Abstract
During phloem unloading, multiple cell-to-cell transport events move organic substances to the root meristem. Whereas the primary unloading event from the sieve elements (SE) to the phloem pole pericycle (PPP) has been characterized to some extent, little is known about post-SE unloading. Here, we report a novel gene, PLM (PHLOEM UNLOADING MODULATOR), in the absence of which plasmodesmata-mediated symplastic transport through the PPP-endodermis interface is specifically enhanced. Increased unloading is attributable to a defect in the formation of the ER-plasma membrane tethers during plasmodesmal morphogenesis, resulting in the majority of pores lacking a visible cytoplasmic sleeve. PLM encodes a putative enzyme required for the biosynthesis of sphingolipids with very long-chain fatty acid (VLCFA). Taken together, our results indicate that post-SE unloading involves sphingolipid metabolism which impacts plasmodesmal ultrastructure. They also raise the question of how and why plasmodesmata with no cytoplasmic sleeve facilitate molecular trafficking.
Plasmodesmata are membrane-lined channels that cross the plant cell wall, connecting neighboring cells to mediate symplastic communication1. Cell-to-cell trafficking of a wide range of molecules via plasmodesmata is involved in the coordination of growth and developmental programs such as cell differentiation, photoassimilate translocation, disease and stress resistance2–9. The typical plasmodesmal structure consists of the plasma membrane (PM) lining the pore and a central rod-like structure, called the desmotubule, derived from the endoplasmic reticulum (ER)10–12. Inside the pore, the ER and the PM are tethered by unidentified spoke-like elements, and this specialized membrane arrangement defines plasmodesmata as a specific type of membrane contact site. The gap between the PM and the desmotubular ER is termed the cytoplasmic sleeve, the presence of which classifies plasmodesmata as type I (no visible sleeve) or type II (clear cytoplasmic sleeve with visible tethering like spokes)13. In addition, plasmodesmata can also be classified as simple (with only a single channel) or branched (with multiple channels joining into a central cavity) based on their morphology14,15. In current models, the cytoplasmic sleeve is assumed to facilitate molecular trafficking through plasmodesmata16,17. The larger the gap between the ER and the PM, the more open the cytoplasmic sleeve and thus the larger the size exclusion limit of the pores. However, this hypothesis was recently challenged by a report that type I plasmodesmata may predominate at cellular interfaces with efficient symplastic trafficking and allow the movement of both micromolecules and macromolecules13.
Plasmodesmata consist of several chemically distinct components. Callose (β-1,3-glucan) accumulates in the cell wall around the pores, and its abundance is known to inversely correlate with trafficking efficiency18–20. Several membrane protein classes, including receptor-like proteins, are known to be associated with plasmodesmata21–23. The membrane lipid environment of the pores is distinct from that of the bulk PM, with an enrichment of complex sphingolipids with saturated VLCFA and a higher ratio of sterols to glycerolipids24. Sterols play a role in the regulation of plasmodesmatal permeability, and inhibiting sterol synthesis affects the targeting of plasmodesmata-localized proteins and ultimately callose homeostasis24,25. However, the precise role of sphingolipids in plasmodesmata remains elusive.
As discussed above, structurally diverse plasmodesmata have been identified and the abundance of the various plasmodesmal forms appears to vary depending on the cellular interface. Recently, another structural variant, funnel plasmodesmata, was identified in the context of phloem unloading26. Transmission electron microscopy revealed large V-shaped structures in SE cell walls, which appear to unload especially large proteins in batches to the neighboring PPP26, indicating a structural adaptation of the symplastic pathway for phloem unloading.
Here, we identify PLM (PHLOEM UNLOADING MODULATOR) as a modulator of symplastic unloading at the interface between the PPP and endodermis in roots. We show that PLM is involved in sphingolipid metabolism. In Arabidopsis, loss-of-function mutants of plm present a defect in the ER-PM tethering transition from type I to type II plasmodesmata, which is correlated with increased trafficking at the PPP-endodermis interface. Our data identify sphingolipids as regulators of plasmodesmal structure and unexpectedly reveal that plasmodesmata with a tight cytoplasmic sleeve are more competent at trafficking than those without a sleeve.
Results
Isolation and identification of the plm mutation, a suppressor of gain-of-function alleles in callose biosynthesis
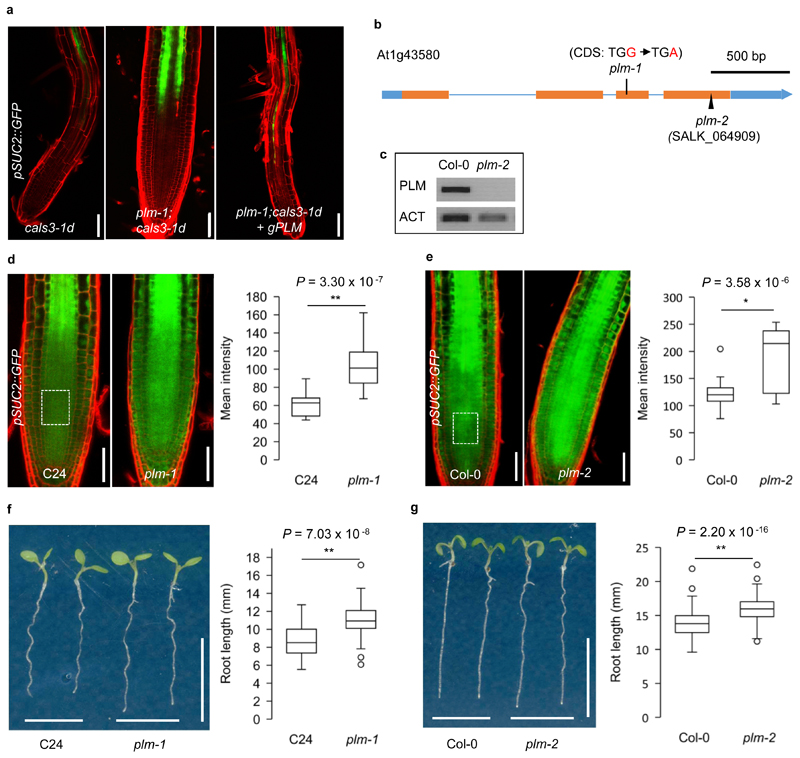
We have previously identified gain-of-function mutants (cals3-d) of the CALLOSE SYNTHASE 3/GLUCAN SYNTHASE-LIKE12 (CALS3/GSL12) gene27. The cals3-d mutants had reduced plasmodesmatal permeability, which in turn resulted in decreased intercellular trafficking, impaired phloem unloading and defective root development in Arabidopsis27. To further investigate the genetic control of symplastic trafficking, a genetic screen for suppressors was performed using ethyl methanesulfonate mutagenized cals3-1d plants expressing pSUC2::GFP27. In addition to numerous intragenic mutants, a putative extragenic suppressor of cals3-1d was eventually identified based on GFP unloading, root growth and subsequent genetic analysis. The suppressor partially rescued the phloem unloading and root phenotypes of cals3-1d. GFP transport was restored up to the transition zone, and seedling growth was similar to wild-type C24 (Fig. 1a and Supplementary Fig.1a). A genetic linkage analysis of F2 individuals derived from a cross between the suppressor and cals3-3d suggested that GFP trafficking and root growth were influenced by a single recessive mutation. By PCR-based positional cloning, the locus, named PLM, was narrowed down to a window of approximately 204 kb in chromosome 1. Subsequent whole-genome resequencing identified a premature STOP codon (TGG to TGA, 717 position of CDS) in exon 3 of the gene At1g43580 (Fig. 1b). When complemented with a 3,705 bp genomic DNA fragment which included 1,508 bp upstream of the gene, the suppressor line (plm-1;cals3-1d) displayed GFP unloading defects and a short primary root similar to cals3-1d (Fig.1a and Supplementary Fig.1a), supporting the identification of At1g43580 as PLM.
Figure 1. Identification and characterization of plm mutants.
a. pSUC2::GFP in the roots of cals3-1d, plm-1;cals3-1d and plm-1;cals3-1d transformed with PLM genomic insert. This experiment was repeated independently five times with similar results. b. Schematic view of the intron-exon structure of the PLM and sites of the mutations/T-DNA insertion. UTRs, white boxes. Exons, blue boxes. Introns, blue lines. c. RT-PCR showing undetectable transcript of PLM in plm-2. ACTIN2 (ACT) was used as the internal control. This experiment was repeated independently three times with similar results.d. Comparison of GFP intensity in the root meristems of wild-type C24 and plm-1 mutant carrying pSUC2::GFP. An area of 50 x 40 μm2 with 50 μm away from QC in the central root meristem (white dash box) was used for the quantification of fluorescence intensity, as well as all the following assays. n =16 (C24) and 37 (plm-1) biologically independent plants. e. Comparison of GFP intensity in the root meristems of wild-type Col-0 and plm-2 mutant carrying pSUC2::GFP. n =13 (Col-0) and 16 (plm-2) biologically independent plants. f. Root length of 5-day-old wild-type C24 and plm-1 seedlings. n=50 (C24), n = 46 (plm-1) biologically independent plants. g. Root length of 5-day-old wild-type Col-0 and plm-2 seedlings. n=213 (Col-0), n = 205 (plm-2) biologically independent plants. In the box plots of (d,e,f) and (g), the boxes indicate the first and third quartiles and the whiskers indicate the minimum and maximum values. The black lines within the boxes indicate the median values. Outliers are shown as dots. Statistically significant differences were determined by two-sided Mann-Whitney-Wilcoxon test (*P<0.05, ** P<0.01). Scale bars, 50 μm (a,d,e), 1 cm (f,g).
To further study the function of PLM, we isolated the plm-1 mutant in the C24 background. Using pSUC2::GFP as a reporter, we found a significant increase in the GFP signal in the root meristem of plm-1 (Fig.1d). To confirm the relationship between the enhanced GFP-trafficking phenotype and the plm mutation, an independent knock-out T-DNA insertion mutant (SALK_064909) in Col-0 was identified, which we named plm-2 (Fig. 1b,c). Like plm-1, plm-2 displayed enhanced GFP-trafficking (Fig. 1e). Furthermore, the plm-2;cals3-3d double mutant with pSUC2::GFP showed a similar GFP trafficking pattern to plm-1;cals3-1d and could also be complemented by genomic PLM and a PLM-YFP fusion (Supplementary Fig. 1b,c). We next characterized the overall phenotype caused by the recessive plm mutations. Both plm-1 and plm-2 displayed an early spurt in root elongation, conferring a slight but significant difference in root elongation for several days (Fig. 1f,g and Supplementary Fig.2a). We also found that the enhanced GFP-unloading phenotype in plm-2 was evident by day 3 after germination (Supplementary Fig. 2f), coinciding with the early root elongation phenotype (Supplementary Fig. 2a). Otherwise, we did not detect other phenotypic differences; for example, general growth and biomass accumulation were similar in the mutants and wild type (Supplementary Fig. 2b,c,d,e). Taken together, our results indicate that PLM may influence the root elongation rate by controlling the extent of phloem unloading soon after germination.
PLM encodes a novel protein involved in sphingolipid biosynthetic pathway
Based on the Arabidopsis database (http://www.Arabidopsis.org), PLM is a single-copy gene that contains 4 exons and 3 introns. It is predicted to encode a sphingomyelin synthase (SMS) family protein with a weak phosphatidic acid phosphatase-related2 domain (PAP2_C) (Fig.2a). Sphingomyelin is an important type of sphingolipid found in organisms ranging from protozoa to mammals28,29. It is synthesized by the transfer of phosphocholine from phosphatidylcholine (PC) to ceramide, yielding diacylglycerol (DAG) as a side product30,31. The reaction is catalyzed by SMS, which is located at the PM and in the Golgi in most mammalian cells32,33,34. However, sphingomyelin has not been detected in plants, as confirmed by our LC-MS analysis in Col-0 Arabidopsis (data not shown). However, plants possess inositol phosphorylceramide synthase (IPCS) enzymes, which also have a PAP2_C domain and employ ceramides as substrates to catalyse the synthesis of IPC in the Golgi34.
Figure 2. Molecular characterization of PLM.
a. Schematic of PLM protein sequence. TM, transmembrane domain. PAP2_C, type 2 phosphatidic acid phosphatase_C terminal. b. Phylogenetic tree of SMSs, IPCSs and PLM homologues from different organisms. Protein accession numbers and sequences are shown in Supplementary Table 2. The tree was constructed by MEGA4.1 using neighbour-joining method. Bootstrap = 500. c. Expression pattern of pPLM::PLM-YFP in roots. The experiment was repeated three times independently with similar results. d. Co-localization of PLM-YFP with RFPer (RFP-HDEL). Signal intensities of YFP (green line) and RFP (red line) were obtained on the dashed white arrow. The experiment was repeated three times independently with similar results. Scale bars, 50 μm (c), 10 μm (d).
Multiple sequence alignment of full-length protein sequences showed that PLM has very low sequence identity (20-26%) with both human SMSs and Arabidopsis IPCSs (Supplementary Fig. 3 and 4). PLM has the typical D1 but no D2 motif of SMSs, while IPCSs lack both32. PLM possesses the three conserved amino acid residues H-H-D in the catalytic site for phosphatidic acid phosphatase activity while displaying low identity for other residues in the D3 and D4 motif 32 (Supplementary Fig. 3). A phylogenetic tree of known and predicted SMSs, IPCSs and PLM homologues from various organisms was generated. It revealed that PLM is distinct from SMSs and IPCSs and might have originated before the separation of Klebsormidiophyceae algae from land plants (Fig.2b and Supplementary Table 2).
A pPLM::PLM-YFP construct was employed to investigate the expression pattern and subcellular localization of PLM. The construct could revert the unloading phenotype of plm-2;cals3-3d back to cals3-3d levels, demonstrating that it was functional (Supplementary Fig.1c). pPLM::PLM-YFP displayed a broad expression pattern in the root (Fig.2c). The tagged protein co-localized with the ER-retained reporter RFPer (RFP-HDEL) indicating that PLM is ER-localized (Fig.2d and Supplementary Fig.5a,b). To further confirm ER-association, we co-expressed UB10::PLM-YFP with the ER-shaping protein reticulon6 (RTNLB6)35. When overexpressed reticulon modifies ER morphology by changing ER cisternae membranes into tubules36. Under these conditions, UB10::PLM-YFP displayed a droplet-like pattern along the ER-strands (Supplementary Fig.5c), confirming that PLM associates with the ER and accordingly change its localization pattern when the ER-shape is modified.
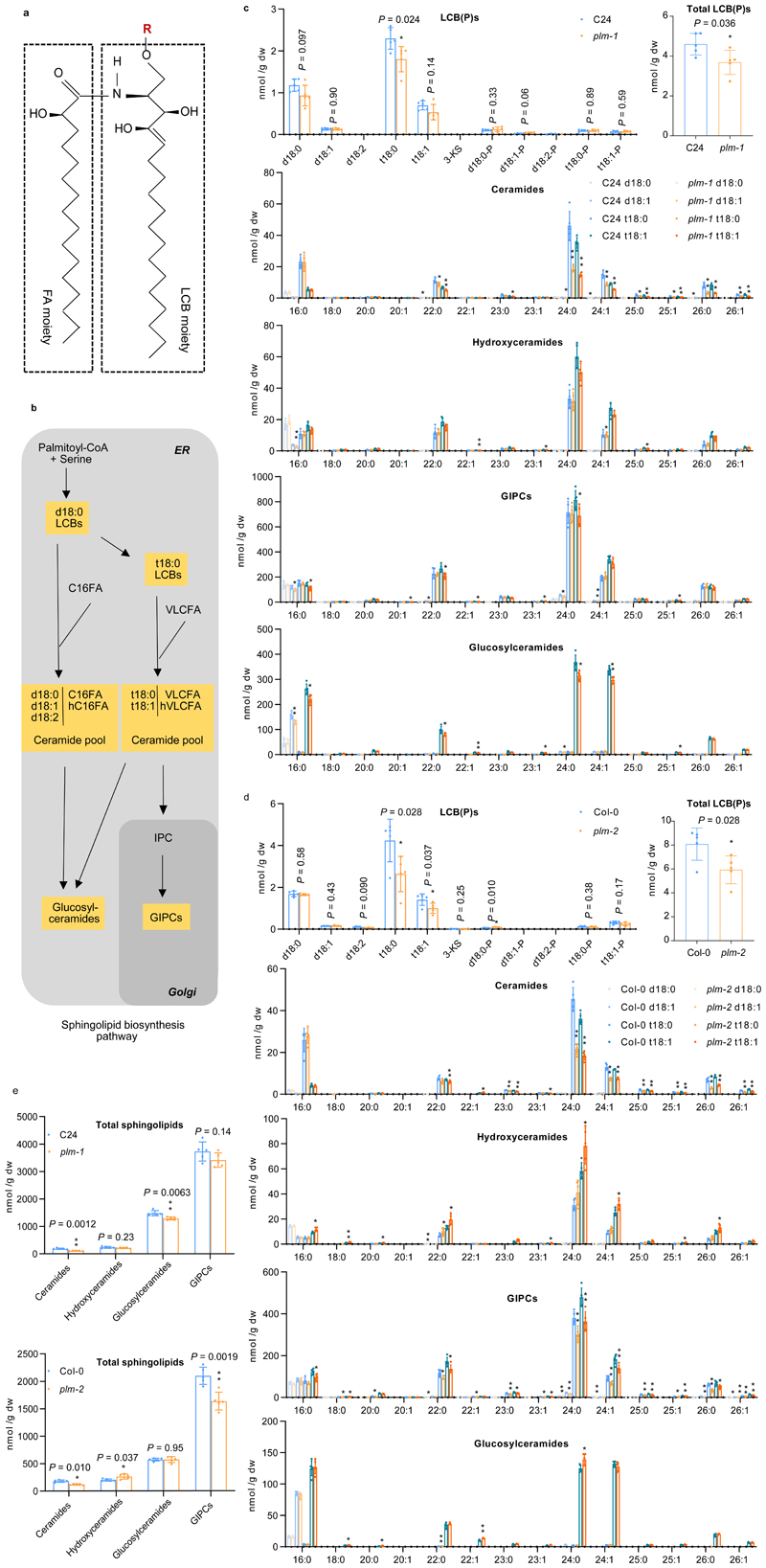
To investigate the biochemical basis for the plm phenotype and the role of PLM in sphingolipid metabolism, a complete sphingolipid analysis was performed on roots of 5-day-old seedlings37. The structural sphingolipids (glucosylceramides, and glycosyl inositol phospho ceramides (GIPCs)) were analyzed along with free long-chain bases (LCBs) and ceramides, thought to act as important sphingolipid precursors (Fig.3a,b). A comparison between plm-1, plm-2 and wild type showed that both plm mutants displayed a strong and consistent reduction in VLCFA-ceramides, especially the main species, t18:0/t18:1 and C24:0/C24:1-ceramides, by 48- 60% (Fig.3c,d and Supplementary Fig.6). A decrease in the trihydroxy LCBs, especially of t18:0 species, was noticeable in both alleles (Fig. 3c,d) although with some variation (Supplementary Fig.6). We also observed a strong reduction (supported by statistical testing) of GIPC species in plm-2 mutant in one set of experiment. Subsequent analyses showed a similar trend for both plm-1 and plm-2 but differences were no longer supported statistically (Fig.3c,d and Supplementary Fig.6).
Figure 3. PLM affects sphingolipid biosynthesis.
a. A diagram of sphingolipid structure. R indicates the substituent of sphingolipid. The FA chain has various carbon number while the LCB moiety are largely synthesized as saturated form (18:0) and desaturated form (18:1) with different OH (d/t) number. b. Simplified schematic representation of sphingolipid biosynthesis pathway in plants. c and d. Free LCB(P), ceramide and complex sphingolipid species characterized by LCB(d18:0, d18:1, t18:0, t18:1) and FA(16:0-26:1) pairings in wild-type C24 and plm-1(c) and Col-0 and plm-2 (d). 3-KS, 3-ketosphinganine. e.Total levels of complex sphingolipid species in wild-type and plm mutants. Measurements are the average of five replicates (n = 5 independent samples). Data are means ± SD.Statistically significant differences are indicated (two-tailed Student’s t-test; *P<0.05. ** P <0.01).
GIPCs possess different head groups that are modified by a number of sugar residues38,39. To investigate whether plm mutant has a depletion of specific GIPC head species, primuline and orcinol staining in addition to MALDI analysis were performed. No differences were found in Col-0 and plm-2 (Supplementary Fig. 7), suggesting a similar composition of GIPC sugar heads. Last, VLCFA-containing hydroxyceramides and VLCFA-containing glucosylceramides were overall unaltered (Fig.3c,d and Supplementary Fig.6).
To investigate whether PLM is specifically involved in sphingolipid metabolism, profiling of polar lipid, DAG and sterol content was performed. No alterations in the amounts of these lipids were detected in plm-2 (Supplementary Fig. 8a,b,c).
Overall, these results indicate that the plm mutation interferes specifically in sphingolipid metabolism, and mostly act early in the sphingolipid biosynthetic pathways on the VLCFA-ceramides with likely consequences on the overall GIPC content.
To further strengthen the difference between PLM and IPCS2, a T-DNA insertion allele of Atipcs2 (SALK_206784) was also included in the sphingolipid profiling. The relative subtle differences observed in ipcs2 mutant (Supplementary Fig. 9) did not match the more significant ones in plm-2. Based on the phylogenetic clustering of plm and its difference from IPCSs in terms of subcellular localization and sphingolipid profiles, PLM is likely to be a new plant-specific enzyme.
plm enhances GFP post-SE unloading from PPP to endodermis
We next focused on the unloading phenotype of plm-2. As described above, plm mutations enhance GFP trafficking from the phloem to the root tip. We therefore explored the unloading process in more detail. GFP expressed by pSUC2::GFP translocates from companion cells to SEs then unloads symplastically into the root meristem from protophloem SEs through the PPP-endodermis interface (Fig. 4a). While wild-type plants presented a strong difference in GFP signal between the unloading domain in the transition zone and the root meristematic zone, plm-2 has a less-defined boundary and a more uniform GFP signal (Fig. 1d,e). GFP unloading in the root tip therefore appears to be enhanced in plm-2. We confirmed this using an inducible version of pSUC2::GFP, which displayed a similar difference between unloading in wild type and plm-2 upon induction (Supplementary Fig. 10a). The unaltered transcript levels of GFP and SUC2 excluded the possibility of regulation of SUC2 expression by PLM (Supplementary Fig.10b). However, the impaired unloading of GFP fusion proteins with bigger size, such as SPORAMIN-GFP and SEOR-YFP, remains same in plm-2 (Supplementary Fig.11). Furthermore, no developmental differences in vasculature were observed in plm-2 based on established markers for phloem (pAPL::YFPer), SE (pCALS7::H2B-YFP) and PPP (pCALS8::YFPer) (Supplementary Fig.12).
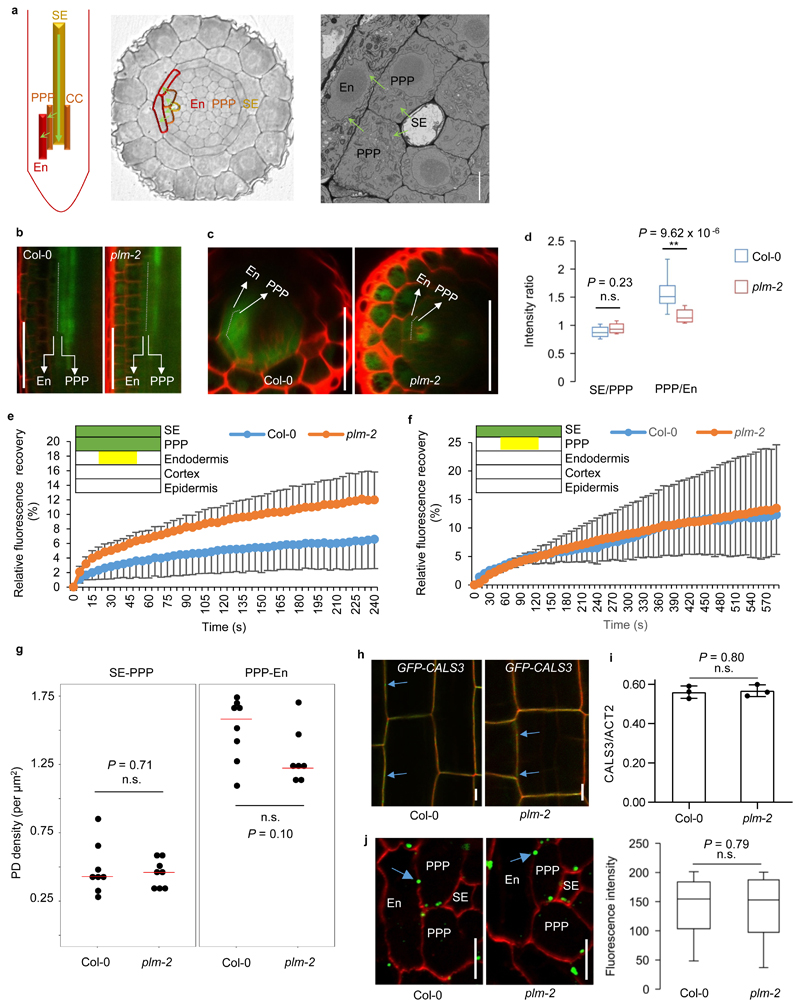
Figure 4. GFP unloading into root tip is enhanced in plm.
a. Schematic view of the unloading path of SE-PPP-endodermis in the root tip. b. GFP signals of PPP and endodermis at the unloading domains in the wild-type Col-0 and plm-2 roots. c. Optical cross sections of (b). The experiments of (b) and (c) were repeated five times independently with similar results. d. Fluorescence intensity ratios of SE/PPP and PPP/endodermis in the unloading domain (3 continuous cells since 200 μm away from QC post SE enucleation). n=13 (Col-0) and 11(plm-2) biologically independent plants. e. FRAP assay showing the recovery of GFP within the endodermis trafficking from PPP post bleaching of endodermis, cortex and epidermal layers in the unloading domain. The yellow box indicates the same region in endodermis described in (d) for GFP recovery assay. n=7 (Col-0) and 9 (plm-2) biologically independent plants. Data are means ± SD. f. FRAP assay showing the recovery of GFP within the PPP trafficking from SE post bleaching of PPP, endodermis, cortex and epidermal layers in the unloading domain. The yellow box indicates the same region in PPP described in (d) for GFP recovery assay. n=4 (Col-0 and plm-2) biologically independent plants. Data are means ± SD. g. Plasmodesmata density assay by SB-EM at the SE-PPP and PPP-endodermis interface in the same region of unloading domain in Col-0 and plm-2. The median values are indicated by red lines. n=8 (n=7 for plm-2 PPP-En) biologically independent samples. h. Localization of CALS3 (blue arrows) in Col-0 and plm-2. The experiment was repeated three times independently with similar results. i. Relative transcript levels of CALS3 in Col-0 and plm-2. Data are based on three independent replicate experiments. Data are means ± SD. j. Immunolocalization of callose at plasmodesmata (blue arrows) at the PPP-endodermis interface in the same region of unloading domain in Col-0 and plm-2. n=102 (Col-0) and 92 (plm-2) independent samples for quantification of fluorescence intensity. In the box plots of (d) and (j), the boxes indicate the first and third quartiles and the whiskers indicate the minimum and maximum values. The black lines within the boxes indicate the median values. Significant differences were determined by two-tailed Mann-Whitney-Wilcoxon test (d, j) and two-tailed two sample t-test (g, i). ** P<0.01. n.s., no significance. En, endodermis. Scale bars, 1 μm (a), 50 μm (b,c), 5 μm (h, j).
Phloem unloading in the root tip occurs from proto-SEs into PPP cells. Further subsequent radial movement into neighboring endodermal cells is tightly regulated and depends on the size of the unloaded macromolecules26 (Fig.4a). In wild-type plants, PPP cells display a stronger GFP signal intensity than endodermal cells (Fig.4b,c). The signal maximum suggests that GFP experiences some form of bottleneck in transport at the PPP-endodermis interface. Differences in the GFP signal between the two layers were reduced in plm-2 (Fig.4b,c,d), indicating that the trafficking restriction that normally occurs at the PPP-endodermis interface is relaxed in the plm-2 mutant. To further explore GFP trafficking across the PPP-endodermis interface, fluorescence recovery after photobleaching (FRAP)40 was performed in the unloading zone. The GFP recovery rate in the endodermis with influx from PPP was faster in plm-2 than in Col-0, while the movement through the SE-PPP interface was not affected (Fig.4e,f). This similar phenomenon was also observed in plm-1 and C24 (Supplementary Fig. 13). These data together suggest that post SE-PPP unloading trafficking is affected in plm mutants, with a specific impact on the PPP-endodermis interface.
To strengthen the link between sphingolipid homeostasis by PLM and enhanced GFP trafficking, we used the ceramide synthase inhibitor FB1, which reduces ceramide and GIPC products41,42. After a 72h treatment at concentrations shown not to affect root growth, the GFP signal in the root tip of Col-0 plants carrying pSUC2::GFP was increased, mimicking the plm mutation (Supplementary Fig.14a,b).
PLM-mediated symplastic trafficking is independent from plasmodesmata density and callose accumulation
We tested multiple hypotheses to determine which change at the PPP-endodermis interface underpins the enhanced GFP trafficking in plm-2. The density of plasmodesmata in the unloading zone was first assessed employing serial block face scanning electron microscopy (SB-EM). There was no increase in the number of plasmodesmata per unit area at the SE-PPP or PPP-endodermis interfaces of plm-2 (Fig. 4g), indicating that the enhanced trafficking is not the result of increased pores.
Since callose is known to modulate symplastic trafficking by regulating plasmodesmata aperture and plm is a suppressor of cals3d, we analyzed the transcript levels of CALS3 in the root and the subcellular localization of the CALS3 protein. Both were unaltered in plm-2 (Fig. 4h,i). To assess the callose level, immunolocalization using a callose antibody on root sections from the unloading domain was performed. No difference in the callose signal at the PPP-endodermis interface (Fig.4j and Supplementary Fig. 15a,b), as well as the whole meristematic zone (Supplementary Fig. 15c) of Col-0 and plm-2 was observed. Moreover, the increased callose accumulation in the early lateral roots of cals3-3d was also observed in plm-2;cals3-3d (Supplementary Fig. 15d). In summary, our results indicate that the plm mutation enhances plasmodesmatal permeability independently of plasmodesmata density and callose regulation.
To further investigate the impact of callose on PLM-mediated trafficking, we carried out an assay of root growth under stress caused by alloxan treatment, which induces callose accumulation and blocks phloem unloading, compromising root growth43. At 28h, 52h and 65h after transferring 5-day old seedlings to alloxan media, plm-2 roots grew longer than wild-type roots, while no difference was observed after transferal to normal media (Supplementary Fig. 16a). Aniline blue staining showed similar increases of callose accumulation in both Col-0 and plm-2 roots after alloxan treatment (Supplementary Fig. 16b). These results suggest that plm plasmodesmata are somewhat resistant to the negative effect of callose deposition. It is possible that the ability of the plm mutation to suppress casl3-d action is based on this characteristic.
PLM regulates plasmodesmal architecture at the PPP-endodermis interface
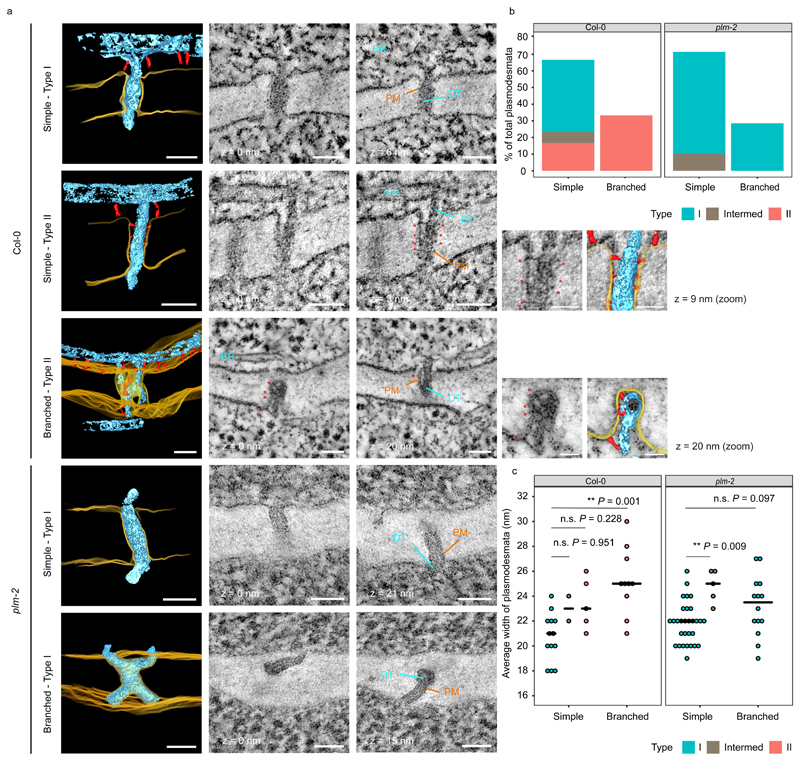
One possible explanation for the observed enhanced symplastic conductivity is an alteration in plasmodesmal ultrastructure. Nicolas et al (2017)13 have shown that type I plasmodesmata (lacking an electron-lucent cytoplasmic sleeve due to close PM-ER contact) are predominant in young root tissue and open to macromolecular trafficking, while type II plasmodesmata (displaying a visible electron-lucent cytoplasmic sleeve with a larger ER-PM gap containing sparse tether-like spoke elements) are more numerous in older tissue. To investigate possible alterations in plasmodesmal ultrastructure, we employed electron tomography13 at the PPP-endodermis and SE-PPP interfaces in Col-0 and plm-2. Detailed examination of our samples showed that while Col-0 plants had an equal proportion of type I and type II plasmodesmata at the PPP-endodermis interface, plm-2 had no type II plasmodesmata (Fig. 5a,b). We also detected a few intermediates, with no clear spokes but partial detachment between the two membranes (Fig. 5b). In terms of morphology, a mixed population of simple and branched plasmodesmata was observed in both Col-0 and plm-2. In Col-0, the branched plasmodesmata were all type II (presumably corresponding to later maturation events), while simple plasmodesmata consisted of both type I and type II (Fig. 5a,b). Defective transition from type I to type II plasmodesmata at the PPP-Endodermis interface in the plm-2 mutant was also demonstrated by measuring the width of the pores (type I being narrower than type II)13. In Col-0, plasmodesmatal width increased between simple type I and branched type II plasmodesmata (Fig. 5c). By contrast, there was no clear difference in the width of simple and branched plasmodesmata in the plm-2 mutant (Fig. 5c). The loss of the width increase between simple and branched plasmodesmata mirrored the depletion of type II plasmodesmata. Overall, the plm mutant therefore seems to be compromised in the transition from type I to type II plasmodesmata at the PPP-Endodermal interface but not from simple to branched structures, implying that independent underlying factors may determine these two morphogenetic processes. Conversely, we still observed type II plasmodesmata at the SE-PPP interface in plm-2 (Supplementary Fig.17), which suggests no defect in type I to type II transition at this interface. This correlates with the data showing unchanged permeability at SE-PPP (Fig. 4f).
Figure 5. plm-2 lacks type II plasmodesmata at the PPP-endodermal interface of the unloading zone.
a. Plasmodesmata representing the various classes detected in Col-0 and plm-2 roots with electron tomography at the PPP-endodermal interface, within the unloading domain. Panels with black background are 3D reconstructions based on 2D image stacks. Note that the 3D view of the branched plasmodesmatata in Col-0 is tilted relative to 2D images for ease of visualisation. Plasma membrane (PM) is rendered in yellow, desmotubule (DT) and ER in light blue, tethers within plasmodesmata and outside of them in red. Selected 2D views are displayed in the neighbouring panels with zooms for tethers within type II plasmodesmata (with a model overlay). Distance to the first 2D view is indicated under or next to each panel. Arrows, symbols and labels indicate various plasmodesmata components and follow the colour scheme of the 3D models (* tether elements within plasmodesmata). Scale bars are of 50nm or 25nm for the zoomed panels. b. Quantification of morphologies and types of plasmodesmata present at PPP-endodermal interface within the unloading domain in Col-0 and plm-2 roots. n=30 (Col-0), n=49 (plm-2) plasmodesmata. The sections with plasmodesmata were acquired from two roots for each genotype. c. Comparison of the widths of plasmodesmata described in panel b. Median values for each group is represented by black horizontal bars. Significant differences in reference to simple (type I) plasmodesmata in Col-0 or plm-2 were determined by two-sided Dunn’s test. The P-values were adjusted with Holm method for multiple comparisons. ** P<0.01, n.s. no significance.
Discussion
In this paper, we establish a link between sphingolipid metabolism, the internal architecture of plasmodesmata and cell-to-cell connectivity. Our results provide evidence for an unexpected role of the cytoplasmic sleeve and ER-PM contacts in plasmodesmata-mediated cell-to-cell trafficking.
The cytoplasmic sleeve of plasmodesmata has been well established as a continuum of symplasm between adjacent cells, and the size exclusion limit of the pores is believed to depend on the conductivity of the cytoplasmic sleeve16,17. Recently Nicolas et al. (2017) postulated a positive correlation between the spatial distribution of type I plasmodesmata (with a very narrow cytoplasmic sleeve) and enhanced trafficking13. Here, we provide genetic support for this model and furthermore demonstrate that type I plasmodesmata are more conductive than type II, which have an “open” cytoplasmic sleeve. The lack of type II plasmodesmata and the higher GFP conductivity at the PPP-endodermis interface in plm (together with the unchanged density of plasmodesmata and callose level) strongly indicate that the narrow cytoplasmic sleeve observed in type I plasmodesmata may facilitate symplastic trafficking. However, how and why type I plasmodesmata favor cell-to-cell transport remains elusive.
Because of the structural homology of PLM with the SMSs and IPCSs, which are involved in sphingolipid metabolism, we profiled sphingolipids and other lipids in the plm-2 mutant. We observed a specific effect of the plm mutation on sphingolipids, whereas phospholipids, DAG or sterols were not affected. Most striking was a massive reduction in trihydroxy VLCFA-ceramides and to some extent a reduction in non-phosphorylated LCBs and VLCFA-GIPC products (Fig. 3). The profile seems unique compared to known sphingolipid biosynthetic mutants. Based on sequence alignment and sphingolipid profiling, it is possible that PLM functions as an enzyme at an unknown synthesis branch for ceramide synthesis in the ER. Alternatively, PLM may indirectly affect the sphingolipid biosynthesis pathway and act as a regulatory element.
The question of how sphingolipids affect plasmodesmata remains unresolved. Sphingolipid-containing VLCFAs and especially GIPCs have been reported to be critical for membrane bending and microdomain formation44–47, and are enriched in plasmodesmata fractions undergoing the type I to type II transition13. One scenario is that by acting early on the sphingolipid biosynthetic pathway and interfering with the pool of ceramide and possibly LCB precursors, plm loss-of function indirectly affects structural sphingolipids at the cellular level and/or more specifically at plasmodesmata. Further work will be required to investigate this latter point, and determine whether plasmodesmata sphingolipid content is altered in the plm mutant background. Alternatively, VLCFA-ceramides may directly regulate plasmodesmata function, the mechanisms of which remain elusive but may involved signalling or membrane shaping function48.
Based on our data, the mode of action of PLM (and sphingolipids) is different from the effect of sterols. Sterol inhibitors resulted in strong and tissue specific defects in callose deposition at PD24 while loss of function of PLM did not alter callose level. Overall, this suggests that the mode of action of different lipid classes on PD likely differ.
In this work, we provide further elements to dissect the phloem unloading pathway. Our previous collaborative work revealed that the SE-PPP route is the predominant unloading pathway26. Solutes such as sugar and macromolecules with the size of GFP can be unloaded from SE into PPPs and then enter the post-SE unloading pathway to the meristem, while larger proteins (for instance, the size of SE OCCLUSION RELATED (SEOR) protein fused to YFP (SEOR-YFP)) remain trapped within the PPP26. Therefore, the post-phloem PPP-endodermis interface has emerged as a key barrier for controlling post-unloading transport to the sinks. The higher conductivity of the PPP-endodermis interface and enhanced GFP input into the root meristem are consistent with the accelerated root elongation seen in young plm seedlings, indicating a transient, rate-limiting role for the PPP-endodermis interface in symplastic trafficking. However, the enhancement of plasmodesmatal permeability by PLM is most likely limited, as larger proteins, are retained in the PPP in plm (Supplementary Fig. 11). This suggests the existence of other regulatory mechanisms which might contribute to the somewhat subtle nature of the phenotype in Arabidopsis.
A unique type of funnel-shaped plasmodesmata has been reported at the SE-PPP interface26, indicating symplastic interfaces are specialized. The observed change in conductivity, based on FRAP, at the PPP-endodermis interface but not at the SE-PPP interface suggests further specialization in the unloading interfaces. Indeed, although PLM is expressed broadly, a slightly higher expression is detected in sink tissues like roots, shoot apex and seeds at early stage (ATTED-II version 9.2, http://atted.jp)49. It is possible that the requirement for VLCFA-containing sphingolipids is quite local. Tissue-specific relevance of VLCFA-containing lipids in the pericycle has been reported in the context of callus generation50.
Finally, we found PLM-like genes in the genomes of virtually both algae and land plants that we investigated (Fig.2b), but how the PLM pathway contributes to the functional diversity of symplastic trafficking events in plants remains to be assessed.
Methods
Plant materials and growth conditions
Arabidopsis thaliana mutants cals3-1d and plm-1 are in the C24 background; cals3-3d and plm-2 are in the Col-0 background. plm-1 was isolated in a genetic screen based on ethyl methanesulfonate mutagenesis of a gain-of-function allele of cals3-1d to identify a second site mutation that could fully or partially rescue root growth and pSUC2::GFP unloading defects. The segregation pattern of identified putative suppressors was analyzed in the M3 generation to determine whether the suppressor was dominant or recessive. Next, suppressors were backcrossed with the pSUC2::GFP line (C24 background), and the phenotypic segregation pattern was studied in the F1 and F2 generations to identify putative extra-genic suppressors. A mapping population was generated by crossing a non-segregating plm suppressor mutant (C24) with cals3-3d (Col-0). A PCR-based positional cloning was used for rough mapping, and whole-genome re-sequencing (Illumina) was used for SNP identification. plm-2 (SALK_064909) was obtained from the Nottingham Arabidopsis Stock Centre. Seedlings were sterilized and grown vertically in Petri dishes on 1/2 Murashige and Skoog (MS) basal salt mixture with 1% agar, 1% sucrose and 0.05% 4-morpholine ethanesulfonic acid (MES) at pH 5.8 in a growth chamber at 23°C under 16h light conditions.
Transgenic work
Transgenic constructs were generated using the Gateway or MultiSite Gateway system (Invitrogen) and were introduced into plants using the floral dip method51. For pPLM::PLM-YFP, a 2.2-kb genomic DNA fragment with a 1.5-kb upstream region was amplified and combined with YFP into the MultiSite destination vector pBm43GW. Analysis was carried out in the F2 or F3 generations of crosses with wild type or mutant plants. Primer sequences are listed in Supplemental Table 1.
Confocal microscopy
Confocal imaging was performed on a Leica TCS SP8. Excitation and emission spectra were, respectively, 484 and 489-505 nm for GFP, 514 and 522-564 nm for YFP, 561 and 580-620 for RFP, and 561 and 600-650 for propidium iodide (PI). PI was used to stain cell contours.
Transcript analysis
For quantification of mRNA, root tips from 5-day-old seedlings were harvested. The total RNA was extracted with a miRNeasy Mini Kit (QIAGEN) and treated with DNase on a column. cDNAs were synthesized from total RNA primed with oligo(dT)18 primers using a First strand synthesis kit (Roche). Semi-quantitative PCR for plm-2 was performed using specific primers across the T-DNA insertion site and ACTIN2 as an internal standard. Quantitative PCRs (qPCRs) were performed using gene-specific primers and real-time PCR mix (Roche) in a LightCycler 480 Real-Time PCR System (Roche) with a standard program for 40 cycles. The levels of expression were calculated relative to ACTIN2. Primer sequences are provided in Supplemental Table 1.
Sphingolipid profile
Sphingolipids were extracted from 10 to 30 mg of lyophilized 5-day-old roots. The molecular species of different sphingolipid classes were analyzed by reverse-phase HPLC coupled to ESI-MS/MS as described in35. P values of statistical analysis for ceramides, hydroxyceramides, GIPCs and glucosylceramides were provided in Supplementary Table 3.
FRAP assay
FRAP was conducted on a Leica TCS SP8 confocal microscope. GFP in the layers outwards of the SE or PPP in the unloading zone was photo-bleached with 476, 488 and 496 nm lasers concurrently with 100% power and 8x zoom with a 25X lens. Subsequent recording of the refilling occurred immediately following photobleaching at 0.75X digital zoom by excitation with the 488 nm laser line, and emission was collected between 505 and 545 nm. The relative recovery profiles were normalized to pre-bleach image with the first image post-bleach set to 0.
SB-EM
Five-day old Col-0 and plm-2 roots were fixed, dehydrated and embedded in Durcupan resin following protocols previously described in52,53. To improve handling, the roots were shortened to a total length of about 600 μm from the tip using a grinding machine (LaboPol-5, Struers Inc). Ground roots were then mounted by the ground side to a 3View pin and trimmed from the opposite end to 240 μm from the root tip using an EM Ultracut UC6i ultramicrotome (Leica Mikrosysteme GmbH) and a diamond knife. The trimming distance was selected based on markers for SE enucleation to get as close as possible to the area of interest. As a control, a histological section was cut and checked after the trimming. The blocks were covered with silver paint (Agar Scientific Ltd.) and platinum-coated using Quorum Q150TS (Quorum Technologies, UK). Images were acquired with an FEG-SEM Quanta 250 (FEI, Hillsboro, OR) using a backscattered electron detector (Gatan Inc.) with 2.5- kV beam voltage, spot size 3 and pressure between 0.1 to 0.3 Torr. Images were collected moving away from the stumped root tip in 40 nm steps. The coordinates of bonding boxes for each dataset were recalculated from root tip for ease of interpretation.
The datasets were processed using the Microscopy Image Browser (MIB)54, run within the Matlab environment (Mathworks Inc.). Plasmodesmata were manually annotated on the cell-cell interfaces of interest. Care was taken to not count the same PD twice, making a conservative estimate. Midlines were drawn across the walls corresponding to the interfaces of interest on multiple slides. Lines were then interpolated and a 3D surface generated using the specially developed Contact Area 3D plugin of MIB. Density was subsequently calculated as the number of plasmodesmata per unit of surface.
Callose immunofluorescence localization
Callose detection was conducted according to27. Roots of 5-day-old seedlings were fixed using 4% formaldehyde (Sigma) and 0.5% glutaraldehyde (Sigma) in 1x PBS buffer at pH7 overnight at 4°C. Roots were then washed, aligned and embedded in 1% low-melting Agarose (Calbiochem). After dehydration by 25%, 50%, 70%, 90%, 96%, 3 × 100% ethanol and resin infiltration (LR White medium grade, Agar scientific), polymerization was conducted overnight at 60°C. Callose immunolocalization was performed on 1 μm sections collected within a 100 μm unloading domain starting from the first enucleated SE using monoclonal antibody to (1→3)-β-glucan (Biosupplies, AU) as the primary antibody (1:1000) and Alexa Fluor Plus 488 goat anti-mouse IgG as a secondary antibody (1:1000). Slides were finally mounted in a 1:1 solution of AF1 antifading agent (Citifluor) and 1x PBS, with the addition of calcofluor as a cell wall counterstain. Images were taken by confocal microscopy (Leica TCS SP8) and processed by Leica Application Suite X (LAS X) and Image J (https://imagej.nih.gov/ij/).
Electron tomography
Sample preparation, sectioning, TEM analysis, tomogram reconstruction and analysis were performed as described13,55.
High pressure freezing-freeze substitution
Root tips from Col-0 and plm-2 mutant plants were frozen with high pressure to achieve vitreous ice in the whole sample. 5-day old seedlings were placed on a glass microscopy slide covered with 20% BSA. Root tips were sampled over 1.5 mm approximately from the root tip in order to have access to the differentiation zone, where phloem enucleation and unloading occur. Roots were then placed in Leica copper membrane carriers with a drop of BSA 20% to serve as cryoprotectant and filler. The loaded membrane carriers were then processed in an EM PACT1 (Leica) high pressure freezer, with no more than 30 seconds separating sampling and freezing. Samples were stored at -196°C in liquid nitrogen until further processing. Samples were then transferred to the well of the AFS2 machine (Leica), precooled to -90°C for the freeze substitution steps. The frozen membrane carriers were incubated in sealed cryotubes with a cryosubstitution cocktail containing 2% osmium tetroxide, 0.1% uranyl acetate and 0.5% glutaraldehyde in pure acetone. The cocktail was left at -90°C for 48h and then temperature was progressively brought up to -50°C (+3°/h). The cryofixation cocktail was then carefully removed and washed out by three consecutive 10-minute long acetone baths. Three pure ethanol baths were then performed, and uncasing of the samples from the copper membrane carriers was performed at the last one. Samples were carefully placed in resin casts (either coffin shaped ones for transversal root sections or cylindrical ones for longitudinal sections). Incubation of the samples in solutions of increasing concentration of HM20 Lowicryl resin in pure ethanol was as follows: 2h in 25% and 50% HM20, overnight in 75% HM20. This was followed by two consecutive 2h baths in 100% HM20. Curing the resin was accomplished by a last incubation in 100% HM20 for 8h before exposing the samples to UV light for 48h using the FSP module of the AFS2 (Leica). The first 24h of UV exposure were done at -50°C, then the temperature was raised to room temperature for the last 24h of UV exposition.
Section collection
Blocks were cut using a EM UC7 ultramicrotome (Leica). Section thickness ranged from 90 to 150nm. Sections were deposited on either formvar filmed and carbon coated slot grid (Electron Microscopy Science) or on 200-mesh copper grids coated with 2% parlodion. Grids with deposited sections were incubated in a solution of 5nm gold fiducial markers on both sides of the grid. The solution is a colloidal gold solution (BBI solutions) mixed with 0.5% BSA in a 1:1 ratio.
Data acquisition
Tilt series were acquired on a FEI TECNAI Spirit 120kV electron microscope equipped with a -70° - +70°C tilting goniometer and an Eagle 4k x 4k bottom camera. A tomography optimized single tilt holder was used (Fichione instruments, model-2020). Acquisitions were done at magnifications between 30 000x and 56 000x with a 1° increment for a range of -65° to +65°. Dual tilt axis tomography was achieved by taking out the sample holder and manually rotating the grid approximately 90° before acquiring the other axis. The batch mode of FEI 3D-explore was used to acquire tilt series automatically on areas of interest.
Tomogram reconstruction
The raw unbinned 4k x 4k data was processed using the IMOD package with the etomo GUI56. Tilt series from both axis were binned down to 2k x 2k to reduce occupancy and processing time for alignment and reconstruction. Alignment was performed using the automatic bead detection/tracking script, and reconstruction was done by weighed back projection, using the SIRT-like filter set at an equivalent of 15 iterations, or by real SIRT, 15 iterations. Then the two tomograms from the orthogonal axis were combined using IMOD again.
Data segmentation
Segmentation was done manually using 3dmod from the IMOD package. The interpolator and diverse drawing tools were used to trace the multiple features of interest. Systematic measurements were taken on all relevant unfiltered tomograms.
GIPC glycosylation assay
5-day-old roots were used for GIPCs extraction. GIPC extraction and analysis were conducted as described in57,58. GIPCs were separated by TLC and visualized by primuline and orcinol. Glycosylated GIPCs were analyzed by MALDI-MS. Peak assignments are based on published data59–62.
Lipid Analysis
Polar lipid extraction and profile were performed according to63,64 using 5-day-old roots. Data were based on 5 replicates.
Transgenic work
For generating pSUC2-XVE::YFP, a 2.1-kb fragment of SUC2 promoter was amplified by primers of iSUC2-F and iSUC2-R, cloned into a P4_P1r plasmid and subsequently recombined into the inducible system65 using multisite gateway cloning.
Aniline blue staining
Aniline blue staining was performed using aniline blue fluorochrome (Biosupplies Australia Pty. Ltd.). A stock solution (0.1 mg/ml in H2O) was diluted 1:3 ratio with 67 mM K3PO4 pH 9.5. Seedlings were incubated in the staining solution for 2 hr at room temperature under vacuum (60 mPa) and imaged in Citifluor/PBS solution (1:2) (Citifluor Ltd.) on confocal using a 405nm UV laser.
Grafting
7-day-old seedlings were used for grafting assay following the protocol described in66 with little modification. Liquid half MS media (2.1g MS basal salt mixture, 1% sucrose and 0.05% MES, pH 5.8) was used for root growing after grafting instead of water. Images of GFP in roots were taken by confocal microscopy at 4 days after grafting.
Transient expression
For transient expression in N. benthamiana, leaves of 3-week-old plants were pressure-infiltrated with GV3101 agrobacterium strains. Agrobacterium were previously transformed using heat chock protocol with the relevant binary plasmids (30 min on ice, 5 min in liquid nitrogen and 5 min at 37°C). Prior to infiltration, agrobacteria cultures were grown in Luria and Bertani medium with appropriate antibiotics at 28°C for 24h then diluted to 1/10 and grown until the culture reached an OD600 of about 0.8. Bacteria were then pelleted and resuspended in water at a final OD600 of 0.4. For expression of more than one protein construction, equal amounts of the water-resuspended bacteria were mixed. Agroinfiltrated N. benthamiana leaves were imaged 3-4 days post infiltration at room temperature. Around 1 x 1 cm leaf pieces were removed from plants and mounted with the lower epidermis facing up onto glass microscope slides.
Supplementary Material
Acknowledgements
This work was supported by Finnish Centre of Excellence in Molecular Biology of Primary Producers (Academy of Finland CoE program 2014-2019, decision #271832), the Gatsby Foundation (GAT3395/PR3), the National Science Foundation Biotechnology and Biological Sciences Research Council grant (BB/N013158/1), University of Helsinki (award 799992091) and the European Research Council Advanced Investigator Grant SYMDEV (No. 323052). This work was also funded as part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. We thank Raymond Wightman for assistance with confocal imaging, Matthieu Bourdon for help in conducting immunolocalization and Katie Abley for help in seed germination assay. The SB-EM imaging was supported by Biocenter Finland (IB, EJ), and we would like to thank Mervi Lindman, Antti Salminen and Matias Veikkolainen for sample preparation for SB-EM. We thank Mary Roth and Ruth Welti for polar lipid analysis, Andrej Shevchenko and Kai Schuchmann for sphingomyelin analysis and Paul Dupree for data interpretation.
Footnotes
Reporting Summary
Further information on the experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data underlying the findings are available from the corresponding authors upon request.
Author Contributions
D.Y., A.P., S.R.Y., E.M.B. and Y.H. designed the experiments. D.Y., A.P., S.R.Y. E.M.B and Y.H. wrote the manuscript with input from other authors. D.Y., S. R. Y., A.V. and J.Y.L generated Arabidopsis lines. S.R.Y., A.V.,L.K. and S.E.S carried out the suppressor screen and gene mapping. D.Y., S.R.Y. and A.P. performed growth phenotype analysis. D.Y., J.E.M. and M.S.G. carried out the sphingolipid profile. A.P., W.J.N. J. D. P. and L. B. carried out the electron tomography analysis with support from E.M.B.. M.K. provided support for FRAP experiment. A.P., E.J. and I.B. carried out the SB-EM analysis. G.M.M. and J.C.M. performed the GIPC glycosylation assay. J. D. P. carried out the transient co-expression assay. The rest of experiments were performed by D.Y.
Competing interests
The authors declare no competing interests.
Contributor Information
Dawei Yan, The Sainsbury Laboratory, University of Cambridge, Cambridge, United Kingdom.
Shri Ram Yadav, Helsinki Institute of Life Science / Institute of Biotechnology, University of Helsinki, 00014 Helsinki, Finland; Department of Biotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
Andrea Paterlini, The Sainsbury Laboratory, University of Cambridge, Cambridge, United Kingdom.
Lysiane Brocard, Bordeaux Imaging Centre, Plant Imaging Plateform, UMS 3420, INRA-CNRS-INSERM-University of Bordeaux, 71 Av Edouard Bourlaux, 33883 Villenave-d’Ornon, France.
Ilya Belevich, Helsinki Institute of Life Science / Institute of Biotechnology, University of Helsinki, 00014 Helsinki, Finland.
Magali S. Grison, Laboratory of Membrane Biogenesis, UMR5200 CNRS, University of Bordeaux, Bordeaux, France
Anne Vaten, Helsinki Institute of Life Science / Institute of Biotechnology, University of Helsinki, 00014 Helsinki, Finland.
Leila Karami, Helsinki Institute of Life Science / Institute of Biotechnology, University of Helsinki, 00014 Helsinki, Finland; Department of Horticulture, Faculty of Agriculture and Natural Resources, Persian Gulf University, Bushehr 7561158818, Bushehr, Iran.
Sedeer el-Showk, Helsinki Institute of Life Science / Institute of Biotechnology, University of Helsinki, 00014 Helsinki, Finland.
Jung-Youn Lee, Department of Plant and Soil sciences, Delaware Biotechnology Institute, University of Delaware, Newark, DE19711, U. S. A..
Michael Knoblauch, School of Biological Sciences, Washington State University, WA, United States.
Eija Jokitalo, Helsinki Institute of Life Science / Institute of Biotechnology, University of Helsinki, 00014 Helsinki, Finland.
Jennifer E. Markham, Department of Biochemistry, University of Nebraska-Lincoln, NE, United States
Emmanuelle M. Bayer, Laboratory of Membrane Biogenesis, UMR5200 CNRS, University of Bordeaux, Bordeaux, France.
Ykä Helariutta, The Sainsbury Laboratory, University of Cambridge, Cambridge, United Kingdom; Helsinki Institute of Life Science / Institute of Biotechnology, University of Helsinki, 00014 Helsinki, Finland.
References
- 1.Lucas WJ. Plasmodesmata - Intercellular Channels for Macromolecular Transport in Plants. Curr Opin Cell Biol. 1995;7:673–680. doi: 10.1016/0955-0674(95)80109-x. [DOI] [PubMed] [Google Scholar]
- 2.Kim I, Zambryski PC. Cell-to-cell communication via plasmodesmata during Arabidopsis embryogenesis. Curr Opin Plant Biol. 2005;8:593–599. doi: 10.1016/j.pbi.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Otero S, Helariutta Y, Benitez-Alfonso Y. Symplastic communication in organ formation and tissue patterning. Curr Opin Plant Biol. 2016;29:21–28. doi: 10.1016/j.pbi.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Lim GH, et al. Plasmodesmata Localizing Proteins Regulate Transport and Signaling during Systemic Acquired Immunity in Plants. Cell Host Microbe. 2016;19:541–549. doi: 10.1016/j.chom.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Yadav SR, Yan D, Sevilem I, Helariutta Y. Plasmodesmata-mediated intercellular signaling during plant growth and development. Front Plant Sci. 2014;5:44. doi: 10.3389/fpls.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benitez-Alfonso Y, et al. Symplastic intercellular connectivity regulates lateral root patterning. Dev Cell. 2013;26:136–147. doi: 10.1016/j.devcel.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Sivaguru M, et al. Aluminum-induced 1-->3-beta-D-glucan inhibits cell-to-cell trafficking of molecules through plasmodesmata. A new mechanism of aluminum toxicity in plants. Plant Physiol. 2000;124:991–1006. doi: 10.1104/pp.124.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tylewicz S, et al. Photoperiodic control of seasonal growth is mediated by ABA acting on cell-cell communication. Science. 2018;360:212–214. doi: 10.1126/science.aan8576. [DOI] [PubMed] [Google Scholar]
- 9.Oparka KJ, et al. Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell. 1999;97:743–754. doi: 10.1016/s0092-8674(00)80786-2. [DOI] [PubMed] [Google Scholar]
- 10.Tilsner J, Amari K, Torrance L. Plasmodesmata viewed as specialised membrane adhesion sites. Protoplasma. 2011;248:39–60. doi: 10.1007/s00709-010-0217-6. [DOI] [PubMed] [Google Scholar]
- 11.Ding B, Turgeon R, Parthasarathy MV. Substructure of Freeze-Substituted Plasmodesmata. Protoplasma. 1992;169:28–41. [Google Scholar]
- 12.Tilsner J, Nicolas W, Rosado A, Bayer EM. Staying Tight: Plasmodesmal Membrane Contact Sites and the Control of Cell-to-Cell Connectivity in Plants. Annu Rev Plant Biol. 2016;67:337–364. doi: 10.1146/annurev-arplant-043015-111840. [DOI] [PubMed] [Google Scholar]
- 13.Nicolas WJ, et al. Architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves. Nat Plants. 2017;3 doi: 10.1038/nplants.2017.82. 17082. [DOI] [PubMed] [Google Scholar]
- 14.Roberts AG, Oparka KJ. Plasmodesmata and the control of symplastic transport. Plant Cell Environ. 2003;26:103–124. [Google Scholar]
- 15.Burch-Smith TM, Stonebloom S, Xu M, Zambryski PC. Plasmodesmata during development: re-examination of the importance of primary, secondary, and branched plasmodesmata structure versus function. Protoplasma. 2011;248:61–74. doi: 10.1007/s00709-010-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz A. Plasmodesmal Widening Accompanies the Short-Term Increase in Symplasmic Phloem Unloading in Pea Root-Tips under Osmotic-Stress. Protoplasma. 1995;188:22–37. [Google Scholar]
- 17.Brunkard JO, Runkel AM, Zambryski PC. The cytosol must flow: intercellular transport through plasmodesmata. Curr Opin Cell Biol. 2015;35:13–20. doi: 10.1016/j.ceb.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Zavaliev R, Ueki S, Epel BL, Citovsky V. Biology of callose (beta-1,3-glucan) turnover at plasmodesmata. Protoplasma. 2011;248:117–130. doi: 10.1007/s00709-010-0247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Storme N, Geelen D. Callose homeostasis at plasmodesmata: molecular regulators and developmental relevance. Front Plant Sci. 2014;5:138. doi: 10.3389/fpls.2014.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui W, Lee JY. Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nature Plants. 2016;2 doi: 10.1038/nplants.2016.34. 16034. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Calvino L, et al. Arabidopsis Plasmodesmal Proteome. Plos One. 2011;6:e18880. doi: 10.1371/journal.pone.0018880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraner ME, Muller C, Sonnewald U. Comparative proteomic profiling of the choline transporter-like1 (CHER1) mutant provides insights into plasmodesmata composition of fully developed Arabidopsis thaliana leaves. Plant J. 2017;92:696–709. doi: 10.1111/tpj.13702. [DOI] [PubMed] [Google Scholar]
- 23.Diao M, et al. Arabidopsis formin 2 regulates cell-to-cell trafficking by capping and stabilizing actin filaments at plasmodesmata. Elife. 2018;7:e36316. doi: 10.7554/eLife.36316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grison MS, et al. Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell. 2015;27:1228–1250. doi: 10.1105/tpc.114.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, et al. Suppressing a Putative Sterol Carrier Gene Reduces Plasmodesmal Permeability and Activates Sucrose Transporter Genes during Cotton Fiber Elongation. Plant Cell. 2017;29:2027–2046. doi: 10.1105/tpc.17.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross-Elliott TJ, et al. Phloem unloading in Arabidopsis roots is convective and regulated by the phloem-pole pericycle. Elife. 2017;6:e24125. doi: 10.7554/eLife.24125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaten A, et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell. 2011;21:1144–1155. doi: 10.1016/j.devcel.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Ullman MD, Radin NS. The enzymatic formation of sphingomyelin from ceramide and lecithin in mouse liver. J Biol Chem. 1974;249:1506–1512. [PubMed] [Google Scholar]
- 29.Elmendorf HG, Haldar K. Plasmodium falciparum exports the Golgi marker sphingomyelin synthase into a tubovesicular network in the cytoplasm of mature erythrocytes. J Cell Biol. 1994;124:449–462. doi: 10.1083/jcb.124.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tafesse FG, Ternes P, Holthuis JCM. The multigenic sphingomyelin synthase family. J Biol Chem. 2006;281:29421–29425. doi: 10.1074/jbc.R600021200. [DOI] [PubMed] [Google Scholar]
- 31.Voelker DR, Kennedy EP. Cellular and Enzymic-Synthesis of Sphingomyelin. Biochemistry-Us. 1982;21:2753–2759. doi: 10.1021/bi00540a027. [DOI] [PubMed] [Google Scholar]
- 32.Huitema K, van den Dikkenberg J, Brouwers JFHM, Holthuis JCM. Identification of a family of animal sphingomyelin synthases. Embo J. 2004;23:33–44. doi: 10.1038/sj.emboj.7600034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yachi R, et al. Subcellular localization of sphingomyelin revealed by two toxin-based probes in mammalian cells. Genes Cells. 2012;17:720–727. doi: 10.1111/j.1365-2443.2012.01621.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang W, et al. An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell. 2008;20:3163–3179. doi: 10.1105/tpc.108.060053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knox K, et al. Putting the Squeeze on Plasmodesmata: A Role for Reticulons in Primary Plasmodesmata Formation. Plant Physiol. 2015;168:1563–1572. doi: 10.1104/pp.15.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tolley N, et al. Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant Journal. 2010;64:411–418. doi: 10.1111/j.1365-313X.2010.04337.x. [DOI] [PubMed] [Google Scholar]
- 37.Markham JE, Jaworski JG. Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:1304–1314. doi: 10.1002/rcm.2962. [DOI] [PubMed] [Google Scholar]
- 38.Markham JE, Li J, Cahoon EB, Jaworski JG. Separation and identification of major plant sphingolipid classes from leaves. J Biol Chem. 2006;281:22684–22694. doi: 10.1074/jbc.M604050200. [DOI] [PubMed] [Google Scholar]
- 39.Gronnier J, Germain V, Gouguet P, Cacas JL, Mongrand S. GIPC: Glycosyl Inositol Phospho Ceramides, the major sphingolipids on earth. Plant Signal Behav. 2016;11:e1152438. doi: 10.1080/15592324.2016.1152438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merrill AH, Wang E, Gilchrist DG, Riley RT. Fumonisins and Other Inhibitors of De-Novo Sphingolipid Biosynthesis. Adv Lipid Res. 1993;26:215–234. [PubMed] [Google Scholar]
- 42.Markham JE, et al. Sphingolipids Containing Very-Long-Chain Fatty Acids Define a Secretory Pathway for Specific Polar Plasma Membrane Protein Targeting in Arabidopsis. Plant Cell. 2011;23:2362–2378. doi: 10.1105/tpc.110.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benitez-Alfonso Y, et al. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. P Natl Acad Sci USA. 2009;106:3615–3620. doi: 10.1073/pnas.0808717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li WM, et al. Depletion of ceramides with very long chain fatty acids causes defective skin permeability barrier function, and neonatal lethality in ELOVL4 deficient mice. Int J Biol Sci. 2007;3:120–128. doi: 10.7150/ijbs.3.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneiter R, et al. A yeast acetyl coenzyme a carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eisenkolb M, Zenzmaier C, Leitner E, Schneiter RA. specific structural requirement for ergosterol in long-chain fatty acid synthesis mutants important for maintaining raft domains in yeast. Mol Biol Cell. 2002;13:4414–4428. doi: 10.1091/mbc.E02-02-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jean-luc C, et al. Revisiting Plant Plasma Membrane Lipids in Tobacco: A Focus on Sphingolipids. Plant physiol. 2016;170:367–384. doi: 10.1104/pp.15.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinto SN, Silva LC, Futerman AH, Prieto M. Effect of ceramide structure on membrane biophysical properties: The role of acyl chain length and unsaturation. BBA-Biomembranes. 2011;1808:2753–2760. doi: 10.1016/j.bbamem.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 49.Obayashi, et al. ATTED-II in 2018: A Plant Coexpression Database Based on Investigation of Statistical Property of the Mutual Rank Index. Plant Cell Physiology. 2018;59:e3. doi: 10.1093/pcp/pcx191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shang BS, et al. Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in Arabidopsis. P Natl Acad Sci USA. 2016;113:5101–5106. doi: 10.1073/pnas.1522466113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 52.Wilke SA, et al. Deconstructing complexity: serial block-face electron microscopic analysis of the hippocampal mossy fiber synapse. J Neurosci. 2013;33:507–522. doi: 10.1523/JNEUROSCI.1600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furuta KM, et al. Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science. 2014;345:933–937. doi: 10.1126/science.1253736. [DOI] [PubMed] [Google Scholar]
- 54.Belevich I, Joensuu M, Kumar D, Vihinen H, Jokitalo E. Microscopy Image Browser: A Platform for Segmentation and Analysis of Multidimensional Datasets. Plos Biol. 2016;14:e1002340. doi: 10.1371/journal.pbio.1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicolas WJ, Bayer E, Brocard L. Electron Tomography to Study the Three-dimensional Structure of Plasmodesmata in Plant Tissues–from High Pressure Freezing Preparation to Ultrathin Section Collection. Bio-protocol. 2018;81:e2681. doi: 10.21769/BioProtoc.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 57.Mortimer JC, et al. Abnormal Glycosphingolipid Mannosylation Triggers Salicylic Acid-Mediated Responses in Arabidopsis. Plant Cell. 2013;25:1881–1894. doi: 10.1105/tpc.113.111500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cacas JL, et al. Revisiting Plant Plasma Membrane Lipids in Tobacco: A Focus on Sphingolipids. Plant Physiol. 2016;170:367–384. doi: 10.1104/pp.15.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bure C, et al. Fast screening of highly glycosylated plant sphingolipids by tandem mass spectrometry. Rapid Commun Mass Spectrom. 2011;25:3131–3145. doi: 10.1002/rcm.5206. [DOI] [PubMed] [Google Scholar]
- 60.Fang L, et al. Loss of Inositol Phosphorylceramide Sphingolipid Mannosylation Induces Plant Immune Responses and Reduces Cellulose Content in Arabidopsis. Plant Cell. 2016;28:2991–3004. doi: 10.1105/tpc.16.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Markham JE, Jaworski JG. Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:1304–1314. doi: 10.1002/rcm.2962. [DOI] [PubMed] [Google Scholar]
- 62.Markham JE, Li J, Cahoon EB, Jaworski JG. Separation and identification of major plant sphingolipid classes from leaves. J Biol Chem. 2006;281:22684–22694. doi: 10.1074/jbc.M604050200. [DOI] [PubMed] [Google Scholar]
- 63.Welti R, et al. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- 64.Xiao S, et al. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell. 2010;22:1463–1482. doi: 10.1105/tpc.110.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuo J, Niu QW, Chua NH. Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- 66.Melnyk CW. Grafting with Arabidopsis thaliana. Methods Mol Biol. 2017;1497:9–18. doi: 10.1007/978-1-4939-6469-7_2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.