Abstract
Purpose-
The purpose of this case-control retrospective discovery study is to identify antigenic bovine pericardium (BP) proteins that stimulate graft-specific humoral immune response in patients implanted with glutaraldehyde fixed bovine pericardial (GFBP) heart valves.
Experimental Design-
Banked serum was collected from age and sex matched patients who had received either a GFBP or mechanical heart valve replacement. Serum IgG was isolated and used to generate poly-polyclonal antibody affinity chromatography columns from each patient. Native and deglycosylated BP protein extracts were separately added to individual patient affinity chromatography columns, with unbound proteins washed through the column. Proteins captured in the affinity chromatography columns were submitted for LC-MS/MS. Differences between GFBP and mechanical heart valve replacement recipients were analyzed with Gaussian linearized modeling.
Results-
Carbohydrate antigens overwhelmed protein capture in the affinity chromatography column, requiring BP protein deglycosylation prior to affinity chromatography. Nineteen BP protein antigens, which stimulated graft-specific IgG production, were identified in patients who received GFBP valve replacements. Identified antigens were significantly over-represented for calcium binding proteins.
Conclusions and Clinical Relevance-
Patients implanted with GFBP valves develop a graft-specific humoral immune response toward BP protein antigens, with 19 specific antigens identified in this work. The molecular functions of over-represented antigens, specifically calcium-binding proteins, may aid in understand the underlying factors that contribute to structural valve deterioration.
Keywords: Antigens, Deglycosylation, Heart Valves, Immunoproteomics, Structural Valve Deterioration
1. Introduction
American Heart Association data indicates that valvular heart disease affects 2.5% of the United States population, resulting in 20,000 deaths and approximately 100,000 heart valve replacements annually [1]. On a global scale, valvular heart disease is estimated to affect over 100 million people with 300,000 replacements performed annually [2]. Heart valve replacements account for 10–20% of all cardiac surgical procedures in the United States [3]. Bovine pericardium (BP) is one of the most commonly utilized heart valve prosthesis leaflet biomaterials, due to its similar function to human valve leaflets [4]. Glutaraldehyde-fixation is utilized to cross-link BP proteins and thereby “mask” antigenic components that would otherwise stimulate a recipient graft-specific adaptive immune response. However, chronic graft-specific adaptive immune responses persist, resulting in leaflet degeneration and calcification [5]. Such structural valve deterioration (SVD) is especially rapid in juvenile patients and young adults (e.g., bicuspid valve replacement patients) due to their increased immune capacity [6]. Consequently, the national heart, lung and blood institute xenotransplantation working group identified xenoantigenicity as the major barrier to expanding use of xenogeneic tissues and organs in clinical practice [7].
Carbohydrate antigens, such as galactose-alpha-1,3-galactose (α-gal), are currently thought to be the most significant immunological hurdle to use of unfixed xenogeneic tissues in heart valve tissue engineering. Humans and old world monkeys exhibit evolutionary loss of the α1,3-galactosyltransferase enzyme necessary for attachment of α-gal to their proteins. This has resulted in an abundance of natural antibodies toward the α-gal epitope [8]. Consequently, a large body of work has been invested into removing or knocking-out α-gal in potential xenogeneic donor species (e.g., porcine). However, α-gal and other carbohydrate antigens (e.g., NeuGc) are far from the only immunogenic components of glutaraldehyde-fixed bovine pericardium (GFBP). Recent advances in immunoproteomic antigen identification, applied to animal models, have demonstrated presence of non-gal antigenic proteins in all subcellular tissue locations of candidate unfixed xenogeneic biomaterials such as BP [4, 9]. Furthermore, valves explanted from human patients with SVD have shown deposition of IgG antibodies, which is independent of α-gal [10]. Antibody deposition has been implicated in leaflet calcification and macrophage infiltration leading to SVD and ultimately valve failure [10]. Finally, previous publications have demonstrated that glutaraldehyde-fixed tissue generates a robust antigen-specific antibody response [11]. However, the specific identities of protein antigens capable of stimulating recipient graft-specific adaptive immune responses in human patients receiving GFBP heart valve prostheses remain largely unknown. Antigen identification is an important step towards furthering our understanding of the mechanisms by which glutaraldehyde-fixed biomaterials degrade, monitoring post-implantation graft-specific immune responses and ultimately to improving their longevity by removing or otherwise modulating the immune response towards specific antigens [12].
We hypothesized that patients implanted with GFBP valves develop a graft-specific humoral immune response toward non-gal BP antigenic proteins. In this manuscript we: (1) Determine IgG response towards both known (e.g., α-gal) and unknown carbohydrate antigens following GFBP valve implantation. (2) Identify specific BP protein antigens responsible for stimulating chronic graft-specific IgG production following GFBP valve replacement. (3) Map those antigens to their respective subcellular locations and molecular functions to identify their potential roles in SVD. This work therefore provides insight into potential mechanisms underlying development of SVD and identifies specific antigens which have potential to serve as biomarkers for monitoring graft-specific immune responses in patients receiving GFBP valves.
2. Methods
All chemicals were purchased from Sigma-Aldrich unless stated otherwise. Each experiment utilized n = 6 patients per group unless otherwise stated.
2.1. Patient enrollment
Patient data was extracted by searching the Mayo Clinic BioBank according to the flow chart (Fig 1). The 56997 patients within the database were searched for ICD code Z95.4, identifying 217 heart valve replacement patients. Eleven patients were manually removed due to meeting the pre-defined exclusion criteria of history of cancer or autoimmune disease. The valve type received was determined for the remaining 206 patients, with 119 patients removed due to use of porcine, homograph or undefined valves. Of the remaining patients, 52 had GFBP valves and 35 had mechanical valves. Patients were first matched by sex, then time with implant was matched within one year, with an average implant time of greater than 10 years. Lastly, patient ages were matched to within two years of each other. Patient serum (n = 6 GFBP, n = 6 mechanical control) was collected via the BioBank at the Mayo Clinic. Healthy control patients were enrolled from the tissue biorepository. All patients were consented and samples collected in accordance with the IRB guidelines at The Mayo Clinic.
Figure 1. Flow Diagram of the Patient Selection Process.
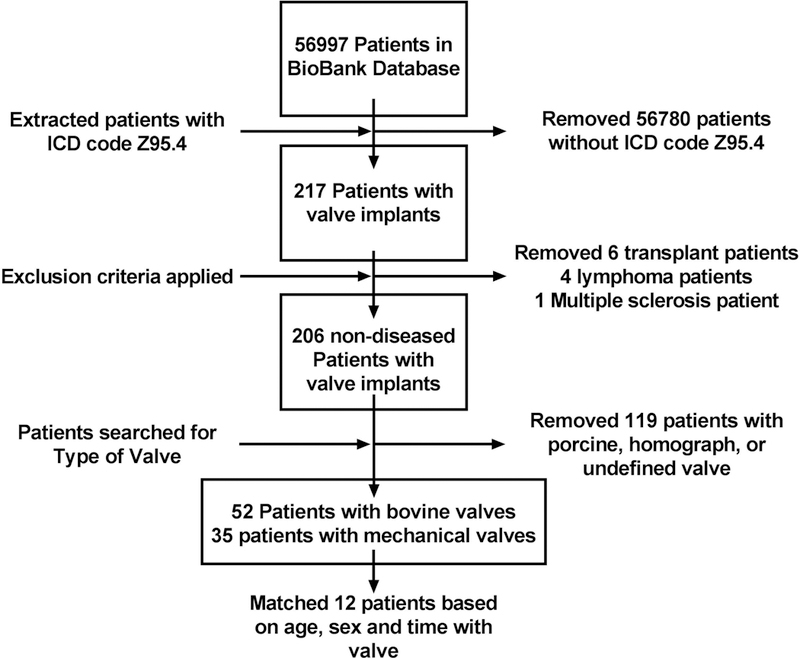
The Mayo Clinic BioBank database was searched for ICD code Z95.4 and patients with a history of transplant, cancer, or autoimmune disease were excluded. The remaining patients were selected for those with mechanical or GFBP valves. The two groups were then matched for sex, time with implant and age within two years.
2.2. Formation of individual patient IgG affinity chromatography columns
Individual patient poly-polyclonal IgG affinity chromatography columns were generated by isolating and crosslinking serum IgG antibodies to Protein G HP SpinTrap columns (GE Healthcare, Pittsburg PA) according to the manufacturer’s recommendations (Fig 2)[9]. Briefly, serum was diluted 1:10 with binding buffer (50 mM Tris, 150 mM NaCl, pH 7.5), and 200 μL was incubated in the column with end-over-end rotation for 30 min. Columns were washed with 400 μL binding buffer, followed by 400 μL of 200 mM triethanolamine and cross-linked with 400 μL of 50 mM Dimethyl pimelimidate dihydrochloride (DMP) in 200 mM triethanolamine for 1 h with end-over-end rotation. Columns were blocked with 400 μL of 100 mM ethanolamine, and unbound antibodies removed with 200 μL of pH 2.9 elution buffer (0.1 M glycine with 2 M urea). Columns were incubated with 200 μL of either hydrophilic or lipophilic protein extract for 1 h with end-over-end rotation. Bound proteins were then eluted using sequential washes with elution buffer at a pH of 5 and 4 for non-specific binding and final specific-binding elution at pH of 2.9. All washes and run-throughs were collected and stored at −80°C for later analysis.
Figure 2. Flow Diagram of Affinity Chromatrography.
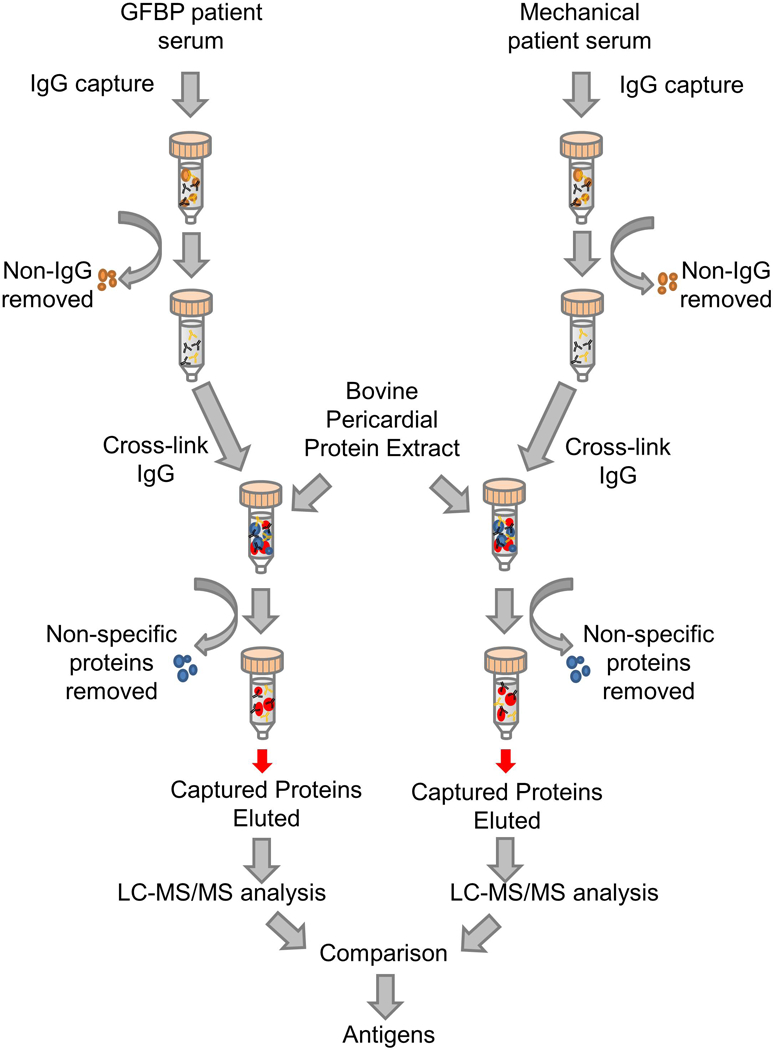
Columns were loaded with serum from patients who had received either a mechanical or GFBP valve. IgG antibodies were captured within the column and non-IgG serum components were washed off the column. The IgG antibodies were crosslinked to the column and then bovine pericardial protein extract was added. Non-specific proteins were washed off the column while captured proteins were eluted separately and submitted for LC-MS/MS analysis. Differences between the two groups were analyzed to determine which proteins were antigenic.
2.3. Native bovine pericardium protein extraction
Protein was extracted from minced bovine pericardium (BP) using a two-step protein extraction process as previously described [13]. Briefly, BP was manually minced and incubated in 1 mL standard extraction solution (10 mM tris–HCl (pH 8.0), 1 mM DTT, 2 mM MgCl2 – 6H2O, 10 mM KCl, 0.5 mM Pefabloc) containing 134 mM NDSB-256. Samples were subjected to mixing at 1,000 rpm in a thermomixter (Eppendorf, Hamburg, Germany), 4°C for 1 h and then centrifuged at 17,000 g, 4°C for 25 min. Supernatant was collected and defined as the hydrophilic protein extract. The insoluble pellet washed twice by resuspension in 1 mL hydrophile extraction solution at 1,400 rpm, 4°C for 30 min and centrifuged at 17,000 g, 4°C for 25 min, with the supernatant discarded. Washed pellets were then incubated in 0.5 mL lipophile extraction solution containing 134 mM NDSB-256 and 1% (w/v) n-dodecyl-β-D-maltoside in standard extraction solution. Samples were subjected to 1,400 rpm, 4°C for 1 h and then centrifuged at 17,000 g, 4°C for 25 min. Supernatant was collected, defined as lipophilic protein extract and stored at −80°C.
2.4. Deglycosylation and α-galactosidase treatment
Hydrophilic and lipophilic protein extracts were deglycosylated using an N- and O-linked deglycosylation kit (Milipore, Burlington, MA) following the non-denaturing protocol for 24 h at 37°C. Deglyclosylated protein extracts were then incubated with 5 U/ml α-Galactosidase for 24 h to remove any residual α-gal, since this known xenoantigen is so ubiquitous.
2.5. Western blot
Eluates from BP proteins captured on IgG affinity chromatography columns (Fig 2) were assessed using one-dimensional SDS-page gels and Western blot as previously described [13]. Briefly, native or deglycosylated BP extracts were loaded onto individual patient IgG affinity chromatography columns. Captured proteins were eluted from the column and run on a 1-DE Nupage gel (Invitrogen), followed by western blotted to nitrocellulose membranes. All blots were probed with the respective individual patient’s post-implant serum (1:100 dilution) and assessed for IgG positivity using peroxidase-conjugated mouse anti-human secondary (1:1000 dilution). All blots were imaged at 1 m exposure time, optical density (OD) of lanes calculated minus background, and compared. All original western blot images are available in the Supporting Information.
2.6. Anti-α-gal ELISA
Anti-α-gal ELISA was modified from a previously described ELISA protocol [14]. 10 μg/mL bovine serum albumin (BSA) linked α-gal (Dextra Laboratories, UK) was used to coat a 96-well plate overnight at 4°C. The plate was blocked for an hour with Pierce Protein-Free Blocking Buffer (ThermoFisher, Waltham, MA) with 0.1% Tween-20 (BioRad, Hercules, CA) to make PFBBT. Bound α-gal was probed with patient serum at a dilution of 1:5000 in PFBBT for 1 h, and positivity was assessed with an HRP conjugated anti-human IgG secondary at a 1:500 dilution in PFBBT for 1 h. Mean absorbance of technical triplicate replicates was calculated for each patient.
2.7. Proteomic analysis
Proteins eluted from individual patient IgG affinity chromatography columns during the specific-binding pH 2.9 elution step were submitted to the Mayo Clinic Proteomics Core and the operator was blinded to the status of each group. Proteins were digested with lysyl endoproteinase Lys-C for 4 h at 25°C, trypsin at 37°C for 20 h, and disulfide bonds reduced with DTT immediately before they were subjected to nanoLC-ESI-MS/MS analysis with an orbitrap mass spectrometer. Tandem mass spectra was extracted and charge state deconvoluted with the database searching software MyriMatch 2.1.138, FreiCore 1.6.209, ProteoWizard 3.0.4140, and ProteoWizard Proteome 3.0.4080. This includes a cRAP database of common laboratory contaminants and an equal number of reverse protein sequences. The fragment mass tolerance was set at 10ppm, searching for trypsin cleavage, with maximum of 3 missed cleavages. The peptide FDR was between 0.66% and 1.54% with the protein FDR between 2.11% and 4.89%. All identified proteins and additional search criteria, including specific bovine sequences utilized to ensure that identified antigens were not human serum contaminants, are included in the supplemental material.
2.8. Statistical analysis
Proteomic data were analyzed at the Mayo Clinic Bioinformatics Core using Gaussian linearized modeling to determine the differential abundance of proteins between patient groups (i.e., GFBP vs. mechanical valve replacement) (Fig. 1). Antigenic proteins were defined as those proteins isolated with statistically greater abundance in IgG affinity chromatography columns generated using serum from GFBP valve replacement patients versus control mechanical valve replacement patients. Western blot OD and ELISA absorbance data were analyzed using paired t-tests in GraphPad. Groups were considered statistically significantly different when p < 0.05.
3. Results
3.1. Carbohydrate antigens overwhelm affinity chromatography capture of protein antigens
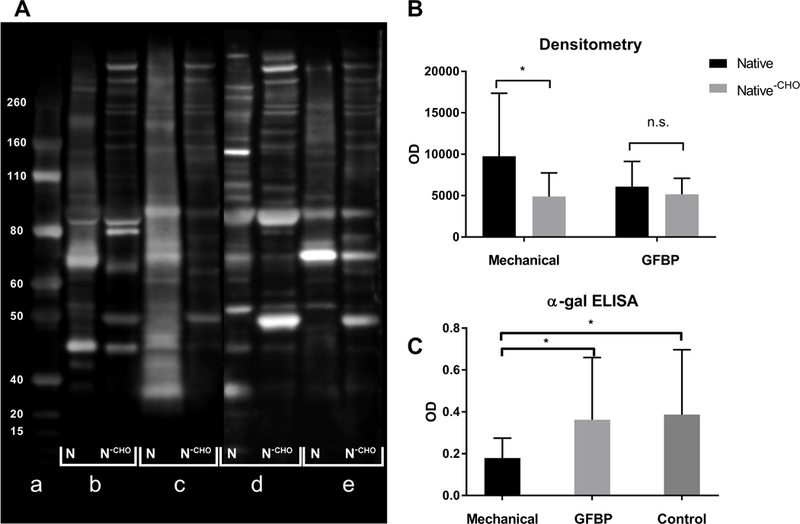
Western blots of native protein extracts captured in affinity chromatography columns, probed with the same patients serum, demonstrated a trend toward higher band intensity in patients implanted with mechanical prostheses than those receiving GFBP valves (9764±7594 vs 6068±3061, p=0.34) (Fig 3B). Eluates from columns loaded with de-glycosylated BP protein extracts resulted in significant reduction of western blots densitometry for mechanical valve patients (9764±7594 vs 4891±2848, p=0.03) whereas GFBP patients showed no statistically significant change (6068±3061 vs 5150±1948, p=0.48) (Fig 3B). Finally, anti-α-gal ELISA showed that mechanical valve patients had significantly higher antibody titers toward α-Gal than GFBP patients (0.362±0.298 vs 0.179±0.09, p=0.03) (Fig 3C). Similarly, healthy control patients had higher anti-α-gal antibody titer than GFBP patients (0.386±0.310 vs 0.179±0.09, p=0.02) (Fig 3C), while there was no difference in anti-α-gal titer between healthy control patients and mechanical valve patients (0.386±0.310 vs 0.362±0.298p > 0.05).
Figure 3. Assessment of Anti-carbohydrate Antibodies in Mechanical Valve and GFBP Valve Patients.
(A) Representative western blot, a: ladder b: native BP protein probed with mechanical valve patient serum c: de-glycosylated BP protein probed with mechanical valve patient serum d: native BP protein probed with GFBP valve patient serum e: de-glycosylated BP protein probed with GFBP valve patient serum. (B) Densitometry analysis of western blots. (C) alpha-gal ELISA for antibodies against the alpha-gal epitope in GFBP valve, Mechanical valve and healthy control patients. All groups have n=6 biological replicates (* p < 0.05).
3.2. GFBP heart valve replacement patients develop graft-specific IgG response towards a greater range of BP antigens than mechanical valve recipients
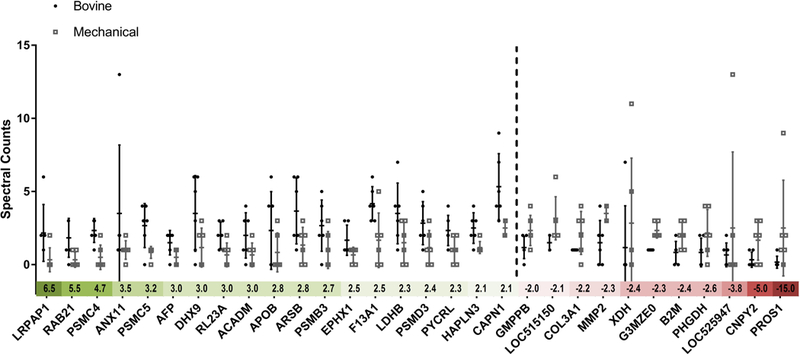
To avoid capture of proteins with carbohydrate post-translational modifications by natural anti-carbohydrate antibodies during IgG affinity chromatography, LC-MS/MS was run with proteins captured from IgG affinity chromatography columns loaded with de-glycosylated and α-Galactosidase treated BP extract. GFBP valve patients had a significant (i.e., ≥ two-fold) increase in abundance of 19 individual proteins compared to mechanical valve patients (p < 0.05) (Fig. 4). Conversely, 11 proteins were found to be significantly increased in abundance in mechanical valve patients compared to GFBP valve patients (Fig. 4). The median fold change in differentially captured protein abundance was 2.8 for GFBP versus only 2.4 for mechanical valve patients. Furthermore, GFBP valve patients had 9 proteins with a greater than 3 fold increase in differential abundance versus only 3 in mechanical valve patients. Finally, the proportion of patients capturing a differentially increased amount of each identified protein was higher for GFBP (i.e., median 86%) than for mechanical valve (i.e., median 66%) patients. Out of the 19 putative GFBP antigens, 13 have literature evidence for their role as antigenic determinants in other diseases (Table 1) [15, 16, 17, 18–22, 23, 24, 25–27]. Conversely, only 5 out of the 11 significant mechanical valve proteins have been previously associated with autoimmune diseases (Table 1) [28–32, 33].
Figure 4. Graphical Representation of Identified Antigen Fold Changes Between Mechanical and GFBP Valve Recipients.
Dotted line differentiates between the BP antigens found to be upregulated in the GFBP group (left) versus the mechanical group (right). The columns represent average spectral counts for each protein with error bars accounting for standard deviation. The colored scale beneath the x-axis indicates the fold change for each protein (green indicating upregulation in GFBP recipients, red in mechanical valve recipients). All groups have n=6 biological replicates and all proteins differences are significant (p < 0.05).
Table 1. Identified Antigens in GFBP Valve and Mechanical Valve Patients.
The table shows the protein name, gene name, accesion number and previous literature evidence of antigenicty is cited in the right hand column. The double black line separates the BP antigens that were found to be significant in GFBP valve patients (top) from those in the mechanical valve patients (bottom).
| Protein Name | Gene Name | Accession Number | Literature Antigenicity |
|---|---|---|---|
| Alpha-2-macroglobulin receptor-associated protein | LRPAP1 | F6RBT3 | [17, 18] |
| Ras-related protein Rab-21 | RAB21 | Q17R06 | |
| 26S protease regulatory subunit 6B | PSMC4 | Q3T030 | [19] |
| Annexin A11 | ANX11 | P27214 | [20–22] |
| 26S protease regulatory subunit 8 | PSMC5 | P62194 | [23] |
| Alpha-fetoprotein | AFP | Q3SZ57 | [24] |
| ATP-dependent RNA helicase A | DHX9 | Q28141 | [25] |
| 60S ribosomal protein L23a | RL23A | Q24JY1 | [26] |
| Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADM | Q3SZB4 | |
| Apolipoprotein B-100 | APOB | E1BNR0 | [27, 28, 35] |
| ARSB protein | ARSB | A6QLZ3 | [29] |
| Proteasome subunit beta type 3 | PSMB3 | P33672 | |
| Epoxide hydrolase 1 | EPHX1 | Q3ZCJ6 | [30, 31] |
| Coagulation factor XIII A chain | F13A1 | F1MW44 | [32] |
| L-lactate dehydrogenase B chain | LDHB | Q5E9B1 | [33] |
| 26S proteasome non-ATPase regulatory subunit 3 | PSMD3 | Q2KJ46 | [34] |
| Pyrroline-5-carboxylate reductase 3 | PYCRL | Q58D08 | |
| Hyaluronan and proteoglycan link protein 3 | HAPLN3 | F6QLF1 | |
| Calpain-1 catalytic subunit | CAPN1 | Q27970 | |
| Mannose-1-phosphate guanyltransferase beta | GMPPB | Q2YDJ9 | |
| Uncharacterized protein | LOC515150 | G3N0S9 | |
| Collagen alpha-1(III) chain | COL3A1 | F1MXS8 | [36] |
| 72 kDa type IV collagenase | MMP2 | F1MKH8 | [37] |
| Xanthine dehydrogenase/oxidase | XDH | F1MUT3 | |
| Uncharacterized protein | G3MZE0 | G3MZE0 | |
| Beta-2-microglobulin | B2M | P01888 | [38] |
| D-3-phosphoglycerate dehydrogenase | PHGDH | Q5EAD2 | [39] |
| Uncharacterized protein | LOC525947 | E1BI82 | |
| Canopy 2 homolog | CNPY2 | Q1LZ72 | |
| Vitamin K-dependent protein S | PROS1 | P07224 | [40–42] |
3.3. Antigens are associated with a wide range of subcellular locations and biological functions
GFBP antigens were identified from a variety of subcellular compartments, specifically, twelve different locations. However, the majority of antigens resided in the cytoplasm (10 antigens), nucleus (5 antigens), proteasome (4 antigens), and secretory (4 antigens) compartments of the cell. The endoplasmic reticulum, endosomes, membranes, golgi apparatus, and cytoskeleton each contained two or three of the identified antigens. One of the membrane antigens was also a transmembrane protein. The rest of the compartments contained only one antigen, which was generally associated with each protein’s more specific function (Fig. 5A). In terms of biological function, the dominant functions of antigenic proteins were hydrolase (5 antigens), metal-binding (5 antigens), calcium-binding (4 antigens), and nucleotide-binding (4 antigens) (Fig. 5B). Compared to the complete bovine proteome all these protein functions are overrepresented. Calcium-binding proteins had the greatest overrepresentation at 4% in the entire proteome versus 21% of identified antigens. Both hydrolase (16% vs 26%) and nucleotide-binding (13% vs 21%) were similarly overrepresented, with metal-binding (19% vs 26%) having the smallest increase in abundance compared to the complete proteome.
Figure 5. Functional Analysis of GBFP Antigens.
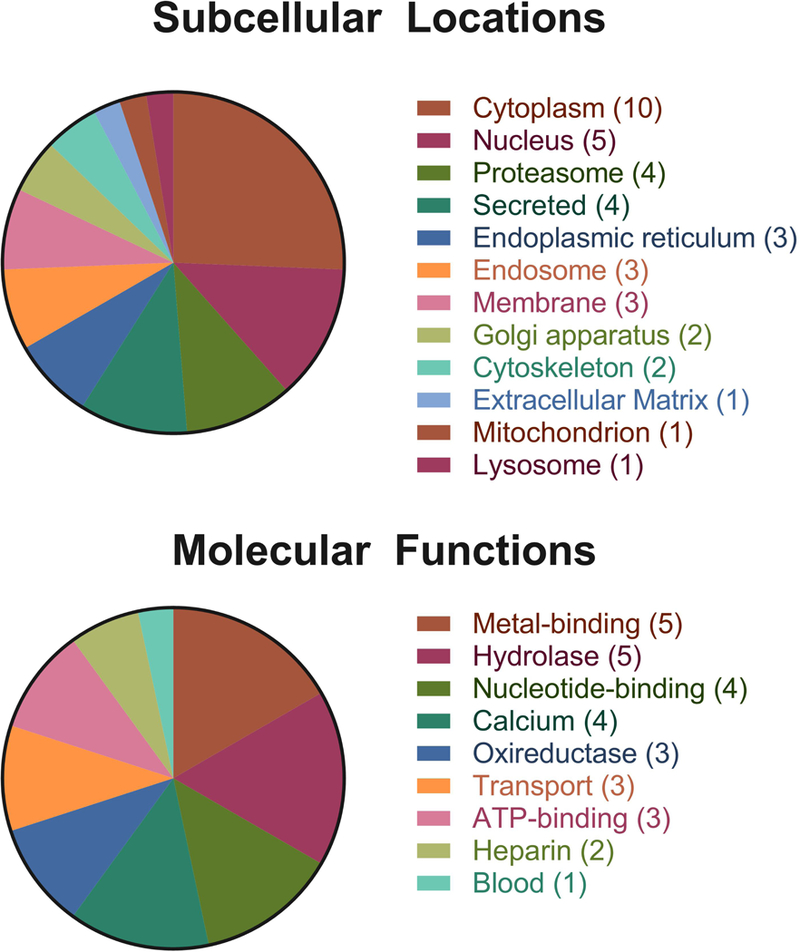
(A) Subcellular locations of GFBP antigens determined by searching UniProt database. (B) Molecular functions of GFBP antigens determined by searching UniProt database.
4. Discussion
Current knowledge about biological heart valve replacements indicates that unknown antigenic components modulate graft-specific adaptive immune activation and progression of SVD. The presented studies demonstrate that GFBP recipient graft-specific humoral immune responses can be leveraged toward identification of antigenic components in current clinically utilized biomaterials (i.e., GFBP). For the first time, the current work identified specific individual BP protein antigens toward which GFBP heart valve recipients develop graft-specific antibodies. Specifically, the results demonstrate that (1) presence of natural antibodies overwhelms detection of antigenic proteins captured using patient specific poly-polyclonal IgG affinity chromatography columns, (2) patients with GFBP valves develop a greater abundance and wider range of graft-specific antibodies toward identified BP protein antigens than patients who receive mechanical valves, and (3) the human immune system is capable of recognizing antigenic proteins from a diverse range of subcellular locations and biological processes despite glutaraldehyde fixation of xenogeneic tissue in GFBP heart valve bioprostheses. The current findings therefore have potential to inform the mechanism, progression and treatment of SVD, as well as informing the development of future BP-based decellularized/antigen removed biomaterials.
Anti-carbohydrate antibodies are more prevalent in mechanical heart valve replacement patients than in those receiving GFBP valves. On initial inspection, the finding that mechanical valve patients have greater response to BP protein extract than GFBP valve patients appears to be counterintuitive, since mechanical heart valve replacement patients should have no incentive to develop an anti-BP graft-specific humoral immune response. However, the protein source in these experiments is native BP and therefore represents a discordant xenogeneic cross-match [8]. Consequently, natural antibodies, directed towards carbohydrate antigens (e.g., α-Gal) represented a potential explanation for these findings [8, 34]. Indeed, the results of deglycosylation experiments and assessment of α-Gal antibody presence support the conclusion that mechanical valve recipients produce a greater amount of anti-BP carbohydrate antigen antibodies than do GFBP recipients. A potential explanation for the higher α-Gal titers in mechanical valve patients could be due to biofilm formation, which is a leading cause of endocarditis in mechanical valve recipients post-implant with a cumulative risk over 5 years of 3.2% to 5.7% [35]. Given that the mean time of implant for mechanical valves was 12.16 ± 3.06 years there is a high cumulative likelihood that these patients could have undergone subclinical chronic exposure to such biofilms. Bacteria express the α-gal epitope on their surface, and therefore exposure to bacterial components in such subclinical biofilms has potential to increase the antibody titer toward this antigen. The alternate hypothesis for the finding of lower anti-carbohydrate antibody presence in GFBP valve recipients is that these patients downregulate production of these natural antibodies. It has been previously demonstrated that even after the fixation process α-Gal epitopes are available on GFBP valve surfaces [36], although explanted valves lack anti-α-Gal antibody deposition [10]. A potential explanation for this previously reported contradictory finding could be that following GFBP valve implantation, patients downregulate anti-α-Gal antibody production. Our western blot findings support this notion as deglycosyation did not significantly decrease GFBP patient antibody binding of BP proteins, indicating that anti-carbohydrate antibodies did not comprise a significant fraction of the anti-BP response in GFBP valve patients. Additionally, healthy control patients had a similar level of anti-α-Gal antibodies to mechanical valve patients, but a significantly higher titer than GFBP valve patients. Consequently, we conclude that GFBP patients’ downregulate production of anti-α-Gal antibodies post-valve replacement. Regardless of underlying pathologic mechanism for the observed differences in anti-carbohydrate antibody presence between GFBP and mechanical valve recipients, for antigen identification experiments deglycosylated samples must be used to overcome the influence of anti-carbohydrate natural antibodies on protein capture in the affinity column.
Removal of the confounding factor of carbohydrate antigens by deglycosylation allowed for individual non-carbohydrate antigenic proteins to be identified in GFBP valve patients. The methodology used to identify xenoantigens has been historically restricted to 2-DE western blots. When investigating non-gal antigens, Boer et al found that a patient implanted with a porcine valve had non-gal antibodies toward the porcine proteins COL6A1 and serum albumin compared to a control patient without a valve implant [37]. However, this work was limited to only one patient due to the difficulties of using 2-DE western blots. The current study utilized a novel, high-throughput poly-polyclonal affinity chromatography method which overcomes the predominant limitations of previous 2-DE immunoproteomic approaches [9]. Many of the antigens identified in the current study have been previously implicated in autoantibody formation in a variety of human diseases [15, 16, 17, 18–22, 23, 24, 25–27]. Furthermore, apolipoprotein B-100 has been previously identified as a bovine xenoantigen in humans implanted with cells cultured in fetal bovine serum [38]. The bovine antigens identified in the current work have had a high degree of sequence homology with their corresponding human proteins, at 89.1%±11.9% via BLAST comparison. Unfortunately, the average sequence homology between human and bovine proteome is currently unknown, and therefore it is not possible to determine if this apparently high sequence homology is truly reflective of high probability of shared epitope regions. However, the possibility that the antigenic epitope for many of the BP antigens identified in the current work are the same as those which stimulate autoantibody production in human diseases is intriguing and requires future epitope mapping studies to resolve.
Analysis of the antigens’ biological function allowed for the deeper examination of potential mechanism which may be involved in development of SVD. A major pitfall in bioprosthetic valves is the calcification of leaflets and subsequent stenosis [39]. The exact mechanism responsible for biological heart valve leaflet calcification is not understood, but significant evidence has implicated both the preservation method (i.e., glutaraldehyde fixation) and residual immunogenicity as being involved in initiation and propagation of the calcification process [40]. Calcium-binding was the most highly overrepresented biological function of antigens identified in GFBP valve recipients. The primary role of these identified calcium-binding proteins is to regulate the amount of free calcium within the cell [41]. Previous studies have documented that antibody binding can interfere with normal protein function [42]. Specifically, in calcium-binding proteins, such antibody binding has potential to induce structural changes preventing calcium from accessing its binding site [43]. The current finding of overrepresentation of calcium-binding proteins in the identified antigens may therefore indicate a potential mechanism for calcium handling dysregulation, initial calcium nidus formation and resultant calcium deposition in SVD. Considering that patients with GFBP valves had elevated levels of antibodies toward calcium binding proteins, this could lead to an imbalance in the normal mechanisms that govern calcium concentrating in the valve leaflet biomaterial. Furthermore, one of the identified antigens (i.e., Hyaluronan and proteoglycan link protein 3) is a matrix protein integral to the process of biomineralization [44]. Antibody-mediated dysfunction of this protein may also therefore contribute to accelerated calcification processes observed in GFBP valve leaflets. Consequently, the current study identified pathways with potential mechanistic links to biomaterial calcification and SVD.
Some antigenic proteins were overrepresented in mechanical valve patients compared to GFBP valve patients, indicating that this control group may develop protein specific antibodies following valve implantation. Given that 5 of the 11 proteins have strong evidence for their involvement in autoimmune disease [28–32, 33], it is possible that mechanical valve patients developed antibodies toward human autoantigens which cross react with the same bovine protein. Indeed, the homology of these 5 proteins with their human counterparts is relatively high at 84.8%±7.9% via BLAST. Intriguingly, the protein that had the greatest antibody response in mechanical valve patients was Vitamin K-dependent protein S, an important cofactor in coagulation. It has been previously demonstrated that patients on chronic anti-coagulant therapy can develop antibodies toward coagulation factors, although the mechanism for this finding is unknown [45]. Furthermore, previous research into thrombin showed that the anti-thrombin antibodies were reactive with both the human and bovine protein [45]. Development of autoantibodies towards self-coagulation factors may therefore explain why the mechanical valve patients in our study were capable of binding the bovine version of Vitamin K-dependent protein S [46]. This hypothesis is further supported by findings from previous studies demonstrating that humans can acquire autoantibodies toward Vitamin K-dependent protein S [32]. Consequently, it is possible that the proteins identified in the control group could in fact be due to an anti-human autoantigen response with cross reactivity to the same protein of bovine origin.
While this paper is the first of its kind, it has distinct limitations. It is a small convenience study and as such has the confounding factors of high baseline biological variability, lack of longitudinal data and potential for unexpected antibody production in the control group. Ideally serum would be collected before and after implantation allowing for a longitudinal study design to reduce the noise associated with a small diverse population. Additionally, the timeframe of this study was aimed at identification of antigens involved in chronic graft-specific immune responses. Recent evidence has shown antibody responses toward GFBP implants may be strongest during the acute phase of the response [14]. Consequently, longitudinal studies incorporating pre-operative, early and late post-operative samples are necessary to define the timeframe and significance of each antigen throughout the implants life expectancy. Although the control group of the present study may not have been as clean as expected, valuable information was generated about this patient cohort. The identification of anti-coagulation autoantibody development in mechanical valve patients has been previously undocumented and warrants future studies to determine its clinical significance. Finally, although our BP protein extraction method has been shown to be superior to harsher sodium dodecyl sulfate-based extraction systems, which interfere with antibody-antigen binding in the affinity chromatography column, as with all proteomic investigations alternative or additional protein extraction steps have potential to identify antigens which were not solubilized by the extraction solutions utilized in this work [9].
The findings of the current study validate the poly-polyclonal affinity chromatography methodology and provides important insights into the antigenic determinants of graft-specific adaptive immune responses in clinical GFBP heart valve replacement recipients. While future studies with prospectively collected samples may discover additional antigens, the fact that 19 antigens were identified in the current study, even with background noise of interpatient variability, indicates the importance of these antigens in stimulating chronic graft-specific immune responses. Several antigens were implicated in the pathogenesis of SVD through calcium regulation. This has potentially far reaching implications in understanding the cause of SVD and requires a deeper exploration into each individual antigens contribution to SVD pathology. Furthermore, the extent to which graft-specific immune response towards individual antigens identified in the current work may be leveraged as biomarkers of SVD remains to be determined. Finally, this study identifies 19 specific BP antigenic components which are recognized by the human immune system and must therefore be removed via decellularization/antigen removal methods in order to overcome graft-specific adaptive immune responses towards next generation xenogeneic heart valve leaflet biomaterials.
Supplementary Material
Statement of clinical relevance.
The National Heart, Lung and Blood Institute states that heart valve replacements are one of the most common cardiovascular procedures. Indeed, 10–20% of all cardiac surgical procedures in the United States are heart valve replacements. Two major types of heart valves have dominated the market, mechanical and biological. While both can replace a diseased valve, neither is capable of truly replicating a fully functioning heart valve without harmful side effects. Bioprosthetic valves are the preferred valve replacement type, due to their superior hemodynamics and avoidance of lifelong anticoagulation. The glutaraldehyde fixation process utilized in bioprosthetic valve production alleviates acute immune system responses towards antigenic tissue components. However, antigen specific chronic immune responses persist ultimately leading to structural valve deterioration (SVD) and failure. One of the most significant immunological hurdles are carbohydrate antigens, such as galactose-alpha-1,3-galactose (α-gal). However, valves explanted from human patients with SVD are also universally strongly positive for deposition of non-α-gal graft-specific IgG antibodies. Currently it is unknown what proteins within biological valves are responsible for eliciting such deleterious graft-specific immune responses. This work utilizes a novel poly-polyclonal affinity chromatography immunoproteomic approach to identify protein antigens which stimulate graft-specific adaptive immune responses in biological heart valve recipients. Identification of such antigenic protein antigens has potential to aid in monitoring of valve deterioration in human subjects. Furthermore, considering the significant interest in generating immunologically-acceptable unfixed biomaterials, understanding which components of xenogeneic tissue are immunogenic and therefore must be removed during biomaterial processing (e.g., decellularization/antigen removal) is critical for generating future unfixed valve bioprostheses.
Acknowledgements
The authors would like to acknowledge funding from the National Institutes of Health (NIH R01HL115205 and T35OD010956–18). Additionally, the authors would like to thank the Proteomics Core at the Mayo Clinic with their funding from the NCI Cancer Center Support Grant 5P30 CA15083–43C1 and Raymond Moore (Bioinformatics Core at Mayo Clinic) for their invaluable guidance in processing and analyzing the proteomic data.
Abbreviations:
- BP
Bovine Pericardium
- SVD
Structural Valve Deterioration
- GFBP
Glutaraldehyde-Fixed Bovine Pericardium
- α-gal
galactose-alpha-1,3-galactose
Footnotes
Disclosures
The authors have no conflicts of interest to declare.
5 References
- [1].Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW, Turner MB, American C Heart Association Statistics, S. Stroke Statistics, Circulation 2015, 131, e29; [DOI] [PubMed] [Google Scholar]; Nkomo VT, Gardin JM, Skelton TN, Gottdiener JS, Scott CG, Enriquez-Sarano M, Lancet 2006, 368, 1005; Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, Carnethon MR, Dai S, de Simone G, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Greenlund KJ, Hailpern SM, Heit JA, Ho PM, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, McDermott MM, Meigs JB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Rosamond WD, Sorlie PD, Stafford RS, Turan TN, Turner MB, Wong ND, Wylie-Rosett J, Circulation 2011, 123, e18. [DOI] [PubMed] [Google Scholar]
- [2].Sun JC, Davidson MJ, Lamy A, Eikelboom JW, Lancet 2009, 374, 565. [DOI] [PubMed] [Google Scholar]
- [3].Maganti K, Rigolin VH, Sarano ME, Bonow RO, Mayo Clin Proc 2010, 85, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Griffiths LG, Choe LH, Reardon KF, Dow SW, Christopher Orton E, Biomaterials 2008, 29, 3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wong ML, Griffiths LG, Acta Biomater 2014, 10, 1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen PC, Sager MS, Zurakowski D, Pigula FA, Baird CW, Mayer JE, del Nido PJ, Emani SM, J Thorac Cardiov Sur 2012, 143, 352. [DOI] [PubMed] [Google Scholar]
- [7].Platt J, DiSesa V, Gail D, Massicot-Fisher J, Circulation 2002, 106, 1043. [DOI] [PubMed] [Google Scholar]
- [8].Cooper DK, Xenotransplantation 1998, 5, 6. [DOI] [PubMed] [Google Scholar]
- [9].Gates KV, Dalgliesh AJ, Griffiths LG, Sci Rep-Uk 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Manji RA, Hara H, Cooper DKC, Xenotransplantation 2015, 22, 406. [DOI] [PubMed] [Google Scholar]
- [11].Human P, Zilla P, Ann Thorac Surg 2001, 71, S385; [DOI] [PubMed] [Google Scholar]; Dahm M, Lyman WD, Schwell AB, Factor SM, Frater RW, J Thorac Cardiovasc Surg 1990, 99, 1082. [PubMed] [Google Scholar]
- [12].Manji RA, Lee W, Cooper DK, Int J Surg 2015. [Google Scholar]
- [13].Wong ML, Wong JL, Athanasiou KA, Griffiths LG, Acta Biomater 2013, 9, 6492. [DOI] [PubMed] [Google Scholar]
- [14].Wong ML, Wong JL, Vapniarsky N, Griffiths LG, Biomaterials 2016, 92, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Strickland DK, Ashcom JD, Williams S, Battey F, Behre E, Mctigue K, Battey JF, Argraves WS, J Biol Chem 1991, 266, 13364; [PubMed] [Google Scholar]; Huang JM, Makker SP, Kidney Int 1995, 47, 432. [DOI] [PubMed] [Google Scholar]
- [16].Molochnikov L, Rabey JM, Dobronevsky E, Bonucelli U, Ceravolo R, Frosini D, Grunblatt E, Riederer P, Jacob C, Aharon-Peretz J, Bashenko Y, Youdim MBH, Mandel SA, Mol Neurodegener 2012, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Iaccarino L, Ghirardello A, Canova M, Zen M, Bettio S, Nalotto L, Punzi L, Doria A, Autoimmunity Reviews 2011, 10, 553; [DOI] [PubMed] [Google Scholar]; Hubers L, Vos H, Schuurman A, Elferink RO, Burgering B, van de Graaf S, Beuers U, J Hepatol 2017, 66, S5; Wang JS, Guo CM, Liu SQ, Qi HB, Yin YL, Liang R, Sun MZ, Greenaway FT, Clin Chim Acta 2014, 431, 164. [Google Scholar]
- [18].Yuan JD, J Immunother Cancer 2016, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Abramsky O, Brenner T, Annals of the New York Academy of Sciences 1983, 417, 108. [DOI] [PubMed] [Google Scholar]
- [20].Valdez BC, Henning D, Busch RK, Woods K, FloresRozas H, Hurwitz J, Perlaky L, Busch H, Nucleic Acids Research 1996, 24, 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ito Y, Hashimoto M, Hirota K, Ohkura N, Morikawa H, Nishikawa H, Tanaka A, Furu M, Ito H, Fujii T, Nomura T, Yamazaki S, Morita A, Vignali DAA, Kappler JW, Matsuda S, Mimori T, Sakaguchi N, Sakaguchi S, Science 2014, 346, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Crowley JT, Drouin EE, Pianta A, Strle K, Wang Q, Costello CE, Steere AC, J Infect Dis 2015, 212, 1841; [DOI] [PMC free article] [PubMed] [Google Scholar]; McLeod O, Silveira A, Fredrikson GN, Gertow K, Baldassarre D, Veglia F, Sennblad B, Strawbridge RJ, Larsson M, Leander K, Gigante B, Kauhanen J, Rauramaa R, Smit AJ, Mannarino E, Giral P, Humphries SE, Tremoli E, de Faire U, Ohrvik J, Nilsson J, Hamsten A, Atherosclerosis 2014, 232, 242; [DOI] [PubMed] [Google Scholar]; Sakamoto N, Tsuji K, Muul LM, Lawler AM, Petricoin EF, Candotti F, Metcalf JA, Tavel JA, Lane HC, Urba WJ, Fox BA, Varki A, Lunney JK, Rosenberg AS, Blood 2007, 110, 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ferla R, Claudiani P, Savarese M, Kozarsky K, Parini R, Scarpa M, Donati MA, Sorge G, Hopwood JJ, Parenti G, Fecarotta S, Nigro V, Sivri HS, Van der Ploeg A, Andria G, Brunetti-Pierri N, Auricchio A, Hum Gene Ther 2015, 26, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Akatsuka T, Kobayashi N, Ishikawa T, Saito T, Shindo M, Yamauchi M, Kurokohchi K, Miyazawa H, Duan HY, Matsunaga T, Komoda T, Morisseau C, Hammock BD, J Autoimmun 2007, 28, 7; [DOI] [PMC free article] [PubMed] [Google Scholar]; Loeper J, De Berardinis V, Moulis Q, Beaune P, Pessayre D, Pompon D, Biological Reactive Intermediates Vi 2001, 500, 121. [DOI] [PubMed] [Google Scholar]
- [25].Lorand L, Velasco PT, Murthy SNP, Lefebvre P, Green D, Blood 1999, 93, 909. [PubMed] [Google Scholar]
- [26].Sabel MS, Liu Y, Griffith KA, He J, Xie X, Lubman DM, Int J Proteomics 2011, 2011, 413742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Parkes JE, Rothwell S, Day PJ, McHugh NJ, Betteridge ZE, Cooper RG, Ollier WE, Chinoy H, Lamb JA, Myositis Genetics C, Arthritis Res Ther 2016, 18, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nakos G, Adams A, Andriopoulos N, Chest 1993, 103, 1051. [DOI] [PubMed] [Google Scholar]
- [29].Helgeland G, Petzold A, Luckman SP, Gilhus NE, Plant GT, Romi FR, Eur Neurol 2011, 65, 53. [DOI] [PubMed] [Google Scholar]
- [30].Falus A, Meretey K, Bohm U, Bozsoky S, Arch Immunol Ther Ex 1979, 27, 647. [PubMed] [Google Scholar]
- [31].Xiang DJ, Xia Q, Chen DX, Feng X, Zhao Y, Liu Y, Liao HY, Liu YM, Li N, Yan HP, Hepatol Res 2011, 41, 867. [DOI] [PubMed] [Google Scholar]
- [32].Boinot C, Planchard D, Giraudeau G, Turhan A, Agius G, Guicheteau M, Thromb Haemostasis 2007, 98, 255; [PubMed] [Google Scholar]; Guermazi S, Hamza M, Dellagi K, Thromb Res 1997, 86, 197. [DOI] [PubMed] [Google Scholar]
- [33].Malnick SDH, Sthoeger ZM, New Engl J Med 1993, 329, 1898. [DOI] [PubMed] [Google Scholar]
- [34].Galili U, Tissue Eng Part B-Re 2015, 21, 231. [DOI] [PubMed] [Google Scholar]
- [35].Litzler PY, Benard L, Barbier-Frebourg N, Vilain S, Jouenne T, Beucher E, Bunel C, Lemeland JF, Bessou JP, J Thorac Cardiovasc Surg 2007, 134, 1025. [DOI] [PubMed] [Google Scholar]
- [36].Naso F, Gandaglia A, Bottio T, Tarzia V, Nottle MB, d’Apice AJF, Cowan PJ, Cozzi E, Galli C, Lagutina I, Lazzari G, Iop L, Spina M, Gerosa G, Xenotransplantation 2013, 20, 252. [DOI] [PubMed] [Google Scholar]
- [37].Boer U, Buettner FFR, Schridde A, Klingenberg M, Sarikouch S, Haverich A, Wilhelmi M, Xenotransplantation 2017, 24. [DOI] [PubMed] [Google Scholar]
- [38].Sakamoto N, Tuji K, Muul LM, Lawler AM, Candotti F, Metcalf JA, Tayel JA, Lane HC, Urba WJ, Fox BA, Varki AP, Lunney JK, Rosenberg AS, J Immunol 2007, 178. [Google Scholar]
- [39].Manji RA, Ekser B, Menkis AH, Cooper DKC, Xenotransplantation 2014, 21, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Brockbank KGM, Song YC, Transplantation 2003, 75, 1133; [DOI] [PubMed] [Google Scholar]; Vesely I, Barber JE, Ratliff NB, Journal of Heart Valve Disease 2001, 10, 471. [PubMed] [Google Scholar]
- [41].Yanez M, Gil-Longo J, Campos-Toimil M, Adv Exp Med Biol 2012, 740, 461. [DOI] [PubMed] [Google Scholar]
- [42].Mian IS, Bradwell AR, Olson AJ, J Mol Biol 1991, 217, 133; [DOI] [PubMed] [Google Scholar]; Shiryaev SA, Remacle AG, Golubkov VS, Ingvarsen S, Porse A, Behrendt N, Cieplak P, Strongin AY, Oncogenesis 2013, 2; Brennan C, Christianson K, Surowy T, Mandecki W, Protein Eng 1994, 7, 509; [DOI] [PMC free article] [PubMed] [Google Scholar]; Wyatt CN, Campbell V, Brodbeck J, Brice NL, Page KM, Berrow NS, Brickley K, Terracciano CMN, Naqvi RU, MacLeod KT, Dolphin AC, J Physiol-London 1997, 502, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Iversen R, Mysling S, Hnida K, Jorgensen TJD, Sollid LM, P Natl Acad Sci USA 2014, 111, 17146; [DOI] [PMC free article] [PubMed] [Google Scholar]; Stearns DJ, Kurosawa S, Sims PJ, Esmon NL, Esmon CT, J Biol Chem 1988, 263, 826. [PubMed] [Google Scholar]
- [44].Rose-Martel M, Smiley S, Hincke MT, J Proteomics 2015, 116, 81. [DOI] [PubMed] [Google Scholar]
- [45].Stricker RB, Lane PK, Leffert JD, Rodgers GM, Shuman MA, Corash L, Blood 1988, 72, 1375. [PubMed] [Google Scholar]
- [46].Stricker RB, Lane PK, Leffert JD, Rodgers GM, Shuman MA, Corash L, Blood 1988, 72, 1375. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


