Abstract
Background
Long noncoding RNA (lncRNA) exerts a potential regulatory role in tumorigenesis. LncRNA NEAT1 expression remains high in osteosarcoma tissues. However, its biological mechanism in osteosarcoma remains unknown.
Methods
In this study, NEAT1 expression in osteosarcoma cells was detected by qRT‐PCR. Proliferative and apoptosis potentials of osteosarcoma cells were determined by CCK‐8 assay and Flow Cytometry, respectively. We identified the potential target of NEAT1 through bioinformatics and dual‐luciferase reporter gene assay. Furthermore, their interaction and functions in regulating the development of osteosarcoma were clarified by Western blot and RIP assay.
Results
Our results demonstrated a high expression of NEAT1 in osteosarcoma tissues and cells. Overexpression of NEAT1 markedly accelerated proliferative and reduced apoptosis potentials of osteosarcoma cells. Besides, NEAT1 could positively regulate the expression of HOXA13 by competing with miR‐34a‐5p.
Conclusion
These results indicated that NEAT1 participated in the development of osteosarcoma as a ceRNA to competitively bind to miR‐34a‐5p and thus mediate HOXA13 expression.
Keywords: apoptosis, long noncoding RNA, osteosarcoma, proliferation
1. INTRODUCTION
Osteosarcoma is common in children and teenagers (Anderson, 2016), whose incidence is second only to lymphoma among malignant tumors in teenagers. Osteosarcoma accounts for about 3%–4% of all pediatric tumors and 30% of malignant bone tumors (Moore & Luu, 2014; Wycislo & Fan, 2015), and it often occurs in the epiphysis and other sites near the knee, proximal tibia, and distal femur (Isakoff, Bielack, Meltzer, & Gorlick, 2015). Many patients with osteosarcoma have varying degrees of lung metastasis when they are first diagnosed (Zhou et al., 2014). If lung metastasis occurs, the 5‐year‐survival rate of these patients will be less than 20% (Friebele, Peck, Pan, Abdel‐Rasoul, & Mayerson, 2015). Currently, the focus of research on osteosarcoma has been extensively and gradually extended to genetic, molecular, and protein levels. For example, Li, Dou, Liu, & He (2017) published a paper Application of Long Noncoding RNAs in Osteosarcoma: Biomarkers and Therapeutic Targets. Zhang et al. (2016) found that the level of long noncoding RNA (lncRNA) MALAT1 in osteosarcoma tissues was higher than that in paired adjacent normal tissues, and it affects the invasion and metastasis abilities of osteosarcoma cells. To further explore the pathogenesis of osteosarcoma, the mechanisms of the occurrence, progression and apoptosis of osteosarcoma were elucidated at the epigenetic level. Our results are of great significance to improve the diagnostic and therapeutic approaches of osteosarcoma.
LncRNAs are noncoding RNAs with more than 200 nucleotides that are capable of regulating gene expressions (Lorenzen & Thum, 2016; Sun, Yang, Xu, & Guo, 2017). They have been widely concerned in recent years due to their complex biological functions. It is reported that certain lncRNAs exert their crucial effects on proliferation, apoptosis, invasion, and infiltration of many types of tumor cells (Chen, Xu, & Zhang, 2017; Mao et al., 2017; Min et al., 2016; Wang et al., 2017). LncRNA Nuclear Enriched Abundant Transcript 1 (NEAT1, NCBI Gene ID: 283131) is an lncRNA with diverse functions in tumorigenesis (Dong et al., 2018; Ghafouri‐Fard & Taheri, 2019; Yang et al., 2017). Previous studies have indicated that NEAT1 participates in the pathogenesis of various diseases, such as nervous system diseases, cardiovascular diseases, and various tumors (Ahmed et al., 2018; Fujimoto et al., 2016; Sunwoo et al., 2017). The promotive role of NEAT1 has been identified in hepatocellular carcinoma (Fang et al., 2017) and ovarian cancer (An, Lv, & Zhang, 2017).
In this study, we examined NEAT1 in osteosarcoma tissues and adjacent noncancerous tissues. Our results verified that NEAT1 was highly expressed in osteosarcoma tissues. The biological role of NEAT1 in the pathological process of osteosarcoma has been pointed out (Li et al., 2018). However, the specific mechanism of NEAT1 involvement in osteosarcoma development still needs to be explored.
2. MATERIALS AND METHODS
2.1. Ethical compliance
The research was approved by the Ethics Committee of The Third Hospital of Hebei Medical University.
2.2. Specimen collection and processing
In this study, 72 pairs of osteosarcoma tissues and matched adjacent normal tissues were collected through surgery and osteosarcoma was confirmed by postoperative pathological examination. Immediately after the fresh tissues from the lesion and the normal tissue adjacent to the tumor were removed by professional physicians using special forceps, these tissues were washed with DEPC, put into the refrigerated tube, and placed in the labeled liquid nitrogen tank for freezing. Subsequently, the clinical data were collected in the department of pathology. Tissue sample collection was approved by the patients and the Ethics Committee. Among these patients, there were 26 females and 46 males aged between 12 and 31, with an average age of 18.3 ± 5.7.
2.3. Cell culture and transfection
Human normal osteoblast cell line hFOB1.19 and osteosarcoma cell lines Saos2, MG63, U2OS, SJSA1, and HOS were purchased from ATCC (Manassas VA). Cells were cultured in DMEM containing 10% FBS (Beyotime, Nantong, China), 100 μg/ml streptomycin, and 100 IU/ml penicillin (Invitrogen, USA), and maintained under 5% CO2 at 37°C. NEAT1 overexpression plasmid, NEAT1 siRNA, miR‐34a‐5p mimics and miR‐34a‐5p inhibitor were all constructed by GenePharma (Shanghai, China). Transfection was performed using Lipofectamine 2000 (Invitrogen, CA).
2.4. RNA extraction and qRT‐PCR
Total RNA was isolated using the TRIzol Reagent (Invitrogen, USA). Using the Reverse Transcription Kit (Takara, Tokyo, Japan), cDNA was obtained from reverse transcription of RNA. Subsequently, an ABI 7900 system (Applied Biosystems, CA) and SYBR Green PCR kit (TaKaRa Biotechnology, Dalian, China) were adopted for qRT‐PCR. With GAPDH as an internal control, the lncRNA expression level was normalized, and 2−ΔCt method was adopted to assess the fold change in lncRNA expression. Primer sequences are displayed below.
NEAT1: F 5′ TCTGTGTGTCAAAGCAAGGC 3′,
R 5′ AGATGCCACTGAATCACCCA 3′,
HOXA13: F: 5′ CTGCCCTATGGCTACTTCGG 3′;
R: 5′ CCGGCGGTATCCATGTACT 3′.
GAPDH: F: 5′ CCGGGAAACTGTGGCGTGATGG 3′,
R: 5′ AGGTGGAGGAGTGGGTGTCGCTGTT 3′,
2.5. Cell proliferation assays
Cells were cultured in 96‐well plates and incubated with CCK‐8 reagent (Beyotime, Nantong, China) for 1 hr. The absorbance at 450 nm was recorded using a TECAN infinite M200 Multimode microplate reader (Tecan, Mechelen, Belgium).
2.6. Cell apoptosis assays
Cells were dyed with Annexin V‐FITC and propidium iodide (PI), and apoptosis was determined by flow cytometry (BD Biosciences, Franklin Lakes, NJ). Apoptosis was assayed through dual‐staining with annexin V‐FITC and propidium iodide (PI; BD Biosciences, Franklin Lakes, NJ). In brief, transfected cells for 48 hr were incubated with Annexin V‐FITC and PI in dark, and subjected to analysis by flow cytometry (BD Biosciences, Franklin Lakes, NJ).
2.7. Subcellular distribution
RNAs in cytoplasm and nucleus were extracted with the PARIS Kit (Life Technologies, USA). Total RNA in each fraction was quantified by qRT‐RCR. GAPDH and U6 were utilized as cytoplasm and nucleus internal references, respectively.
2.8. Dual‐luciferase reporter gene assay
Wild‐type plasmids NEAT1‐WT and HOXA13‐WT, as well as mutant‐type plasmids NEAT1‐MUT and HOXA13‐MUT were constructed. HOS and Saos2 cells seeded into 24‐well plates were co‐transfected with 50 nmol/L miR‐34a‐5p mimics or a negative control and wild‐type or mutant‐type plasmid using Lipofectamine 2000. 5 ng of pRL‐SV40 was added per 80 ng of plasmid. Dual‐luciferase reporter assay kit (Promega, Madison, WI) was used for determining luciferase intensity on a microplate reader.
2.9. RNA‐binding protein immunoprecipitation (RIP)
The Magna Nuclear RIP™ (Native) Nuclear RIP Kit (Millipore, Bedford, MA) was used for RIP assay. Cells were lysed in complete RIPA buffer containing protease inhibitor cocktail and an RNase inhibitor. Cell extract was incubated with RIP buffer containing magnetic beads conjugated to human anti‐AGO2 antibody (Millipore) or IgG control. Immunoprecipitated RNA was obtained from protein digestion. Finally, the purified RNA was quantified by qRT‐PCR. Anti‐NEAT1 used for RIP assay was purchased from Abcam (Cambridge, MA).
2.10. Western blot
Protein samples were extracted and quantified by BCA, separated by SDS‐PAGE gel electrophoresis, and blocked with 5% skim milk. Membranes were then incubated with the primary antibodies (rabbit anti‐human IgG antibodies against HOXA13 and GAPDH) and corresponding secondary antibodies. Bands were exposed, and the color was developed by chemiluminescence.
2.11. Statistical processing
SPSS 20.0 software and GraphPad Prism 6.0 were utilized for statistical analysis. Quantitative data were represented as mean ± SD. Measurement data were analyzed by the t test, whereas data not conformed to the normal distribution were subjected to nonparametric test. The value of p < 0.05 indicated statistical significance.
3. RESULTS
3.1. NEAT1 expression and function in osteosarcoma
NEAT1 expression in osteosarcoma tissues and paired paracancerous tissues was detected by qRT‐PCR. The results showed that NEAT1 was highly expressed in osteosarcoma tissues (Figure 1a). We then examined the relative expression of NEAT1 in osteosarcoma cells (Saos2,MG63, U2OS, SJSA1, and HOS) and human normal osteoblast cells hFOB1.19 by qRT‐PCR. The above results indicate that NEAT1 is highly expressed in osteosarcoma cells, which is consistent with the results found by Wang, Yu, Fan, & Luo (2017). In particular, HOS cells expressed the highest level, while Saos2 cells expressed the lowest level of NEAT1 among the selected osteosarcoma cell lines, which were chosen for the subsequent experiments (Figure 1b). Additionally, the prognostic value of NEAT1 expression was determined for overall survival in osteosarcoma patients by the Kaplan–Meier analysis, suggesting that high expression level of NEAT1 is significantly correlated with shorter overall survival (Figure 1c and d).
Figure 1.
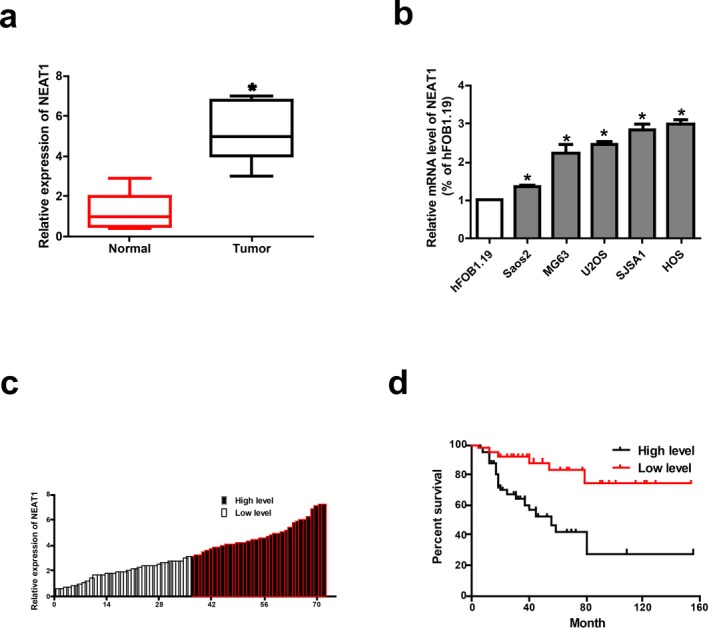
High expression of NEAT1 in osteosarcoma. (a) NEAT1 is highly expressed in osteosarcoma tissues. (b) NEAT1 expression in osteosarcoma cell lines (Saos2, MG63, U2OS, SJSA1, HOS) and human normal osteoblast cell line hFOB1.19 detected by qRT‐PCR. (c) The heights of the columns in the chart represented the log2‐transformed fold changes (osteosarcoma tissues/normal tissues) in NEAT1 expression in 72 patients with osteosarcoma. (d) Correlation between NEAT1 expression and overall survival of patients with osteosarcoma detected by Kaplan–Meier analysis. Data are presented as mean ± SD. *p < 0.05
3.2. NEAT1 facilitates cell proliferation and inhibits apoptosis signaling
After knock‐down of NEAT1 in HOS cell line and overexpression of NEAT1 in Saos2 cell line, we calculate their transfection efficiency (Figure 2a). Then, CCK‐8 assay indicated that NEAT1 downregulation markedly decreased the proliferative ability of osteosarcoma cells. NEAT1 overexpression accelerated the proliferative rate of osteosarcoma cells (Figure 2b). In addition, cell apoptosis experiment showed that overexpression of NEAT1 reduced the apoptosis capacity of osteosarcoma cells, and NEAT1 downregulation induced the apoptosis capacity of osteosarcoma cells (Figure 2c and d). Taken together, these results indicate that NEAT1 may exert regulatory effects on apoptosis and proliferative potentials of osteosarcoma cells.
Figure 2.
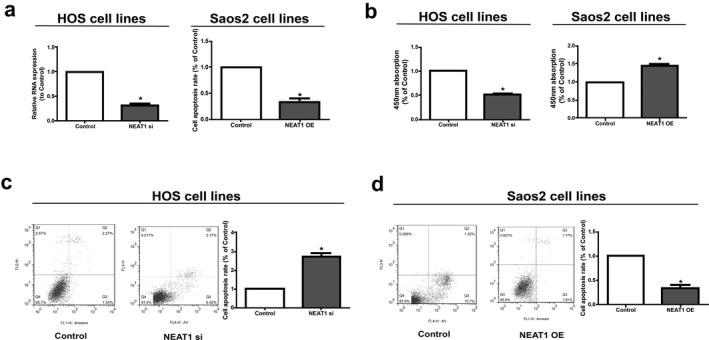
Regulatory effect of NEAT1 on proliferation and apoptosis of osteosarcoma cells. (a) After knock‐down of NEAT1 in HOS cell line and overexpression of NEAT1 in Saos2 cell line, we calculate their transfection efficiency. (b) CCK‐8 assay showed proliferation of HOS transfected with NEAT1 siRNA and Saos2 cells transfected with NEAT1 overexpression vector. (c and d) Cell apoptosis detected by BD Biosciences FACS Calibur Flow Cytometry. Data are presented as mean ± SD. *p < 0.05
3.3. Subcellular distribution of NEAT1
Subcellular distribution of lncRNA determines its biological function. To confirm the cellular localization of NEAT1, we isolated osteosarcoma cells into cytoplasmic and nuclear fractions, with GAPDH and U6 as controls, respectively. QRT‐PCR results showed that 69.5% and 71.05% of NEAT1 were distributed in the cytoplasmic fraction of HOS and Saos2 cells, respectively (Figure 3a). We may conclude that NEAT1 participates in the development of osteosarcoma through post‐transcriptional regulation.
Figure 3.
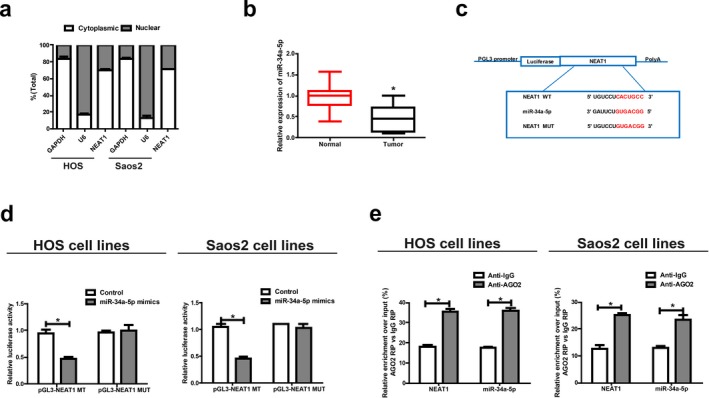
NEAT1 directly interacts with miR‐34a‐5p. (a) Cytoplasmic and nuclear levels of NEAT1 in HOS and Saos2 cells analyzed by qRT‐PCR. (b) MiR‐34a‐5p expression in osteosarcoma tissues. (c) Bioinformatics evidence of binding of miR‐34a‐5p onto 3′‐UTR of NEAT1. (d) Dual‐luciferase reporter gene assay in HOS and Saos2 cells after transfection with negative control or miR‐34a‐5p mimics, renilla luciferase vector pRL‐SV40 and the reporter constructs. (e) RIP experiments for the amount of NEAT1 and miR‐34a‐5p in HOS and Saos2 cells. Data are presented as mean ± SD. *p < 0.05
3.4. NEAT1 is targeted by miR‐34a‐5p
Given that NEAT1 was primarily located in the cytoplasmic fraction, we hypothesized that NEAT1 may act as a ceRNA in the development of osteosarcoma. QRT‐PCR data revealed that miR‐34a‐5p expression was lower in osteosarcoma tissues, which was contrary to the expression trend of NEAT1 (Figure 3b). Through RegRNA, Starbase prediction, we found that sequences in miR‐34a‐5p that were highly matched to NEAT1 3′UTR. Based on these binding sequences, we constructed pGL3‐NEAT1‐WT and pGL3‐NEAT1‐MUT (Figure 3c). Luciferase activity was obviously downregulated in HOS and Saos2 cells co‐transfected with NEAT1 WT and miR‐34a‐5p mimics, while it did not change after transfection with NEAT1 MUT (Figure 3d). RIP analysis was carried out to elucidate whether NEAT1 was involved in RNA‐containing ribonucleoprotein complex. QRT‐PCR results showed that NEAT1 was enriched in anti‐AGO2 antibody compared with in controls. Similar results were yielded in miR‐34a‐5p (Figure 3e). The above results imply that miR‐34a‐5p can bind to NEAT1 in vitro.
3.5. NEAT1 regulates HOXA13, the target gene of miR‐34a‐5p
To investigate the potential role of miR‐34a‐5p in the development of osteosarcoma, we screened out the target genes of miR‐34a‐5p by bioinformatics prediction (TargetScan, Starbase, RegRNA). Finally, HOXA13 was selected for further analyses. After construction of luciferase plasmids pGL3‐ HOXA13‐WT and pGL3‐ HOXA13‐MUT, they were cotransfected with miR‐34a‐5p mimics or NC in HOS and Saos2 cells, respectively (Figure 4a). Luciferase activity of the WT reporter was inhibited, while MUT reporter group did not change (Figure 4b). The above results indicate that HOXA13 is a potential target gene of miR‐34a‐5p. Subsequently, HOXA13 expression in osteosarcoma tissues and paired paracancerous tissues was determined by qRT‐PCR. The mRNA levels of HOXA13 were remarkably elevated in osteosarcoma tissues compared to paracancerous tissues (Figure 4c). Western blot analysis revealed the same result at the protein level (Figure 4d).
Figure 4.
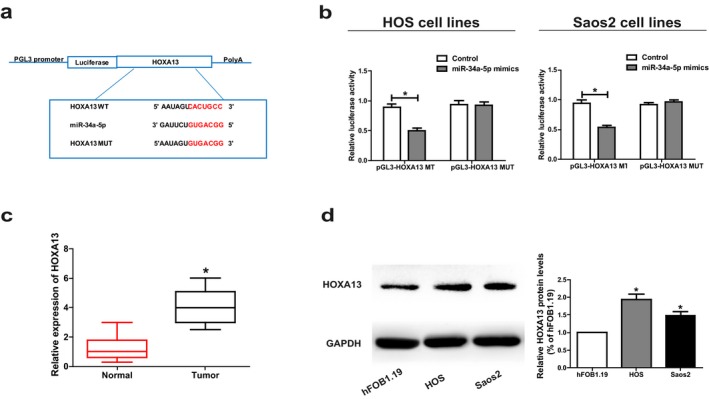
HOXA13 is the direct target of miR‐34a‐5p. (a) The putative miRNA‐binding sites in the HOXA13 sequence. (b) Direct target sites confirmed by dual‐luciferase reporter gene assay. (c) HOXA13 expression in osteosarcoma tissues. (d) Protein level of HOXA13 in hFOB1.19, HOS and Saos2 cell lines detected by Western Blot. Data are presented as mean ± SD. *p < 0.05
To elucidate whether NEAT1 regulated HOXA13 expression via targeting miR‐34a‐5p, we detected expression levels of HOXA13 in osteosarcoma cells after altering NEAT1 or miR‐34a‐5p expressions. Transfection with miR‐34a‐5p inhibitor in HOS cells upregulated HOXA13 expression, which was reversed by cotransfection with miR‐34a‐5p inhibitor and NEAT1 siRNA (Figure 5a and b). Furthermore, transfection with miR‐34a‐5p mimics in Saos2 cells inhibited HOXA13 expression, which was reversed by cotransfection with miR‐34a‐5p mimics and NEAT1 overexpression plasmid (Figure 5c and d). Subsequently, Saos2 cells were transfected with NEAT1 overexpression plasmid and its mutant overexpression plasmid, followed by the determination of HOXA13 expression. Both qRT‐PCR and Western blot results showed that overexpression of wild‐type NEAT1 upregulated HOXA13 expression in osteosarcoma cells, whereas mutant‐type NEAT1 did not disrupt base pairing between NEAT1 and miR‐34a‐5p (Figure 5e and f). To sum up, our findings confirm that NEAT1 positively regulates the expression of HOXA13 by directly binding to miR‐34a‐5p.
Figure 5.
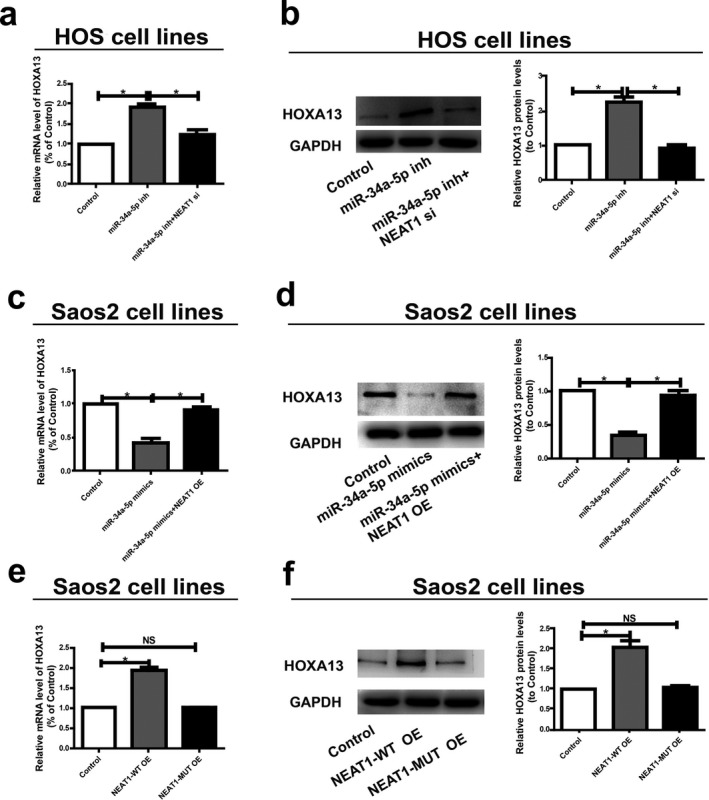
NEAT1/miR‐34a‐5p axis is critical for the expression of HOXA13. (a) MiR‐34a‐5p inhibitor with or without NEAT1 siRNA is transfected into HOS cells and the mRNA level of HOXA13 is evaluated by qRT‐PCR. (b) Western blot analysis of HOXA13 protein level following treatment of HOS cells with miR‐34a‐5p inhibitor or NEAT1 siRNA. GAPDH is used as control. (c) Saos2 cells are transfected with miR‐34a‐5p with or without NEAT1 overexpress plasmid and qRT‐PCR is applied to detect the relative mRNA levels of HOXA13 compared with controls. (d) Relative protein level of HOXA13 when transfected with miR‐34a‐5p mimics and reversed by NEAT1 expression plasmid. (e) Relative mRNA level of HOXA13 when transfected with NEAT1‐WT overexpression plasmid or NEAT1‐MUT overexpression plasmid. (f) Relative protein level of HOXA13 when transfected with NEAT1‐WT overexpression plasmid or NEAT1‐MUT overexpression plasmid. Data are presented as mean ± SD. *p < 0.05, ns, no significantly difference
3.6. NEAT1/miR‐34a‐5p axis regulates behaviors of osteosarcoma cells
We next explored whether miR‐34a‐5p could affect proliferative and apoptosis potentials of HOS and Saos2 cells. Downregulation of miR‐34a‐5p in HOS cells markedly promoted proliferative and inhibited apoptosis potentials compared to controls, which were partially reversed by cotransfection with miR‐34a‐5p inhibitor and NEAT1 siRNA (Figure 6a and c). In addition, overexpression of miR‐34a‐5p inhibited proliferative and promoted apoptosis potentials of Saos2 cells, and were partially reversed by NEAT1 overexpression (Figure 6b and d). Based on the above results, NEAT1/miR‐34a‐5p axis exerts great effects on regulating behaviors of osteosarcoma cells.
Figure 6.
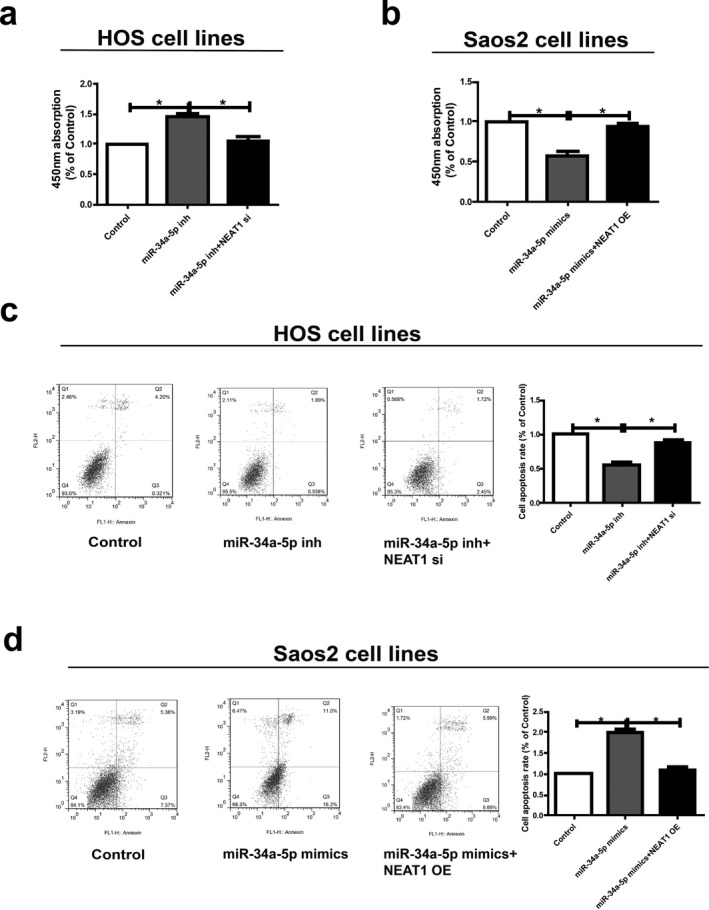
NEAT1 regulates cell function through miR‐34a‐5p. (a and b) Determination of the proliferation of HOS and Saos2 cells via CCK‐8 assay. (c and d) The apoptosis ability with respect to changes of HOS and Saos2 cell lines after different transfection. Data are presented as mean ± SD. *p < 0.05
4. DISCUSSION
Osteosarcoma is ranked among the leading causes of cancer‐related death in the pediatric age group. The cancer's low prevalence and its large tumor heterogeneity make it difficult to obtain meaningful progress in patient's survival (Botter, Neri, & Fuchs, 2014). LncRNA has been shown to be a regulator of various cellular processes. Dysregulated lncRNAs have been identified to be related to the disease development (Yu, Chuang, & Kuo, 2018). NEAT1 has been confirmed to participate in cell proliferation (Wang, Wang, Zhang, Deng, & Long, 2017; Zhu et al., 2019), so we hypothesized that NEAT1 may be involved in the pathogenesis of osteosarcoma.
Our study showed a higher expression of NEAT1 in osteosarcoma cells relative to normal osteoblast cells. In addition, downregulation of NEAT1 expression remarkably reduced proliferative and induced apoptosis capacities, suggesting that NEAT1 was an important regulator in the growth of osteosarcoma cells as an oncogene. Therefore, explorations on the effect of NEAT1 on accelerating growth of osteosarcoma cells are of great significance for in‐depth studies of the occurrence and development of osteosarcoma.
Through separation of cytoplasm and nucleus, we confirmed that NEAT1 was mainly distributed in cell cytoplasm, indicating that NEAT1 may serve as a ceRNA. Subsequently, RIP and dual‐luciferase reporter gene assay clarified that NEAT1 could bind to miR‐34a‐5p. So far, miR‐34a‐5p has been proved to be lowly expressed in ovarian cancer and glioma (Ding, Wu, Tao, & Peng, 2017; Xu et al., 2018). Our study demonstrated that miR‐34a‐5p was downregulated in osteosarcoma cells, and transfection with miR‐34a‐5p mimics promoted apoptosis and suppressed proliferative capacities of osteosarcoma cells, which could be reversed by NEAT1 overexpression. We believed that both NEAT1 and miR‐34a‐5p may participate in the development and progression of osteosarcoma.
HOX genes play a fundamental role in the development of the vertebrate central nervous system, axial skeleton, limbs, gut, urogenital tract and external genitalia, but mutations in two of the 39 human HOX genes (HOXD13 mutated in synpolydactyly, and HOXA13 mutated in hand‐foot‐genital syndrome) have been shown to cause congenital malformations (Goodman & Scambler, 2001; Grier et al., 2005). Studies have shown that HOXA13 exerts an important regulatory effect on organ development, cell differentiation, and tumorigenesis (Luo, Rhie, Lay, & Farnham, 2017). Quagliata et al. (2018) declared that high expression of HOXA13 is correlated with poorly differentiated hepatocellular carcinomas and modulates sorafenib response in in vitro models. Hu, Chen, Cheng, Li, & Zhang (2017) suggested that dysregulated expression of homebox gene HOXA13 is related to the poor prognosis in bladder cancer. Accordingly, our study verified that upregulated NEAT1 increased expression of HOXA13, the target gene of miR‐34a‐5p, further leading to abnormal proliferation and apoptosis of osteosarcoma cells.
To sum up, NEAT1 functioned as a competitive endogenous RNA to regulate HOXA13 expression by sponging miR‐34a‐5p, thus regulating the development of osteosarcoma.
CONFLICT OF INTEREST
None.
Ji S, Wang S, Zhao X, Lv L. Long noncoding RNA NEAT1 regulates the development of osteosarcoma through sponging miR‐34a‐5p to mediate HOXA13 expression as a competitive endogenous RNA. Mol Genet Genomic Med. 2019;7:e673 10.1002/mgg3.673
REFERENCES
- Ahmed, A. S. I. , Dong, K. , Liu, J. , Wen, T. , Yu, L. , Xu, F. , … Zhang, W. (2018). Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells. Proceedings of the National Academy of Sciences of the United States of America, 115, E8660–E8667. 10.1073/pnas.1803725115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, J. , Lv, W. , & Zhang, Y. (2017). LncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR‐194. OncoTargets and Therapy, 10, 5377–5390. 10.2147/OTT [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M. E. (2016). Update on survival in Osteosarcoma. Orthopedic Clinics of North America, 47, 283–292. 10.1016/j.ocl.2015.08.022 [DOI] [PubMed] [Google Scholar]
- Botter, S. M. , Neri, D. , & Fuchs, B. (2014). Recent advances in osteosarcoma. Current Opinion in Pharmacology, 16, 15–23. 10.1016/j.coph.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Xu, D. , & Zhang, T. (2017). Inhibition of proliferation and invasion of hepatocellular carcinoma cells by lncRNA‐ASLNC02525 silencing and the mechanism. International Journal of Oncology, 51, 851–858. 10.3892/ijo.2017.4069 [DOI] [PubMed] [Google Scholar]
- Ding, N. , Wu, H. , Tao, T. , & Peng, E. (2017). NEAT1 regulates cell proliferation and apoptosis of ovarian cancer by miR‐34a‐5p/BCL2. Onco Targets Therapy, 10, 4905–4915. 10.2147/OTT [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, P. , Xiong, Y. , Yue, J. , Hanley, S. J. B. , Kobayashi, N. , Todo, Y. , & Watari, H. (2018). Long non‐coding RNA NEAT1: A novel target for diagnosis and therapy in human tumors. Frontier in Genetics, 9, 471 10.3389/fgene.2018.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, L. , Sun, J. , Pan, Z. , Song, Y. , Zhong, L. , Zhang, Y. , … Huang, P. (2017). Long non‐coding RNA NEAT1 promotes hepatocellular carcinoma cell proliferation through the regulation of miR‐129‐5p‐VCP‐IkappaB. American Journal of Physiology. Gastrointestinal and Liver Physiology, 313, G150–G156. 10.1152/ajpgi.00426.2016 [DOI] [PubMed] [Google Scholar]
- Friebele, J. C. , Peck, J. , Pan, X. , Abdel‐Rasoul, M. , & Mayerson, J. L. (2015). Osteosarcoma: A meta‐analysis and review of the literature. American Journal of Orthopedics, 44, 547–553. [PubMed] [Google Scholar]
- Fujimoto, A. , Furuta, M. , Totoki, Y. , Tsunoda, T. , Kato, M. , Shiraishi, Y. , … Wardell, C. P. (2016). Whole‐genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nature Genetics, 48, 500–509. 10.1038/ng.3547 [DOI] [PubMed] [Google Scholar]
- Ghafouri‐Fard, S. , & Taheri, M. (2019). Nuclear Enriched Abundant Transcript 1 (NEAT1): A long non‐coding RNA with diverse functions in tumorigenesis. Biomedicine & Pharmacotherapy, 111, 51–59. 10.1016/j.biopha.2018.12.070 [DOI] [PubMed] [Google Scholar]
- Goodman, F. R. , & Scambler, P. J. (2001). Human HOX gene mutations. Clinical Genetics, 59, 1–11. 10.1034/j.1399-0004.2001.590101.x [DOI] [PubMed] [Google Scholar]
- Grier, D. G. , Thompson, A. , Kwasniewska, A. , McGonigle, G. J. , Halliday, H. L. , & Lappin, T. R. (2005). The pathophysiology of HOX genes and their role in cancer. The Journal of Pathology, 205, 154–171. 10.1002/(ISSN)1096-9896 [DOI] [PubMed] [Google Scholar]
- Hu, H. , Chen, Y. , Cheng, S. , Li, G. , & Zhang, Z. (2017). Dysregulated expression of homebox gene HOXA13 is correlated with the poor prognosis in bladder cancer. Wiener Klinische Wochenschrift, 129, 391–397. 10.1007/s00508-016-1108-4 [DOI] [PubMed] [Google Scholar]
- Isakoff, M. S. , Bielack, S. S. , Meltzer, P. , & Gorlick, R. (2015). Osteosarcoma: Current treatment and a collaborative pathway to success. Journal of Clinical Oncology, 33, 3029–3035. 10.1200/JCO.2014.59.4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Dou, P. , Liu, T. , & He, S. (2017). Application of long noncoding RNAs in osteosarcoma: Biomarkers and therapeutic targets. Cellular Physiology and Biochemistry, 42, 1407–1419. 10.1159/000479205 [DOI] [PubMed] [Google Scholar]
- Li, P. , Huang, R. , Huang, T. , Cheng, S. , Chen, Y. , & Wang, Z. (2018). Long non‐coding RNA NEAT1 promotes proliferation, migration and invasion of human osteosarcoma cells. International Journal of Medical Sciences, 15, 1227–1234. 10.7150/ijms.25662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen, J. M. , & Thum, T. (2016). Long noncoding RNAs in kidney and cardiovascular diseases. Nature Reviews Nephrology, 12, 360–373. 10.1038/nrneph.2016.51 [DOI] [PubMed] [Google Scholar]
- Luo, Z. , Rhie, S. K. , Lay, F. D. , & Farnham, P. J. (2017). A prostate cancer risk element functions as a repressive loop that regulates HOXA13. Cell Reports, 21, 1411–1417. 10.1016/j.celrep.2017.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Z. , Li, H. , Du, B. , Cui, K. , Xing, Y. , Zhao, X. , & Zai, S. (2017). LncRNA DANCR promotes migration and invasion through suppression of lncRNA‐LET in gastric cancer cells. Bioscience Reports, 37. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Min, L. , Hong, S. , Duan, H. , Zhou, Y. , Zhang, W. , Luo, Y. , … Tu, C. (2016). Antidifferentiation noncoding RNA regulates the proliferation of osteosarcoma cells. Cancer Biotherapy and Radiopharmaceuticals, 31, 52–57. 10.1089/cbr.2015.1888 [DOI] [PubMed] [Google Scholar]
- Moore, D. D. , & Luu, H. H. (2014). Osteosarcoma. Cancer Treatment and Research, 162, 65–92. 10.1007/978-3-319-07323-1 [DOI] [PubMed] [Google Scholar]
- Quagliata, L. , Quintavalle, C. , Lanzafame, M. , Matter, M. S. , Novello, C. , di Tommaso, L. , … Piscuoglio, S. (2018). High expression of HOXA13 correlates with poorly differentiated hepatocellular carcinomas and modulates sorafenib response in in vitro models. Laboratory Investigation, 98, 95–105. 10.1038/labinvest.2017.107 [DOI] [PubMed] [Google Scholar]
- Sun, W. , Yang, Y. , Xu, C. , & Guo, J. (2017). Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genetics, 216, 105–110. 10.1016/j.cancergen.2017.06.003 [DOI] [PubMed] [Google Scholar]
- Sunwoo, J. S. , Lee, S. T. , Im, W. , Lee, M. , Byun, J. I. , Jung, K. H. , … Kim, M. (2017). Altered expression of the long noncoding RNA NEAT1 in Huntington's disease. Molecular Neurobiology, 54, 1577–1586. 10.1007/s12035-016-9928-9 [DOI] [PubMed] [Google Scholar]
- Wang, M. W. , Liu, J. , Liu, Q. , Xu, Q. H. , Li, T. F. , Jin, S. , & Xia, T. S. (2017). LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. European Review for Medical and Pharmacological Sciences, 21, 4613–4622. [PubMed] [Google Scholar]
- Wang, Q. , Wang, W. , Zhang, F. , Deng, Y. , & Long, Z. (2017). NEAT1/miR‐181c regulates osteopontin (OPN)‐mediated synoviocyte proliferation in osteoarthritis. Journal of Cellular Biochemistry, 118, 3775–3784. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Yu, Y. , Fan, S. , & Luo, L. (2017). Knockdown of long non‐coding RNA NEAT1 inhibits proliferation and invasion and induces apoptosis of osteosarcoma by inhibiting miR‐194 expression. Yonsei Medical Journal, 58, 1092–1100. 10.3349/ymj.2017.58.6.1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wycislo, K. L. , & Fan, T. M. (2015). The immunotherapy of canine osteosarcoma: A historical and systematic review. Journal of Veterinary Internal Medicine, 29, 759–769. 10.1111/jvim.12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Zhang, Y. , Qi, L. , Ding, L. , Jiang, H. , & Yu, H. (2018). NFIX circular RNA promotes glioma progression by regulating miR‐34a‐5p via notch signaling pathway. Frontiers in Molecular Neuroscience, 11, 225 10.3389/fnmol.2018.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. , Li, Z. , Li, Y. , Xu, R. , Wang, Y. , Tian, Y. , & Chen, W. (2017). Long non‐coding RNA NEAT1 overexpression is associated with poor prognosis in cancer patients: A systematic review and meta‐analysis. Oncotarget, 8, 2672–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C. Y. , Chuang, C. Y. , & Kuo, H. C. (2018). Trans‐spliced long non‐coding RNA: An emerging regulator of pluripotency. Cellular and Molecular Life Sciences, 75, 3339–3351. 10.1007/s00018-018-2862-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. H. , Kang, X. H. , Lu, P. , Miao, Z. H. , Sun, G. S. , Cao, X. J. , & Cao, F. (2016). Expression of long non‐coding RNA MALAT1 in osteosarcoma and its effect on invasiveness and metastatic potential of osteosarcoma cells. Zhonghua Bing Li Xue Za Zhi, 45, 561–565. [DOI] [PubMed] [Google Scholar]
- Zhou, W. , Hao, M. , Du, X. , Chen, K. , Wang, G. , & Yang, J. (2014). Advances in targeted therapy for osteosarcoma. Discovery Medicine, 17, 301–307. [PubMed] [Google Scholar]
- Zhu, L. , Yang, N. , Li, C. , Liu, G. , Pan, W. , & Li, X. (2019). Long noncoding RNA NEAT1 promotes cell proliferation, migration, and invasion in hepatocellular carcinoma through interacting with miR‐384. Journal of Cellular Biochemistry, 120, 1997–2006. 10.1002/jcb.27499 [DOI] [PMC free article] [PubMed] [Google Scholar]


