Abstract
We report a case of Mycoplasma hominis ventriculitis in a preterm neonate that was successfully identified with 16S ribosomal ribonucleic acid (16s rRNA) sequencing and whole genome sequencing (WGS) after failure to detect the pathogen with conventional diagnostic methods. The infant required doxycycline with subsequent clearance of the infection and no evidence of drug toxicity.
Keywords: Mycoplasma hominis, meningitis, neonate, whole genome sequencing, 16S rRNA sequencing
INTRODUCTION
Mycoplasma hominis colonizes the pharynx and respiratory tract of both sexes and the genitourinary tract of 10–20% of females (1). Neonates can become colonized via passage through the birth canal but invasive infection is rare. Infections in neonates typically present as conjunctivitis, sepsis, meningitis, pneumonia, or subcutaneous abscess after forceps delivery in both term and preterm infants. Features include onset of symptoms from hours to months after birth, clinical decompensation despite empiric therapy for meningitis, and negative cerebrospinal fluid (CSF) testing on routine cultures. In a review of 29 cases, neurologic sequelae and mortality, attributed to delay in diagnosis and lack of adequate treatment, were reported in 28% (2).
Although culture is considered the gold standard, this bacteria can be difficult to identify with standard microbiologic techniques. Newer molecular diagnostic tools may allow characterization of isolates such as M. hominis that are difficult to identify. Recognition of the organism as a potential pathogen in this patient population is important, as M. hominis typically is resistant to antibiotics often used to treat other Mycoplasma infections. Delays in pathogen identification can postpone recognition of the organism’s distinct susceptibility profile and implementation of appropriate antibiotic therapy although there is a lack of consensus on optimal management of disease due to its relative rarity as well.
We describe a preterm infant with ventriculitis caused by a slowly growing organism ultimately identified as M. hominis. Challenges in identification at the species level and unfamiliarity with this organism’s distinct resistance pattern led to delayed initiation of active therapy. Whole genome sequencing (WGS) sequencing successfully identified the organism and allowed evaluation for the presence of genetic markers of antimicrobial resistance.
CASE REPORT
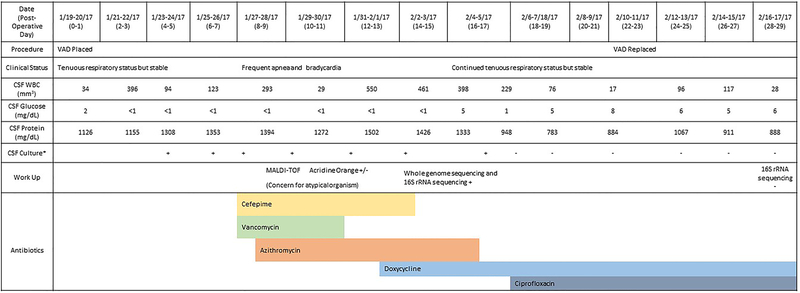
An infant male was born at 24 weeks and 3 day gestation via vaginal delivery due to premature rupture of membranes and clinical chorioamnionitis with a birth weight of 720 grams at a community hospital. The 30-year-old gravida 5 parity 1 mother was group B streptococcus unknown but the remainder of her laboratory tests including human immunodeficiency virus, syphilis, gonorrhea/chlamydia, and hepatitis B antigen testing were negative. He required transfer to a quaternary pediatric hospital at 5 weeks of life for posthemorrhagic hydrocephalus due to bilateral grade 4 intraventricular hemorrhage (IVH) visualized on both head ultrasound and magnetic resonance imaging (MRI) of the brain. The infant received a ventricular access device (VAD) at 5.5 weeks of life with daily reservoir taps for hydrocephalus management. At the time of VAD placement, there were 34 white blood cells (WBC) and 975 red blood cells (RBC) cells/mm3 in the CSF with a glucose of 2 mg/dL and protein of 1126 mg/dL. The team theorized that the hypoglycorrhachia and elevated protein at the time of VAD placement was likely related to the high cell burden from the bleed and subsequently altered metabolism (3). Daily CSF cultures were sent (Figure 1).
Figure 1.
Clinical Timeline *All cultures obtained on the indicated dates grew after 3–5 days of inclubation.
On VAD post-operative day 8, the infant developed frequent apnea and bradycardia requiring bagged ventilation and was started on cefepime and vancomycin for possible sepsis. CSF cell counts showed WBC 40 cells/mm3, RBC 2200 cells/mm3, protein 1219 mg/dL, and glucose <1 mg/dL. A peripheral blood count had WBC count of 28 K/mcL with 70% neutrophils and no bands, hemoglobin of 12.1 gm/dL, and platelets of 195 K/mcL. Urine and blood cultures were negative. Post-operative day 7, the microbiology laboratory noted a fine haze on the blood agar plate from a CSF specimen obtained 3 days prior. No organism was seen on Gram stain, and matrix assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS), identified possible Ureaplasma or Mycoplasma species in the context of clinical suspicion for other atypical bacteria such as Chlamydia or Legionella species. The organism had minimal fluorescence by acridine orange staining. WGS was then performed on the isolate on-site. DNA was extracted from a scraping of the organism using the NucliSENS® easyMag® platform. Library generation and sequencing was performed with the Illumina Nextera XT kit and Illumina NextSeq500 DNA sequencer to a depth of greater than 100-fold coverage of the genome. Identification of the organism was performed by aligning sequence reads to a microbial genome which revealed that >90% of reads were assigned to M. hominis (4, 5). Samples were simultaneously sent to University of Washington for 16S rRNA sequencing which identified the isolate as M. hominis shortly after the WGS results were available.
CSF cultures continued to be positive for M. hominis on post-operative days 4, 6, 8, and 10–14. Empiric cefepime was stopped after 5 days and vancomycin was discontinued after 2 days. With concern for an atypical organism, azithromycin was started on post-operative day 9, and doxycycline was added on post-operative day 13 for better CSF penetration. CSF cultures were negative on post-operative day 15 and 16, but again positive on post-operative day 17.
Due to the organism’s slow growth in culture, we were unable to perform antimicrobial sensitivity testing. Instead, the genome sequence of the isolate was interrogated for mutations. Raw sequence reads were assembled to gyrA, gyrB, parC and parE and aligned to a reference database of antibiotic resistance genes to identify known resistance determinants using BLAST® (6). None of the mutations known to cause resistance to tetracyclines, or quinolones were identified whereas the sequence of the M. hominis 23S rRNA gene contained the mutations known to confer lincosamide and macrolide resistance. Those mutations are G2063A and C2618U (numbering is based on the 23S rRNA sequence AF443616.3 from an M. hominis isolate with known macrolide resistance) (7). An additional C1548T substitution in the 23S rRNA gene was identified, which has not thus far been correlated with antibiotic susceptibility.
Based on these results and more research on the intrinsic resistance patterns of the organism, azithromycin was replaced with ciprofloxacin on post-operative day 18. On that same day, CSF cultures became negative and subsequent 16S rRNA analysis failed to amplify bacterial DNA. The CSF WBC peaked on post-operative day 14 at 1144 cells/mm3 with cell counts, protein, and glucose that slowly improved over the next 2 weeks but did not completely normalize, thought to be due to his massive and evolving IVH. The VAD was replaced on post-operative day 22 when the patient, although persistently tenuous from a respiratory standpoint, was overall stable. He completed a course of ciprofloxacin and doxycycline for 42 days after replacement of the VAD with no relapses after discontinuation of antimicrobials. A ventriculoperitoneal shunt was placed at 4 months of life (2.5 weeks after completion of antibiotic course). An MRI of the brain at 5-months-old showed adequate position of a ventricular catheter, cystic encephalomalacia, white matter volume loss, and evolving IVH. He required tracheostomy at 7 months of age due to his chronic lung disease as well as gastrostomy tube at 14 months for dysphagia. At the time of publication, the child is 24-months-old and is in the custody of grandparents.
DISCUSSION
M. hominis are small, pleomorphic bacteria that lack a cell wall, making detection difficult as they cannot be visualized on Gram stain. Bacterial culture is slow (2–5 days) and, although possible with standard media, is enhanced with arginine-enriched plates. When isolated, M. hominis colonies have a characteristic “fried egg” shaped appearance. MALDI-TOF MS allows for rapid identification of pathogens but has limitations as it requires growth of the organism and its presence in the database for identification. Molecular techniques such as real-time PCR and 16S rRNA sequencing are becoming more common although the tests are not extensively available. The sensitivity of detection of pathogens in the CSF with 16S rRNA can be as high as 75–98% although the field is rapidly expanding with multiple ambiguities on the sensitivity and specificity depending on the technique used (8). Bacterial WGS also holds promise as a means to characterize clinical isolates. Currently, it is utilized to assess strain relatedness during epidemiologic investigation but is not widely adapted to routine clinical care as the turnaround time for sequencing and analysis is typically too slow to provide clinically actionable information.
M. hominis has been reported to be susceptible to tetracyclines, lincosamides, chloramphenicol, and fluoroquinolones but the use of these agents is discouraged in infants due to toxic side effects (9). M. hominis is intrinsically resistant to antimicrobials such as beta-lactams and glycopeptides which are often used in empiric management of meningitis. Furthermore, in vitro data and studies of genitourinary infections in adults have revealed that M. hominis, unlike most other Mycoplasma, is generally resistant to macrolides (9, 10). Increasing resistance to tetracyclines and fluoroquinolones has been reported in pregnant women as well (10). The medical teams initiated azithromycin as empiric therapy when the organism was identified as a Mycoplasma or Ureaplasma. After the organism was identified at the species level and the organism continued to grow despite macrolide therapy, doxycycline was added for better CNS penetration and coverage but we did not see the desired response. WGS data was then used to interrogate the organism’s genome for markers of antimicrobial resistance. Reads were assembled and mutations known to cause fluoroquinolone resistance were found to be absent. Based on these results, azithromycin was changed to ciprofloxacin. CSF cultures became negative on day 0 of quinolone therapy and day 6 of tetracycline treatment and remained so with a prolonged course of doxycycline and ciprofloxacin. It is unclear whether doxycycline alone would have been sufficient as testing sent the same day as ciprofloxacin initiation showed the CSF had cleared. Two case reports demonstrated successful recovery with dual therapy with moxifloxacin and doxycycline in a preterm infant with IVH (11) and with monotherapy with gentamicin in a full-term 4-month-old infant (12). Interestingly, there are reports of neonates with M. hominis who recovered without long-term defects despite not receiving antimicrobial therapy (13).
We present a rare case of meningitis in a neonate due to Mycoplasma hominis where WGS guided the diagnosis and management. Molecular techniques such as 16S rRNA and WGS may be faster and more effective in diagnosing invasive infections from isolates of this organism than bacterial cultures alone. Selection of antimicrobial agents can be challenging as few practitioners have experience with the organism, there is a lack of phenotypic data, and known resistance patters differ significantly from other Mycoplasma species. This case highlights not only the importance of M. hominis as a pathogen in neonates but also how clinical knowledge of resistance patterns can impact care and the potential uses of WGS to analyze genetic determinants of antibiotic resistance. We and others found that tetracyclines and fluoroquinolones can used without apparent toxicities (11).
SOURCES OF SUPPORT:
Felicia Scaggs Huang, Joel Mortensen, Jesse Skoch, Mary Staat, and Joshua Schaffzin have nothing to disclose.
Heidi Andersen receives funding from NIH (T32ES010957–14) at the University of Cincinnati
This work was supported by funding from Dr. Haslam’s Precision Metagenomic Core Laboratory.
REFERENCES
- 1.Chua K, Ngeow YF, Ng K, Chye JK, Lim C. Ureaplasma urealyticum and Mycoplasma hominis isolation from cervical secretions of pregnant women and nasopharyngeal secretions of their babies at delivery. Singapore medical journal. 1998;39(7):300–2. [PubMed] [Google Scholar]
- 2.Hata A, Honda Y, Asada K, Sasaki Y, Kenri T, Hata D. Mycoplasma hominis meningitis in a neonate: case report and review. Journal of Infection. 2008;57(4):338–43. [DOI] [PubMed] [Google Scholar]
- 3.Mathew OP, Bland HE, Pickens JM, James EJ. Hypoglycorrhachia in the survivors of neonatal intracranial hemorrhage. Pediatrics. 1979;63(6):851–4. [PubMed] [Google Scholar]
- 4.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome biology. 2014;15(3):R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome research. 2008;18(5):821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacterial Antimicrobial Resistance Reference Gene Database PubMed. [cited 2018 July 9]. Available from: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA313047.
- 7.Pereyre S, Gonzalez P, De Barbeyrac B, Darnige A, Renaudin H, Charron A, et al. Mutations in 23S rRNA account for intrinsic resistance to macrolides in Mycoplasma hominis and Mycoplasma fermentans and for acquired resistance to macrolides in M. hominis. Antimicrobial agents and chemotherapy. 2002;46(10):3142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srinivasan L, Pisapia JM, Shah SS, Halpern CH, Harris MC. Can broad-range 16S ribosomal ribonucleic acid gene polymerase chain reactions improve the diagnosis of bacterial meningitis? A systematic review and meta-analysis. Annals of emergency medicine. 2012;60(5):609–20.e2. [DOI] [PubMed] [Google Scholar]
- 9.Krausse R, Schubert S. In‐Vitro activities of tetracyclines, macrolides, fluoroquinolones and clindamycin against Mycoplasma hominis and Ureaplasma ssp. isolated in Germany over 20 years. Clinical Microbiology and Infection. 2010;16(11):1649–55. [DOI] [PubMed] [Google Scholar]
- 10.Redelinghuys MJ, Ehlers MM, Dreyer AW, Lombaard HA, Kock MM. Antimicrobial susceptibility patterns of Ureaplasma species and Mycoplasma hominis in pregnant women. BMC infectious diseases. 2014;14(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watt KM, Massaro MM, Smith B, Cohen-Wolkowiez M, Benjamin DK Jr, Laughon MM. Pharmacokinetics of moxifloxacin in an infant with Mycoplasma hominis meningitis. The Pediatric infectious disease journal. 2012;31(2):197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdul-Wahab OM, Dahish AAA, Elawad ME. Mycoplasma Hominis. Bahrain Medical Bulletin. 2017;39(2). [Google Scholar]
- 13.Huang Y-F, Mu X-P. Mycoplasma hominis Meningitis in a Full-Term Neonate: Rapid Recovery without Specific Treatment. The Indian Journal of Pediatrics. 2016;9(83):1030–1. [DOI] [PubMed] [Google Scholar]