Abstract
Prostate cancer (PCa) initiation and progression requires activation of numerous oncogenic signaling pathways. Nuclear-cytoplasmic transport of oncogenic factors is mediated by Karyopherin proteins during cell transformation. However, the role of nuclear transporter proteins in PCa progression has not been well defined. Here, we report that the KPNB1, a key member of Karyopherin beta subunits, is highly expressed in advanced prostate cancers. Further study showed that targeting KPNB1 suppressed the proliferation of prostate cancer cells. The knockdown of KPNB1 reduced nuclear translocation of c-Myc, the expression of downstream cell cycle modulators, and phosphorylation of regulator of chromatin condensation 1 (RCC1), a key protein for spindle assembly during mitosis. Meanwhile, CHIP assay demonstrated the binding of c-Myc to KPNB1 promoter region, which indicated a positive feedback regulation of KPNB1 expression mediated by the c-Myc. In addition, NF-κB subunit p50 translocation to nuclei was blocked by KPNB1 inhibition, which led to an increase in apoptosis and a decrease in tumor sphere formation of PCa cells. Furthermore, subcutaneous xenograft tumor models with a stable knockdown of KPNB1 in C42B PCa cells validated that the inhibition of KPNB1 could suppress the growth of prostate tumor in vivo. Moreover, the intravenously administration of importazole, a specific inhibitor for KPNB1, effectively reduced PCa tumor size and weight in mice inoculated with PC3 PCa cells. In summary, our data established the functional link between KPNB1 and PCa prone c-Myc, NF-kB, and cell cycle modulators. More importantly, inhibition of KPNB1 could be a new therapeutic target for PCa treatment.
Keywords: KPNB1, prostate cancer, cell cycle, tumorigenesis, c-Myc, NF-kB, RCC1
Introduction
Prostate cancer is the most common cancer type diagnosed and the second-leading cause of cancer deaths in men. Comparing to traditional cytotoxic agents, targeted drug therapy based on the unique molecular and cellular signatures that distinguish tumor cells from their normal counterparts is preferred for its reduced, dose-limiting toxicities to normal cells.
Increasing evidences show that the nuclear trafficking plays a critical role in multiple cell processes, including mitosis, migration as well as cell transformation. The crucial nuclear trafficking process is mediated by a series of finely choreographed protein-protein interactions in which Karyopherin superfamily members play a vital role. Consisted of a group of soluble nuclear transporters, the Karyopherin proteins include both alpha and beta subunits. The alpha subunits containing NLS (nuclear localization signal) recognition domain enable them to function as adaptor proteins to bind to the NLS of cargo proteins. On the other hand, the Karyopherin beta subunits normally interact with the complex of alpha subunit and cargo proteins to facilitate nuclear import. However, Karyopherin beta subunits can directly interact with cargo proteins as well [1]. Increasing evidences have shown that aberrant expression of Karyopherin proteins due to the dysregulated promoter activity of Karyopherin proteins is associated with cancer initiation and progression [2]. Among them, our previous study supports that KPNA4, a member of Karyopherin alpha family, is associated with the skeletal metastasis of prostate cancer through the regulation of NF-κB signaling [3]. The increased expression of Karyopherin proteins provides a plausible mechanism which helps tumor cells to cope with the extra metabolic demands to maintain abnormally high rate of mitosis. In particular, more Karyopherin protein could facilitate more frequent entries of oncogenic transcription factors such as c-Myc[4], Snail[5] and c-Jun[6].
The nuclear localization of NF-κB proteins through Karyopherins alpha subunits, including KPNA2, KPNA3, and KPNA4 has been widely reported [7, 8]. As a receptor of the alpha units, KPNB1 can also regulate NF-κB nuclear translocation [9, 10], as well as the expression level of the downstream genes [11]. Another study indicates that KPNB1 may directly interact with epidermal growth factor receptor (EGFR), not just indirectly cooperating through alpha units[12]. Correspondingly, an organic inhibitor intervening the interaction between KPNB1 and EGFR significantly suppresses invasion of lung cancer[12]. Targeting KPNB1 as an anti-cancer strategy has been reported in several cancer types, including cervical cancer[13], lung cancer[14], breast cancer[15] and ovarian cancer[16]. However, the functional role of KPNB1 in prostate cancer growth has not been studied.
Here, we firstly proved the clinical relevance of KPNB1 overexpression in prostate cancer by immunohistochemistry of human tissue microarrays and screening of multiple human prostate cancer datasets. We found that KPNB1 was significantly upregulated in advanced prostate cancer samples. Targeting KPNB1 can efficiently suppress prostate cancer cell growth by prompting cell cycle arrest at G1 phase and increasing cell death. Further study revealed that inhibition of KPNB1 could cause remarkable reductions of Cyclin D1 and CDK1/Cyclin B1 complex expression, and the phosphorylation of a key regulator of mitosis, RCC1. We further demonstrated the existence of a regulatory loop between KPNB1 and c-Myc in PCa cells. In addition, targeting KPNB1 largely blocked the nuclear translocation of NF-κB p50, which led to a significant induction of cell apoptosis and reduction of tumor sphere formation of PCa cells. We also demonstrated that KPNB1 could be a targeted drug therapy for prostate cancer through the application of shRNA oligos of KPNB1 and importazole, a specific inhibitor that alters the interaction of KPNB1 with RanGTP [17]. Both stable knockdown of KPNB1 and intravenous administration of nanoparticle packaged importazole achieved a promising therapeutic outcome on prostate cancer progression in xenograft mouse models.
Results
Overexpression of KPNB1 in prostate carcinoma
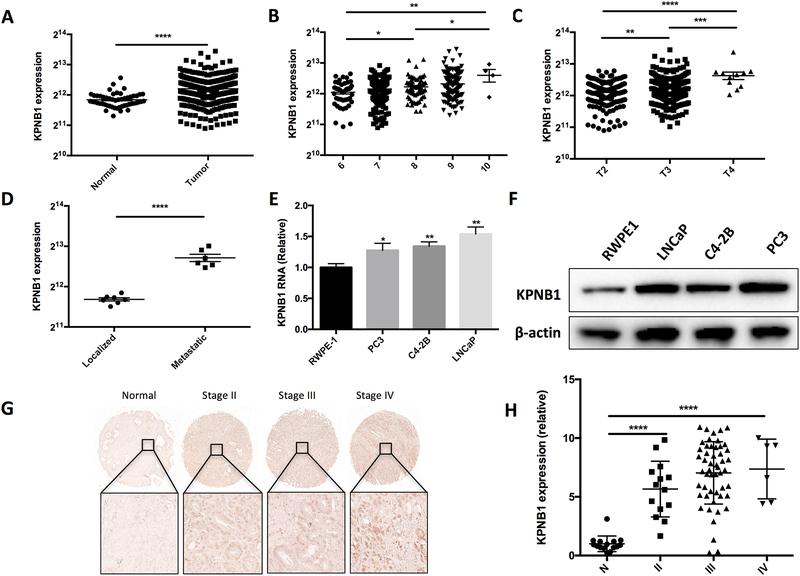
To study the expression profile of KPNB1 in human prostate cancer, we analyzed a TCGA dataset that containing 498 cases of prostate adenocarcinoma and 52 cases of normal subjects as control. The analysis revealed that KPNB1 was highly expressed in PCa samples (Fig 1A). Further analysis based on the classified stages of prostate cancer confirmed that KPNB1 expression levels were positively associated with the PCa progression indicated by the Glison scores (Fig 1B) as well as the pathological stages (Fig 1C). In other words, the KPNB1 expression levels were higher in the more advanced prostate cancer samples. We further compared the expression levels of KPNB1 in the localized and metastatic prostate cancer tissue samples included in a GEO (Gene Expression Omnibus) dataset. We found that the KPNB1 expression levels in metastatic PCa samples were much higher than those localized PCa samples (Fig 1D). Samples from another GEO dataset also confirmed that KPNB1 expression levels were greater in invasive prostate tumor samples than prostate intraepithelial neoplasia (PIN) samples in mouse (Supplementary fig. 1). We also measured the expression levels of KPNB1 in human PCa cell lines using qPCR and western blotting techniques. The results showed overexpression of KPNB1 at both RNA (Fig 1E) and protein (Fig 1F) levels in PCa cell lines in comparison to normal human epithelia cell line, RWPE1. To verify the findings of the TCGA and GEO datasets, we assessed the expression of KPNB1 using a human PCa tissue microarray (TMA) slides. Immunohistochemistry (IHC) result confirmed a significant elevation of KPNB1 expression in PCa samples in comparison to the normal counterparts (adjacent prostate tissues) (Fig 1G and 1H).
Figure 1. KPNB1 expression is associated with human prostate cancer progression.
A) The expression level of KPNB1 in human prostate cancer samples (N=498) and normal prostate tissues (N=52). Samples are from TCGA dataset. B) The expression level of KPNB1 human prostate cancer samples with different Gleason scores. Samples are from TCGA dataset. C) The expression level of KPNB1 human prostate cancer samples with different pathological stages. Samples are from TCGA dataset. D) The expression level of KPNB1 of localized and metastasis human prostate cancer samples. Samples are from GEO dataset. E) qPCR results showing relative expression level of KPNB1 RNA in indicated cell lines. FTH1 was used as internal control. F) Immunoblotting results showing protein level of KPNB1 in indicated cell lines. β-actin was used as internal control. G) Representative images of IHC staining of KPNB1 on human prostate cancer TMA slides. H) Statistical results of KPNB1 positive cells on the TMA slides. *p <0.05, **p<0.01, *** p<0.001, ****p<0.0001.
KPNB1 inhibition induced cell cycle arrest in prostate carcinoma cells
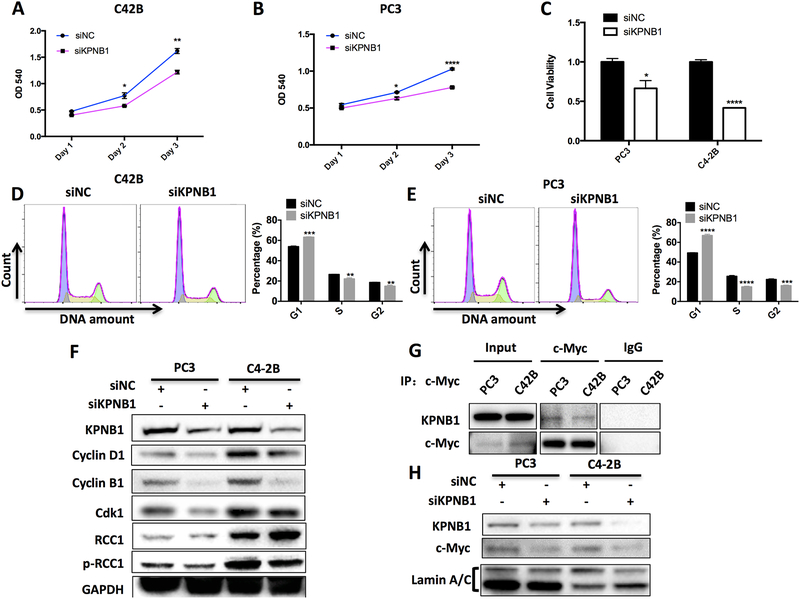
The overexpression of KPNB1 in PCa indicates that it could be functionally involved in the progression of PCa. To assess the potential function of KPNB1 in prostate cancer progression, we applied KPNB1 siRNA to knockdown KPNB1 expression in two human PCa cell lines, C42B and PC3. As measured by crystal violet staining, knockdown of KPNB1 significantly reduced cell growth in both cell lines (Fig 2A and 2B). Moreover, MTT assay revealed that knockdown of KPNB1 decreased the cell viability in both cell lines (Fig 2C). To test if the impaired cell growth by KPNB1 knockdown is contributed by cell cycle arrest, we performed flow cytometry after PI staining to examine the cell cycle phase distribution. As expected, the knockdown of KPNB1 induced the increase of percentage of cells in G1 phase and a significant decreased percentage of cells in S and G2/M phases in both PCa cell lines (Fig 2D and 2E). Consistently, western blotting analysis revealed that KPNB1 knockdown induced the reduction of cell cycle regulatory proteins including Cyclin D1, CDK1 and Cyclin B1. The phosphorylation of RCC1 but not the expression of total RCC1, was also downregulated with the knockdown of KPNB1 (Fig 2F).
Figure 2. KPNB1 knockdown inhibits PCa growth.
A and B) Crystal violet staining showing the growth of indicated cell lines with KPNB1 knockdown. Both cell lines were transfected with KPNB1 siRNA (50 nM) or control siRNA for 24 hours, 48 hours and 72 hours respectively. C) MTT assay showing the relative cell viabilities of indicated cell lines that were transfected with KPNB1 siRNA (50 nM) or control siRNA for 48 hours. D and E) Flow cytometry showing the cell cycle stage distribution of indicated cell lines that were transfected with KPNB1 siRNA (50 nM) or control siRNA for 48 hours. F) Immunoblotting results showing protein level of KPNB1, Cyclin D1, Cyclin B1, CDK1, RCC1 and pRCC1 of indicated cell lines that were transfected with KPNB1 siRNA (50 nM) or control siRNA for 48 hours. GAPDH was used as internal control. G) Immunoblotting results showing physical interaction between KPNB1 and c-Myc proteins by immunoprecipitation assay in both of C42B and PC3 cell lines. H) Immunoblotting results showing KPNB1 and c-Myc from nuclear extracts of C42B and PC3 that were transfected with KPNB1 siRNA (50 nM) or control siRNA for 48 hours. *p <0.05, **p<0.01, *** p<0.001, ****p<0.0001.
Previously, we have described the regulatory role of c-Myc oncogene in PCa cells [18]. In addition, an assay of large-scale identification of c-MYC-associated proteins has suggested c-Myc as a potential cargo protein of KPNB1[19]. Thus, we performed co-IP assay to determine if KPNB1 interacts with c-Myc in PCa. Indeed, immunoblotting results from both PC3 and C42B cells exhibited that KPNB1 bound with c-Myc (Fig 2G). To investigate if KPNB1 mediates nuclear translocation of c-Myc, we collected the nuclear extracts from PC3 and C42B cells transfected with KPNB1 siRNA. Immunoblotting results suggested that the knockdown of KPNB1 attenuate c-Myc’s nuclear localization in PC3 and C42B cells (Fig 2H). Interestingly, cytoplasmic c-Myc is decreased as well following KPNB1 knockdown (Supplementary Fig. 2), probably due to the impaired NF-κB translocation [10, 20].
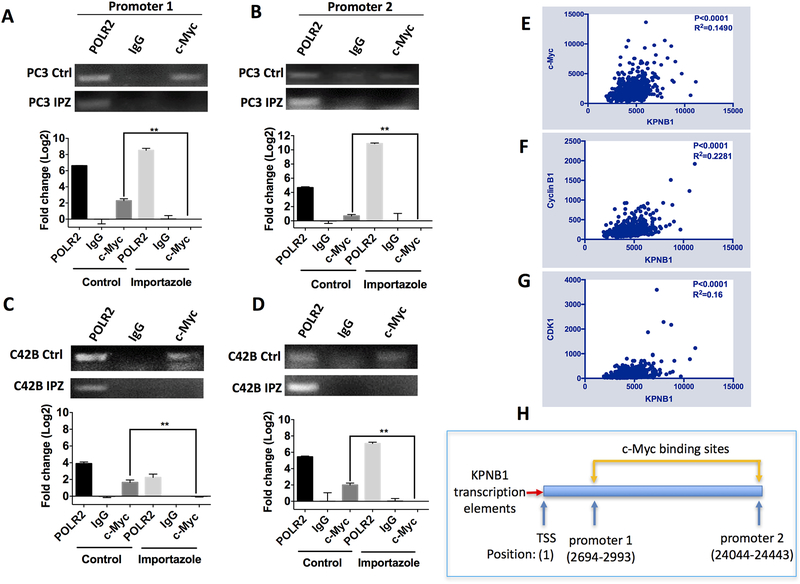
On the other hand, a Myc-driven murine PCa model implied that c-Myc could also directly regulate KPNB1 expression [21]. Our results did indicate that KPNB1 expression reduced when c-Myc was knockdown by siRNA (Supplementary Fig 3). It is highly plausible to speculate that there is a positive feed-back regulation between KPNB1 and c-Myc. To test this possibility, c-Myc-bound genomic DNA were pulled down using a CHIP assay kit in PC3 and C42B cells treated with or without KPNB1 inhibitor, importazole. Importazole is a small molecule inhibitor of KPNB1 which specifically blocks KPNB1-mediated nuclear import [17]. As demonstrated by the qPCR results, c-Myc did bind to the two tested promoter regions of KPNB1 in PCa cells. Inhibiting KPNB1 importing activity using importazole significantly blocked the binding of c-Myc to KPNB1 promoter region in PC3 cells (Fig 3A–B). Similar results were observed in C42B cells (Fig 3C–D). The interaction between KPNB1 and c-Myc in PC3 and C42B cells revealed by the CHIP assays were further supported by TCGA dataset. A positive relationship between the expression levels of KPNB1 and c-Myc was exhibited in the dataset including 498 PCa cases (Fig 3E). In addition, the RNA levels of Cyclin B1 (Fig 3F) and CDK1 (Fig 3G) were also positively associated with KPNB1 levels in the same dataset. Interestingly, there was no association between KPNB1 and Cyclin D1 or KPNB1 and RCC1 (Supplementary Fig. 4) which resonated with our observation that RCC1’s phosphorylation but not the expression level was downregulated by KPNB1 (Fig 2F).
Figure 3. c-Myc binds to the promoter regions of KPNB1.
A-D) qPCR showing the interaction of c-Myc with promoter regions of KPNB1. The indicated PCa cell lines were treated with importazole (20 μM) for 24 hours, DMSO was used as vehicle control. DNA of the treated cells were precipitated and purified by c-Myc antibody for CHIP assay. Polymerase II (POLR 2) antibody and control IgG antibody were used as positive and negative control, respectively. Relative amount of precipitated promoter DNA was analyzed using qPCR, and the PCR product was revealed by running an agarose gel. E-G) Analysis of the TCGA dataset to show the correlation of KPNB1 with c-Myc, Cyclin B1 and CDK1 in human prostate cancer samples. Correlations between indicated genes were analyzed by computing Pearson correlation coefficients. P value and R squared were revealed as well. H) Schematic model indicating the promoter region (c-Myc binding sites) of KPNB1. **p<0.01.
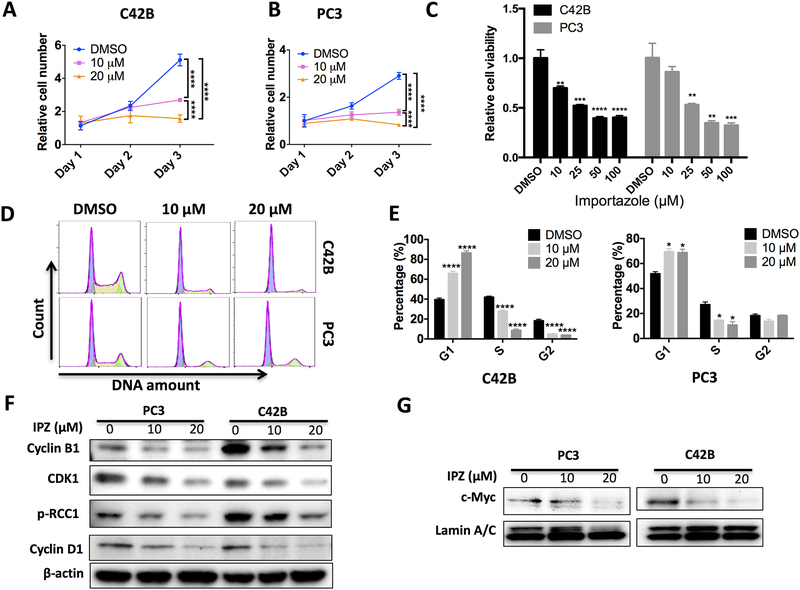
Next, we assessed whether inhibition of KPNB1 importing activity with importazole can inhibit PCa cell growth. Consistent with the effects of KPNB1 siRNA, importazole inhibited PCa cell growth in C42B (Fig 4A), PC3 (Fig 4B) and Myc-CaP mouse PCa cells (Supplementary Fig 5). The cell viability measured with MTT assay demonstrated a dose dependent manner of importazole’s inhibitory effects in these PCa cells (Fig 4C). Importazole was also able to induce cell cycle arrest in human PCa cells (Fig 4D–E). Immunoblotting experiments showed that importazole inhibited the expression of Cyclin D1, CDK1, Cyclin B1, and phosphorylation of RCC1 in a dose dependent manner (Fig 4F), as well as the nuclear importing of c-Myc in both PC3 and C42B cell lines (Fig 4G). Of note, the half lethal dose of importazole in non-malignant prostatic epithelial RWPE1 cells, was double amount of that in PCa cell lines (Supplementary Fig 6). These results suggested that PCa cells were more sensitive to KPNB1 inhibition when compared to non-tumor prostate cells.
Figure 4. Inhibition of KPNB1 using importazole suppresses PCa growth.
A and B) Crystal violet staining showing the growth of indicated cell lines that were treated with 10 μM or 20 μM of importazole for 24 hours, 48 hours or 72 hours. DMSO was used as vehicle control. C) MTT assay showing relative cell viabilities of PC3 or C42B that was treated with importazole of indicated concentrations for 48 hours. DMSO was used as vehicle control. D and E) Flow cytometry showing the cell cycle stage distribution of indicated cell lines that were treated with 10 μM or 20 μM of importazole for 48 hours. DMSO was used as vehicle control. F) Immunoblotting results showing protein level of Cyclin B1, CDK1, pRCC1 and Cyclin D1 of indicated cell lines that were treated with DMSO, 10 μM or 20 μM of importazole for 48 hours. β-actin was used as internal control. G) Immunoblotting results showing nuclear distribution of c-Myc of indicated cell lines that were treated with 10 μM or 20 μM of importazole for 48 hours. DMSO was used as vehicle control. Lamin A/C was used as loading control of nuclear protein. *p <0.05, **p<0.01, *** p<0.001, ****p<0.0001.
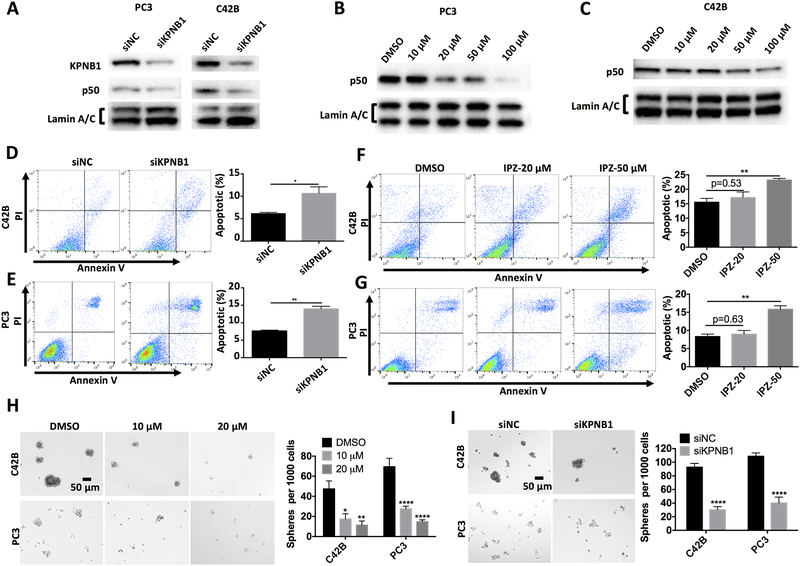
KPNB1 inhibition blocked nuclear importing of NF-κB in PCa cell
KPNB1 is a key factor mediating the nuclear importing of NF-κB subunits, which preludes the activation of NF-κB signaling. Considering that NF-κB activation has been functionally linked to pro-proliferation and anti-apoptosis in cancer cells, we assessed whether inhibition of KPNB1 can reduce NF-κB activation in PCa cells by measuring the nuclear localization of NF-κB subunits. Our data confirmed that inhibiting KPNB1 with either KPNB1 siRNA or importazole was sufficient to block the nuclear importing of NF-κB subunit p50 (Fig 5A–C) in PCa cell lines. The blockade of p50 nuclear translocation by either KPNB1 siRNA (Fig 5D–E) or importazole (Fig 5F–G) led to increased apoptosis in C42B and PC3 PCa cell cultures. Importazole also suppressed the total expression levels of p50 (Supplementary Fig 7).
Figure 5. Inhibition of KPNB1 using importazole induces apoptosis and suppresses sphere formation of PCa through NF-κB signaling.
A) Immunoblotting results showing nuclear NF-κB p50 of PC3 or C42B that was transfected with KPNB1 siRNA (50 nM) or control siRNA for 48 hours. Lamin A/C was used as internal control. B and C) Immunoblotting results showing nuclear NF-κB p50 of PC3 or C42B that was treated with indicated concentrations of importazole or DMSO. Lamin A/C was used as internal control. D and E) Flow cytometry showing the apoptosis of PC3 or C42B that was transfected KPNB1 siRNA (50 nM) or control siRNA for 48 hours. Annexin V positive cells were counted as apoptotic population. F and G) Flow cytometry showing the apoptosis of PC3 or C42B that was treated with indicated concentrations of importazole or DMSO. Annexin V positive cells were counted as apoptotic population. Sphere formation assay of PC3 or C42B that was H) treated with indicated concentrations of importazole or DMSO, or I) transfected with KPNB1 siRNA (50 nM) or control siRNA for 1 week. *p <0.05, **p<0.01, ****p<0.0001.
NF-κB has been known as a key transcriptional factor in tumorigenesis[22]. We speculated that KPNB1 inhibition could reduce the tumorigenesis of PCa cells. We conducted sphere formation assay with C42B and PC3 cells and confirmed that inhibiting KPNB1 through either importazole treatment (Fig 5H) or siRNA transfection (Fig 5I) effectively suppressed the sphere formation in both types of PCa cells.
KPNB1 inhibition suppressed PCa growth in vivo
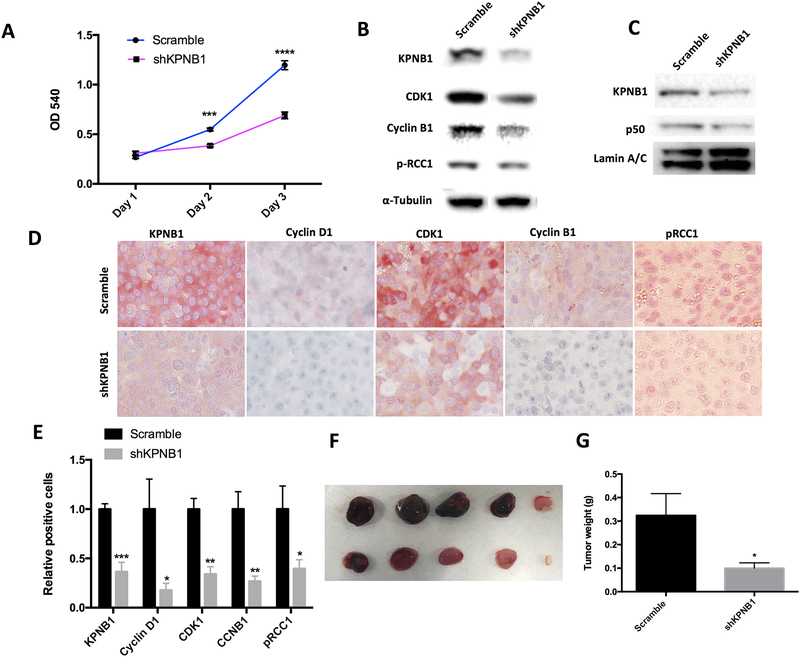
In order to validate the function of KPNB1 in vivo, we generated a KPNB1 stable knockdown cell line, C42B-shKPNB1, using retrovirus and observed a significant inhibition of cell growth in C42B-shKPNB1 in comparison to C42B cells infected with vector control (C42B-scramble) (Fig 6A). The knockdown efficiency of KPNB1 reached 90% reduction at protein level which was accompanied with decreased levels of cell cycle modulators including CDK1, Cyclin B1 and pRCC1 (Fig 6B). Consistent to previous results on p50 nuclear localization, C42B-shKPNB1 nuclear extract showed less p50 proteins than that of C42B-scramble cells (Fig 6C). Next, we compared the tumor burden yield in xenograft model using athymic nude mice bearing subcutaneous implant with either C42B-scramble or C42B-shKPNB1 cells. Six-week later, tumor burdens were dissected and the immune-staining of the tumor sections supported the western blotting results that the protein levels of KPNB1, Cyclin D1, CDK1, Cyclin B1, and phosphorylated RCC1 were downregulated in C42B-shKPNB1 tumors (Fig 6D–E). The weights of tumors in mice bearing C42B-shKPNB1 cells were significantly less than those bearing C42B-scramble cells by about 70% (Fig 6F–G). Consistently, decreased Ki67 and increased Caspase 3 were observed in the C42B-shKPNB1 tumor tissue by IHC staining (Supplementary Fig 8). Orthotropic prostate cancer model showed a similar trend of tumor suppression by KPNB1 knockdown (Supplementary Fig 9).
Figure 6. KPNB1 knockdown attenuates PCa tumor growth in vivo.
A) Crystal violet staining showing the growth of C42B-scramble or C42B-shKPNB1 cell line. B) Immunoblotting results showing protein level of KPNB1, Cyclin B1, CDK1, pRCC1 of C42B-scramble and C42B-shKPNB1 cell lines. α-Tubulin was used as internal control. C). Immunoblotting results showing NF-κB p50 within nuclear of C42B-scramble and C42B-shKPNB1. Lamin A/C was used as internal control. D) IHC staining of KPNB1, Cyclin D1, Cyclin B1, CDK1, pRCC1 of tumor sections. E) Statistical results of positive stained cells of tumor sections. F and G) Tumor burdens of C42B-scramble and C42B-shKPNB1 through subcutaneous injection were dissected and weighted. *p <0.05, **p<0.01, *** p<0.001.
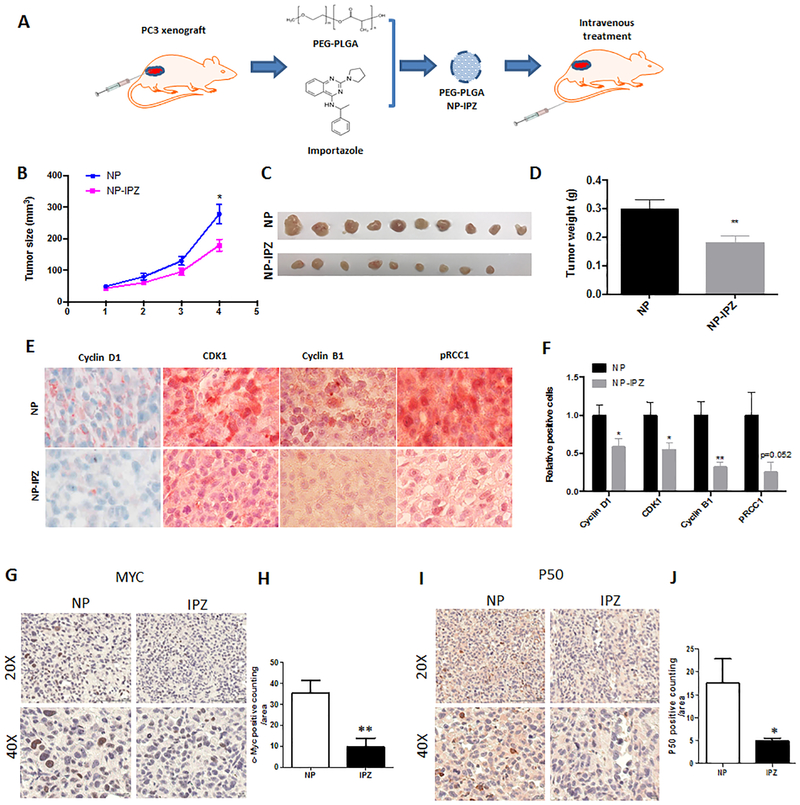
To explore the therapeutic efficacy of importazole in vivo, we assembled importazole powder into a delivery vehicle using PEG-PLGA based lipid nanoparticles. The tumor-bearing mice were subcutaneously inoculated with 1 million PC3 cells and a week later, they were randomly assigned to receive the treatments, either importazole containing particles (NP-IPZ) or empty particles (NP), through tail intravenous injection (Fig 7A). The tumor sizes and weights demonstrated significant difference as early as 3 weeks of treatments. Mice received 6 times of importazole treatment exhibited a significant inhibition of PCa tumor growth (Fig 7B–D). IHC staining of the tumor sections verified that KPNB1 inhibition by importazole can suppress the expression of Cyclin D1, CDK1 and Cyclin B1, as well as the phosphorylation of RCC1 (Fig 7E–F). Additionally, c-Myc and p50 expressions of the PC3 tumor sections were also reduced in mice treated with importazole (Fig 7G–J). Similar to C42B-shKPNB1 tumor, decreased Ki67 and increased Caspase 3 were observed in the importazole treated mice (Supplementary Fig 10A–B). Importantly, in comparison to control, importazole treatment did not change the body weight of mice (Supplementary Fig 10C).
Figure 7. Importazole suppresses PCa tumor growth in vivo.
A) Schematic model showing the procedure of importazole treatment in vivo. One million PC3 cells were inoculated to nude mice through subcutaneous injection. After one week, importazole was encapsulated into PEG-PLGA nanoparticle and injected intravenously twice a week for 3 weeks. B) Tumor growth curve of indicated groups. Tumor sizes of both groups were measured weekly. Tumor burdens of indicated groups were C) dissected and D) weighted. E) IHC staining of Cyclin D1, Cyclin B1, CDK1, pRCC1 of tumor sections. F) Statistical results of positive stained cells of tumor sections. IHC staining of G-H) c-Myc and I-J) p50 using tumor tissue form mice treated with NP-IPZ or NP vehicle. Positive cells were quantified using Image J software. *p <0.05, **p<0.01.
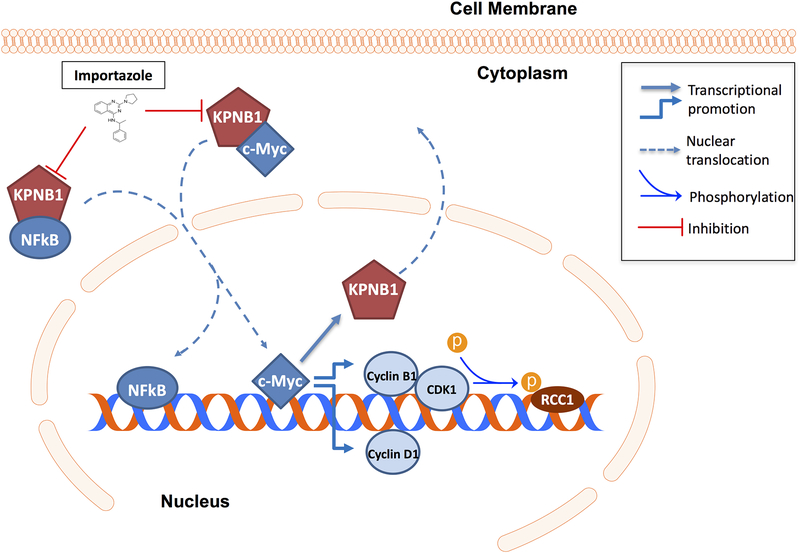
In conclusion, our study demonstrated an important role of KPNB1 in PCa growth through its regulation of c-Myc, cell cycle modulators and NF-κB. We further validated its potential as a therapeutic target for prostate cancer treatment in vivo. As depicted in the diagram (Fig 8), KPNB1 overexpression in prostate cancer activates NF-κB and c-Myc which leads to the dysregulation of Cyclin D1, CDK1/Cyclin B1 and p-RCC1 to promote multiple processes favoring tumorigenesis. Targeting KPNB1 with importazole could effectively inhibit prostate cancer growth in vitro and in vivo.
Figure 8. Schematic model of the role of KPNB1 in PCa progression.
Dysregulation of KPNB1 in PCa facilitates the nuclear translocation of NF-κB p50, by which it can promote tumorigenesis and cell survival PCa cells. In addition, KPNB1-c-Myc positive feedback loop increases the expression level of Cyclin D1, Cyclin B1 and CDK1. The complex of Cyclin B1 and CDK1 can further enhances phosphorylation level of RCC1, by which it promotes PCa cell proliferation through accelerating cell cycle and mitosis.
Discussion
Several Karyopherin proteins have been proposed as a new anti-cancer targets by studies from ours and other groups [3, 23–25]. The pro-cancer function of alpha members of Karyopherin proteins such as KPNA2 [4, 26, 27], KPNA4[28] and KPNA7[29, 30] indicates the fundamental role the Karyopherin family mediated nuclear trafficking in tumorigenesis of prostate carcinoma. One possible reason of this phenomenon is that increased metabolism rate in transformed cells requires enhanced nucleocytoplasmic transport efficiency [31]. In addition, dysregulation of Karyopherin may contribute to tumor progression by transporting key oncogenic signaling molecules across nuclear membrane into the nucleus [32]. As a receptor of Karyopherin alpha subunits, it is not a surprise that KPNB1 is involved in multiple cell processes by importing transcription factors like NF-κB, and cell signaling proteins [9, 33]. Here, we demonstrated that KPNB1 is overexpressed in PCa, including patient tissues and cell lines. The differential fold change between RNA and protein levels indicates a post-transcriptional regulation mechanism might be involved in the dysregulation of KPNB1 [34]. Also, KPNB1 inhibition led to cell cycle arrest through reducing the expression of several key cell cycle modulators including Cyclin D1, Cyclin B1, CDK1 and RCC1 phosphorylation.
A live-cell imaging RNAi screen has identified that KPNB1 regulates post-mitotic assembly mechanisms in human cells through its cooperation with PP2A-B55alpha [35]. Our current study demonstrated that phosphorylation of RCC1, a key nucleotide-exchange factor for the Ran GTPase, was a downstream molecule regulated by KPNB1. RCC1 plays pivotal roles in mitosis and nuclear-envelope assembly[36]. Localization of RCC1 to chromosomes has been demonstrated to be critical for proper mitotic spindle assembly in human cells[37]. Previous study has revealed that targeting RCC1 using Latcripin-13 domain could induce apoptosis and cell cycle arrest of human lung carcinoma [38]. Affinity of RCC1 to chromosome relies on its phosphorylation status[39], which can be regulated by CDK/Cyclin B1 complex[40]. In line with that, we demonstrated that the inhibition of KPNB1 led to decreased expression of CDK1/Cyclin B1 accompanied with suppression of RCC1 phosphorylation but not its expression. The analysis of human TCGA dataset also support that KPNB1 level is not associated with the expression of RCC1.
Amplification of the c-MYC oncogene is a key event during PCa development [41, 42]. Accelerated cell cycle in transformed cells upon c-Myc enforced gene expression has been described in many models [43]. We have previously reported that targeting c-Myc overexpression helped to prevent the initiation and progression of prostate cancer [18]. In the present study, we further revealed that the dysregulated cell cycle in PCa could be driven by the KPNB1-c-Myc positive feedback loop. In which, overexpression of c-Myc directly enhanced KPNB1 expression by binding to the promoter region of KPNB1. Subsequently, the upregulated KPNB1 accelerated the nuclear importing of c-Myc to further stimulate the expression of c-Myc regulated genes including KPNB1 itself and collectively favored cell cycle progression. These evidences revealed a concealed regulatory loop between c-Myc and KPNB1 in promoting cancer growth.
Constitutive activation of NF-κB contributes to oncogenesis and tumor progression via induction of anti-apoptotic signaling has been widely reported[44]. Previous studies provided evidences indicating that KPNB1 functions as a mediator to transduce NF-κB signaling into the nuclei [9, 11, 45]. Our study could add new evidence to support a critical role of KPNB1 in the nuclear importing of NF-κB subunit p50. The regulation of c-Myc and NF-κB by KPNB1 indicates the multifaceted functions of KPNB1 in tumor growth. Indeed, in addition to halting cell division process, we proved that the inhibition of KPNB1 also stimulated PCa cell apoptosis and suppressed its tumorigenesis.
Of note, NF-κB activation requires the NLS-recognition domain of KPNA3 and KPNA4 [7, 8]. Therefore, nuclear transduction of NF-κB mediated by KPNB1 probably needs its co-interaction with the alpha subunits. To date, KPNB1 is known to be the most significant Karyopherin beta subunit in transporting NF-κB [9]. Other members of Karyopherin beta family, such as XPO7 and IPO8, are still able to bind to NF-κB in a NLS-independent manner [9]. The increases of KPNB1 in malignant cells indicate a specific need of KPNB1 in tumor while normal cells may not heavily rely on KPNB1 for NF-κB importing. Thus, it is likely that specific inhibition of KPNB1 does not affect the normal physiological activation of NF-κB in normal cells. As mentioned earlier, the half lethal dose of importazole in non-malignant prostatic epithelial RWPE1 cells, was double amount of that in PCa cell lines (Supplementary Fig 6) suggesting that non-tumor prostate cells were less sensitive to KPNB1 inhibition when compared to PCa cells. In view of this, targeting KPNB1 may cause minimum toxicity in normal cells expressing baseline level of KPNB1.
To date, multiple proteins and small molecule inhibitors have been discovered to target KPNB1 directly [17, 46–48]. The antitumor effects of compounds targeting the epigenetic regulator of KPNB1 has been evaluated previously in malignant peripheral nerve sheath tumors [49]. However, to our best knowledge, our study is the first to assess the inhibitory effects of importazole on tumor growth in vivo. Importazole specifically inhibits the function of KPNB1 by altering its interaction with RanGTP and our results strongly support that the inhibition of KPNB1 with importazole can effectively inhibit prostate cancer progression. Interestingly, importazole directly reduced the mRNA levels of RCC1 which was not observed in knockdown KPNB1 expression by siRNA or shRNA. Considering the critical role of RCC1 for chromatin stabilization and mitosis [50], reduction of RCC1 expression may directly lead to cell cycle arrest and cell death which makes importazole more potent as an anti-cancer agent. Similarly, importazole not only abolishes the nuclear localization, but also diminishes the total expression level of NF-κB p50. It is possible that importazole inhibits cancer growth through other mechanisms in addition to inhibiting KPNB1 activity. Future studies are required to explore the mechanisms. The selection of a more convenient and effective delivery method for importazole is also important for its potential clinical application.
In summary, our study revealed the role of KPNB1 overexpression in prostate cancer development. Targeting KPNB1 using importazole might be a promising method of therapeutic intervention for cancers with KPNB1 overexpression.
Materials and methods
Cell culture
Human prostate cancer cell lines PC3 and LNCaP were purchased from the American Type Culture Collection (ATCC). Human epithelial cell line RWPE-1 was provided by Dr. Susan Logan (New York University Langone Medical Center) and cultured in using a Keratinocyte-Serum Free Medium kit (Gibco, Gaithersburg, MD, USA). Human prostate cancer cell line C42B originally derived from metastatic human PCa lesions in the spine of an athymic mouse [51, 52] was a gift from Dr. Laurie McCauley (University of Michigan School of Dentistry). PC3, LNCaP and C42B were cultured using RPMI medium 1640 (Thermo fisher scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA, USA) and 1% penicillin-streptomycin (Thermo fisher scientific). The Myc-CaP cell line derived from prostate carcinoma of a Hi-Myc transgenic mouse [53] was a gift from Charles Sawyers (Howard Hughes Medical Institute and Memorial Sloan-Kettering Cancer Center) and cultured using DMEM (Thermo fisher scientific) medium with high glucose (4.5 g/L) supplemented with 10% FBS and 1% penicillin-streptomycin (Thermo fisher scientific). All cell lines were maintained in a 37℃ chamber (Sanyo, Osaka, Japan) containing 5% CO2.
Human samples analysis
The human prostate cancer datasets were downloaded from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases. The KPNB1, Cyclin D1, CDK1, Cyclin B1 and RCC1 expression values, pathological stages of individual patients were extracted using an R program. The samples were then distributed into different groups based on the pathological stage classification or specific gene expression level. Bar graphs, the Kaplan-Meier survival curves and Ozone correlation graphs were generated using Prism GraphPad software (GraphPad Software, Inc., La Jolla, CA, USA).
Human PCa tissue microarray (TMA)
Prostate adenocarcinoma test tissue arrays were purchased from US Biomax, Inc (Derwood, MD 20855, USA). Array PR956b contains 40 cases with matched normal adjacent tissue and metastatic bones, with TNM, Gleason scores, PSA level and survival data. Array T195b contains 12 cases including TNM, clinical stage and pathology grade.
Real-time PCR
Total RNA was extracted using RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). cDNA was generated using TaqMan reverse transcription reagents (Life technologies, Carlsbad, CA, USA). SYBR green reagents (Life technologies) was applied to performed qPCR on CFX384 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). The sequences of primers used in this study were listed in the supplementary materials (Supplementary table 1).
Immuno-blotting
Total protein was purified using Radioimmunoprecipitation assay (RAPI) buffer (Thermo fisher scientific). Concentration of protein samples were determined by Pierce BCA protein assay kit (Thermo fisher scientific). Protein samples mixed with loading buffer and NuPAGE sample reducing reagent (Thermo fisher scientific) were heat-shocked at 85℃ for 2 minutes before loaded into SDS-PAGE gel (Thermo fisher scientific). Electrophoresis was performed on a Novex mini-cell device (Invitrogen, Carlsbad, CA, USA) for 90 munities. The gel was then attached to a PVDF membrane (Thermo fisher scientific) to transfer the protein. After blocked in 1% BSA buffer for 1 hour, the membrane was incubated in diluted antibodies solution overnight at 4 ℃. Anti-KPNB1, anti-Cyclin B1 primary antibodies were purchased from Novus biologicals (Littleton, CO, USA). Anti-Cyclin D1 primary antibody was purchased from Santa Cruz Biotechnology. Anti-CDK1, anti-RCC1, anti-pRCC1, anti-β-actin, anti-Ki67, and anti-caspase 3 primary antibodies and all secondary antibodies were purchased from Cell signaling technology (Danvers, MA, USA). Targeted protein was imaged using SuperSignal West substrate (Thermo fisher scientific).
Plasmid construction, virus packaging and infection
ShRNA oligos of KPNB1 are purchased from Integrated DNA Technologies, Inc. (Coralville, IA,USA). Annealed double strand oligos were cloned into pSUPER.retro.puro vector (OligoEngine, Seattle, WA, USA) according to the provided manual for the expression of shRNA. The reconstructed plasmids were amplified in E.coli (New England Biolabs, Ipswich, MA, USA) and purified using plasmid miniprep kit (Thermo fisher scientific).
Virus was produced using Gryphon retroviral packaging cell lines (Allele Biotechnology, San Diego, CA, USA). In brief, 10 μg of plasmid was transfected to Grypho cell lines in a 6-cm dish. In 48 hours, supernatant that contains virus particles was collected. For virus infection, 1 ml of the collected virus supernatant supplemented with 3 μl of polybrene (EMD Millipore, Billerica, MA, USA) at 5 mg/ml was added to the culture medium of C42B in a 6-cm plate. In 24 hours, virus supernatant was replaced with fresh medium. Infected cells were selected using puromycin (Sigma-Aldrich-Aldrich, St. Louis, MO, USA).
Cell cycle assay
PC3 or C42B cells were seeded in a 6-wells plate (0.8 million cells per well) and transfected with siRNA (Invitrogen) for 48 hours or treated with Importazole (Selleckchem, Houston, TX, USA) for 24 hours. The post-treatment cells were collected and fixed with 70% ethanol. After 2 washes with PBS, the cells were stained with propidium iodide (PI) solution (20 μg/ml PI (Roche, Basel, Switzerland) in PBS containing 0.1% Triton-100 (Sigma-Aldrich-Aldrich) and 0.2 mg/ml DNAse-free RNAse (Thermo fisher scientific)) and incubated at 37℃ for 15 minutes. The stained cells were washed with PBS for once and re-suspended in 500 μl PBS. Flow cytometry was performed on a BD FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). The results were interpreted and displayed using Flowjo software (Flowjo, Ashland, OR, USA).
CHIP assay
Six to eight million of PC3 or C42B cells were harvested for the chromatin immunoprecipitation (ChIP) assay. EpiQuik Chromatin Immunoprecipitation kit (Epigentek, Farmingdale, NY, USA) was used for the extraction of nuclear complex and immunoprecipitation of c-Myc binding DNA. c-Myc antibody was purchased from Santa Cruz Biotechnology (Dallas, Texas, USA). Promoter region of KPNB1 was predicted using Promoter Scan software (https://www-bimas.cit.nih.gov/molbio/proscan/). KPNB1 promoter in c-Myc associated DNA fragment was evaluated using qPCR.
MTT assay
PC3 or C42B cells were seeded in a 96-wells plate (10,000 cells per well). After treatment, culture medium was replaced with 100 μl fresh medium. 10 μl of 12mM MTT (Thermo fisher scientific) stock solution was added to each well, 10 μl of MTT stock solution mixed with 100 μl of fresh medium was used as negative control. After incubation at 37℃ for 4 hours, remove all but 25 μl medium from each well and add 50 μl DMSO with thoroughly mixture by pipetting. After incubation at 37℃ for another 10 minutes, the samples were mixed again. Absorbance was read at 540 nm to assess cell viability.
Crystal violet staining
For the assessment of siRNA effect, PC3 or C42B cells were seeded in a 96-wells plate (10,000 cells per well). After 24 hours, 48 hours and 72 hours’ post-transfection, cells were fixed with formalin (Fisher scientific, Hampton, NH, USA) for 5 minutes and stained with 0.05% crystal violet solution (Sigma-Aldrich) for 30 minutes. After staining, the cells were washed with tap water twice and plate was drained for a couple minutes. 300 μl of methanol was added to each well to solubilize the dye. Absorbance was read at 540 nm by taking an aliquot of 100 μl soluble dye to another plate.
Xenograft model
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at New York University Medical Center and in compliance with ethical regulations.
Athymic nude mouse (Charles River, Malvern, PA, US) at age of 6–8 weeks were purchased and distributed randomly to different groups. For C42B cell line, 2 million of C42B-scramble or C42B-shKPNB1 cells were injected subcutaneously to each of the mouse. After 6 weeks, tumors from both groups of mice were dissected to measure the volume and weight, as well as IHC staining. For PC3 cell line, 1 million of PC3 cells were injected subcutaneously to each mouse of both groups. From day 4 post-injection, 100 μl of empty nanoparticles (NP) or Importazole encapsulated nanoparticles (NP-IPZ) was injected intravenously to the mice twice a week for 4 weeks. The tumor volume was measured weekly. After 4 weeks, tumors were harvested for weight measurement and histologic analysis.
Encapsulation of importazole in biodegradable nanoparticles
Importazole was encapsulated into PLGA-PEG as described by Elgogary et al [54]. Briefly, 7.2 mg of Importazole and 60 mg of the PLGA-PEG powder (Creative PEGWorks, Chapel Hill, NC, USA) were dissolved in 144 μl of DMSO (Sigma-Aldrich) or 1.356 ml of dichloromethane (DCM, Sigma-Aldrich) respectively and mixed together immediately. 1 ml of the mixture was added into 5ml of 1% sucrose fatty acid ester (TCI America, Portland, OR, USA) and emulsified using a probe sonicator for 10 minutes on an ice-water bath. The emulsion was poured into another 40 ml 0.5% sucrose ester solution and mixed using magnetic stirring at 700 rpm for 1 hour. The solvent was transferred to centrifuge tubes for speed vacuum. After 2 hours of evaporation, the remaining solvent was collected and centrifuged at 2000×g for 15 minutes. The NP-IPZ remains in the supernatant and was pelleted at 20,000×g for 25 minutes. NP-IPZ pellets was re-suspended in 2ml PBS for intravenous injection. Empty nanoparticle (without Importazole) was prepared in the same way.
Immunohistochemistry
Immunohistochemistry was carried out on TMA sections or 10% neutral buffered formalin fixed, paraffin-embedded xenograft tumor tissues. Briefly, formalin fixed, paraffin embedded sections for Cleaved Caspase-3 labeling were deparaffinized and antigen retrieved for 20 minutes. Endogenous peroxidase activity was blocked with hydrogen peroxide. KPNB1, Cyclin D1, CDK1, Cyclin B1, pRCC1, Ki67 or Cleaved Caspase-3 antibody was diluted in Cell Signaling diluent and incubated for three hours. Above primary antibodies (Cell Signaling Technology) were further detected by using Avidin-Biotin Complex (ABC) method following NYU Experimental Pathology Immunohistochemistry Core Laboratory protocol. All slides were washed in distilled water, counterstained with hematoxylin, dehydrated and mounted with permanent media. Appropriate positive and negative controls were run in parallel to the experimental sections.
Statistical analysis
An analysis of the relationship between KPNB1 levels and clinical PCa stages or Gleason scores was carried out using the nonparametric one-way ANOVA test. The results revealed statistically significant differences in KPNB1 expression in PCa with different Gleason scores (R sqare=0.0603; p < 0.0001). We also examined the extent to which the TNM scores were associated with KPNB1 expression using the nonparametric one-way ANOVA test. Our results revealed significant differences in KPNB1 expression as a function of TNM (R square=0.0568; p < 0.0001). We used GraphPad Prism software (GraphPad Software, La Jolla, CA, USA) to statistically analyze the remaining experimental outcomes. We expressed the data as means ± SEMs of at least three independent determinations. Statistical significance was determined using the unpaired t-test. Results with p values less than 0.05(*), 0.01(**), 0.001(***) or 0.0001(****) were considered to be statistically significant.
Study approval
All of the animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee (IACUC) of New York University Medical Center following the guidelines for the proper care and use of animals for research purpose.
Supplementary Material
Acknowledgments
This study was partly supported by NYU CSCB Pilot Study Award to J.Y., National Institutes of Health grants R01CA180277, R01DE025992 and R01DE027074 to X.L. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclosure Statement: The authors declare no conflicts of interest.
References
- 1.Ghildyal R, Ho A, Wagstaff KM, Dias MM, Barton CL, Jans P et al. Nuclear import of the respiratory syncytial virus matrix protein is mediated by importin beta1 independent of importin alpha. Biochemistry 2005; 44: 12887–12895. [DOI] [PubMed] [Google Scholar]
- 2.van der Watt PJ, Ngarande E, Leaner VD. Overexpression of Kpnbeta1 and Kpnalpha2 importin proteins in cancer derives from deregulated E2F activity. PLoS One 2011; 6: e27723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J, Lu C, Wei J, Guo Y, Liu W, Luo L et al. Inhibition of KPNA4 attenuates prostate cancer metastasis. Oncogene 2017; 36: 2868–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang L, Wang HY, Li JD, Wang JH, Zhou Y, Luo RZ et al. KPNA2 promotes cell proliferation and tumorigenicity in epithelial ovarian carcinoma through upregulation of c-Myc and downregulation of FOXO3a. Cell Death Dis 2013; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L, Zhou Y, Cao XP, Lin JX, Zhang L, Huang ST et al. KPNA2 promotes migration and invasion in epithelial ovarian cancer cells by inducing epithelial-mesenchymal transition via Akt/GSK-3beta/Snail activation. J Cancer 2018; 9: 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aggarwal A, Agrawal DK. Importins and Exportins Regulating Allergic Immune Responses. Mediat Inflamm 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagerlund R, Kinnunen L, Kohler M, Julkunen I, Melen K. NF-{kappa}B is transported into the nucleus by importin {alpha}3 and importin {alpha}4. J Biol Chem 2005; 280: 15942–15951. [DOI] [PubMed] [Google Scholar]
- 8.Cai Y, Shen Y, Gao L, Chen M, Xiao M, Huang Z et al. Karyopherin Alpha 2 Promotes the Inflammatory Response in Rat Pancreatic Acinar Cells Via Facilitating NF-kappaB Activation. Dig Dis Sci 2016; 61: 747–757. [DOI] [PubMed] [Google Scholar]
- 9.Liang P, Zhang H, Wang G, Li S, Cong S, Luo Y et al. KPNB1, XPO7 and IPO8 mediate the translocation ofNF-kappaB/p65 into the nucleus. Traffic 2013; 14: 1132–1143. [DOI] [PubMed] [Google Scholar]
- 10.Yan W, Li R, He J, Du J, Hou J. Importin beta1 mediates nuclear factor-kappaB signal transduction into the nuclei of myeloma cells and affects their proliferation and apoptosis. Cell Signal 2015; 27: 851–859. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, Zhao Y, Xia N, Zhang W, Tang Z, Wang C et al. KPNbeta1 promotes palmitate-induced insulin resistance via NF-kappaB signaling in hepatocytes. J Physiol Biochem 2015; 71: 763–772. [DOI] [PubMed] [Google Scholar]
- 12.Ha S, Jeong J, Oh J, Rhee S, Ham SW. A Small Organic Molecule Blocks EGFR Transport into the Nucleus by the Nonclassical Pathway Resulting in Repression of Cancer Invasion. Chembiochem 2017. [DOI] [PubMed] [Google Scholar]
- 13.Angus L, van der Watt PJ, Leaner VD. Inhibition of the nuclear transporter, Kpnbeta1, results in prolonged mitotic arrest and activation of the intrinsic apoptotic pathway in cervical cancer cells. Carcinogenesis 2014; 35: 1121–1131. [DOI] [PubMed] [Google Scholar]
- 14.Martens-de Kemp SR, Nagel R, Stigter-van Walsum M, van der Meulen IH, van Beusechem VW, Braakhuis BJ et al. Functional genetic screens identify genes essential for tumor cell survival in head and neck and lung cancer. Clin Cancer Res 2013; 19: 1994–2003. [DOI] [PubMed] [Google Scholar]
- 15.Kuusisto HV, Jans DA. Hyper-dependence of breast cancer cell types on the nuclear transporter Importin beta1. Biochim Biophys Acta 2015; 1853: 1870–1878. [DOI] [PubMed] [Google Scholar]
- 16.Kodama M, Kodama T, Newberg JY, Katayama H, Kobayashi M, Hanash SM et al. In vivo loss-of-function screens identify KPNB1 as a new druggable oncogene in epithelial ovarian cancer. Proc Natl Acad Sci U S A 2017; 114: E7301–E7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M et al. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem Biol 2011; 6: 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akinyeke T, Matsumura S, Wang X, Wu Y, Schalfer ED, Saxena A et al. Metformin targets c-MYC oncogene to prevent prostate cancer. Carcinogenesis 2013; 34: 2823–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koch HB, Zhang R, Verdoodt B, Bailey A, Zhang CD, Yates JR 3rd et al. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle 2007; 6: 205–217. [DOI] [PubMed] [Google Scholar]
- 20.Qin ZH, Chen RW, Wang Y, Nakai M, Chuang DM, Chase TN. Nuclear factor kappaB nuclear translocation upregulates c-Myc and p53 expression during NMDA receptor-mediated apoptosis in rat striatum. J Neurosci 1999; 19: 4023–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 2003; 4: 223–238. [DOI] [PubMed] [Google Scholar]
- 22.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer 2002; 2: 301–310. [DOI] [PubMed] [Google Scholar]
- 23.Laurila E, Vuorinen E, Savinainen K, Rauhala H, Kallioniemi A. KPNA7, a nuclear transport receptor, promotes malignant properties of pancreatic cancer cells in vitro. Exp Cell Res 2014; 322: 159–167. [DOI] [PubMed] [Google Scholar]
- 24.Winkler J, Ori A, Holzer K, Sticht C, Dauch D, Eiteneuer EM et al. Prosurvival function of the cellular apoptosis susceptibility/importin-alpha1 transport cycle is repressed by p53 in liver cancer. Hepatology 2014; 60: 884–895. [DOI] [PubMed] [Google Scholar]
- 25.Yang LL, Hu BY, Zhang YX, Qiang SL, Cai J, Huang W et al. Suppression of the nuclear transporter-KPN beta 1 expression inhibits tumor proliferation in hepatocellular carcinoma. Med Oncol 2015; 32. [DOI] [PubMed] [Google Scholar]
- 26.Alshareeda AT, Negm OH, Green AR, Nolan CC, Tighe P, Albarakati N et al. KPNA2 is a nuclear export protein that contributes to aberrant localisation of key proteins and poor prognosis of breast cancer. Brit J Cancer 2015; 112: 1929–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altan B, Yokobori T, Mochiki E, Ohno T, Ogata K, Ogawa A et al. Nuclear karyopherin-2 expression in primary lesions and metastatic lymph nodes was associated with poor prognosis and progression in gastric cancer. Carcinogenesis 2013; 34: 2314–2321. [DOI] [PubMed] [Google Scholar]
- 28.Wang HJ, Tao T, Yan W, Feng Y, Wang YZ, Cai JQ et al. Upregulation of miR-181s reverses mesenchymal transition by targeting KPNA4 in glioblastoma. Sci Rep-Uk 2015; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vuorinen EM, Rajala NK, Rauhala HE, Nurminen AT, Hytonen VP, Kallioniemi A. Search for KPNA7 cargo proteins in human cells reveals MVP and ZNF414 as novel regulators of cancer cell growth. Bba-Mol Basis Dis 2017; 1863: 211–219. [DOI] [PubMed] [Google Scholar]
- 30.Shen SH, Gui TT, Ma CC. Identification of molecular biomarkers for pancreatic cancer with mRMR shortest path method. Oncotarget 2017; 8: 41432–41439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stelma T, Chi A, van der Watt PJ, Verrico A, Lavia P, Leaner VD. Targeting nuclear transporters in cancer: Diagnostic, prognostic and therapeutic potential. IUBMB Life 2016; 68: 268–280. [DOI] [PubMed] [Google Scholar]
- 32.Mahipal A, Malafa M. Importins and exportins as therapeutic targets in cancer. Pharmacol Ther 2016; 164: 135–143. [DOI] [PubMed] [Google Scholar]
- 33.Lu T, Bao Z, Wang Y, Yang L, Lu B, Yan K et al. Karyopherinbeta1 regulates proliferation of human glioma cells via Wnt/beta-catenin pathway. Biochem Biophys Res Commun 2016; 478: 1189–1197. [DOI] [PubMed] [Google Scholar]
- 34.Zhang P, Garnett J, Creighton CJ, Al Sannaa GA, Igram DR, Lazar A et al. EZH2-miR-30d-KPNB1 pathway regulates malignant peripheral nerve sheath tumour cell survival and tumourigenesis. J Pathol 2014; 232: 308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz MH, Held M, Janssens V, Hutchins JR, Hudecz O, Ivanova E et al. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol 2010; 12: 886–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF, Macara IG. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nature Cell Biology 2007; 9: 596–U203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore WJ, Zhang CM, Clarke PR. Targeting of RCC1 to chromosomes is required for proper mitotic spindle assembly in human cells. Curr Biol 2002; 12: 1442–1447. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Wan XY, Gao YF, Zhong MT, Sha L, Liu B et al. Latcripin-13 domain induces apoptosis and cell cycle arrest at the G1 phase in human lung carcinoma A549 cells. Oncol Rep 2016; 36: 441–447. [DOI] [PubMed] [Google Scholar]
- 39.Hood FE, Clarke PR. RCC1 isoforms differ in their affinity for chromatin, molecular interactions and regulation by phosphorylation. J Cell Sci 2007; 120: 3436–3445. [DOI] [PubMed] [Google Scholar]
- 40.Hutchins JR, Moore WJ, Hood FE, Wilson JS, Andrews PD, Swedlow JR et al. Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Curr Biol 2004; 14: 1099–1104. [DOI] [PubMed] [Google Scholar]
- 41.Gurel B, Iwata T, Koh CM, Jenkins RB, Lan FS, Van Dang C et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Modern Pathol 2008; 21: 1156–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and Prostate Cancer. Genes Cancer 2010; 1: 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta 2015; 1849: 506–516. [DOI] [PubMed] [Google Scholar]
- 44.Nakanishi C, Toi M. Nuclear factor-kappa B inhibitors as sensitizers to anticancer drugs. Nature Reviews Cancer 2005; 5: 297–309. [DOI] [PubMed] [Google Scholar]
- 45.Gagne B, Tremblay N, Park AY, Baril M, Lamarre D. Importin beta 1 targeting by hepatitis C virus NS3/4A protein restricts IRF3 and NF-kappa B signaling of IFNB1 antiviral response. Traffic 2017; 18: 362–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lundberg L, Pinkham C, Baer A, Amaya M, Narayanan A, Wagstaff KM et al. Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan Equine Encephalitis Virus replication. Antiviral Res 2013; 100: 662–672. [DOI] [PubMed] [Google Scholar]
- 47.Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu SC, Hung MC. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J Cell Biochem 2006; 98: 1570–1583. [DOI] [PubMed] [Google Scholar]
- 48.van der Watt PJ, Chi A, Stelma T, Stowell C, Strydom E, Carden S et al. Targeting the Nuclear Import Receptor Kpnbeta1 as an Anticancer Therapeutic. Mol Cancer Ther 2016; 15: 560–573. [DOI] [PubMed] [Google Scholar]
- 49.Zhang PY, Yang XB, Ma XY, Ingram DR, Lazar AJ, Torres KE et al. Antitumor effects of pharmacological EZH2 inhibition on malignant peripheral nerve sheath tumor through the miR-30a and KPNB1 pathway. Mol Cancer 2015; 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hadjebi O, Casas-Terradellas E, Garcia-Gonzalo FR, Rosa JL. The RCC1 superfamily: from genes, to function, to disease. Biochim Biophys Acta 2008; 1783: 1467–1479. [DOI] [PubMed] [Google Scholar]
- 51.Thalmann GN, Anezinis PE, Chang SM, Zhau HE, Kim EE, Hopwood VL et al. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer research 1994; 54: 2577–2581. [PubMed] [Google Scholar]
- 52.Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. International journal of cancer Journal international du cancer (Research Support, U.S. Gov’t, P.H.S.) 1994; 57: 406–412. [DOI] [PubMed] [Google Scholar]
- 53.Watson PA, Ellwood-Yen K, King JC, Wongvipat J, Lebeau MM, Sawyers CL. Context-dependent hormone-refractory progression revealed through characterization of a novel murine prostate cancer cell line. Cancer research 2005; 65: 11565–11571. [DOI] [PubMed] [Google Scholar]
- 54.Elgogary A, Xu Q, Poore B, Alt J, Zimmermann SC, Zhao L et al. Combination therapy with BPTES nanoparticles and metformin targets the metabolic heterogeneity of pancreatic cancer. Proc Natl Acad Sci U S A 2016; 113: E5328–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.