Abstract
The chromosome 8q24.21 locus, which contains the proto-oncogene c-MYC, long non-coding RNA PVT1, and microRNAs (miRs), is the most commonly amplified region in human prostate cancer. A long-range interaction of genetic variants with c-MYC or long non-coding PVT1 at this locus contributes to the genetic risk of prostate cancer. At this locus is a cluster of genes for six miRs (miR-1204, −1205, −1206, −1207–3p, −1207–5p, and −1208), but their functional role remains elusive. Here, the copy numbers and expressions of miRs-1204~1208 were investigated using quantitative PCR for prostate cancer cell lines and primary tumors. The data revealed that copy numbers and expression of miR-1205 were increased in both castration-resistant prostate cancer cell lines and in primary tumors. In castration-resistant prostate cancer specimens, the copy number at the miR-1205 locus correlated with expression of miR-1205. Furthermore, functional analysis with an miR-1205 mimic, an miR-1205 inhibitor, and CRISPR/Cas9 knockout revealed that, in human prostate cancer cells, miR-1205 promoted cell proliferation and cell cycle progression and inhibited hydrogen peroxide-induced apoptosis. In these cells, miR-1205 downregulated expression of the Egl-9 family hypoxia inducible factor 3 (EGLN3) gene and targeted a site in its 3’-untranslated region to downregulate its transcriptional activity. Thus, by targeting EGLN3, miR-1205 has an oncogenic role and may contribute to the genetic risk of castration-resistant prostate cancer.
Keywords: microRNA, prostate cancer, EGLN3, cell proliferation
INTRODUCTION
Based on statistics for 2016, prostate cancer has the highest incidence rate (180,890 new cases, 21% of all male cases) in the United States 1. Although death rates for prostate cancer are down about 50% as a result of improvements in early detection and treatment, it is still the second leading cause of death (26,120 cases) for American men 1. However, the genetic mechanism underlying the development and progression of prostate cancer remains poorly understood. It is therefore important to identify the genetic risk factors for prostate cancer. Genome-wide association studies (GWAS) showed evidence for an association of prostate cancer risk with independent genetic variants on chromosome 8q24, conferring prostate cancer susceptibility at this locus 2–5. Nevertheless, at the locus with these genetic variants, no specific gene has been identified as responsible for risk of prostate cancer. At 8q24.21, there is a long-range interaction of these genetic variants with c-MYC or long non-coding RNAs (lncRNAs) in a tissue-specific manner, including prostate 6–8, suggesting master genetic factors at 8q24.21 that contribute to this genetic risk.
The locus at chromosome 8q24.21, the most commonly amplified region in human prostate cancer 9–11, contains the oncogene c-MYC and, adjacent to it, the gene for lncRNA PVT1 12–15 (Figure 1a). In human prostate cancers, c-MYC is a commonly amplified oncogene 16, 17. Both copy number alterations and expression of PVT1 are elevated in various human cancers, including prostate cancer 8, 18. In 8q24-amplified human cancer cells, a gain of PVT1 expression is required for high c-MYC protein levels 12. In most cancers, the copy number of PVT1 increases with high c-MYC copies, suggesting that co-expression of PVT1 and c-MYC is a characteristic of human cancers 12, and that c-MYC and PVT1 contribute to the genetic risk of prostate cancer. Functional analyses show that PVT1 and c-MYC promoters compete for enhancer contact in cis and that the PVT1 promoter inhibits c-MYC expression, but silencing of this promoter enhances breast cancer cell competition and growth 19, 20. However, other analyses show that, in triple-negative breast cancer cells, depletion of PVT1 inhibits tumor growth through KLF5/beta-catenin signaling 21 and that, in gastric cancer cells, PVT1 promotes angiogenesis through activation of the STAT3/VEGFA axis 22. Thus, the functional role of PVT1 in cancer cells remains elusive.
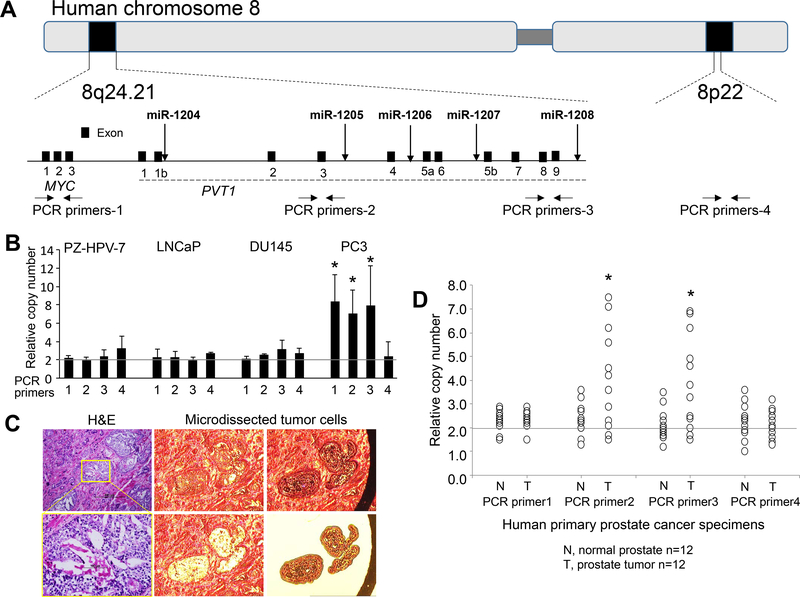
Figure 1. DNA copy numbers for chromosome 8q24.21 in human prostate cancer cells.
(a) Diagram of the position of human miRs-1204~1208, coding gene c-MYC, and long non-coding RNA PVT1 at 8q24.21 and a reference locus at 8q22. Down-arrows indicate loci of miRs-1204~1208. Horizontal arrows indicate the loci for design of PCR primers. (b) Relative DNA copy number of 8q24.21 and 8p22 loci against multiple independent loci in the genome determined by a multicopy reference assay of human prostate cancer cell lines. Data are presented as means ± SD. * p < 0.05 by two-tailed t-test for PCR primers 1–3 vs. primer 4. (c) Representative images of laser capture microdissection of tumor cells in prostate cancer tissues. Left panels: H&E staining; Right and middle panels: laser capture microdissection of tumor cells from target tissues. (d) Relative DNA copy numbers of 8q24.21 and 8p22 loci in primary castration-resistant prostate cancer specimens. Data are presented as the means ± SD. * p < 0.05 by two-tailed t-test in T group vs. N group. T, micro-dissected prostate cancer cells; N, micro-dissected normal prostate epithelial cells. All experiments were repeated three times.
A cluster of six microRNAs (miR-1204, −1205, −1206, −1207–3p, −1207–5p, and −1208) is located at the PVT1 locus of 8q24.2114, 15 (Figure 1a), but no functional role for any of these miRNAs has been found for prostate cancer cells. The mature forms of these miRNAs are differentially expressed in various cancer cell lines 15. In colon cancer cells, there is a p53-dependent induction of miR-1204 23 but, in nasopharyngeal carcinoma cells, downregulation of miR-1204 24. In breast cancers, miR-1204 targets the vitamin D receptor (VDR) gene to promote the epithelial-mesenchymal transition and metastasis 25. In the pleural mesothelioma cell line HP10, there is > 3-fold increased expression of miR-1205 as compared to other malignant mesothelioma cell lines 26. In ovarian cancers, overexpression of miR-1207 promotes cancer stem cell-like traits by activating the Wnt/β-catenin signaling pathway 27, and PVT1-derived miR-1207–5p promotes breast cancer cell growth by targeting STAT6 28. miR-1207–3p is under-expressed in prostate cancer cell lines; it inhibits cell proliferation and migration, and induces apoptosis via targeting of FNDC1 29.
Thus, characterization of these miRNAs would provide new insights into the genetic risk of prostate cancer associated with 8q24. In the present effort, we measured the copy numbers and expression levels of miRs-1204~1208 and assessed their potential roles in prostate cancer cells. We further addressed the functional role and target genes of miR-1205 and its regulatory mechanism in these cells.
RESULTS
Somatic amplification and overexpression of miR-1205 at 8q24.21 in human castration-resistant prostate cancer cells
GWAS analyses reveal common germline genetic variants at 8q24 that are associated with various cancers, especially prostate cancer 2–5. For prostate cancer, somatic amplification at 8q24, including the c-MYC locus on 8q24.21, shows copy number gains, and these gains are implicated in tumor progression, lymph node metastasis, and tumor recurrence 11, 30. At 8q24, long-range enhancers interact with c-MYC and PVT1 at 8q24.21 (Figure 1a) and contribute to the genetic risk of prostate cancer 6, 31, 32. In the present study, using PCR quantitative copy number assays, we identified, in the human castration-resistant prostate cancer cell line PC3, the somatic DNA amplification of 8q24.21 loci, including the c-MYC locus (PCR primer-1 region) and PVT1 and the locus for miRs-1205~1208 (PCR primers-2~3 regions), but this amplification was not evident in the human castration-resistant cell line DU145, in the human androgen-sensitive cell line LNCaP, or in the human normal prostate epithelial cell line, PZ-HPV-7 transformed by transfection with human papillomavirus 18 33 (Figure 1b). Since there may be linkage disequilibrium between the c-MYC locus and PVT1 and the miRs-1205~1208 locus, in PC3 cells, miRs-1205~1208 may be passengers amplified with c-MYC. To address this question, using laser-capture microdissection in combination with PCR quantitative copy number assays, we found 12 primary prostate cancer tissues without c-MYC amplification from 30 castration-resistant prostate cancers (Figures 1c-d and Table 1). Using the 12 micro-dissected prostate cancer tissues, we identified DNA amplification of the miRs-1205~1208 locus (PCR primers-2~3 regions) in prostate cancer cells from more than 50% of cases (7/12; PCR primers-2 region, p = 0.013; PCR primer-3 region, p = 0.007) (Figure 1d), suggesting that amplification of miRs-1205~1208 is a frequent event and may be independent of c-MYC in about 23% (7/30) of all cases. In addition, we assessed the DNA copy numbers of the miRs-1205~1208, c-MYC, and PVT1 loci using various public datasets. As shown in Supplementary Figures S1a-b, there was amplification of miR-1205 in 8% of all cases and 13% of metastatic cases, but c-MYC-independent amplification of miR-1205 only in a few cases. These data suggest that amplification of miR-1205 is a frequent event in prostate cancer, especially metastatic prostate cancer, but MYC-independent miR-1205 amplification is an infrequent event. Of note, DNA amplification at the miR-1205 locus was associated with poor survival of patients with prostate cancer (Supplementary Figures S1c-d).
Table 1.
The clinicopathological characteristics of the prostate cancer patients
| Categories | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 | Case 10 | Case 11 | Case 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age at the time of diagnosis | 75 | 72 | 77 | 65 | 72 | 69 | 80 | 76 | 75 | 62 | 72 | 65 |
| Year of diagnosis | 2012 | 2012 | 2012 | 2014 | 2014 | 2014 | 2015 | 2015 | 2015 | 2013 | 2014 | 2015 |
| Gleason score | 3+3 | 3+4 | 3+4 | 4+3 | 4+4 | 4+5 | 4+3 | 3+3 | 3+4 | 3+4 | 4+4 | 4+4 |
| Tumor stage (TNM) | T2N0M0 | T3N0M0 | T2N0M0 | T3N1M0 | T2N0M0 | T3N2M0 | T3N0M0 | T3N1M0 | T2N0M0 | T2N0M0 | T3N0M0 | T3N1M0 |
Next, we assessed the association of somatic DNA amplification with expressions of miRs-1204, −1205, −1206, −1207–3p, −1207–5p, and −1208 at 8q24.21 in prostate cancer cells. Except for miR-1207–5p, all miRNAs were detected in LNCaP, DU145, and PC3 cells. The highest expression of the miR-1205 was in PC3 cells, and higher expressions of miR-1204 and miR-1205 were present in DU145 cells, but there were lower expressions of all miRs-1204~1208 in LNCaP and PZ-HPV-7 cells (Figure 2a). In the 12 micro-dissected prostate cancer tissues, expression of only miR-1205 was increased in castration-resistant prostate cancer cells compared with expression in adjacent normal prostate epithelial cells (p = 0.011) (Figure 2b). Likewise, expression analysis of miR-1205 as normalized to U6 was validated by an additional RNA reference, miR-16–5p (Supplementary Figure S2). In addition, for prostate cancer samples, there were significant associations of expression of miR-1205 with the host gene PVT1 as determined with the TCGA dataset (Supplementary Figure S3). For castration-resistant prostate cancer cells, the DNA copy number at the miR-1205 locus (PCR-primer 2 region) correlated with expression of miR-1205 (r = 0.760, p = 0.004) but not in matched adjacent normal prostate epithelial cells (r = 0.412, p = 0.138) (Supplementary Figure S4). Likewise, both amplification and overexpression of miR-1205 was present in PC3 cells (Figures 1b and 2a), but, in DU145 cells, there was overexpression without amplification of miR-1205. To investigate the potential regulatory mechanism of miR-1205 in DU145 cells, direct bisulfite-treated sequencing was performed to determine the DNA methylation status at the CpG island in the promoter region of miR-1205 (Supplementary Figure S5a). DNA hypermethylation at the CpG island was evident in LNCaP and PZ-HPV-7 cells but not in DU145 and PC3 cells (Supplementary Figures S5b-c), suggesting epigenetic regulation in the transcription of miR-1205.
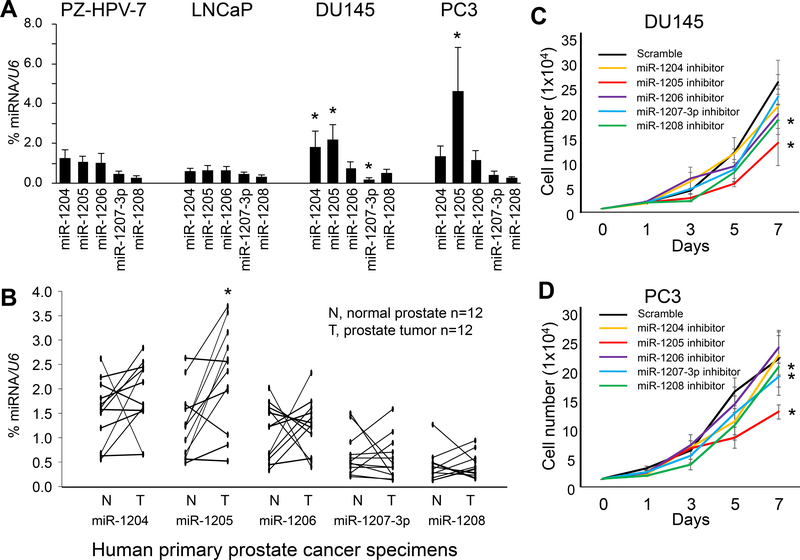
Figure 2. Expression levels of miRs-1204~1208 and their effect on cell proliferation in human prostate cancer cells.
(a) Quantification of expression of miRs-1204~1208 by qPCR as a percentage of U6 expression in PZ-HPV-7, LNCaP, DU145, and PC3 cells. Data are presented as means ± SD. * p < 0.05 by two-tailed t-test for each miRNA in PC3, DU145 or LNCaP vs. PZ-HPV-7 cells. (b) Quantification of miRs-1204~1208 expression by qPCR as a percentage of U6 expression in primary castration-resistant prostate cancer specimens. * p < 0.05 by Wilcoxon signed-rank test of T group vs. N group. T, micro-dissected prostate cancer cells; N, micro-dissected normal prostate epithelial cells. (c, d) Proliferation of DU145 and PC3 cells after transfection with an miR-1204~1208 inhibitor for 7 days. Data are presented as means ± SD. * p < 0.05 by two-way ANOVA test vs. scramble group. All experiments were repeated three times.
miR-1205 facilitates cell proliferation and cell cycle progression in human prostate cancer cells
To determine the role of miRs-1204~1208, a scrambled control or an miRNA inhibitor was transiently transfected into DU145 and PC3 cells. Compared with the scrambled control, the miR-1205 inhibitor reduced cell proliferation in DU145 (p = 0.002, Figure 2c) and PC3 cells (p < 0.001, Figure 2d). To validate the role of miR-1205 in prostate cancer cells, a scrambled control or an miR-1205 mimic was transiently transfected into LNCaP cells, which showed low miR-1205 expression (Figure 2a). For LNCaP cells, transfection of an miR-1205 mimic promoted cell proliferation compared with that of a scrambled control (p = 0.042, Figure 3a). Likewise, we validated this observation for LNCaP C4–2 cells, which are androgen receptor-positive, castration-resistant prostate cancer cells (Supplementary Figures S6a-b). Furthermore, to determine whether miR-1205 is related to androgen resistance, LNCaP cells were treated with 5α-dihydrotestosterone (5α-DHT) for 7 days following transfection with an miR-1205 mimic. As shown in Supplementary Figure S6c, cell proliferation was increased after 5α-DHT stimulation in cells transfected with the scramble miRNA. Although transfection with an miR-1205 mimic promoted cell proliferation, 5α-DHT stimulation did not enhance proliferation of cells transfected with miR-1205, suggesting castration resistance. In addition, we assessed VCaP cell growth under serum-starved conditions in the absence of androgen for 5 days following transfection with the miR-1205 mimic or scramble control miRNA. As shown in Supplementary Figure S6d, transfection with the miR-1205 mimic promoted cell proliferation in serum-free medium without androgen as compared to transfection with scramble control miRNA, suggesting promotion of androgen-independent cell growth by miR-1205.
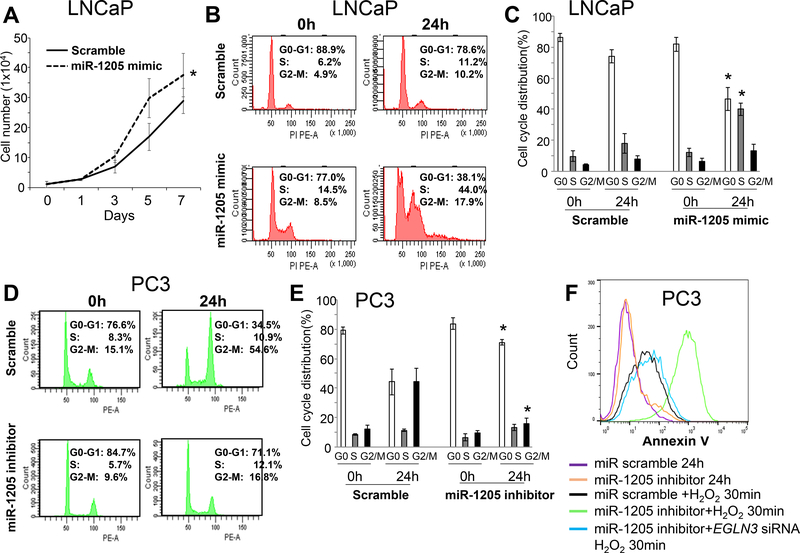
Figure 3. Effect of miR-1205 on cell proliferation, cell cycle progression, and apoptosis in human prostate cancer cells.
(a) Cell proliferation after transfection of LNCaP cells with an miR-1205 mimic for 7 days. Data are presented as means ± SD. * p < 0.05 by two-way ANOVA test vs. scramble group. (b) Cell cycle progression in LNCaP cells after starvation for 48 hours monitored by PI staining and flow cytometry at 0 and 24 hours after transfection with a scrambled control or an miR-1205 mimic. (c) Quantitative cell cycle data for LNCaP cells presented as means ± SD of triplicates. * p < 0.05 by one-way ANOVA followed by protected least-significant difference test vs. scramble group. (d) Cell cycle progression of PC3 cells after starvation for 48 hours monitored by PI staining at 0 and 24 hours after transfection with a scrambled control or an miR-1205 inhibitor. (e) Quantitative cell cycle data for PC3 cells presented as the means ± SD of triplicates. * p < 0.05 by one-way ANOVA followed by protected least-significant difference test vs. scramble group. (f) Apoptosis of PC3 cells was detected by Annexin V staining and flow cytometry after exposure to H2O2 (0.1 mM) for 30 minutes (min) following transfection with an miRNA scramble control, an miR-1205 inhibitor, or an miR-1205 inhibitor plus an EGLN3 siRNA for 24 hours (h). All experiments were repeated three times.
Next, the effect of miR-1205 on cell cycle progression of LNCaP cells (expressing low miR-1205) and PC3 cells (expressing high miR-1205) was addressed. To arrest the cells at the G0-G1 phase, they were starved under serum-free conditions for 48 hours before transfection with an miR-1205 mimic or miR-1205 inhibitor. After 24 hours of serum stimulation, approximately 10% of the LNCaP cells transfected with the scrambled miR were at the S or G2-M phases, whereas 44% and 18% of cells transfected the miR-1205 mimic entered the S and G2-M phases, respectively (Figures 3b-c). Likewise, > 65% of PC3 cells transfected with the scrambled miR were at either the S or G2-M phases, whereas < 30% PC3 cells transfected with the miR-1205 inhibitor entered the S or G2-M phases (Figures 3d-e). However, after transfection of PC3 cells for 7 days, the miR-1205 inhibitor had little influence on apoptosis (Figure 3f).
miR-1205 downregulates the expression of the Egl-9 family hypoxia inducible factor 3 (EGLN3) gene in human prostate cancer cells
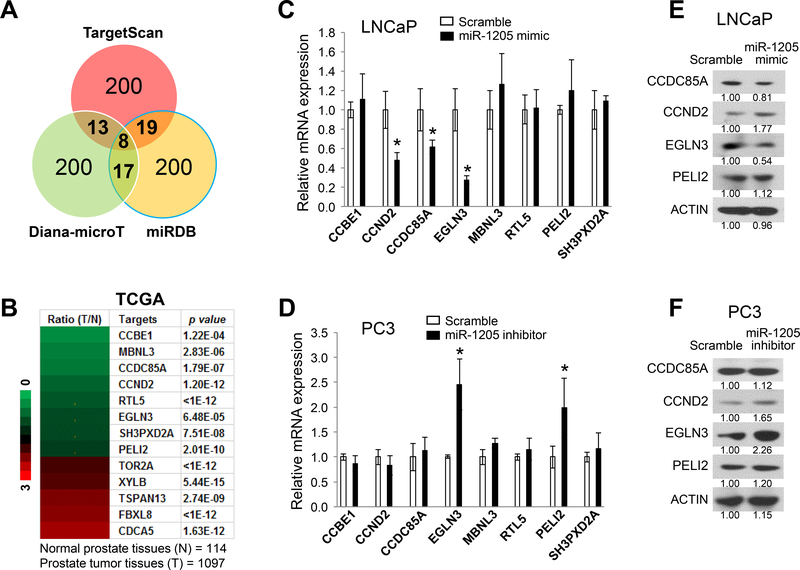
The potential target genes of miR-1205 were predicted using the public miRNA databases TargetScan 34, Diana-microT 35, and miRDB 36. Due to a different scoring algorithm for each database, we selected the top 200 candidate targets of miR-1205 from each. As shown in Figure 4a and Supplementary Table S1, we predicted 57 candidate targets as identified in at least two databases. Furthermore, we analyzed the mRNA expression of these candidate target genes in the Cancer Genome Atlas (TCGA) dataset. Of the 57 candidate genes, 8 were downregulated and 5 were upregulated (>1.5-fold change, p < 0.001) in prostate cancer tissues compared with normal prostate tissues (Figure 4b). To validate the effect of miR-1205 on the downregulated candidate genes, we transiently transfected a scrambled control or an miR-1205 mimic into LNCaP cells (with low expression of miR-1205). Quantitative PCR (qPCR) analysis showed downregulation of CCND2 (p = 0.0113), CCDC85A (p = 0.0457), and EGLN3 (p = 0.005) expressions after transfection with the miR-1205 mimic (Figure 4c). Likewise, we transiently transfected the scrambled control or miR-1205 inhibitor into PC3 cells (with high expression of miR-1205). Upregulation of EGLN3 (p = 0.008) and PELI2 (p = 0.0410) expressions were evident after transfection with the inhibitor (Figure 4d). However, there was a change of protein levels only for EGLN3 after transfection with the mimic or inhibitor of miR-1205 in LNCaP cells (Figure 4e) and PC3 cells (Figure 4f).
Figure 4. Identification of miR-1205 target genes in human prostate cancer cells.
(a) Prediction of miR-1205 target genes by multiple algorithms using three miRNA databases: TargetScan (www.targetscan.org), Diana-microT (diana.imis.athena-innovation.gr), and miRDB (mirdb.org). A total of 57 candidates of miR-1205 target genes were identified in at least two databases. (b) Heat map of mRNA expression levels of the predicted miR-1205 target genes in prostate cancer tissues (T) compared with that in normal prostate tissues (N) from the TCGA dataset. A total of 13 predicted target genes were changed (>1.5-fold change, p < 0.001, T vs. N), including 8 downregulated genes (green color) and 5 upregulated genes (red color). (c) Relative expression levels of downregulated candidate genes were determined by qPCR in LNCaP cells after treatment with a scrambled control or an miR-1205 mimic. Data are presented as means ± SD. * p < 0.05 by two-tailed t-test for the miR-1205 mimic group vs. the scramble group. (d) Relative expression levels of downregulated candidate genes were determined by qPCR in LNCaP cells after treatment with a scrambled control or an miR-1205 inhibitor. Data are presented as means ± SD. * p < 0.05 by two-tailed t-test for the miR-1205 inhibitor group vs. the scramble group. (e) Protein expression in LNCaP cells measured by Western blotting at 48 hours after transfection with a scrambled control or an miR-1205 mimic. (f) Protein expression measured by Western blotting in PC3 cells at 48 hours after transfection with a scrambled control or an miR-1205 inhibitor. All experiments were repeated three times.
miR-1205 promotes cell proliferation through EGLN3 in human prostate cancer cells
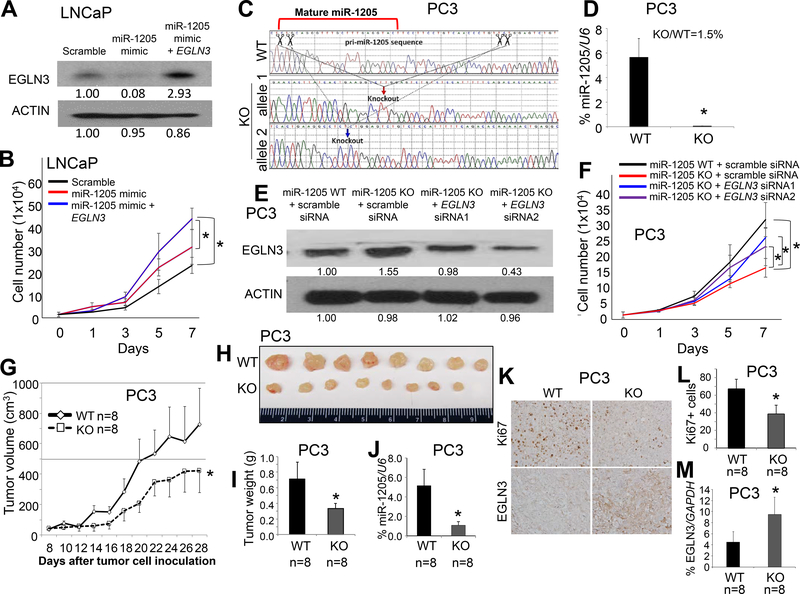
To determine the basal expression of miR-1205 and EGLN3 protein, we performed expression analysis for PZ-HPV-7 cells as compared to LNCaP, DU145, and PC3 cells. The expression of miR-1205 was similar in PZ-HPV-7 and LNCaP cells but lower than in DU145 and PC3 cells (Figure 2a). In contrast, protein expression of EGLN3 was higher in PZ-HPV-7 and LNCaP cells than in PC3 and DU145 cells (Supplementary Figure S7a). Of note, in the cell lines, expression of miR-1205 appeared to be conversely associated with expression of EGLN3, supporting an interaction between miR-1205 and EGLN3 (Figure 2a and Supplementary Figure S7a). To determine if EGLN3 is responsible for miR-1205-induced cell proliferation, we transiently transfected a scrambled control, miR-1205 mimic, or miR-1205 mimic plus exogenous EGLN3 into LNCaP cells (with low expression of miR-1205). The effective transfection of exogenous EGLN3 and the effect of the miR-1205 mimic on expression of EGLN3 was validated by Western blotting (Figure 5a). After transfection, cell proliferation was induced by the miR-1205 mimic (p < 0.001, miR-1205 mimic vs. scrambled control), but this induction was partially blocked by exogenous EGLN3 (p = 0.007, miR-1205 mimic + EGLN3 vs. miR-1205 mimic) (Figure 5b). Furthermore, using CRISPR/Cas9 technology, miR-1205 was knocked out in PC3 cells (with high expression of miR-1205), and the miR-1205 knockout cells were confirmed by Sanger sequencing (Figure 5c) and qPCR (Figure 5d). The scrambled control or either of two EGLN3 small interfering RNAs (siRNAs) were transiently transfected into the miR-1205 knockout PC3 cells, and the effect of miR-1205 knockout and EGLN3 siRNAs on the expression of EGLN3 was validated by Western blotting (Figure 5e). Cell proliferation was reduced in miR-1205 knockout PC3 cells compared with that in wild-type PC3 cells (p < 0.001, miR-1205 wild-type vs. knockout), but knockdown of EGLN3 partially rescued cell proliferation in miR-1205 knockout PC3 cells (p = 0.026 for EGLN3 siRNA1 and p = 0.019 for EGLN3 siRNA2 vs. scrambled control) (Figure 5f). Since EGLN3 appears to control cell migration and motility 37, 38, we also tested the effect of miR-1205 on cell migration in PC3 cells by scratch assays and Transwell assays, but there was no difference in cell migration between miR-1205 knockout and wild-type PC3 cells (Supplementary Figures S8a-c). We performed immunofluorescence assays with F-actin and anti-EGLN3 antibody in miR-1205 WT and KO PC3 cells, and miR-1205 KO increased the expression of EGLN3, but F-actin staining was not appreciably changed in miR-1205 KO cells compared with miR-1205 WT cells (Supplementary Figure S8d). In addition, to determine the effect of miR-1205 on tumor growth, miR-1205 wild-type or knockout PC3 cells were subcutaneously injected into immunodeficient nude mice. Xenograft tumor growth (p < 0.001) and weights (p < 0.001) were reduced in mice with miR-1205 knockout PC3 cells compared with wild-type PC3 cells (Figures 5g-i). A decreased expression of Ki67 but increased expressions of EGLN3 protein and mRNA was evident in miR-1205 knockout xenograft tumors compared with that in the WT xenograft tumors (Figures 5j-m).
Figure 5. Effects of miR-1205 and its target gene EGLN3 on growth of human prostate cancer cells.
(a) Protein expression of EGLN3 was measured by Western blotting in LNCaP cells at 48 hours after transfection with a scrambled control, an miR-1205 mimic, or an miR-1205 mimic and the pcDNA6-EGLN3-coding sequence (without the 3’-UTR). (b) Proliferation of LNCaP cells after transfection for 7 days. Data are presented as means ± SD. * p < 0.05 by two-way ANOVA test between the two groups. (c) Sanger sequencing of CRISPR/Cas9 editing PC3 cells. Down-arrows indicate knockout sites. WT, wild-type; KO, knockout. (d) Quantification of miR-1205 expression by qPCR as a percentage of U6 expression in PC3 cells. Data are presented as means ± SD. * p < 0.05 by two-tailed t-test for the miR-1205 KO group vs. WT group. (e) Protein expression of EGLN3 was measured by Western blotting of miR-1205 WT or KO PC3 cells at 48 hours after transfection with a scrambled control or an miR-1205 inhibitor or both an miR-1205 inhibitor and EGLN3 siRNA. (f) Proliferation of PC3 cells after transfection for 7 days. Data are presented as means ± SD. * p < 0.05 by two-way ANOVA test between two groups. (g) Tumor growth in nude mice subcutaneously injected with miR-1205 WT or KO PC3 cells. Data are presented as means ± SD of the tumor volumes. * p < 0.05 by two-way ANOVA test between WT and KO groups. Representative images (h) and weights (i) of xenograft tumors at day 28 after injection. (j) Quantification of expression of miR-1205 by qPCR as a percentage of U6 expression in micro-dissected xenograft prostate cancer cells. (k) Representative immunohistochemical staining of Ki67 and EGLN3 in xenograft prostate tumor tissues. Top panels: Ki67 staining; bottom panels: EGLN3 staining. (l) The percentage of Ki67+ cells as an indicator of proliferating cells among the xenograft prostate tumor tissues. At least five 40x fields for each mouse were counted. (m) Quantification of mRNA expression of EGLN3 by qPCR as a percentage of GAPDH expression in micro-dissected xenograft prostate cancer cells. Data are presented as means ± SD. * p < 0.05 by two-tailed t-test between WT and KO groups. All experiments were repeated three times.
The tumor suppressive function of EGLN3 is independent of HIF1α and NF-κB 39, 40, and lack of mRNA expression of EGLN3 causes an absence of response to hypoxia 41. EGLN3 induces apoptosis in cancer cells under normoxia 42, 43, but loss of EGLN3 enhances cancer cell proliferation and survival in hypoxic microenvironments 39, 40. Hydrogen peroxide (H2O2) is a non-radical reactive oxygen species (ROS) molecule that is induced by hypoxia 44, 45. Thus, to address whether inhibition of miR-1205 induces apoptosis upon oxidative stress, the PC3 cells were treated with H2O2 for induction of apoptosis. At 24 hours after transfection with an miR-1205 inhibitor, apoptosis was not evident in the cells, but apoptosis was elevated upon H2O2 stimulation for 30 minutes in cells treated with an miR-1205 inhibitor as compared to that in cells treated with an miRNA scramble control (Figure 3f). Of note, in the cells exposed to H2O2, hypoxia-mediated apoptosis induced by the miR-1205 inhibitor was reduced by EGLN3 siRNA, suggesting that inhibition of the miR-1205-EGLN3 axis promotes apoptosis through oxidative stress.
Post-transcriptional regulation of the EGLN3 gene by miR-1205 in human prostate cancer cells
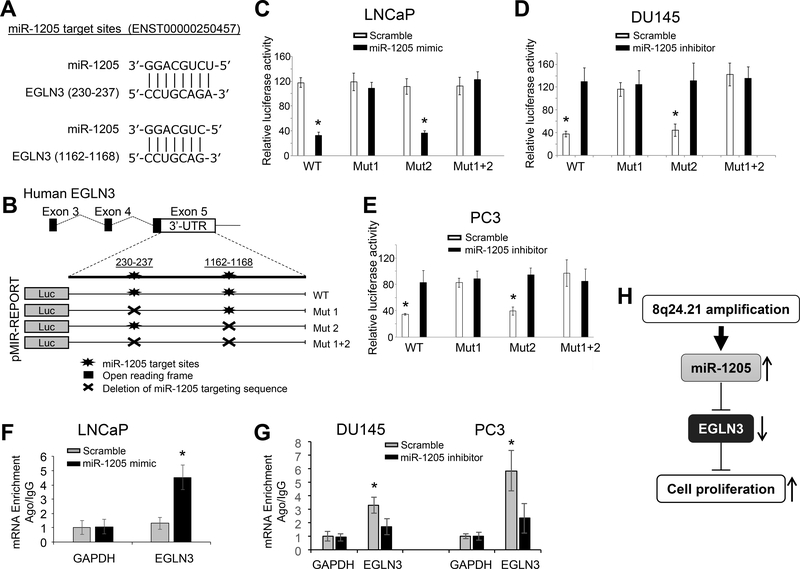
We predicted the miR-1205-targeting sequences in the 3’-untranslated region (3’-UTR) of EGLN3 (Figure 6a). Sequence alignment analysis revealed two potential miR-1205-targeting sites (Figures 6a-b). The post-transcriptional regulation of EGLN3 was characterized by targeting of miR-1205 using a pmiR-luciferase reporting system in prostate cancer cells. In LNCaP cells (with low expression of miR-1205), transfection of the miR-1205 mimic reduced the luciferase activity of the wild-type EGLN3 3’-UTR (3.6-fold, p < 0.001), but deletion of the first miR-1205-targeting sequence (CCUGCAGA) in the EGLN3 3’-UTR rescued the luciferase activity of EGLN3 3’-UTR in LNCaP cells transfected with the miR-1205 mimic (Figure 6c). The post-transcriptional regulation of EGLN3 was verified by targeting of miR-1205 in DU145 and PC3 cells (with high expression of miR-1205). Transfection of the miR-1205 inhibitor induced the luciferase activity of the wild-type EGLN3 3’-UTR in DU145 cells (3.4-fold, p = 0.003) and PC3 cells (2.4-fold, p = 0.002), but deletion of the first miR-1205-targeting sequence reversed the suppression of luciferase activity of the EGLN3 3’-UTR in DU145 and PC3 cells transfected with the miR-1205 inhibitor (Figures 6d-e). To test whether miR-1205 directly interacts with EGNL3 mRNA, we performed an miRNA-mRNA interaction analysis using miRNA target immunoprecipitation (IP) with Argonate proteins (Ago1/2/3) upon transfection of LNCaP, DU145, and PC3 cells with the miR-1205 mimic or inhibitor. Ago IP analysis showed that EGLN3 mRNA, but not GAPDH mRNA, directly bound with Ago1/2/3 proteins in the presence of miR-1205 as compared to scramble control miRNA (Figures 6f-g), supporting a direct interaction of miR-1205 with EGLN3 mRNA in prostate cancer cells. In addition, we interrogated the TCGA dataset for a correlation of miR-1205 with EGLN3 mRNA in primary prostate cancers. However, we did not find a strong correlation between miR-1205 with EGLN3 mRNA (Pearson correlation r = 0.138; Supplementary Figure S9a). To test whether miR-1205 is related to EGLN3 in castration-resistant prostate cancer, we selected 10 additional samples of castration-resistant prostate cancers to obtain micro-dissected tumor cells, which were used to determine the correlation of expression between miR-1205 and EGLN3. As shown in Supplementary Figure S9b, there was a negative correlation of expression between miR-2015 and EGLN3 (Pearson correlation, r = −0.482; p = 0.023) in these samples, supporting the presence of a functional miR-1205-EGLN3 axis in castration-resistant prostate cancer.
Figure 6. Molecular mechanism for the post-transcriptional regulation of EGLN3 by miR-1205 in prostate cancer cells.
(a) Sequencing alignment of mature human miR-1205 with the 3’-UTR of EGLN3. (b) Construction of the WT-pMIR-EGLN3 3’-UTR reporter vector or Mut-pMIR-EGLN3 3’-UTR reporter vector with deletion of the miR-1205 targeting sequence. These vectors were transfected into prostate cancer cells in conjunction with either a scrambled control or an miR-1205 mimic or inhibitor. (c) Quantification of luciferase activity in LNCaP cells co-transfected with the pMIR-EGLN3 3’-UTR reporter vector and a scrambled control or an miR-1205 mimic. (d, e) Quantification of luciferase activity for DU145 and PC3 cells co-transfected with the pMIR-EGLN3 3’-UTR reporter vector and a scrambled control or an miR-1205 inhibitor. (f, g) Quantification of Ago-IP EGLN3 mRNA in the presence or absence of miR-1205 for LNCaP, DU145 and PC3 cells. Ago1/2/3-precipitated miRNA/mRNA complexes were treated with proteinase K, and extracted mRNAs were converted to cDNA and quantified by qPCR. The Ago-IP values were normalized by a negative control IgG-IP. (h) Diagram of an miR-1205-EGLN3 axis and its induction of proliferation in prostate cancer cells. Data are presented as means ± SD. * p < 0.05 by two-tailed t-test vs. control group. WT, wild-type; Mut, deletion of the miR-1205 targeting sequence. All experiments were repeated three times.
DISCUSSION
In the present study, we found somatic DNA amplification at 8q24.21 in human prostate cancer cells, particularly castration-resistant prostate cancer cells. In primary castration-resistant prostate cancer specimens, this amplification positively correlated with expression of miR-1205. Our functional analysis revealed that, in human prostate cancer cells, miR-1205 promoted cell proliferation and cell cycle progression but inhibited hydrogen peroxide-induced apoptosis. Further, in human prostate cancer cells, miR-1205 downregulated the expression of EGLN3, which was implicated in the miR-1205-induced cell proliferation. In human prostate cancer cells, miR-1205 targeted a specific site in the 3’-UTR of EGLN3 to downregulate its transcriptional activity. Thus, in these cells, we identified a potentially functional miR-1205-EGLN3 axis (Figure 6h).
EGLN3 (also known as PHD3, HPH1, and SM-20) is a member of the Caenorhabditis elegans gene egl-9 (EGLN) family of prolyl hydroxylases. The EGLN3 protein catalyzes hydroxylation of HIF1α, resulting in its ubiquitination and degradation and subsequent decrease in activity 46. In HEK293T cells, EGLN3 inhibits NF-κB activity through inhibition of IKKγ ubiquitination 47, and, in PC3 cells, lack of mRNA expression of EGLN3 causes an absence of response to hypoxia 41. The growth-suppressive function of EGLN3 is exerted through suppression of EGFR signaling, but, in gliomas, this function is independent to HIF1α and NF-κB 39, 40. In hypoxic microenvironments, loss of EGLN3 in tumor cells enhances cell proliferation and survival 39, 40. Functional analyses suggest that, in fibroblasts, neurons, and myoblasts, EGLN3 facilitates p53-induced growth arrest, apoptosis, and differentiation 48–50. Although miR-1205 promotes cell proliferation by targeting of EGLN3, which may be relevant to suppression of cell cycle progression, other studies indicate that EGLN3 enhances cell cycle progression in cancer cells 51, 52. However, there is evidence that, in cancer cells, EGLN3 promotes p53 protein stability through de-ubiquitination, leading to G1 cell cycle arrest and apoptosis, which further supports a tumor suppressive role of EGLN3 53. Bioinformatics analysis of the public datasets showed low mRNA expression of EGLN3 in primary prostate cancer tissues (Supplementary Figure S10a); this low expression of EGLN3 is more evident in cases with low Gleason scores (Supplementary Figure S10b) and is associated with disease relapse (Supplementary Figures S10c-d) but not with patient survival, whereas high expression of miR-1205 is associated with poor survival (Supplementary Figure S10e). Thus, these data suggest that miR-1205 induces cell proliferation, cell cycle progression, and inhibition of apoptosis, at least partially through EGLN3, indicating the existence of a functional miR-1205-EGLN3 axis in prostate cancer cells.
At 8q24.21, the miR-1205 locus (with PCR-primer 2) was amplified in the PC3 (castration-resistant) prostate cancer cell line and in primary castration-resistant prostate cancers. Since DNA amplification was associated with overexpression of miR-1205 in castration-resistant prostate cancer specimens, miR-1205 may contribute to the genetic risk of castration-resistant prostate cancer. However, although this cell line does not show amplification at the miR-1205 locus, overexpression of miR-1205 was evident in DU145 cells, a castration-resistant prostate cancer line, suggesting that different mechanisms underlie the regulation of miR-1205 expression in DU145 and PC3 cells. Furthermore, our data showed the DNA amplification of both PCR primer 2 and 3 loci, including miR-1205 and miR-1207, in castration-resistant prostate cancer cells. Although miR-1205 and miR-1207 may be amplified together at the same locus, there was no difference in expression levels of miR-1207–3p between benign and malignant tissues, suggesting that, in prostate cancer cells, a selective transcription of miR-1205 in the amplified locus includes an epigenetic regulatory mechanism. Indeed, DNA hypermethylation at the CpG island in the promoter region of miR-1205 was identified in normal prostate epithelial cells and androgen-sensitive prostate cancer cells but not in castration-resistant prostate cancer cells, supporting an epigenetic regulation in transcription of miR-1205.
Luciferase reporter assays are commonly used to measure functional miRNA-mRNA interactions. However, our mutation analysis showed that only one of the two binding sites was possibly involved in miR-1205-mediated regulation of EGLN3 stability. Mutation of the second seed sequence did not have any effect despite having a 100% homologous binding sequence. However, our miR-1205 target IP analysis supported a direct interaction of miR-1205 with EGLN3 mRNA in prostate cancer cells. In addition, although 100 nM of an miRNA mimic is commonly used for transient transfection, this concentration led to supraphysiological expression of miR-1205 in the cells (Supplementary Figure S11), resulting in a limitation of these results. However, all such results were validated by knockdown or knockout of miR-1205, which should alleviate this limitation.
In conclusion, in castration-resistant prostate cancer cells, DNA amplification at 8q24.21 is associated with overexpression of miR-1205, which may contribute to the genetic risk of prostate cancer. Functional analyses indicate that EGLN3, a target of miR-1205, is associated with miR-1205-mediated growth promotion, and suggest the existence of a functional miR-1205-EGLN3 axis in prostate cancer cells. However, miR-1205 targets, other than EGLN3, may contribute to miR-1205-induced cell proliferation. Although miR-1205 enhances cell cycle progression in these cells, the molecular mechanism underlying this process remains unknown.
MATERIALS AND METHODS
Cell lines, antibodies, and reagents
Prostate cancer cell lines, PZ-HPV-7, LNCaP, LNCaP C4–2, VCaP, DU145, and PC3, were obtained from the American Type Culture Collection (Manassas, VA, USA). Cell lines were authenticated by examination of morphology and growth characteristics and were confirmed to be mycoplasma-free. A short tandem-repeat analysis for DNA fingerprinting was also used to verify the cell lines 54. Cells were maintained in Dulbecco’s Modified Eagle’s medium (DMEM for DU145 and PC3) and Roswell Park Memorial Institute 1640 medium (RPMI-1640 for LNCaP) supplemented with 10% fetal bovine serum or keratinocyte serum-free medium (KSFM for PZ-HPV-7) (Thermo Fisher Scientific, Waltham, MA, USA). Specific primary antibodies were used to detect the following proteins: Ki67 (ab15580, ABCAM, Cambridge, MA, USA), CCDC85A (aas80681c, 1:1,000, Antibody Verify, Las Vegas, NV, USA), CCND2 (ab81359, 1:2,000, ABCAM), EGLN3 (PHD3) (aas02524c, 1:1,000, Antibody Verify), and PELI2 (aas76488c, 1:1,000, Antibody Verify). EGLN3 siRNAs and miRNA mimic and inhibitor are listed in Supplementary Table S2. These small RNAs (nmol/L) were transfected with Lipofectamine RNAiMAX (Thermo Fisher Scientific) into cells according to the protocol of the manufacturer.
Human tissue specimens
Formalin-fixed and paraffin-embedded prostate cancer specimens were obtained from the Department of Pathology, Jilin University Second Hospital. The tumor and tumor-adjacent normal specimens were collected from 12 patients with castration-resistant prostate cancer who underwent primary surgery between January 2012 and June 2015. All had histologically confirmed prostate adenocarcinoma with information on tumor stage (Tumor-Node-Metastasis) and grade (Gleason) and had received hormone therapy (Table 1). This study, involving the use of human prostate tumor specimens, was approved by the Institutional Review Board of the Jilin University Second Hospital. For all specimens, informed consent was obtained from all subjects in accordance with the requirements of the Institutional Review Board.
Laser capture microdissection
Formalin-fixed and paraffin-embedded tissue sections (8-μm) of human prostate specimens were cut and transferred to glass slides not coated with polylysine. The slides were stained with Harris hematoxylin for 50 seconds and eosin for 30 seconds and then dried in a laminar flow hood for 5 to 10 min prior to microdissection. For analysis of gene copy number and gene expression, target tissue sections were laser-capture micro-dissected using the Arcturus PixCell II system (Thermo Fisher Scientific) with an Olympus IX-50 microscope to obtain prostate epithelial and cancer cells (5 × 103), as described previously 55–57. RNA was extracted with PicoPure RNA extraction kits (Thermo Fisher Scientific) and amplified by RT-PCR. Genomic DNA was extracted from micro-dissected tissue using PicoPure DNA Isolation kits (Thermo Fisher Scientific). The concentrations were measured with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific).
Quantification of gene copy numbers
Quantitative analysis of copy numbers was conducted by QIAGEN qBiomarker Copy Number PCR assays (QIAGEN, Germantown, MD, USA) based on a 7500 Real Time PCR System (Thermo Fisher Scientific) as previously described 58, 59. All qBiomarker Copy Number PCR Assays were designed for unique regions of the genome. A qBiomarker Multicopy Reference Copy Number PCR Assay (MRef, QIAGEN) was included in each assay. The reference assay recognizes a stable sequence that appears in the human genome more than 40 times and whose copy number is not affected or minimally affected by local genomic changes. Relative gene copy numbers for each specimen were calculated as 2 × (tumor copy number/MRef copy number) as described previously 59.
Quantitative PCR of gene expression
For miRNA expression, 5 μl of RNA in 20-μl reactions was reverse-transcribed using miScript II RT Kits (QIAGEN) according to the manufacturer’s protocol. Then, 2 μl of cDNA was used as a template for real-time PCR using a LightCycler 480 Real Time PCR System (Roche Applied Sciences, Indianapolis, IN) with miScript SYBR Green PCR kits (QIAGEN) at 95°C for 2 min, followed by 50 cycles of 95°C for 15 sec and 60°C for 1 min. The relative quantities of miRNA were determined by the comparative method (2−ΔCt) with a U6 reference, as described previously 54, 60. For mRNA expression of coding genes, relative expression levels were determined by qPCR with SYBR Green Dye (Promega, Madison, WI, USA) using the comparative method (2−ΔΔCt) against endogenous GAPDH in accordance with the manufacturer’s protocol. The qPCR primer sequences are listed in Supplementary Table S2.
miRNA/mRNA IP assay
miRNA/mRNA IP was performed by using miRNA Target IP kits (Active Motif, Carlsbad, CA USA) according to the accompanying protocol. Briefly, cells (1.5 × 107) were transfected with scrambled control or miRNA mimic or inhibitor (50 nmol/L) using Lipofectamine RNAiMAX (Thermo Fisher Scientific) for 24 hours. Transfected cells of each sample were lysed in 150 μL of complete lysis buffer and incubated on ice for 5 min and then at −80°C for 2 hours. Protein G magnetic beads (50 μL) were mixed with 200 μL of BSA solution for 10 min, and then the tubes were placed on a magnet to pellet beads. After removal of the supernatant, 100 μL of wash buffer containing 5 μg of anti-Ago1/2/3 antibody or negative control anti-IgG antibody was incubated for 30 min. The lysate, mixed with 1000 μL IP buffer, was added to the protein G magnetic beads, incubated for overnight at 4 °C, and treated with proteinase K to digest protein for 30 min at 55 °C. Purification and detection of mRNA and miRNA are described above. The results of Ago-IP were normalized to that of negative control IgG-IP.
Cell proliferation, cell cycle progression, and apoptosis
Cells (1 × 105) were transfected with scrambled control or miRNA mimic or inhibitor (100 nmol/L) using Lipofectamine RNAiMAX (Thermo Fisher Scientific) for 48 hours. After transfection, cell morphology, viability, and numbers were monitored microscopically at 0, 1, 3, 5, and 7 days. At 0 and 24 hours after transfection, apoptosis was assessed by flow cytometry based on cell binding to Annexin V (BD Biosciences, San Jose, CA, USA). For apoptosis induction by H2O2, cells were treated with 0.1 mM H2O2 for 30 minutes following transfection with an miRNA scramble control, an miR-1205 inhibitor or an miR-1205 inhibitor plus an EGLN3 siRNA for 24 hours. After starvation of cells for 48 hours, cell-cycle progression at 0 and 24 hours was determined by propidium iodide (PI) (50 μg/ml, BD Biosciences) staining and flow cytometry, as described previously 57, 61.
Western blots
Western blotting was performed as previously described 56, 61. Briefly, cell lysates containing 50–100 μg of protein were resolved by electrophoresis on 10% SDS-polyacrylamide gels. Proteins were transferred to Hybond-P membranes, which were incubated in 5% non-fat dry milk for 1 hour and overnight at 4°C in 0.25% non-fat milk containing antibodies. The membranes were incubated for 1 hour at room temperature in 0.25% non-fat milk with an anti-rabbit or mouse IgG HRP-linked secondary antibody (Cell Signaling, Danvers, MA, USA). To ensure equal loading of proteins, the membranes were stripped under the same conditions as described above. They were then incubated with enhanced chemiluminescence reagents and exposed to X-ray film for 1–5 min.
Luciferase assay
The pMIR-REPORT luciferase reporter vectors (Thermo Fisher Scientific) were used to construct DNA fragments from the 3’-UTR of EGLN3 (Transcript: EGLN3–201 ENST00000250457) by Spe I and Mlu I (Promega) digestion according to the manufacturer’s protocol. The PCR primer sequences for DNA construction and mutagenesis are listed in Supplementary Table S2. Luciferase activity was measured as described previously 56, 62, 63. Briefly, cells were plated at a density of 5 × 104 per well into 24-well plates and then transiently co-transfected using Lipofectamine 3000 (Thermo Fisher Scientific) with luciferase gene reporter vectors (pMIR-EGLN3–3’UTR) and miR-1205 mimic or inhibitor (100 nmol/L) according to the protocol of the manufacturer. After transient transfections for 48 h, cells were washed twice with ice-cold PBS and were lysed in 1 x lysis buffer (Promega) for 15 min on a shaker. The luciferase activity was assessed with a Veritas Microplate Luminometer (Turner BioSystems, Sunnyvale, CA, USA) using a Dual Luciferase Assay System (Promega).
Site-directed mutagenesis
Site-directed mutagenesis of the EGLN3 3’-UTR-luciferase reporter plasmid was accomplished following the protocols from the Site-Directed Mutagenesis System (Thermo Fisher Scientific). The primers for mutagenesis are shown in Supplementary Table S2. The miR-1205 targeting motif sequences (230 to 237: CCUGCAGA and 1162 to 1168: CCUGCAG after stop coding sequence) in the pMIR-EGLN3–3’UTR construct were mutagenized to generate the targeting motif deletions.
Establishment of miR-1205 knockout cells
The miR-1205 single guide RNAs (sgRNAs) were designed using the online CRISPR design tool (Benchling, San Francisco, CA, USA, https://benchling.com). The pri-miR-1205 region was selected to be targeted by CRISPR/Cas9 editing. A ranked list of sgRNAs was generated with specificity and efficiency scores. In two flanking sites of the mature miR-1205 sequence, pair sgRNAs with more than 30% specificity and efficiency scores were selected. The pair of oligos for two targeting sites was annealed and ligated to the Bbs I-digested pSpCas9(BB)-2A-GFP (PX458) vector (Addgene, Cambridge, MA, USA). The pX458 plasmid containing each target sgRNA sequence or pX458 empty vector was transfected into PC3 cells with Lipofectamine 3000 (Thermo Fisher Scientific). After flow cytometry sorting with GFP, 100 GFP+ cells were seeded into each well of a 96-well plate. After selection of single colonies, miR-1205 knockout colonies were determined by Sanger sequencing with isolated genomic DNA, and miR-1205 expression levels in each clone were validated by qPCR. All sgRNAs were accessed using the online off-target searching tool (Cas-OFFinder, Daejeon, South Korea, http://www.rgenome.net/cas-offinder) 64. To avoid an off-target effect, potential off-target regions were selected and subjected to PCR and Sanger sequence analysis. The sgRNAs and primers for CRISPR design and DNA construct are shown in Supplementary Table S2.
Xenogeneic transplantation
BALB/c nude mice were purchased from Beijing Institute for Experimental Animals (Beijing, China). All experiments were conducted in accordance with accepted standards of animal care and approved by the Institutional Animal Care and Use Committee of Jilin University Second Hospital. miR-1205 wild-type or miR-1205 knockout PC3 cells (100 μl, 5 × 106) were subcutaneously injected into the left flanks of 8-week-old male nude mice. Xenograft tumor growth was observed for 28 days after tumor cell injection. Tumor sizes and weights were measured as described previously 55, 56.
Prediction of miR-1205 target genes
The source of potential miR-1205 target genes was selected from three miRNA databases, TargetScan34, Diana-micro T35, and miRDB 36. Briefly, the top 200 candidate target genes of miR-1205 were selected from each database due to a different scoring algorithm between them (Supplementary Table S1). All selected target genes were identified in at least two databases.
Datasets, analysis of gene expression data, and annotation
The TCGA Data Portal was used to download the data from samples of prostate adenocarcinomas (n=1097) and normal prostate controls (n=114). The RNAseqV2 level 3 data were used before statistical analysis. Gene-level normalized expression data were used in Partek Genomic Suite (PGS, St. Louis, MO, USA) for additional normalization, statistics, and annotation. False discovery rate (FDR) corrections (Benjamini-Hochberg methods) were applied for the purpose of testing multiple hypotheses.
Statistical analyses
Continuous variables were summarized using mean, standard deviation (SD), and median values. For each group, the distribution of data was evaluated using a one-sample Kolmogorov-Smirnov test. In samples with normal distributions, the means of the variables were compared using a two-tailed t-test between two groups. In samples with non-normal distributions, the medians of the variable between two groups were compared by a Mann–Whitney U test. Analysis of variance (ANOVA), one- and two-way, were used to test for overall differences, followed by a protected least significant difference test for differences between groups. All data were entered into an access database using Excel 2016 and analyzed with SPSS (version 24; IBM, Armonk, NY, USA), and StatView (version 5.0.1, SAS Institute Inc., Cary, NC, USA).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Donald L Hill for editorial assistance in preparing this manuscript. This work was supported by grants from the National Institutes of Health/National Cancer Institute (CA118948, CA179282, and CA013148 to LW and RL), the Department of Defense (BC160808, PC130594, and PC140308 to LW and RL), the Mike Slive Foundation for Prostate Cancer Research (LW), and the Natural Science Foundation of China (No. 31571126, 31571342, and 81772757 to XL, BL and RC). Results are based, in part, upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016; 66: 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet 2007; 39: 631–637. [DOI] [PubMed] [Google Scholar]
- 3.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet 2007; 39: 645–649. [DOI] [PubMed] [Google Scholar]
- 4.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet 2008; 40: 316–321. [DOI] [PubMed] [Google Scholar]
- 5.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet 2008; 40: 310–315. [DOI] [PubMed] [Google Scholar]
- 6.Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci U S A 2010; 107: 9742–9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim T, Cui R, Jeon YJ, Lee JH, Lee JH, Sim H et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc Natl Acad Sci U S A 2014; 111: 4173–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer KB, Maia AT, O’Reilly M, Ghoussaini M, Prathalingam R, Porter-Gill P et al. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet 2011; 7: e1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cher ML, MacGrogan D, Bookstein R, Brown JA, Jenkins RB, Jensen RH. Comparative genomic hybridization, allelic imbalance, and fluorescence in situ hybridization on chromosome 8 in prostate cancer. Genes Chromosomes Cancer 1994; 11: 153–162. [DOI] [PubMed] [Google Scholar]
- 10.Visakorpi T, Kallioniemi AH, Syvanen AC, Hyytinen ER, Karhu R, Tammela T et al. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res 1995; 55: 342–347. [PubMed] [Google Scholar]
- 11.Van Den Berg C, Guan XY, Von Hoff D, Jenkins R, Bittner, Griffin C et al. DNA sequence amplification in human prostate cancer identified by chromosome microdissection: potential prognostic implications. Clin Cancer Res 1995; 1: 11–18. [PubMed] [Google Scholar]
- 12.Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014; 512: 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huppi K, Pitt JJ, Wahlberg BM, Caplen NJ. The 8q24 gene desert: an oasis of non-coding transcriptional activity. Front Genet 2012; 3: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck-Engeser GB, Lum AM, Huppi K, Caplen NJ, Wang BB, Wabl M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology 2008; 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huppi K, Volfovsky N, Runfola T, Jones TL, Mackiewicz M, Martin SE et al. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res 2008; 6: 212–221. [DOI] [PubMed] [Google Scholar]
- 16.Sato K, Qian J, Slezak JM, Lieber MM, Bostwick DG, Bergstralh EJ et al. Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. J Natl Cancer Inst 1999; 91: 1574–1580. [DOI] [PubMed] [Google Scholar]
- 17.Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res 2007; 67: 8504–8510. [DOI] [PubMed] [Google Scholar]
- 18.Bawa P, Zackaria S, Verma M, Gupta S, Srivatsan R, Chaudhary B et al. Integrative Analysis of Normal Long Intergenic Non-Coding RNAs in Prostate Cancer. PLoS One 2015; 10: e0122143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho SW, Xu J, Sun R, Mumbach MR, Carter AC, Chen YG et al. Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell 2018; 173: 1398–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The PVT1 Promoter Suppresses MYC Transcription to Reduce Cell Growth. Cancer Discov 2018. [Google Scholar]
- 21.Tang J, Li Y, Sang Y, Yu B, Lv D, Zhang W et al. LncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene 2018. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, Du P, Cui P, Qin Y, Hu C, Wu J et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene 2018. [DOI] [PubMed] [Google Scholar]
- 23.Barsotti AM, Beckerman R, Laptenko O, Huppi K, Caplen NJ, Prives C. p53-Dependent induction of PVT1 and miR-1204. J Biol Chem 2012; 287: 2509–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng X, Cao P, Li J, He D, Han S, Zhou J et al. MiR-1204 sensitizes nasopharyngeal carcinoma cells to paclitaxel both in vitro and in vivo. Cancer Biol Ther 2015; 16: 261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Bi L, Wang Q, Wen M, Li C, Ren Y et al. miR-1204 targets VDR to promotes epithelial-mesenchymal transition and metastasis in breast cancer. Oncogene 2018. [DOI] [PubMed] [Google Scholar]
- 26.Riquelme E, Suraokar MB, Rodriguez J, Mino B, Lin HY, Rice DC et al. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J Thorac Oncol 2014; 9: 998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu G, Liu A, Zhu J, Lei F, Wu S, Zhang X et al. MiR-1207 overexpression promotes cancer stem cell-like traits in ovarian cancer by activating the Wnt/beta-catenin signaling pathway. Oncotarget 2015; 6: 28882–28894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan C, Chen Y, Kong W, Fu L, Liu Y, Yao Q et al. PVT1-derived miR-1207–5p promotes breast cancer cell growth by targeting STAT6. Cancer Sci 2017; 108: 868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das DK, Naidoo M, Ilboudo A, Park JY, Ali T, Krampis K et al. miR-1207–3p regulates the androgen receptor in prostate cancer via FNDC1/fibronectin. Exp Cell Res 2016; 348: 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato H, Minei S, Hachiya T, Yoshida T, Takimoto Y. Fluorescence in situ hybridization analysis of c-myc amplification in stage TNM prostate cancer in Japanese patients. Int J Urol 2006; 13: 761–766. [DOI] [PubMed] [Google Scholar]
- 31.Sotelo J, Esposito D, Duhagon MA, Banfield K, Mehalko J, Liao H et al. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci U S A 2010; 107: 3001–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wasserman NF, Aneas I, Nobrega MA. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res 2010; 20: 1191–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weijerman PC, Konig JJ, Wong ST, Niesters HG, Peehl DM. Lipofection-mediated immortalization of human prostatic epithelial cells of normal and malignant origin using human papillomavirus type 18 DNA. Cancer Res 1994; 54: 5579–5583. [PubMed] [Google Scholar]
- 34.Shi Y, Yang F, Wei S, Xu G. Identification of Key Genes Affecting Results of Hyperthermia in Osteosarcoma Based on Integrative ChIP-Seq/TargetScan Analysis. Med Sci Monit 2017; 23: 2042–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Min H, Yoon S. Got target? Computational methods for microRNA target prediction and their extension. Exp Mol Med 2010; 42: 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015; 43: D146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo W, Lin B, Wang Y, Zhong J, O’Meally R, Cole RN et al. PHD3-mediated prolyl hydroxylation of nonmuscle actin impairs polymerization and cell motility. Mol Biol Cell 2014; 25: 2788–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi H, Zheng B, Wu Y, Tang Y, Wang L, Gao Y et al. Ubiquitin ligase Siah1 promotes the migration and invasion of human glioma cells by regulating HIF-1alpha signaling under hypoxia. Oncol Rep 2015; 33: 1185–1190. [DOI] [PubMed] [Google Scholar]
- 39.Garvalov BK, Foss F, Henze AT, Bethani I, Graf-Hochst S, Singh D et al. PHD3 regulates EGFR internalization and signalling in tumours. Nat Commun 2014; 5: 5577. [DOI] [PubMed] [Google Scholar]
- 40.Henze AT, Garvalov BK, Seidel S, Cuesta AM, Ritter M, Filatova A et al. Loss of PHD3 allows tumours to overcome hypoxic growth inhibition and sustain proliferation through EGFR. Nat Commun 2014; 5: 5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Place TL, Fitzgerald MP, Venkataraman S, Vorrink SU, Case AJ, Teoh ML et al. Aberrant promoter CpG methylation is a mechanism for impaired PHD3 expression in a diverse set of malignant cells. PLoS One 2011; 6: e14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S, Nakamura E, Yang H, Wei W, Linggi MS, Sajan MP et al. Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell 2005; 8: 155–167. [DOI] [PubMed] [Google Scholar]
- 43.Schlisio S Neuronal apoptosis by prolyl hydroxylation: implication in nervous system tumours and the Warburg conundrum. J Cell Mol Med 2009; 13: 4104–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 2009; 8: 579–591. [DOI] [PubMed] [Google Scholar]
- 45.Vergara R, Parada F, Rubio S, Perez FJ. Hypoxia induces H2O2 production and activates antioxidant defence system in grapevine buds through mediation of H2O2 and ethylene. J Exp Bot 2012; 63: 4123–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaelin WG Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 2008; 30: 393–402. [DOI] [PubMed] [Google Scholar]
- 47.Fu J, Taubman MB. EGLN3 inhibition of NF-kappaB is mediated by prolyl hydroxylase-independent inhibition of IkappaB kinase gamma ubiquitination. Mol Cell Biol 2013; 33: 3050–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lipscomb EA, Sarmiere PD, Crowder RJ, Freeman RS. Expression of the SM-20 gene promotes death in nerve growth factor-dependent sympathetic neurons. J Neurochem 1999; 73: 429–432. [DOI] [PubMed] [Google Scholar]
- 49.Madden SL, Galella EA, Riley D, Bertelsen AH, Beaudry GA. Induction of cell growth regulatory genes by p53. Cancer Res 1996; 56: 5384–5390. [PubMed] [Google Scholar]
- 50.Gerber SA, Yatsula B, Maier CL, Sadler TJ, Whittaker LW, Pober JS. Interferon-gamma induces prolyl hydroxylase (PHD)3 through a STAT1-dependent mechanism in human endothelial cells. Arterioscler Thromb Vasc Biol 2009; 29: 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hogel H, Rantanen K, Jokilehto T, Grenman R, Jaakkola PM. Prolyl hydroxylase PHD3 enhances the hypoxic survival and G1 to S transition of carcinoma cells. PLoS One 2011; 6: e27112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hogel H, Miikkulainen P, Bino L, Jaakkola PM. Hypoxia inducible prolyl hydroxylase PHD3 maintains carcinoma cell growth by decreasing the stability of p27. Mol Cancer 2015; 14: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez J, Herrero A, Li S, Rauch N, Quintanilla A, Wynne K et al. PHD3 Regulates p53 Protein Stability by Hydroxylating Proline 359. Cell Rep 2018; 24: 1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang G, Zhang W, Li B, Stringer-Reasor E, Chu C, Sun L et al. MicroRNA-200c and microRNA- 141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast Cancer Res 2017; 19: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu R, Yi B, Wei S, Yang WH, Hart KM, Chauhan P et al. FOXP3-miR-146-NF-kappaB Axis and Therapy for Precancerous Lesions in Prostate. Cancer Res 2015; 75: 1714–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, Liu R, Li W, Chen C, Katoh H, Chen GY et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell 2009; 16: 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang W, Yi B, Wang C, Chen D, Bae S, Wei S et al. Silencing of CD24 Enhances the PRIMA-1-Induced Restoration of Mutant p53 in Prostate Cancer Cells. Clin Cancer Res 2016; 22: 2545–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma L, Chung WK. Quantitative analysis of copy number variants based on real-time LightCycler PCR. Curr Protoc Hum Genet 2014; 80: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo Y, Li B, Zhang G, He Y, Bae JH, Hu F et al. Integrated Oncogenomic Profiling of Copy Numbers and Gene Expression in Lung Adenocarcinomas without EGFR Mutations or ALK Fusion. J Cancer 2018; 9: 1096–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao S, Wang Y, Wang M, Li Z, Zhao Z, Wang RX et al. MicroRNA-155, induced by FOXP3 through transcriptional repression of BRCA1, is associated with tumor initiation in human breast cancer. Oncotarget 2017; 8: 41451–41464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Liu R, Ye P, Wong C, Chen GY, Zhou P et al. Intracellular CD24 disrupts the ARF-NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation. Nat Commun 2015; 6: 5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu R, Liu C, Chen D, Yang WH, Liu X, Liu CG et al. FOXP3 Controls an miR-146/NF-kappaB Negative Feedback Loop That Inhibits Apoptosis in Breast Cancer Cells. Cancer Res 2015; 75: 1703–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu R, Wang L, Chen G, Katoh H, Chen C, Liu Y et al. FOXP3 up-regulates p21 expression by site-specific inhibition of histone deacetylase 2/histone deacetylase 4 association to the locus. Cancer Res 2009; 69: 2252–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014; 30: 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.