sjl1Δ SJL2 sjl3Δ cells (YCS157; top) and sjl1Δ sjl2ts sjl3Δ cells (YCS176; bottom) at 26°C (left column) and 38°C (right column). Cells were grown to early log phase, shifted to the appropriate temperature for 2 h, fixed with 3.7% formaldehyde, and labeled with rhodamine-phalloidin to visualize actin filaments. (B) Quantitative comparisons of PtdIns(4,5)P2 levels synthesized by wild-type (SEY6210), sjl1Δ sjl2Δ MSS4 (AAY219.2), mss4ts (AAY202), and sjl1Δ sjl2Δ mss4ts (AAY219) cells at 26°C are shown. These data represent the means ± SD of at least two independent experiments. (C) Actin cytoskeletal organization in sjl1Δ sjl2Δ MSS4 cells (AAY219.2; left) and sjl1Δ sjl2Δ mss4ts cells (AAY219; right) at 26°C.
We further examined whether the actin defects observed in yeast synaptojanin mutants were directly due to increased levels of PtdIns(4,5)P2 or due to indirect effects of sjl null mutations. Previous work (Srinivasan et al., 1997; Stolz et al., 1998a) and our own studies have demonstrated that sjl1Δ sjl2Δ mutant cells possess increased PtdIns(4,5)P2 levels (Figure 3B) and defects in organization of the actin cytoskeleton (Figure 3C). Thus, we created an sjl1Δ sjl2Δ mss4ts triple mutant strain. MSS4 encodes the major yeast PtdIns(4)P 5-kinase (Desrivieres et al.,1998). The mss4ts allele used in this study conferred a weak defect in PtdIns(4,5)P2 synthesis even at the permissive temperature of 26°C compared with cells expressing wild-type MSS4 (Figure 3 B; Table 2). At the nonpermissive temperature, mss4ts cells displayed an approximately sevenfold decrease in PtdIns(4,5)P2 synthesis compared with MSS4 cells identically treated (Table 2). Importantly, at the permissive temperature, sjl1Δ sjl2Δ mss4ts triple mutant cells synthesized PtdIns(4,5)P2 at levels similar to wild-type cells (Figure 3B; Table 2).
To visualize actin, sjl1Δ sjl2Δ and sjl1Δ sjl2Δ mss4ts cells grown at the permissive temperature were fixed, stained with rhodamine-phalloidin, and observed by fluorescence microscopy. Whereas sjl1Δ sjl2Δ cells displayed random distribution of actin patches in both mother and daughter cells, proper organization of the actin cytoskeleton was restored in sjl1Δ sjl2Δ mss4ts cells, with actin cables in mother cells properly aligned toward cortical actin patches in the bud and septum (Figure 3C). These results indicated that increased cellular PtdIns(4,5)P2 levels directly affected organization of the actin cytoskeleton in sjl1Δ sjl2Δ cells, because sjl1Δ sjl2Δ mss4ts cells possessed normal PtdIns(4,5)P2 levels at the permissive temperature.
sjl2ts Cells Exhibit Impaired Rates of Endocytosis and an Aberrant Cell Surface Morphology
PIs have previously been implicated in intracellular trafficking and membrane dynamics (Simonsen et al., 2001). We investigated whether endocytic trafficking was affected in sjl2ts cells when incubated at the nonpermissive temperature. To do this, we examined rates of transport of the lipophilic dye FM4-64 to the vacuole. This vital dye is internalized from the plasma membrane into punctate intracellular compartments and ultimately is delivered to the vacuole membrane. Accordingly, SJL2 and sjl2ts cells were grown at the permissive temperature and then further incubated at the permissive or nonpermissive temperatures. These cells were pulse-labeled with FM4-64 and CMAC (CMAC stains the vacuole lumen) for 15 min, washed, and chased in media not containing FM4-64 or CMAC for either 0 or 30 min at the appropriate temperature. At the permissive temperature, SJL2 and sjl2ts cells displayed similar rates of transport of FM4-64 to the vacuole membrane (Stefan, Audhya, and Emr, unpublished results).
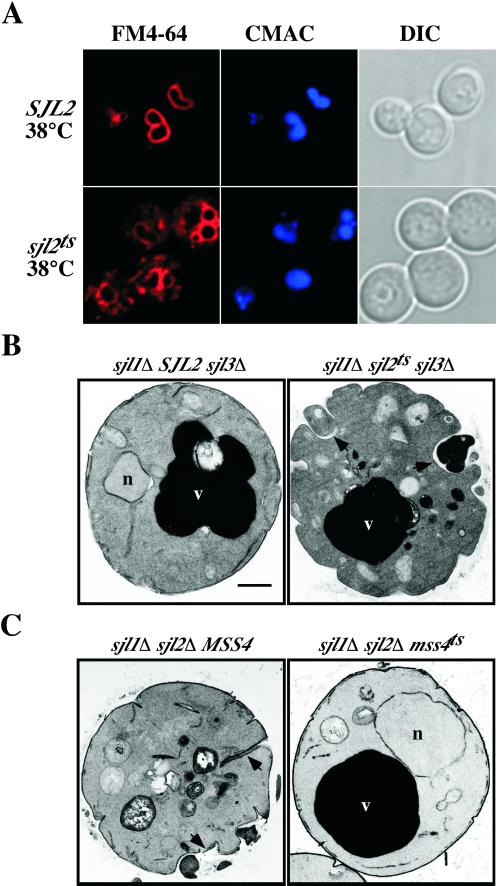
After a chase at restrictive temperature for 30 min, most SJL2 cells (>75%) displayed FM4-64 fluorescence only in the vacuole membrane (Figure 4A, top). In contrast, sjl2ts cells clearly displayed defects in endocytic delivery of FM4-64 to the vacuole (Figure 4A, bottom). In >80% of these cells, FM4-64 fluorescence accumulated in intracellular, punctate structures clearly distinct from the vacuole, as defined by CMAC staining. These results indicated that yeast synaptojanin activity is required for efficient transport of FM4-64 to the vacuole membrane and suggested that the endocytic defect correlated with increases in PtdIns(4,5)P2 levels because the effect was observed only after a shift to the nonpermissive temperature.
Figure 4.
Synaptojanin PI 5-Pase activity is required for normal endocytic transport and plasma membrane morphology. (A) sjl1Δ SJL2 sjl3Δ cells (YCS157; top) or sjl1Δ sjl2ts sjl3Δ cells (YCS176; bottom) were incubated at 38°C for 90 min and then labeled with the vital dyes FM4-64 and CMAC for 15 min. After labeling, cells were chased for 30 min at the nonpermissive temperature. The left column shows cells under fluorescent illumination in the rhodamine channel (FM4-64). The middle column shows cells as observed by CMAC fluorescence (CMAC) to visualize the lumen of vacuoles. The right column shows cells as observed by Nomarski optics (DIC). (B) Ultrastructure of sjl1Δ SJL2 sjl3Δ cells (YCS157; left) and sjl1Δ sjl2ts sjl3Δ cells (YCS176; right) at the restrictive temperature. YCS157 and YCS176 were grown to early log phase, shifted to 38°C for 90 min, fixed with 3% glutaraldehyde, and processed for electron microscopy. (C) Ultrastructure of sjl1Δ sjl2Δ MSS4 cells (AAY219.2; left) and sjl1Δ sjl2Δ mss4ts cells (AAY219; right) at 26°C. (B and C) Arrowheads indicate abnormal invaginations at the cell surface; v, vacuole; n, nucleus; bar, 0.5 μm.
At the nonpermissive temperature, sjl2ts cells displayed large, aberrant cell surface structures stained with FM4-64 at the 0-min chase time (Stefan, Audhya, and Emr, unpublished results), likely corresponding to large invaginations at the cell surface previously observed in sjl1Δ sjl2Δ mutants (Srinivasan et al., 1997; Singer-Kruger et al., 1998; Stolz et al., 1998a). To determine whether the yeast synaptojanins play a primary role in regulating membrane dynamics at the cell surface, SJL2 and sjl2ts cells were examined at the ultrastructural level. Electron microscopy revealed that sjl2ts cells were similar to SJL2 cells at the permissive temperature, except for the presence of slightly fragmented vacuoles in some cells (Stefan, Audhya, and Emr, unpublished results). However, sjl2ts cells formed large, abnormal invaginations at the plasma membrane (indicated by arrowheads in Figure 4B) in contrast to cells expressing SJL2 at the restrictive temperature.
To determine whether the abnormal cell surface morphology observed in yeast synaptojanin mutants was directly due to increased levels of PtdIns(4,5)P2 or caused by indirect effects of sjl null mutations, we examined sjl1Δ sjl2Δ and sjl1Δ sjl2Δ mss4ts cells at the ultrastructural level. Electron microscopy revealed that, whereas >70% of sjl1Δ sjl2Δ cells displayed large, abnormal invaginations at the cell surface (indicated by arrowheads in Figure 4C), normal morphology of the plasma membrane was restored in >90% sjl1Δ sjl2Δ mss4ts cells grown at the permissive temperature (Figure 4C). These results indicated that increased cellular PtdIns(4,5)P2 levels directly affected membrane dynamics at the cell surface, because sjl1Δ sjl2Δ mss4ts cells possessed normal PtdIns(4,5)P2 levels at the permissive temperature (Figure 3B).
The 5-Pase Activity of the Yeast Synaptojanins Regulates TGN/Endosomal Sorting via Clathrin-coated Vesicles
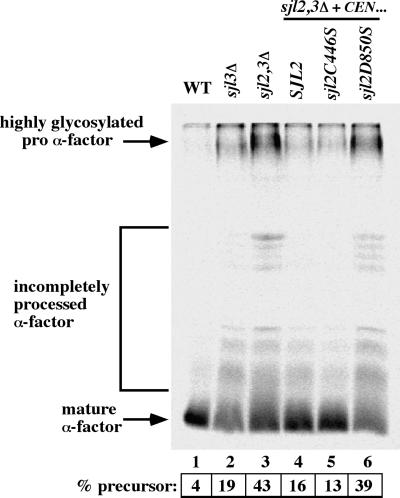
Sjl3p has been implicated in clathrin-dependent transport between the TGN and endosomes (Bensen et al., 2000). To address whether SacI-like PPIPase or 5-Pase activities were involved in the control of this pathway, we took advantage of the sjl2C446S and sjl2D850S alleles that inactivate the SacI-like or 5-Pase activities of Sjl2p, respectively. These sjl2 alleles were expressed in sjl2Δ sjl3Δ cells and examined for effects on processing of secreted α-factor. Maturation of precursor α-factor is mediated by the protease Kex2p. Kex2p cycles between the TGN and endosomes in a clathrin-dependent manner. If clathrin is impaired, Kex2p becomes mislocalized and inefficient maturation of α-factor precursor occurs (Bensen et al., 2000). Thus, the extent of α-factor maturation serves as a measure of clathrin function at the TGN. To monitor α-factor maturation, cells expressing various SJL2 alleles were labeled with [35S]methionine, and secreted α-factor was immunoprecipitated from the medium.
Whereas wild-type cells secreted fully mature pheromone, sjl3Δ cells displayed a slight impairment in α-factor processing (19% precursor form; Figure 5, lanes 1 and 2), consistent with previously published observations (Bensen et al., 2000). Moreover, although deletion of SJL2 had no effect on α-factor processing (Bensen et al., 2000), deletion of SJL2 and SJL3 conferred additive effects upon α-factor maturation, because sjl2Δ sjl3Δ cells secreted ∼40% of α-factor in the precursor form (Figure 5, lane 3). In sjl2Δ sjl3Δ cells expressing wild-type SJL2 from a LEU2-marked CEN plasmid, α-factor processing was similar to that in sjl3Δ cells (16% precursor form), as expected. Importantly, sjl2Δ sjl3Δ cells expressing sjl2C446S from a CEN plasmid did not display impaired α-factor processing, compared with sjl3Δ cells (13% precursor form; Figure 5, lane 5). In contrast, sjl2Δ sjl3Δ cells expressing sjl2D850S secreted the precursor form of α-factor at levels nearly identical to that of sjl2Δ sjl3Δ cells (Figure 5, lane 6). Taken together, these results indicated that the 5-Pase activity of Sjl2p regulated clathrin-dependent protein sorting between the TGN and endosomes.
Figure 5.
Inactivation of SJL2-encoded PI 5-Pase activity affects α-factor maturation. Wild-type (WT; SEY6210; lane 1), sjl3Δ (JGY131; lane 2), sjl2Δ sjl3Δ (JGY132; lane 3), SJL2 sjl3Δ (JGY132 carrying pRS416SJL2; lane 4), sjl2C446S sjl3Δ (JGY132 carrying pRS416sjl2C446S; lane 5), and sjl2D850S sjl3Δ (JGY132 carrying pRS416sjl2D850S; lane 6) cells were metabolically labeled at 26°C for 45 min, and secreted α-factor was immunoprecipitated from the culture supernatants. Samples were analyzed by SDS-PAGE and fluorography. Precursor levels of α-factor were quantified by phosphoimage analysis. The levels of secreted precursor α-factor indicated are the means of two independent experiments; SD < 15%.
We investigated whether the effects of inactivating the yeast synaptojanins were specific for clathrin-mediated protein transport pathways from the TGN. Accordingly, we examined roles for the yeast synaptojanins in additional protein-trafficking pathways, including two TGN to vacuole pathways and the secretory pathway. First, we determined whether the yeast synaptojanins were required for vacuole protein sorting and transport. Using the sjl1Δ sjl2Δ sjl3Δ strains expressing either the SJL2 or sjl2ts alleles, we examined the transport and processing of two vacuolar proteins, CPY and ALP. However, even after an extended preshift to the nonpermissive temperature, neither CPY nor ALP transport was affected in sjl2ts cells compared with SJL2 control cells (Stefan, Audhya, and Emr, unpublished results). Finally, we investigated a role for the yeast synaptojanins in secretion. By performing pulse-chase experiments, we tested whether Hsp150p, a high-molecular-weight glycoprotein that is rapidly secreted, was affected upon inactivation of the yeast synaptojanins. However, no significant impairment of Hsp150p glycosylation or secretion was observed in sjl2ts cells after shift to the nonpermissive temperature compared with cells expressing SJL2 (Stefan, Audhya, and Emr, unpublished results). Taken together, these results indicated that inactivation of the yeast synaptojanins does not confer rapid, nonspecific pleiotropic effects on protein transport from the Golgi complex.
The Yeast Syanptojanins Control the Intracellular Distribution of PtdIns(4,5)P2
Because our results indicated that the yeast synaptojanins control several cellular processes by regulating PtdIns(4,5)P2, we wished to localize this PI in vivo. For this purpose, we took advantage of the PH domain from PLCδ, which has been shown to bind PtdIns(4,5)P2 in vitro (Kavran et al., 1998). We fused two tandem copies of the PLCδ PH domain to GFP to create a PtdIns(4,5)P2-specific FLARE, GFP-2×PH(PLCδ). In wild-type cells, GFP-2×PH(PLCδ) fluorescence was observed at the plasma membrane and weakly in the cytosol but not on intracellular compartments (Figure 6A). To demonstrate the specificity of this FLARE for PtdIns(4,5)P2 in vivo, we expressed GFP-2×PH(PLCδ) in mss4ts mutant cells. At the permissive temperature, GFP-2×PH(PLCδ) localized to the plasma membrane similar to wild-type cells (Stefan, Audhya, and Emr, unpublished results). On shift to the nonpermissive temperature, GFP fluorescence became diffuse in mss4ts cells, mainly throughout the cytosol, indicating that recruitment of GFP-2×PH(PLCδ) to the plasma membrane was dependent on Mss4p activity (Figure 6A).
Figure 6.
The steady-state localization of a PtdIns(4,5)P2-specific FLARE at the plasma membrane in yeast is dependent on Mss4p and synaptojanin-like activities. (A) Wild-type (SEY6210; left) or mss4ts cells (AAY202; right) expressing a GFP-2×PH(PLCδ) fusion were incubated at 38°C for 45 min and observed by fluorescent microscopy using a DeltaVision deconvolution system. (B) sjl1Δ SJL2 sjl3Δ cells (YCS157; left) or sjl1Δ sjl2ts sjl3Δ cells (YCS176; right) expressing GFP-2×PH(PLCδ) were incubated for 0 or 60 min at the nonpermissive temperature. (C) sjl1Δ sjl2ts sjl3Δ cells (YCS176) expressing GFP-2×PH(PLCδ) incubated at the nonpermissive temperature for 30 min were killed by the addition of NaN3 and NaF to inhibit further internalization from the plasma membrane, stained with FM4-64 on ice, and observed by fluorescent microscopy. DIC, Nomarski optics.
Next, we examined the role of the yeast synaptojanins in regulating the cellular location of PtdIns(4,5)P2. In SJL2 cells expressing GFP-2×PH(PLCδ), GFP fluorescence was observed at the plasma membrane at both the permissive and nonpermissive temperatures (Figure 6B, left). Likewise, localization of GFP-2×PH(PLCδ) was restricted to the plasma membrane in sjl2ts cells at the permissive temperature (Figure 6B, top right). Interestingly, GFP fluorescence at the plasma membrane became punctate in sjl2ts cells expressing GFP-2×PH(PLCδ) when shifted to the nonpermissive temperature (Figure 6B, bottom right). Moreover, at the nonpermissive temperature, GFP-2×PH(PLCδ) was observed on intracellular compartments as well as the plasma membrane in sjl2tscells (Figure 6B, bottom right). To confirm that these structures were indeed intracellular compartments, sjl2ts cells expressing GFP-2×PH(PLCδ), which had been incubated at the nonpermissive temperature for 30 min, were killed by the addition of NaN3 and NaF to inhibit further internalization from the plasma membrane and stained with FM4-64 on ice. Under these conditions, punctate intracellular compartments containing GFP fluorescence were observed that clearly did not colocalize with FM4-64 fluorescence at the plasma membrane (Figure 6C). However, colocalization was observed between GFP and FM4-64 fluorescence on punctate, intracellular compartments in metabolically active sjl2ts cells expressing GFP-2×PH(PLCδ) after a brief labeling and chase with FM4-64 at the nonpermissive temperature, suggesting that these structures may be endocytic compartments (Stefan, Audhya, and Emr, unpublished results). Taken together, these results indicated that the yeast synaptojanins are essential to restrict the steady-state accumulation of PtdIns(4,5)P2 to the plasma membrane.
Our results using the PtdIns(4,5)P2-specific FLARE indicated that the yeast synaptojanins control the distribution of PtdIns(4,5)P2 within cells. As a control, we utilized another PH domain from the mammalian protein FAPP1 that specifically bound PtdIns(4)P in vitro (Dowler et al., 2000). This PH domain was fused to GFP to create a PtdIns(4)P-specific FLARE, GFP-PH(FAPP1), and expressed in yeast. In wild-type cells expressing GFP-PH(FAPP1), GFP fluorescence was observed on punctate, intracellular compartments but not at the plasma membrane (Figure 7). Next, we examined the localization of GFP-PH(FAPP1) in various yeast PI kinase mutants. Interestingly, GFP-PH(FAPP1) was diffuse throughout the cytosol in pik1ts cells (Audhya et al., 2000) when incubated at the nonpermisive temperature. Thus, PIK1-encoded PtdIns 4-kinase activity was necessary for the punctate localization of GFP-PH(FAPP1) (Figure 7). In contrast, Mss4p activity was not required for the localization of GFP-PH(FAPP1) to puncta, because GFP-PH(FAPP1) displayed wild-type localization in mss4ts cells at the nonpermissive temperature (Figure 7). Moreover, we found that the localization of GFP-PH(FAPP1) did not change in sjl2ts cells at the nonpermissive temperature (Stefan, Audhya, and Emr, unpublished results), providing additional evidence for the PI-binding specificities of the FLAREs used in these studies.
Figure 7.
The intracellular localization of a PtdIns(4)P-specific FLARE to the Golgi is dependent on Pik1p activity. Wild-type cells (SEY6210), pik1ts cells (AAY104), mss4ts cells (AAY202), or arf1Δ cells (6210arf1Δ) expressing GFP-PH(FAPPI) were incubated at 26°C (left column) or 38°C (right column) for 20 min as indicated and observed by fluorescent microscopy using a DeltaVision deconvolution system. DIC, Nomarski optics.
Because Pik1p has been implicated in transport from the Golgi (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000), we examined whether the structures that contained GFP-PH(FAPP1) corresponded to Golgi compartments. To do this, we expressed GFP-PH(FAPP1) in arf1Δ mutant cells. In arf1Δ cells, atypical membrane ring structures accumulate that contain known Golgi proteins (Gaynor et al., 1998). Similarly, GFP-PH(FAPP1) localized to ring-like structures in arf1Δ cells (Figure 7, arrows), suggesting that GFP-PH(FAPP1) recognized PtdIns(4)P generated by Pik1p on intracellular Golgi compartments.
DISCUSSION
In this study, we examined the consequences of inactivating the yeast synaptojanins by generating a strain that expresses only a temperature-sensitive SJL2 allele. Impairment of the yeast synaptojanins resulted in effects on the actin cytoskeleton, endocytic transport, cell surface morphology, and clathrin-dependent transport between the TGN and endosomes, consistent with previously published reports. Although sjl2ts cells accumulated increased levels of both PtdIns(3,5)P2 and PtdIns(4,5)P2, our studies indicated that these phenotypes correlated with accumulation of PtdIns(4,5)P2. Moreover, we found that PtdIns(4,5)P2 accumulated on intracellular compartments, as well as the plasma membrane when the yeast synaptojanins were inactivated, providing the first demonstration that the yeast synaptojanins control the subcellular localization of PtdIns(4,5)P2. This spatial control of PtdIns(4,5)P2 was essential for normal cell morphology and membrane transport.
The Yeast Synaptojanins Regulate Cellular PtdIns(3,5)P2 and PtdIns(4,5)P2 Levels
By measuring total cellular PI levels, our data confirmed that the yeast synaptojanins regulated both PtdIns(3,5)P2 and PtdIns(4,5)P2 levels in vivo. In sjl2ts cells, PtdIns(3,5)P2 levels were increased (twofold) at both the permissive and restrictive temperatures. More importantly, a specific increase in PtdIns(4,5)P2 levels was observed in sjl2ts cells at the restrictive temperature (2.2-fold; see Figure 2), consistent with our initial results demonstrating a requirement for PI 5-Pase activity for viability.
The physiological consequences of inactivating the yeast synaptojanins observed in this study were specifically due to increases in PtdIns(4,5)P2. Thus, an interesting question arises as to why Sjl2p, Sjl3p, and mammalian synaptojanin possess both PPIPase and 5-Pase activities. Previous in vitro studies have indicated that the 5-Pase and Sac1 domains of yeast and mammalian synaptojanins act sequentially to dephosphorylate PtdIns(4,5)P2 to PtdIns(4)P and PtdIns, respectively (Guo et al., 1999). However, our results suggested that the Sac1 and 5-Pase domains of Sjl2p may have separate functions as well, because sjl2ts cells displayed increases in both PtdIns(3,5)P2 and PtdIns(4,5)P2 cellular levels. Consistent with this, previous work has indicated overlapping roles for the Sac1 domain-containing proteins Sjl2p, Sjl3p, and Sac1p in the control of PtdIns(4)P cellular levels (Foti et al., 2001).
Coordinate Control of PtdIns(4,5)P2, Actin, and Cell Surface Morphology by PI 5-Pases
Previous studies have implicated PIs in the regulation of the actin cytoskeleton (Janmey, 1994). Our studies confirmed that synaptojanin activity is required for proper actin organization in yeast. Consistent with this, previous work has shown that Sjl2p and Sjl3p partially colocalize with actin patches (Ooms et al., 2000). The defects in actin cytoskeletal organization observed in synaptojanin-deficient cells specifically correlated with increases in PtdIns(4,5)P2 levels, because sjl2ts cells displayed defects in the actin cytoskeleton only at the nonpermissive temperature (Figure 3A). Others have shown that Mss4p, the major yeast PtdIns(4)P 5-kinase, was required for proper actin organization (Desrivieres et al., 1998). Thus, both increases and decreases in cellular PtdIns(4,5)P2 levels affected actin organization in yeast. Interestingly, we found that restoration of normal cellular PtdIns(4,5)P2 levels in sjl1Δ sjl2Δ mss4ts cells rescued the actin cytoskeletal defects observed in sjl1Δ sjl2Δ cells (see Figure 3, B and C), indicating that cellular PtdIns(4,5)P2 levels directly affected organization of the actin cytoskeleton. Consistent with this, a previous study showed that overexpression of a mammalian PI 5-Pase corrected the lipid and actin defects observed in cells lacking Sjl1p, Sjl2p, and Sjl3p (O'Malley et al., 2001).
Thus, in a simple model, Mss4p and the yeast synaptojanins control cellular PtdIns(4,5)P2 levels, which in turn govern the actin cytoskeleton by recruiting and/or regulating actin-binding proteins. Accordingly, PtdIns(4,5)P2 has been shown to control actin polymerization by binding several actin regulatory proteins, such as N-WASP, profilin, cofilin, and capping proteins (Schafer et al., 1996; Higgs and Pollard, 2000; Prehoda et al., 2000; Rohatgi et al., 2000). Thus, both increases and decreases in cellular PtdIns(4,5)P2 levels may confer effects upon actin polymerization and organization in vivo. We were not able to detect significant changes in the relative amounts of filamentous actin in pellet fractions made from lysates of SJL2 and sjl2ts cells after incubation at the nonpermissive temperature (Stefan, Audhya, and Emr, unpublished results). Thus, the yeast synaptojanins may not regulate cellular levels of filamentous actin per se but rather control the organization of actin filaments in vivo, similar to a previously published study using various mutants and conditions known to affect the arrangement of yeast actin patches and cables (Karpova et al., 1998). Additional biochemical experiments will be required to more directly examine the effects of inactivating the yeast synaptojanins on PtdIns(4,5)P2-regulated actin regulatory proteins.
Several mammalian studies have shown that alterations in intracellular PtdIns(4,5)P2 levels have varying effects on the actin cytoskeleton. Overexpression of PI 5-Pases has been shown to concurrently reduce PtdIns(4,5)P2 levels, formation of actin stress fibers, and the attachment of existing filaments to the plasma membrane (Sakisaka et al., 1997; Raucher et al., 2000). Likewise, overexpression of type I PI 5-kinase induced dramatic changes in the actin cytoskeleton. These included stabilization of comet-like actin tails associated with vesicles (Rozelle et al., 2000), either induction or inhibition of plasma membrane ruffling (Honda et al., 1999; Yamamoto et al., 2001), and stabilization of thick actin stress fibers (Yamamoto et al., 2001). Thus, extensive evidence including our work has indicated that both increases and decreases in cellular PtdIns(4,5)P2 levels can confer effects on actin cytoskeletal organization in vivo.
Previous studies have implicated PIs in the regulation of membrane dynamics and organelle morphology (Odorizzi et al., 2000). Accordingly, a previous study has demonstrated that PtdIns(4,5)P2 controls tension between the plasma membrane and actin cytoskeleton (Raucher et al., 2000). Our studies demonstrated that the yeast synaptojanins control plasma membrane dynamics by regulating cellular PtdIns(4,5)P2 levels. Importantly, we found that restoration of normal cellular PtdIns(4,5)P2 levels in sjl1Δ sjl2Δ mss4ts cells rescued the cell surface morphology effects observed in sjl1Δ sjl2Δ cells (see Figure 4 C).
The cell surface morphology defects observed in yeast synaptojanin mutants are likely because of additive effects on PtdIns(4,5)2-mediated control of multiple pathways. Besides effects on actin and plasma membrane dynamics, inactivation of the yeast synaptojanins confers rapid effects on cell wall structure (Srinivasan et al., 1997; Stolz et al., 1998a; Stefan, Audhya, and Emr, unpublished results). The thickened cell wall phenotype observed in yeast synaptojanin mutants may be due to improper localization of chitin synthases at the plasma membrane (Ziman et al., 1998). Alternatively, PtdIns(4,5)P2 could regulate yeast cell surface morphology through the PKC1 protein kinase pathway, which controls the actin cytoskeleton and cell wall biosynthetic enzymes after heat shock or other extracellular stimuli (Helliwell et al., 1998; Delley and Hall, 1999). Previous studies implicated PtdIns(4,5)P2 in the yeast PKC1 protein kinase pathway, because mss4ts cells are unable to repolarize their actin cytoskeletons and undergo cell lysis at restrictive temperatures (Desrivieres et al., 1998). Accordingly, the yeast synaptojanins may act as regulators of the PKC1 pathway, because sjl2ts cells display actin defects and cell wall thickening at the nonpermissive temperature. Additional experiments will be needed to examine the precise roles of the yeast synaptojanins in each of these pathways. Nonetheless, our studies suggest that the yeast synaptojanins coordinately control actin cytoskeletal organization, endocytosis, and cell wall synthesis by regulating PtdIns(4,5)P2 levels at the plasma membrane.
The Role of PI 5-Pases in Clathrin Function
PIs have been implicated in intracellular trafficking (Simonsen et al., 2001). Accordingly, previous work has suggested that mammalian synaptojanin regulates the association of clathrin with membranes (Hill et al., 2001). Mammalian synaptojanin has been proposed to act in the uncoating step of endocytic clathrin-coated vesicles (Cremona et al., 1999) and has been demonstrated to physically interact with clathrin coats (Haffner et al., 2000). Likewise, the yeast synaptojanins have been implicated in endocytosis (Singer-Kruger et al., 1998) and have been shown to genetically interact with clathrin (Bensen et al., 2000). Our results indicated that regulation of PtdIns(4,5)P2 by the yeast synaptojanins controls two clathrin-mediated transport pathways, endocytosis (Figure 4A) and sorting between the TGN and endosomes (Figure 5). Although α-factor processing was affected in synaptojanin-deficient cells, we did not observe effects on additional transport pathways from the TGN. Thus, improper accumulation of PtdIns(4,5)P2 may specifically affect clathrin-mediated trafficking between the TGN and endosomes rather than conferring general effects on transport from the Golgi.
Spatial Control of PtdIns(4,5)P2 by the Yeast Syanptojanins
Because the accumulation of PtdIns(4,5)P2 was detrimental to several cellular processes, we developed a convenient and reliable method to visualize this PI in yeast. For this purpose, we used the PH domain from PLCδ that specifically binds PtdIns(4,5)P2 in vitro (Kavran et al., 1998). The PtdIns(4,5)P2-specific FLARE used in this study, GFP-2×PH(PLCδ), localized to the plasma membrane in a manner dependent on Mss4p activity but not on intracellular compartments (Figure 6A). Likewise, GFP-2×PH(PLCδ) was restricted to the plasma membrane in sjl2ts cells at the permissive temperature (Figure 6B). This result indicated that Sjl2p 5-Pase activity was sufficient to maintain the steady-state distribution of PtdIns(4,5)P2 at the plasma membrane and not in intracellular compartments. However, GFP-2×PH(PLCδ) was observed in intracellular compartments as well as the plasma membrane in sjl2ts cells at the nonpermissive temperature (Figure 6, B and C), indicating that the yeast synaptojanins were essential to restrict the steady-state accumulation of PtdIns(4,5)P2 to the plasma membrane. Although these experiments did not address whether the PtdIns(4,5)P2 that accumulated intracellularly in sjl2ts cells was synthesized at the plasma membrane or in intracellular compartments, previous work demonstrated that Mss4p localized primarily to the plasma membrane and not on internal structures (Homma et al., 1998).
Whereas our initial studies indicate that the phenotypes observed in synaptojanin-deficient cells correlate with an increase in total cellular PtdIns(4,5)P2 levels, our studies uising GFP-2×PH(PLCδ) demonstrate that the yeast synaptojanins are critical for the spatial control of PtdIns(4,5)P2. Thus, certain phenotypes observed in synaptojanin-deficient cells may be due to the inappropriate localization of PtdIns(4,5)P2 rather than slight increases of this lipid at the plasma membrane. Although overall PtdIns(4,5)P2 levels are increased only twofold in synaptojanin-deficient cells, the relative concentration of PtdIns(4,5)P2 on certain intracellular membranes may be increased by more than an order of magnitude (see Figure 6). Consistent with this idea, although sjl2Δ sjl3Δ cells do not display an increase in total cellular PtdIns(4,5)P2 levels (Stolz et al., 1998a), we find that the GFP-2×PH(PLCδ) reporter is present on intracellular structures in these cells (Stefan, Audhya, and Emr, unpublished results). Accordingly, PtdIns(4,5)P2-interacting proteins may become mislocalized in synaptojanin-deficient cells, resulting in the titration of these PtdIns(4,5)P2 effector molecules away from their normal site of function at the plasma membrane. Alternatively, mislocalization of PtdIns(4,5)P2 may result in the recruitment of effectors to new membrane sites, where they could interfere with the normal function of these membranes. These effects could explain the lethality of cells lacking synaptojanin-like activity. The future identification of these effectors (e.g., various PH and/or ENTH domain-containing proteins) will allow us to directly test this hypothesis.
Previous studies using various PI-specific FLAREs have suggested that distinct PI isoforms are compartmentalized within cells (Balla et al., 2000). Our studies using a PtdIns(4,5)P2-specific FLARE indicated that PtdIns(4,5)P2 accumulated on the plasma membrane in wild-type yeast cells. Other studies utilizing PtdIns(3)P-specific FLAREs have demonstrated that PtdIns(3)P was present in endosomal compartments (Burd and Emr, 1998; Gillooly et al., 2000). Several recent reports have suggested that Pik1p, a PtdIns 4-kinase, regulates transport from the Golgi in yeast (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000). Thus, we fused GFP to a PH domain that specifically binds PtdIns(4)P in vitro (Dowler et al., 2000). We found that this PtdIns(4)P-specific FLARE, GFP-PH(FAPP1), localized to punctate, intracellular compartments in a manner dependent on Pik1p activity (Figure 7). Moreover, GFP-PH(FAPP1) localized to ring-like structures in arf1Δ cells (Figure 7), perhaps corresponding to the abnormal ring-like Golgi compartments previously observed in arf1Δ cells by electron microscopy and fluorescence experiments (Gaynor et al., 1998). Taken together, these results suggested that PtdIns(4)P generated by the PtdIns 4-kinase Pik1p localized to yeast Golgi compartments, consistent with a role for Pik1p in transport from the Golgi. Accordingly, Pik1p has previously been localized to Golgi structures (Walch-Solimena and Novick, 1999).
Overall, these studies using GFP-based FLAREs with distinct PI-binding specificities provide additional evidence for the compartmentalization of particular PI species at discrete membrane sites. More importantly, our studies provide the first evidence that the yeast synaptojanins not only control PtdIns(4,5)P2 cellular levels but also are essential for the proper spatial control of this PI isoform. Future studies using synaptojanin-deficient cells will likely reveal various effects on the activity and/or localization of several downstream effector proteins that bind PtdIns(4,5)P2.
ACKNOWLEDGMENTS
We are indebted to Dr. John York for generously providing SJL1 and SJL2 plasmids and helpful discussions. We also thank Dr. Mark Lemmon and Dr. Dario Alessi for generously providing plasmids encoding the PLCδ PH domain or the FAPP1 PH domain, respectively. We thank Ingrid Niesman and Tammie McQuistan for assisting with the electron microscopic analysis (Immunoelectron Microscopy Core B of Program Project grant CA58689 headed by M. Farquhar) and Steven Padilla and Perla Arcaira for helpful technical assistance. We also thank members of the Emr laboratory, Pietro De Camilli, Jeremy Thorner, Sean Munro, and Greg Payne for useful discussions. This work was supported by grant CA58689 from the National Institutes of Health (to S.D.E.). C.J.S. is a fellow of the American Cancer Society supported by the Holland Peck Charitable Fund. S.D.E. is an investigator of the Howard Hughes Medical Institute.
Abbreviations used:
- ALP
alkaline phosphatase
- CEN, centromeric
CPY, carboxypeptidase Y
- FLARE
fluorescent lipid-associated reporter
- FM4-64
N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide
- 5-FOA
5-fluoroorotic acid
- FYVE domain
for Fab1, YGL023, Vps27, and EEA1
- GFP
green fluorescent protein
- 5-Pase
phosphoinositide 5-phosphatase
- PH
pleckstrin homology
- PI
phosphoinositide
- PLCδ
phospholipase C δ1
- PPIPase
polyphosphoinositide phosphatase
- PtdIns
phosphatidylinositol
- PtdIns(3)P
phosphatidylinositol 3-phosphate
- PtdIns(4)P
phosphatidylinositol 4-phosphate
- PtdIns(3,5)P2
phosphatidylinositol (3,5)-bisphosphate
- PtdIns(4,5)P2
phosphatidylinositol (4,5)-bisphosphate
- Sjl
synaptojanin-like
- TGN
trans-Golgi network
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–10-0476. Article and publication date are at www.molbiolcell.org/cgi/10.1091/mbc.01–10-0476.
REFERENCES
- Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T, Bondeva T, Varnai P. How accurately can we image inositol lipids in living cells? Trends Pharmacol Sci. 2000;21:238–241. doi: 10.1016/s0165-6147(00)01500-5. [DOI] [PubMed] [Google Scholar]
- Bensen ES, Costaguta G, Payne GS. Synthetic genetic interactions with temperature-sensitive clathrin in Saccharomyces cerevisiae: roles for synaptojanin-like Inp53p and dynamin-related Vps1p in clathrin-dependent protein sorting at the trans-Golgi network. Genetics. 2000;154:83–97. doi: 10.1093/genetics/154.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho RJ, Teruel M, Dierckman R, Anderson R, Wells A, York JD, Meyer T, Grinstein S. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J Cell Biol. 2000;151:1353–1368. doi: 10.1083/jcb.151.7.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- Cremona O, Di Paolo G, Wenk MR, Luthi A, Kim WT, Takei K, Daniell L, Nemoto Y, Shears SB, Flavell RA, McCormick DA, De Camilli P. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 1999;99:179–188. doi: 10.1016/s0092-8674(00)81649-9. [DOI] [PubMed] [Google Scholar]
- Czech MP. PIP2 and PIP3: complex roles at the cell surface. Cell. 2000;100:603–606. doi: 10.1016/s0092-8674(00)80696-0. [DOI] [PubMed] [Google Scholar]
- Delley PA, Hall MN. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J Cell Biol. 1999;147:163–174. doi: 10.1083/jcb.147.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrivieres S, Cooke FT, Parker PJ, Hall MN. MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J Biol Chem. 1998;273:15787–15793. doi: 10.1074/jbc.273.25.15787. [DOI] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD. SacI lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell. 2001;12:2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Gaullier JM, Simonsen A, D'Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- Gaynor EC, Chen CY, Emr SD, Graham TR. ARF is required for maintenance of yeast Golgi and endosome structure and function. Mol Biol Cell. 1998;9:653–670. doi: 10.1091/mbc.9.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Haffner C, Di Paolo G, Rosenthal JA, de Camilli P. Direct interaction of the 170 kDa isoform of synaptojanin 1 with clathrin and with the clathrin adaptor AP-2. Curr Biol. 2000;10:471–474. doi: 10.1016/s0960-9822(00)00446-2. [DOI] [PubMed] [Google Scholar]
- Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Schmidt A, Ohya Y, Hall MN. The Rho1 effector Pkc1, but not Bni1, mediates signaling from Tor2 to the actin cytoskeleton. Curr Biol. 1998;8:1211–1214. doi: 10.1016/s0960-9822(07)00511-8. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD. Activation by Cdc42 and PIP2 of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J Cell Biol. 2000;150:1311–1320. doi: 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E, van Der Kaay J, Downes CP, Smythe E. The role of dynamin and its binding partners in coated pit invagination and scission. J Cell Biol. 2001;152:309–323. doi: 10.1083/jcb.152.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma K, Terui S, Minemura M, Qadota H, Anraku Y, Kanaho Y, Ohya Y. Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J Biol Chem. 1998;273:15779–15786. doi: 10.1074/jbc.273.25.15779. [DOI] [PubMed] [Google Scholar]
- Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA, Kanaho Y. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Meyer T. Subcellular targeting by membrane lipids. Curr Opin Cell Biol. 2001;13:146–152. doi: 10.1016/s0955-0674(00)00191-5. [DOI] [PubMed] [Google Scholar]
- Janmey PA. Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly. Annu Rev Physiol. 1994;56:169–191. doi: 10.1146/annurev.ph.56.030194.001125. [DOI] [PubMed] [Google Scholar]
- Karpova TS, McNally JG, Moltz SL, Cooper JA. Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J Cell Biol. 1998;142:1501–1517. doi: 10.1083/jcb.142.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J Biol Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- Majerus PW, Kisseleva MV, Norris FA. The role of phosphatases in inositol signaling reactions. J Biol Chem. 1999;274:10669–10672. doi: 10.1074/jbc.274.16.10669. [DOI] [PubMed] [Google Scholar]
- Martin TF. PI(4,5)P2 regulation of surface membrane traffic. Curr Opin Cell Biol. 2001;13:493–499. doi: 10.1016/s0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- Meili R, Ellsworth C, Lee S, Reddy TB, Ma H, Firtel RA. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci. 2000;25:229–235. doi: 10.1016/s0968-0004(00)01543-7. [DOI] [PubMed] [Google Scholar]
- O'Malley CJ, McColl BK, Kong AM, Ellis SL, Wijayaratnam AP, Sambrook J, Mitchell CA. Mammalian inositol polyphosphate 5-phosphatase II can compensate for the absence of all three yeast SacI-like-domain-containing 5-phosphatases. Biochem J. 2001;355:805–817. doi: 10.1042/bj3550805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms LM, McColl BK, Wiradjaja F, Wijayaratnam AP, Gleeson P, Gething MJ, Sambrook J, Mitchell CA. The yeast inositol polyphosphate 5-phosphatases Inp52p and Inp53p translocate to actin patches following hyperosmotic stress: mechanism for regulating phosphatidylinositol 4,5-bisphosphate at plasma membrane invaginations. Mol Cell Biol. 2000;20:9376–9390. doi: 10.1128/mcb.20.24.9376-9390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehoda KE, Scott JA, Mullins RD, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- Raucher D, Stauffer T, Chen W, Shen K, Guo S, York JD, Sheetz MP, Meyer T. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell. 2000;100:221–228. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4048. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4,5-cbisphosphate. J Cell Biol. 2000;150:1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozelle AL, Machesky LM, Yamamoto M, Driessens MH, Insall RH, Roth MG, Luby-Phelps K, Marriott G, Hall A, Yin HL. Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr Biol. 2000;10:311–320. doi: 10.1016/s0960-9822(00)00384-5. [DOI] [PubMed] [Google Scholar]
- Sakisaka T, Itoh T, Miura K, Takenawa T. Phosphatidylinositol 4,5-bisphosphate phosphatase regulates the rearrangement of actin filaments. Mol Cell Biol. 1997;17:3841–3849. doi: 10.1128/mcb.17.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Maniatis T, Fritsch EF. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, Bourne HR. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence LW. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1979. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Wurmser AE, Emr SD, Stenmark H. The role of phosphoinositides in membrane transport. Curr Opin Cell Biol. 2001;13:485–492. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- Singer-Kruger B, Nemoto Y, Daniell L, Ferro-Novick S, De Camilli P. Synaptojanin family members are implicated in endocytic membrane traffic in yeast. J Cell Sci. 1998;111:3347–3356. doi: 10.1242/jcs.111.22.3347. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Seaman M, Nemoto Y, Daniell L, Suchy SF, Emr S, De Camilli P, Nussbaum R. Disruption of three phosphatidylinositol-polyphosphate 5-phosphatase genes from Saccharomyces cerevisiae results in pleiotropic abnormalities of vacuole morphology, cell shape, and osmohomeostasis. Eur J Cell Biol. 1997;74:350–360. [PubMed] [Google Scholar]
- Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer TP, Ahn S, Meyer T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- Stolz LE, Huynh CV, Thorner J, York JD. Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics. 1998a;148:1715–1729. doi: 10.1093/genetics/148.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz LE, Kuo WJ, Longchamps J, Sekhon MK, York JD. INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem. 1998b;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- Tsujishita Y, Guo S, Stolz LE, York JD, Hurley JH. Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase. Cell. 2001;105:379–389. doi: 10.1016/s0092-8674(01)00326-9. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K, Gunn-Moore F, Oatey PB, Tavare JM, Cullen PJ. Nerve growth factor- and epidermal growth factor-stimulated translocation of the ADP-ribosylation factor-exchange factor GRP1 to the plasma membrane of PC12 cells requires activation of phosphatidylinositol 3-kinase and the GRP1 pleckstrin homology domain. Biochem J. 1998;335:139–146. doi: 10.1042/bj3350139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiradjaja F, Ooms LM, Whisstock JC, McColl B, Helfenbaum L, Sambrook JF, Gething MJ, Mitchell CA. The yeast inositol polyphosphate 5-phosphatase Inp54p localizes to the endoplasmic reticulum via a C-terminal hydrophobic anchoring tail: regulation of secretion from the endoplasmic reticulum. J Biol Chem. 2001;276:7643–7653. doi: 10.1074/jbc.M010471200. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Hilgemann DH, Feng S, Bito H, Ishihara H, Shibasaki Y, Yin HL. Phosphatidylinositol 4,5-bisphosphate induces actin stress-fiber formation and inhibits membrane ruffling in CV1 cells. J Cell Biol. 2001;152:867–876. doi: 10.1083/jcb.152.5.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Tsung M, Hamamoto S, Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]