Abstract
Background
Colorectal cancer is the third most common cancer worldwide. Recently, an increasing number of evidences suggest that genetic susceptibility plays an important role in the occurrence of colorectal cancer. This study aimed to better understand the influence of MIR17HG polymorphisms on colorectal cancer susceptibility in the Chinese Han population.
Methods
We recruited 514 patients with colorectal cancer and 510 healthy controls to investigate the association between polymorphisms of MIR17HG and risk of colorectal cancer in the Chinese Han population. Genotyping was performed with the Agena MassARRAY platform. We used the χ 2 test to compare the distributions of single nucleotide polymorphisms (SNPs) allele and genotypes frequencies between cases and controls. Odds ratios and 95% confidence intervals were calculated by logistic regression analysis to evaluate the association under genetic models. Linkage disequilibrium between the five SNPs was assessed using the Haploview software.
Results
Overall analysis found that rs7336610 and rs1428 and haplotype CTAGA were significantly associated with increased risk of colorectal cancer. However, we found rs7318578 was associated with a decreased risk of colorectal cancer in the dominant model. Stratification analysis showed that rs7336610, rs7318578, and rs1428 were also associated with rectal cancer risk. Gender stratification analysis found that rs7336610, rs7318578, rs17735387, and rs1428 were significantly associated with colorectal cancer risk in males.
Conclusion
In conclusion, this study indicated that the polymorphisms of MIR17HG were associated with colorectal cancer risk. Therefore, our findings may provide new insights into the development of colorectal cancer. Further association and functional studies are of great importance to confirm these results and to define the potential biological mechanism of colorectal cancer.
Keywords: case–control study, colorectal cancer, MIR17HG, single nucleotide polymorphisms, susceptibility
1. INTRODUCTION
Colorectal cancer is the third most common cancer worldwide and a major causes of cancer related morbidity and mortality (Bray et al., 2018). In China, the incidence and mortality of colorectal cancer have a rapid increase during the past few decades (Chen et al., 2016). The colorectal cancer occurrence and progression are comprehensive, multifactorial, and multistep process which caused by the interaction of environmental and genetic factors. However, the mechanism of colorectal carcinogenesis remains still not fully understood. Although obesity, sedentary behavior, and a high‐meat, high‐calorie, fat‐rich, fiber‐deficient diet, alcohol consumption, and tobacco smoking were found to be major risk factors for the development of colorectal cancer (Bishehsari, Mahdavinia, Vacca, Malekzadeh, & Mariani‐Costantini, 2014; Marley & Nan, 2016), only a fraction of individuals exposed to the same risk factors develop colorectal cancer during their lifetime, suggesting that other factors were associated with the development of colorectal cancer. The single nucleotide polymorphism (SNP) is the most common form of human genetic variations and are significantly associated with many cancers risk (Geng et al., 2015; Hu et al., 2019; Tian et al., 2018). Recently, an increasing number of evidence suggests that genetic susceptibility plays an important role in the occurrence of colorectal cancer (Duan et al., 2014; Su et al., 2015; Wang et al., 2015; Zhang, Li, Du, et al., 2014b).
The miR‐17‐92a‐1 cluster host gene (MIR17HG) located on chromosome 13q31.3 in the third intron of an open reading frame termed the c13orf25 (chromosome 13 open reading frame 25) gene. The miR‐17‐92 cluster transcript is about 800 nucleotides and encompasses six miRNAs (miR‐17, miR‐18a, miR‐19a, miR‐20a, miR‐19b‐1, and miR‐92a‐1) (Tanzer & Stadler, 2004). All members of miR‐17‐92 cluster were overexpressed in colorectal cancer, pointing to a key role of miR‐17‐92 cluster in colorectal cancer carcinogenesis (Koga et al., 2010). It has been reported that the MIR17HG overexpression is associated with poor prognosis in colorectal cancer (Yu et al., 2012; Zhang, Li, Zhou, Xiao, & Zhou, 2014a; Zhou, Zhang, Liu, Xia, & Tian, 2013). Furthermore, functional studies have confirmed the pivotal role of members of the MIR17HG in the development, progression, and aggressiveness of colorectal cancer (Ma et al., 2016; Tsuchida et al., 2011; Zhang, Li, Zhou, et al., 2014a).
However, few association studies on polymorphisms of MIR17HG and colorectal cancer risk has been reported (Sun et al., 2017). To better understand the influence of MIR17HG polymorphisms on colorectal cancer susceptibility in the Chinese Han population. In this study, we recruited 514 patients with colorectal cancer and 510 healthy controls to investigate the association between polymorphisms (rs72640334, rs7336610, rs7318578, rs17735387, and rs1428) of MIR17HG and risk of colorectal cancer in the Chinese Han population.
2. MATERIALS AND METHODS
2.1. Study subjects
In this case–control study, we recruited of 514 colorectal cancer cases and 510 healthy control subjects from the Shaanxi Province Cancer Hospital. All cases were patients newly diagnosed with histologically confirmed colorectal cancer who were admitted to the hospital and without restrictions of age, sex, or disease stage. The patients who had received radiotherapy or chemotherapy were excluded in the study. The controls were randomly selected from the general health check‐up center at the same time period. The case and control subjects were unrelated ethnic Han Chinese and these subjects had no history of cancer. The characteristics of all subjects were taken from patients’ medical records by well‐trained interviewers.
2.2. Ethics statement
This study protocol was approved by the Ethics Committee of the Shaanxi Province Cancer Hospital and was conducted in accordance with the principles of the Declaration of Helsinki. All subjects provided written informed consent before the collection of blood samples.
2.3. DNA isolation
We used venipuncture into ethylene diamine tetraacetic acid‐coated blood vacutainer collection tubes to collect peripheral blood samples from each subject and then stored at −20°C for further DNA isolation. The GoldMag‐Mini Whole Blood Genomic DNA Purification Kit (GoldMag. Co. Ltd., Xi'an, China) was used to extract genomic DNA from blood samples following the manufacturer's instructions (Liu et al., 2017). The purity and concentration of the isolated DNA were analyzed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) by absorbance measurements at 260 and 280 nm.
2.4. SNPs selection and genotyping
We selected the tagSNPs of MIR17HG with the minor allele frequency (MAF) greater than 0.05 in global population from the 1,000 Genome Projects. As a result, five tagSNPs (rs72640334, rs7336610, rs7318578, rs17735387, and rs1428) were selected using a pairwise Tagger method with r 2 > 0.8 to capture other SNPs. Primer sequences of amplification and extension for the polymorphisms of MIR17HG were designed using the Agena Bioscience Assay Design Suite V2.0 software (https://agenacx.com/online-tools/). Genotyping was performed with the Agena MassARRAY platform with iPLEX gold chemistry (Agena Bioscience, San Diego, CA) according to the standard protocol recommended by the manufacturer. Data management and analysis were performed using the Agena Bioscience TYPER software, version 4.0.
2.5. Statistical analysis
The differences between the cases and controls in demographic characteristics were evaluated by Student's t test (for age) and Pearson's χ 2 test (for gender). We used the chi‐square test to assess whether the genotype frequencies of SNPs among the control group was consistent with Hardy–Weinberg equilibrium (HWE). We compared the distributions of allele frequencies of SNPs between cases and controls using the χ 2 test. The association analyses were conducted under the codominant, dominant, recessive, and additive genetic models. Logistic regression analysis was carried out to calculate odds ratios (ORs) and its 95% confidence intervals (CIs) with the adjustment of gender and age (Dai et al., 2019). Pair‐wise linkage disequilibrium (LD) between the five SNPs was assessed using the Haploview software (version 4.2) (Barrett, Fry, Maller, & Daly, 2005). All two‐sided p values less than 0.05 were considered statistically significant. The statistical analyses were performed using the Statistical Package of the PLINK software (version 1.07) (Purcell et al., 2007) and Social Sciences (SPSS) software version 20.0 (SPSS Inc., Chicago, IL) and Microsoft Excel (Microsoft Corp., Redmond, WA).
3. RESULTS
3.1. Characteristics of study subjects
The demographic characteristics of participants are described in Table 1. Among the 1,024 participants, 514 were patients with colorectal cancer (228 females and 286 males) and 510 were healthy controls (224 females and 286 males). The mean age of the cases was 60.27 years old compared with 60.13 years old in controls, which revealed no statistically difference (p = 0.847). Furthermore, there was no significant difference in sex distribution (p = 0.839). Among the patients, the number of cases with colon cancer, rectal cancer, and other were 217 (42.2%), 244 (47.5%), and 53 (10.3%), respectively. The tumor stage for I–II, III–IV and missing were 146 (28.4%), 212 (41.2%), and 156 (30.4%), respectively.
Table 1.
Basic characteristics of study objects
| Characteristics | Case (%) | Control (%) | p | |
|---|---|---|---|---|
| Number | 514 | 510 | ||
| Age | Mean ± SD (years) | 60.27 ± 11.81 | 60.13 ± 10.61 | 0.847 |
| Gender | Female | 228 (50.3) | 224 (49.4) | 0.839 |
| Male | 286 (50.0) | 286 (50.0) | ||
| Tumor stage | I–II | 146 (28.4) | ||
| III–V | 212 (41.2) | |||
| Missing | 156 (30.4) | |||
| LN metastasis | No | 188 (36.6) | ||
| Yes | 170 (33.1) | |||
| Missing | 156 (30.3) | |||
| Tumor type | Colon cancer | 217 (42.2) | ||
| Rectal cancer | 244 (47.5) | |||
| Other | 53 (10.3) | |||
p values were calculated by Student's t test for age and χ 2 test for gender.
p < 0.05 indicates statistical significance.
SD: standard deviation; LN: lymph node
3.2. Allele models analysis
Genotypes frequencies distributions of the five SNPs of MIR17HG among the healthy control were consistent with the HWE in this study, which indicated a representative distribution of the subjects in the regional population. The frequencies of rs7336610 minor allele T and rs1428 minor allele A among the patients with colorectal cancer were significantly different from those among the control subjects (p = 0.007; p = 0.008, respectively), as shown in Table 2. The results revealed that individuals carrying the allele T of rs7336610 and allele A of rs1428 were associated with significantly increased risk of colorectal cancer (OR = 1.27, 95% CI: 1.07–1.51; OR = 1.27, 95% CI: 1.06–1.51, respectively). However, no statistically significant association was found between the other four MIR17HG polymorphisms and colorectal cancer risk in the allele model.
Table 2.
Association with between polymorphisms of MIR17HG and colorectal cancer risk
| SNP‐ID | Chr | Position |
Alleles A/B |
MAF | OR (95% CI) | p | |
|---|---|---|---|---|---|---|---|
| Case | Control | ||||||
| rs72640334 | 13 | 91352674 | A/C | 0.084 | 0.096 | 0.86 (0.64–1.17) | 0.341 |
| rs7336610 | 13 | 91352883 | T/C | 0.554 | 0.495 | 1.27 (1.07–1.51) | 0.007 |
| rs7318578 | 13 | 91353215 | C/A | 0.263 | 0.297 | 0.85 (0.70–1.03) | 0.089 |
| rs17735387 | 13 | 91353800 | A/G | 0.164 | 0.188 | 0.85 (0.68–1.07) | 0.157 |
| rs1428 | 13 | 91354516 | A/C | 0.555 | 0.496 | 1.27 (1.06–1.51) | 0.008 |
SNP: Single nucleotide polymorphism; Chr: chromosome; A: Minor alleles; B: Major alleles; MAF: Minor allele frequency; OR: Odds ratio; 95% CI: 95% Confidence interval
p values were calculated from χ 2 test ( two sided).
p < 0.05 was considered statistically significant.
3.3. Genetic models analysis
Then, we further assessed the association between the five SNPs of MIR17HG and colorectal cancer risk under the four genetic models (codominant, dominant, recessive, and additive) by logistic regression analysis adjusting for gender and age (Table 3). Compared to the wild homozygous genotype CC of rs7336610, individuals carrying rs7336610 TT genotype were associated with a significantly increased risk of colorectal cancer before and after adjusting for gender and age (OR = 1.57, 95% CI: 1.12–2.20, p = 0.009; Table 3). The SNP rs7336610 was also found to be associated with an increased risk of colorectal cancer in the dominant model (TT + TC vs. CC: OR = 1.44, 95% CI: 1.08–1.92, p = 0.013) and the additive model (OR = 1.25, 95% CI: 1.06–1.48, p = 0.010) before and after adjusting for gender and age. However, we found rs7318578 was associated a decreased risk of colorectal cancer in the dominant model before adjusting for gender and age (CC + CA vs. AA: OR = 0.78, 95% CI: 0.61–1.00, p = 0.049).
Table 3.
Genetics model analysis of association between MIR17HG polymorphisms and colorectal cancer risk
| SNP‐ID | Model | Genotype | Case | Control | OR (95% CI) | p | Adjusted OR (95% CI) | Adjusted p |
|---|---|---|---|---|---|---|---|---|
| rs72640334 | Codominant | CC | 426 | 416 | 1.00 | 1.00 | ||
| CA | 75 | 90 | 0.81 (0.58–1.14) | 0.228 | 0.82 (0.58–1.14) | 0.234 | ||
| AA | 5 | 4 | 1.22 (0.33–4.58) | 0.768 | 1.23 (0.33–4.60) | 0.763 | ||
| Dominant | CC | 426 | 416 | 1.00 | 1.00 | |||
| AA + CA | 80 | 94 | 0.83 (0.60–1.15) | 0.268 | 0.83 (0.60–1.16) | 0.275 | ||
| Recessive | CC + CA | 501 | 506 | 1.00 | 1.00 | |||
| AA | 5 | 4 | 1.26 (0.34–4.73) | 0.729 | 1.27 (0.34–4.75) | 0.725 | ||
| Additive | – | – | – | 0.86 (0.64–1.17) | 0.344 | 0.87 (0.64–1.17) | 0.352 | |
| rs7336610 | Codominant | CC | 109 | 142 | 1.00 | 1.00 | ||
| TC | 240 | 230 | 1.36 (1.00–1.85) | 0.051 | 1.36 (1.00–1.85) | 0.051 | ||
| TT | 165 | 137 | 1.57 (1.12–2.20) | 0.009 | 1.57 (1.12–2.20) | 0.009 | ||
| Dominant | CC | 109 | 142 | 1.00 | 1.00 | |||
| TT + TC | 405 | 367 | 1.44 (1.08–1.92) | 0.013 | 1.44 (1.08–1.92) | 0.013 | ||
| Recessive | CC + TC | 349 | 372 | 1.00 | 1.00 | |||
| TT | 165 | 137 | 1.28 (0.98–1.68) | 0.069 | 1.28 (0.98–1.68) | 0.070 | ||
| Additive | – | – | – | 1.25 (1.06–1.48) | 0.010 | 1.25 (1.06–1.48) | 0.010 | |
| rs7318578 | Codominant | AA | 277 | 245 | 1.00 | 1.00 | ||
| CA | 199 | 227 | 0.78 (0.60–1.00) | 0.052 | 0.78 (0.60–100) | 0.054 | ||
| CC | 35 | 38 | 0.81 (0.50–1.33) | 0.413 | 0.82 (0.50–1.33) | 0.417 | ||
| Dominant | AA | 277 | 245 | 1.00 | 1.00 | |||
| CC + CA | 234 | 265 | 0.78 (0.61–1.00) | 0.049 | 0.78 (0.61–1.00) | 0.051 | ||
| Recessive | AA + CA | 476 | 472 | 1.00 | 1.00 | |||
| CC | 35 | 38 | 0.91 (0.57–1.47) | 0.709 | 0.91 (0.57–1.47) | 0.713 | ||
| Additive | – | – | – | 0.84 (0.69–1.02) | 0.083 | 0.84 (0.69–1.03) | 0.087 | |
| rs17735387 | Codominant | GG | 361 | 335 | 1.00 | 1.00 | ||
| GA | 137 | 158 | 0.80 (0.61–1.06) | 0.119 | 0.80 (0.61–1.06) | 0.116 | ||
| AA | 16 | 17 | 0.87 (0.43–1.76) | 0.704 | 0.87 (0.43–1.75) | 0.695 | ||
| Dominant | GG | 361 | 335 | 1.00 | 1.00 | |||
| AA + GA | 153 | 175 | 0.81 (0.62–1.06) | 0.119 | 0.81 (0.62–1.05) | 0.116 | ||
| Recessive | GG + GA | 498 | 493 | 1.00 | 1.00 | |||
| AA | 16 | 17 | 0.93 (0.47–1.87) | 0.842 | 0.93 (0.46–1.87) | 0.837 | ||
| Additive | – | – | – | 0.85 (0.68–1.07) | 0.159 | 0.85 (0.67–1.07) | 0.155 | |
| rs1428 | Codominant | CC | 109 | 142 | 1.00 | 1.00 | ||
| CA | 239 | 228 | 1.37 (1.00–1.86) | 0.048 | 1.37 (1.00–1.86) | 0.048 | ||
| AA | 165 | 138 | 1.56 (1.11–2.18) | 0.010 | 1.56 (1.11–2.19) | 0.010 | ||
| Dominant | CC | 109 | 142 | 1.00 | 1.00 | |||
| AA + CA | 404 | 366 | 1.44 (1.08–1.92) | 0.013 | 1.44 (1.08–1.92) | 0.013 | ||
| Recessive | CC + CA | 348 | 370 | 1.00 | 1.00 | |||
| AA | 165 | 138 | 1.27 (0.97–1.66) | 0.081 | 1.27 (0.97–1.67) | 0.081 | ||
| Additive | – | – | – | 1.24 (1.05–1.47) | 0.011 | 1.24 (1.05–1.47) | 0.011 |
SNP: Single nucleotide polymorphism; OR: Odds ratio; 95% CI: 95% Confidence interval
p < 0.05 was considered statistically significant.
In addition, the genotypes CA and AA of rs1428 were significantly associated with increased risk of colorectal cancer, compared to the CC genotype before and after adjusting for gender and age (CA vs. CC: OR = 1.37, 95% CI: 1.00–1.86, p = 0.048; AA vs. CC: OR = 1.56, 95% CI: 1.11–2.19, p = 0.010). In both the dominant and additive models, there were significant association between rs1428 and the risk of colorectal cancer before and after adjusting for gender and age (dominant AA + CA vs. CC: OR = 1.44, 95% CI: 1.08–1.92, p = 0.013; additive: OR = 1.24, 95% CI: 1.05–1.47, p = 0.011). However, no significant association between the other SNPs of MIR17HG (rs72640334 and rs17735387) and colorectal cancer risk under the four genetic models.
3.4. Tumor type stratification analysis
Statistical analysis based on stratification of tumor type revealed that the SNP rs7336610 was remarkably increased risk of rectal cancer after adjusting for gender and age (allele T vs. C: OR = 1.32, 95% CI: 1.06–1.64, p = 0.013; TC vs. CC: OR = 1.56, 95% CI: 1.05–2.33, p = 0.028; TT vs. CC: OR = 1.72, 95% CI: 1.12–2.66, p = 0.014; dominant TT + TC vs. CC: OR = 1.62, 95% CI: 1.12–2.36, p = 0.011; additive: OR = 1.30, 95% CI: 1.05–1.60, p = 0.017) (Table 4). The SNP rs1428 was also found to be associated with a higher risk of rectal cancer after adjusting for gender and age (allele A vs. C: OR = 1.31, 95% CI: 1.06–1.63, p = 0.014; CA vs. CC: OR = 1.57, 95% CI: 1.05–2.33, p = 0.027; dominant AA + CA vs. CC: OR = 1.62, 95% CI: 1.12–2.36, p = 0.011; additive: OR = 1.29, 95% CI: 1.05–1.60, p = 0.018). The SNP rs7318578 had a significantly lower risk of rectal cancer (CA vs. AA: OR = 0.71, 95% CI: 0.51–0.98, p = 0.038; dominant CC + CA vs. AA: OR = 0.73, 95% CI: 0.54–1.00, p = 0.047). However, no association was found between the SNPs of MIR17HG and colon cancer risk.
Table 4.
Tumor type stratification analysis of association between MIR17HG polymorphisms and colorectal cancer risk
| SNP–ID | Model | Genotype | Rectal cancer | Colon cancer | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95% CI) | p | Case | Control | OR (95% CI) | p | |||
| rs7336610 | Allele | C | 213 | 514 | 1.00 | 199 | 514 | 1.00 | ||
| T | 275 | 504 | 1.32 (1.06–1.64) | 0.013 | 235 | 504 | 1.20 (0.96–1.51) | 0.106 | ||
| Codominant | CC | 47 | 142 | 1.00 | 50 | 142 | 1.00 | |||
| TC | 119 | 230 | 1.56 (1.05–2.33) | 0.028 | 99 | 230 | 1.23 (0.82–1.84) | 0.310 | ||
| TT | 78 | 137 | 1.72 (1.12–2.66) | 0.014 | 68 | 137 | 1.42 (0.92–2.19) | 0.119 | ||
| Dominant | CC | 47 | 142 | 1.00 | 50 | 142 | 1.00 | |||
| TT + TC | 197 | 367 | 1.62 (1.12–2.36) | 0.011 | 167 | 367 | 1.30 (0.90–1.89) | 0.168 | ||
| Recessive | CC + TC | 166 | 372 | 1.00 | 149 | 372 | 1.00 | |||
| TT | 78 | 137 | 1.28 (0.91–1.78) | 0.152 | 68 | 137 | 1.24 (0.87–1.75) | 0.229 | ||
| Additive | – | – | – | 1.30 (1.05–1.60) | 0.017 | – | – | 1.19 (0.96–1.47) | 0.120 | |
| rs7318578 | Allele | A | 359 | 717 | 1.00 | 311 | 717 | 1.00 | ||
| C | 125 | 303 | 0.82 (0.65–1.05) | 0.119 | 121 | 303 | 0.92 (0.72–1.18) | 0.516 | ||
| Codominant | AA | 135 | 245 | 1.00 | 111 | 245 | 1.00 | |||
| CA | 89 | 227 | 0.71 (0.51–0.98) | 0.038 | 89 | 227 | 0.88 (0.63–1.22) | 0.434 | ||
| CC | 18 | 38 | 0.86 (0.47–1.56) | 0.618 | 16 | 38 | 0.93 (0.50–1.75) | 0.828 | ||
| Dominant | AA | 135 | 245 | 1.00 | 111 | 245 | 1.00 | |||
| CC + CA | 107 | 283 | 0.73 (0.54–1.00) | 0.047 | 105 | 265 | 0.88 (0.64–1.22) | 0.449 | ||
| Recessive | AA + CA | 224 | 1.00 | 200 | 472 | 1.00 | ||||
| CC | 18 | 38 | 1.00 (0.56–1.79) | 0.994 | 16 | 38 | 0.99 (0.54–1.82) | 0.979 | ||
| Additive | – | – | – | 0.82 (0.64–1.05) | 0.113 | – | – | 0.92 (0.71–1.19) | 0.538 | |
| rs1428 | Allele | C | 212 | 512 | 1.00 | 199 | 512 | 1.00 | ||
| A | 274 | 504 | 1.31 (1.06–1.63) | 0.014 | 235 | 504 | 1.20 (0.96–1.50) | 0.113 | ||
| Codominant | CC | 47 | 142 | 1.00 | 50 | 142 | 1.00 | |||
| CA | 118 | 228 | 1.57 (1.05–2.33) | 0.027 | 99 | 228 | 1.24 (0.83–1.86) | 0.288 | ||
| AA | 78 | 138 | 1.71 (1.11–2.64) | 0.015 | 68 | 138 | 1.41 (0.91–2.17) | 0.125 | ||
| Dominant | CC | 47 | 142 | 1.00 | 50 | 142 | 1.00 | |||
| AA + CA | 196 | 366 | 1.62 (1.12–2.36) | 0.011 | 167 | 366 | 1.30 (0.90–1.89) | 0.162 | ||
| Recessive | CC + CA | 165 | 370 | 1.00 | 149 | 370 | 1.00 | |||
| AA | 78 | 138 | 1.27 (0.91–1.77) | 0.161 | 68 | 138 | 1.22 (0.86–1.73) | 0.256 | ||
| Additive | – | – | – | 1.29 (1.05–1.60) | 0.018 | – | – | 1.18 (0.95–1.47) | 0.127 | |
SNP: Single nucleotide polymorphism; OR: Odds ratio; 95% CI: 95% Confidence interval
OR and 95% CI were calculated using logistic regression adjusted with age and gender.
p < 0.05 was considered statistically significant.
3.5. Gender stratification analysis
Gender stratification analysis found rs7336610 (allele T vs. C: OR = 1.33, 95% CI: 1.06–1.68, p = 0.015; dominant TT + TC vs. CC: OR = 1.48, 95% CI: 1.02–2.15, p = 0.039; additive: OR = 1.30, 95% CI: 1.04–1.63, p = 0.021) and rs1428 (allele A vs. C: OR = 1.33, 95% CI: 1.06–1.68, p = 0.015; AA vs. CC: OR = 1.70, 95% CI: 1.08–2.65, p = 0.021; dominant AA + CA vs. CC: OR = 1.50, 95% CI: 1.03–2.17, p = 0.034; additive: OR = 1.30, 95% CI: 1.04–1.63, p = 0.021) were significantly associated with increased risk of colorectal cancer after adjusting for age in males (Table 5). Moreover, rs7318578 (CA vs. AA: OR = 0.66, 95% CI: 0.47–0.94, p = 0.020; dominant CC + CA vs. AA: OR = 0.70, 95% CI: 0.51–0.98, p = 0.036) and rs17735387 (GA vs. GG: OR = 0.67, 95% CI: 0.46–0.96, p = 0.031) were found to be associated with reduced risk of colorectal cancer after adjusting for age in males. However, no association was found between the SNPs of MIR17HG and colorectal cancer risk among females.
Table 5.
Gender stratification analysis of association between MIR17HG polymorphisms and colorectal cancer risk
| SNP‐ID | Model | Genotype | Male | Female | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95% CI) | p | Case | Control | OR (95% CI) | p | |||
| rs7336610 | Allele | C | 263 | 304 | 1.00 | 261 | 236 | 1.00 | ||
| T | 309 | 268 | 1.33 (1.06–1.68) | 0.015 | 195 | 210 | 0.84 (0.65–1.09) | 0.192 | ||
| Codominant | CC | 66 | 88 | 1.00 | 76 | 67 | 1.00 | |||
| TC | 131 | 128 | 1.36 (0.91–2.04) | 0.130 | 109 | 102 | 0.94 (0.61–1.44) | 0.770 | ||
| TT | 89 | 70 | 1.70 (1.08–2.65) | 0.021 | 43 | 54 | 0.70 (0.42–1.18) | 0.177 | ||
| Dominant | CC | 66 | 88 | 1.00 | 76 | 67 | 1.00 | |||
| TT + TC | 220 | 198 | 1.48 (1.02–2.15) | 0.039 | 152 | 156 | 0.86 (0.58–1.27) | 0.443 | ||
| Recessive | CC + TC | 197 | 216 | 1.00 | 185 | 169 | 1.00 | |||
| TT | 89 | 70 | 1.39 (0.97–2.01) | 0.077 | 43 | 54 | 0.73 (0.46–1.14) | 0.166 | ||
| Additive | – | – | – | 1.30 (1.04–1.63) | 0.021 | – | – | 0.85 (0.65–1.09) | 0.199 | |
| rs7318578 | Allele | A | 413 | 394 | 1.00 | 340 | 323 | 1.00 | ||
| C | 155 | 178 | 0.83 (0.64–1.07) | 0.155 | 114 | 125 | 0.87 (0.64–1.17) | 0.342 | ||
| Codominant | AA | 152 | 128 | 1.00 | 125 | 117 | 1.00 | |||
| CA | 109 | 138 | 0.66 (0.47–0.94) | 0.020 | 90 | 89 | 0.95 (0.65–1.40) | 0.803 | ||
| CC | 23 | 20 | 0.97 (0.51–1.84) | 0.920 | 12 | 18 | 0.62 (0.29–1.35) | 0.231 | ||
| Dominant | AA | 152 | 128 | 1.00 | 125 | 117 | 1.00 | |||
| CC + CA | 132 | 158 | 0.70 (0.51–0.98) | 0.036 | 102 | 107 | 0.90 (0.62–1.30) | 0.563 | ||
| Recessive | AA + CA | 261 | 266 | 1.00 | 215 | 206 | 1.00 | |||
| CC | 23 | 20 | 1.17 (0.63–2.19) | 0.618 | 12 | 18 | 0.64 (0.30–1.36) | 0.242 | ||
| Additive | – | – | – | 0.82 (0.63–1.07) | 0.145 | – | – | 0.87 (0.64–1.17) | 0.346 | |
| rs17735387 | Allele | G | 477 | 457 | 1.00 | 382 | 371 | 1.00 | ||
| A | 95 | 115 | 0.79 (0.59–1.07) | 0.127 | 74 | 77 | 0.93 (0.66–1.32) | 0.699 | ||
| Codominant | GG | 203 | 181 | 1.00 | 158 | 154 | 1.00 | |||
| GA | 71 | 95 | 0.67 (0.46–0.96) | 0.031 | 66 | 63 | 1.02 (0.67–1.53) | 0.944 | ||
| AA | 12 | 10 | 1.07 (0.45–2.54) | 0.876 | 4 | 7 | 0.54 (0.15–1.88) | 0.331 | ||
| Dominant | GG | 203 | 181 | 1.00 | 158 | 154 | 1.00 | |||
| AA + GA | 83 | 105 | 0.70 (0.50–1.00) | 0.051 | 70 | 70 | 0.97 (0.65–1.44) | 0.872 | ||
| Recessive | GG + GA | 274 | 276 | 1.00 | 224 | 217 | 1.00 | |||
| AA | 12 | 10 | 1.21 (0.51–2.86) | 0.660 | 4 | 7 | 0.53 (0.15–1.86) | 0.325 | ||
| Additive | – | – | – | 0.80 (0.59–1.07) | 0.135 | – | – | 0.92 (0.65–1.32) | 0.661 | |
| rs1428 | Allele | C | 263 | 302 | 1.00 | 260 | 238 | 1.00 | ||
| A | 309 | 266 | 1.33 (1.06–1.68) | 0.015 | 194 | 210 | 0.85 (0.65–1.10) | 0.211 | ||
| Codominant | CC | 66 | 88 | 1.00 | 76 | 68 | 1.00 | |||
| CA | 131 | 126 | 1.39 (0.93–2.07) | 0.112 | 108 | 102 | 0.94 (0.62–1.44) | 0.788 | ||
| AA | 89 | 70 | 1.70 (1.08–2.65) | 0.021 | 43 | 54 | 0.71 (0.42–1.19) | 0.194 | ||
| Dominant | CC | 66 | 88 | 1.00 | 76 | 68 | 1.00 | |||
| AA + CA | 220 | 196 | 1.50 (1.03–2.17) | 0.034 | 151 | 156 | 0.86 (0.58–1.28) | 0.464 | ||
| Recessive | CC + CA | 197 | 214 | 1.00 | 184 | 170 | 1.00 | |||
| AA | 89 | 70 | 1.38 (0.96–2.00) | 0.086 | 43 | 54 | 0.73 (0.47–1.15) | 0.181 | ||
| Additive | – | – | – | 1.30 (1.04–1.63) | 0.021 | – | – | 0.85 (0.66–1.10) | 0.218 | |
SNP: Single nucleotide polymorphism; OR: Odds ratio; 95% CI: 95% Confidence interval
OR and 95% CI were calculated using logistic regression adjusted with age and gender.
p < 0.05 was considered statistically significant.
3.6. LD and haplotype analysis
The results of pair‐wise LD analysis with these five SNPs are shown in Figure 1. We observed one haplotype block composed of rs72640334, rs7336610, rs7318578, rs17735387, and rs1428. Overall analysis found that the distributions of the frequency of the haplotype CTAGA were significantly different between colorectal cancer and control groups (p = 0.007); and the haplotype CTAGA was significantly associated with an increased colorectal cancer risk after adjusting for gender and age (OR = 1.26, 95% CI: 1.06–1.49) (Table 6). Statistical analysis found that the haplotype CTAGA was also associated with high risk of rectal cancer after adjusting for gender and age (OR = 1.29, 95% CI: 1.04–1.59, p = 0.018). Furthermore, in logistic regression analysis adjusted for age and gender, the haplotype CTAGA in MIR17HG was associated with high risk of colorectal cancer among males (OR = 1.32, 95% CI: 1.06–1.65, p = 0.015).
Figure 1.
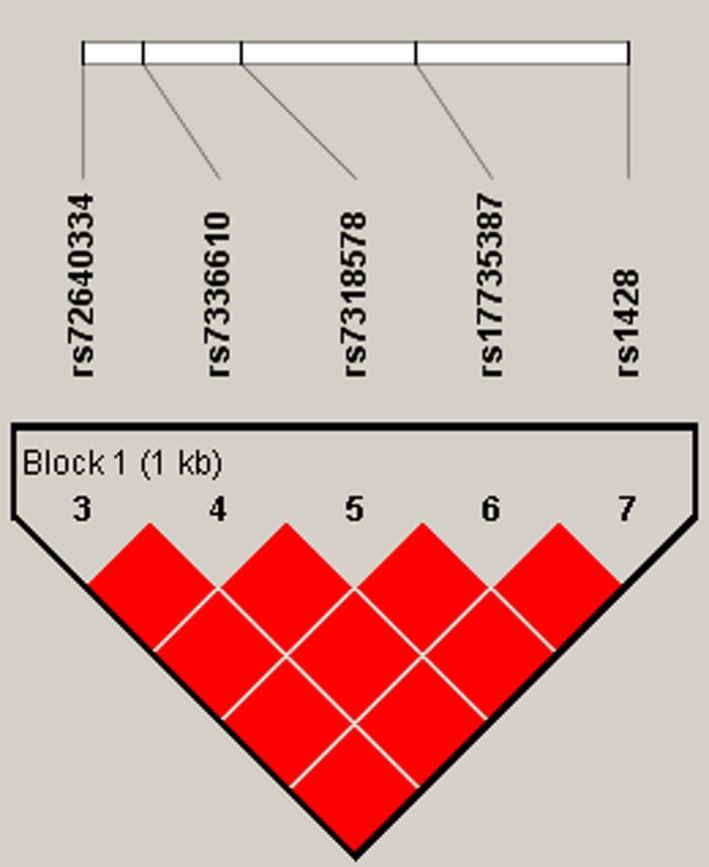
Haplotype block map for the five SNPs in the MIR17HG gene. Standard color frame is used to show LD pattern. One block in the figure showed higher LD. Bright red represents very strong LD
Table 6.
Haplotype analysis of association between MIR17HG polymorphisms and colorectal cancer risk
| SNP‐ID | Haplotype | Overall | Rectal cancer | Male | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95% CI) | p | Case | Control | OR (95% CI) | p | Case | Control | OR (95% CI) | p | ||
| rs72640334| | CTAGA | 0.558 | 0.496 | 1.26 (1.06–1.49) | 0.007 | 0.564 | 0.496 | 1.29 (1.04–1.59) | 0.018 | 0.544 | 0.469 | 1.32 (1.06–1.65) | 0.015 |
| rs7336610| | CCAAC | 0.834 | 0.812 | 1.17 (0.93–1.47) | 0.183 | 0.846 | 0.812 | 1.27 (0.95–1.70) | 0.113 | 0.832 | 0.799 | 1.24 (0.92–1.67) | 0.160 |
| rs7318578| | ACCGC | 0.917 | 0.904 | 1.17 (0.86–1.58) | 0.323 | 0.918 | 0.904 | 1.18 (0.81–1.73) | 0.394 | 0.905 | 0.897 | 1.09 (0.74–1.61) | 0.657 |
| rs17735387| | CCCGC | 0.824 | 0.799 | 1.18 (0.94–1.49) | 0.148 | 0.823 | 0.799 | 1.19 (0.89–1.58) | 0.250 | 0.827 | 0.792 | 1.28 (0.94–1.75) | 0.118 |
| rs1428 | CCAGC | 0.983 | 0.981 | 1.12 (0.58–2.18) | 0.739 | 0.977 | 0.981 | 0.82 (0.38–1.74) | 0.598 | 0.981 | 0.981 | 0.99 (0.42–2.32) | 0.980 |
SNP: Single nucleotide polymorphism; OR: Odds ratio; 95% CI: 95% Confidence interval
OR and 95% CI were calculated using logistic regression adjusted with age and gender.
p < 0.05 was considered statistically significant.
4. DISCUSSION
In this case–control study, we investigated the association between the polymorphisms of MIR17HG and colorectal cancer risk in the Chinese Han population. Overall analysis indicated that rs7336610 and rs1428 were associated with increased risk of colorectal cancer; but rs7318578 was associated with a decreased risk of colorectal cancer under the dominant model. Stratification analysis showed that rs7336610, rs7318578, rs17735387, and rs1428 were associated with colorectal cancer risk. Moreover, haplotype analysis confirmed that the haplotype CTAGA was significantly associated with an increased risk of colorectal cancer.
MIR17HG is located at humans chromosome 13q31, a genomic region frequently amplified in a large spectrum of human cancers including colorectal cancer. According to UALCAN database (http://ualcan.path.uab.edu/cgi-bin/ualcan-res.pl), we found that the expression of MIR17HG is significantly different between normal and colon and rectal cancer tissues (Figures 2 and 3; Chandrashekar et al., 2017). The overexpression of miR‐17‐92 cluster is not only involved in the progression of colorectal adenoma to adenocarcinoma but also related to poor survival of colorectal cancer (Diosdado et al., 2009; Yu et al., 2012). Previous study demonstrated that miR‐17‐92 suppressed colorectal cancer progression by inhibiting tumor angiogenesis in a genetically engineered mouse model, indicating the presence of cellular context‐dependent pro‐ and anti‐cancer effects of miR‐17‐92 (Ma et al., 2016). It also found that higher levels of miR‐17‐92 contribute to inhibition of tumor growth and metastasis in a mouse tumor model (Jiang et al., 2014). Recent research identified that the miR‐17‐92 cluster was a crucial player in the development of the immune system (Kuo, Wu, & Yang, 2018). Previous study indicated that MIR17HG copy numbers would seem to be related to response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer (Molinari et al., 2016). Moreover, it has been reported that the miR‐92 upregulation in plasma may be used as a noninvasive molecular marker for colorectal cancer screening, with a sensitivity of 89% and a specificity of 70% (Ng et al., 2009). These findings suggest that miR‐17‐92 cluster play a pivotal role in the of development colorectal cancer.
Figure 2.
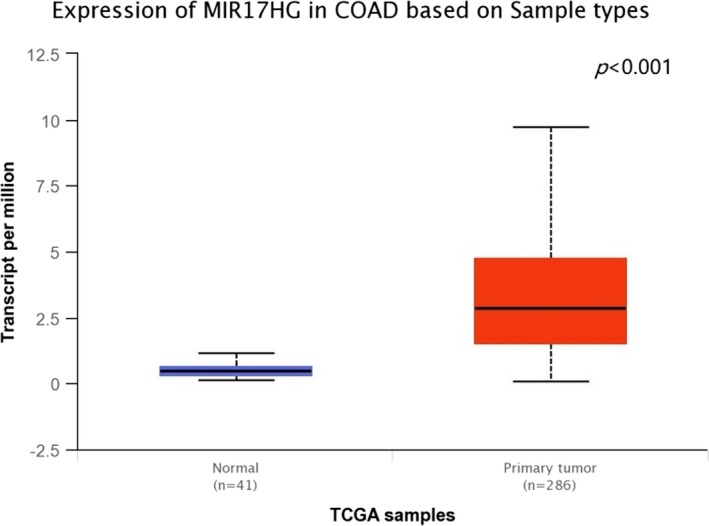
The expression of MIR17HG between normal and colon adenocarcinoma tissues from the UALCAN database. COAD: colon adenocarcinoma
Figure 3.
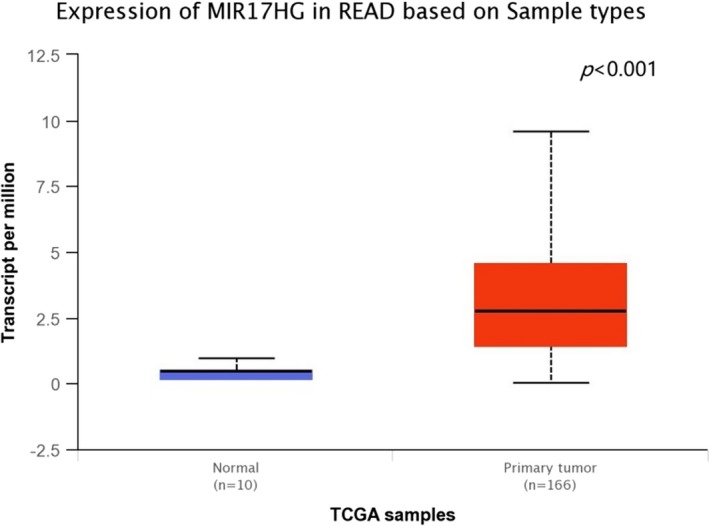
The expression of MIR17HG between normal and rectum adenocarcinoma tissues from the UALCAN database. READ: Rectum adenocarcinoma
Previous study reported that two functional polymorphisms (rs9588884 and rs982873) in the promoter region of miR‐17‐92 cluster are associated with a decreased risk of colorectal cancer (Sun et al., 2017). It has been reported that the SNP rs9515692 in the promoter region of miR‐17‐92 was a protective factor for the susceptibility of systemic lupus erythematosus (Wang et al., 2018). Statistical analysis of allele frequencies in cases and controls in the Genomics Research Centre Breast Cancer population for rs7336610 showed significance; and haplotypic analysis of results showed that the AC haplotype of rs4824505/rs7336610 are associated with risk of breast cancer development (Chacon‐Cortes, Smith, Lea, Youl, & Griffiths, 2015). In this study, we investigated the association between the polymorphisms of MIR17HG and colorectal cancer risk in the Chinese Han population. The results indicated that the two SNPs (rs7336610 and rs1428) of MIR17HG were associated with increased colorectal cancer risk, but the two SNPs (rs7318578, rs17735387) of MIR17HG were associated with decreased colorectal cancer risk in the Chinese Han population. To date, no association study was carried out to investigate the association of SNPs (rs72640334, rs7336610, rs7318578, rs17735387, and rs1428) of MIR17HG and colorectal cancer risk. Therefore, further association study with a large sample is needed to confirm these results.
There are some potential limitations in this study must be considered. First, only subjects of Chinese Han descent were included in this study, additional studies included different ethnic populations should be conducted to confirm these results. Second, data were not available for some risk factors (e.g., cigarette smoking, alcohol consumption), which prevented our further gene‐environment interaction analysis. Third, functional studies were not performed in this study. More detailed data are required to create a comprehensive understanding of the MIR17HG in colorectal cancer tumorigenesis.
5. CONCLUSIONS
In conclusion, this study provides the first evidence that the polymorphisms (rs7336610, rs7318578, rs17735387, and rs1428) of MIR17HG were associated with colorectal cancer risk. Therefore, our findings may provide new insights into the development of colorectal cancer. Further association and functional studies are of great importance to confirm these results and help us to define the potential biological mechanism of colorectal cancer.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
ACKNOWLEDGMENTS
The work was supported by the China‐Nepal Friendship Medical Research Laboratory of Prof. Rajiv Kumar Jha in Xi'an Medical University (No. 18LJM01). We are grateful to the patient and control individuals for their participation in the study.
Chen P, Bai Y, Li Y, et al. Association between polymorphisms of MIR17HG and risk of colorectal cancer in the Chinese Han population. Mol Genet Genomic Med. 2019;7:e667 10.1002/mgg3.667
Contributor Information
Hongli Zhu, Email: zhuyjw1971@nwu.edu.cn.
Chao Chen, Email: chaochenxd898@gmail.com.
REFERENCES
- Barrett, J. C. , Fry, B. , Maller, J. , & Daly, M. J. (2005). Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics, 21(2), 263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Bishehsari, F. , Mahdavinia, M. , Vacca, M. , Malekzadeh, R. , & Mariani‐Costantini, R. (2014). Epidemiological transition of colorectal cancer in developing countries: Environmental factors, molecular pathways, and opportunities for prevention. World Journal of Gastroenterology, 20(20), 6055–6072. 10.3748/wjg.v20.i20.6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, F. , Ferlay, J. , Soerjomataram, I. , Siegel, R. L. , Torre, L. A. , & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68(6), 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Chacon‐Cortes, D. , Smith, R. A. , Lea, R. A. , Youl, P. H. , & Griffiths, L. R. (2015). Association of microRNA 17–92 cluster host gene (MIR17HG) polymorphisms with breast cancer. Tumour Biology: the Journal of the International Society for Oncodevelopmental Biology and Medicine, 36(7), 5369–5376. 10.1007/s13277-015-3200-1 [DOI] [PubMed] [Google Scholar]
- Chandrashekar, D. S. , Bashel, B. , Balasubramanya, S. A. H. , Creighton, C. J. , Ponce‐Rodriguez, I. , Chakravarthi, B. V. S. K. , & Varambally, S. (2017). UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia, 19(8), 649–658. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Zheng, R. , Baade, P. D. , Zhang, S. , Zeng, H. , Bray, F. , … He, J. (2016). Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians, 66(2), 115–132. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- Dai, Z. , Tian, T. , Wang, M. , Yang, T. , Li, H. , Lin, S. , … … N. (2019). Genetic polymorphisms of estrogen receptor genes are associated with breast cancer susceptibility in Chinese women. Cancer Cell International, 19, 11 10.1186/s12935-019-0727-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diosdado, B. , van de Wiel, M. A. , Terhaar Sive Droste, J. S. , Mongera, S. , Postma, C. , Meijerink, W. J. H. J. , … Meijer, G. A. (2009). MiR‐17‐92 cluster is associated with 13q gain and c‐myc expression during colorectal adenoma to adenocarcinoma progression. British Journal of Cancer, 101(4), 707–714. 10.1038/sj.bjc.6605037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, X. , Li, X. , Lou, H. , Geng, T. , Jin, T. , Liang, P. , … Chen, C. (2014). Genetic association of PLCE1, C11orf92‐C11orf93, and NOC3L with colorectal cancer risk in the Han population. Tumour Biology: the Journal of the International Society for Oncodevelopmental Biology and Medicine, 35(3), 1813–1817. 10.1007/s13277-013-1242-9 [DOI] [PubMed] [Google Scholar]
- Geng, T.‐T. , Xun, X.‐J. , Li, S. , Feng, T. , Wang, L.‐P. , Jin, T.‐B. , & Hou, P. (2015). Association of colorectal cancer susceptibility variants with esophageal cancer in a Chinese population. World Journal of Gastroenterology, 21(22), 6898–6904. 10.3748/wjg.v21.i22.6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, D. , Liu, Q. , Lin, X. , Zhang, H. , Lin, J. , Zheng, X. , & Peng, F. (2019). Association of RAGE gene four single nucleotide polymorphisms with the risk, invasion, metastasis and overall survival of gastric cancer in Chinese. Journal of Cancer, 10(2), 504–509. 10.7150/jca.26583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H. , Wang, P. , Wang, Q. , Wang, B. , Mu, J. , Zhuang, X. , … Zhang, H.‐G. (2014). Quantitatively controlling expression of miR‐17~92 determines colon tumor progression in a mouse tumor model. The American Journal of Pathology, 184(5), 1355–1368. 10.1016/j.ajpath.2014.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, Y. , Yasunaga, M. , Takahashi, A. , Kuroda, J. , Moriya, Y. , Akasu, T. , … Matsumura, Y. (2010). MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prevention Research, 3(11), 1435–1442. 10.1158/1940-6207.CAPR-10-0036 [DOI] [PubMed] [Google Scholar]
- Kuo, G. , Wu, C. Y. , & Yang, H.‐Y. (2018). MiR‐17‐92 cluster and immunity. Journal of the Formosan Medical Association, 1118, 2–6. 10.1016/j.jfma.2018.04.013 [DOI] [PubMed] [Google Scholar]
- Liu, D. i. , Wang, M. , Tian, T. , Wang, X.‐J. , Kang, H.‐F. , Jin, T.‐B. , … Dai, Z.‐J. (2017). Genetic polymorphisms (rs10636 and rs28366003) in metallothionein 2A increase breast cancer risk in Chinese Han population. Aging, 9(2), 547–555. 10.18632/aging.101177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H. , Pan, J.‐S. , Jin, L.‐X. , Wu, J. , Ren, Y.‐D. , Chen, P. , … Han, J. (2016). MicroRNA‐17~92 inhibits colorectal cancer progression by targeting angiogenesis. Cancer Letters, 376(2), 293–302. 10.1016/j.canlet.2016.04.011 [DOI] [PubMed] [Google Scholar]
- Marley, A. R. , & Nan, H. (2016). Epidemiology of colorectal cancer. International Journal of Molecular Epidemiology and Genetics, 7(3), 105–114. [PMC free article] [PubMed] [Google Scholar]
- Molinari, C. , Salvi, S. , Foca, F. , Teodorani, N. , Saragoni, L. , Puccetti, M. , … Calistri, D. (2016). miR‐17‐92a‐1 cluster host gene (MIR17HG) evaluation and response to neoadjuvant chemoradiotherapy in rectal cancer. OncoTargets and Therapy, 9, 2735–2742. 10.2147/OTT.S105760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, E. K. , Chong, W. W. , Jin, H. , Lam, E. K. , Shin, V. Y. , Yu, J. , … Sung, J. J. (2009). Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut, 58(10), 1375–1381. 10.1136/gut.2008.167817 [DOI] [PubMed] [Google Scholar]
- Purcell, S. , Neale, B. , Todd‐Brown, K. , Thomas, L. , Ferreira, M. A. R. , Bender, D. , … Sham, P. C. (2007). PLINK: A tool set for whole‐genome association and population‐based linkage analyses. American Journal of Human Genetics, 81(3), 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Q. , Wang, Y. , Zhao, J. , Ma, C. , Wu, T. , Jin, T. , & Xu, J. (2015). Polymorphisms of PRLHR and HSPA12A and risk of gastric and colorectal cancer in the Chinese Han population. BMC Gastroenterology, 15, 107 10.1186/s12876-015-0336-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, R. , Liang, Y. , Yuan, F. , Nie, X. , Sun, H. , Wang, Y. , … Zhang, L. (2017). Functional polymorphisms in the promoter region of miR‐17‐92 cluster are associated with a decreased risk of colorectal cancer. Oncotarget, 8(47), 82531–82540. 10.18632/oncotarget.19753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer, A. , & Stadler, P. F. (2004). Molecular evolution of a microRNA cluster. Journal of Molecular Biology, 339(2), 327–335. 10.1016/j.jmb.2004.03.065 [DOI] [PubMed] [Google Scholar]
- Tian, T. , Wang, M. , Zheng, Y. , Yang, T. , Zhu, W. , Li, H. , … Zhou, L. (2018). Association of two FOXP3 polymorphisms with breast cancer susceptibility in Chinese Han women. Cancer Management and Research, 10, 867–872. 10.2147/CMAR.S158433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida, A. , Ohno, S. , Wu, W. , Borjigin, N. , Fujita, K. , Aoki, T. , … Kuroda, M. (2011). miR‐92 is a key oncogenic component of the miR‐17‐92 cluster in colon cancer. Cancer Science, 102(12), 2264–2271. 10.1111/j.1349-7006.2011.02081.x [DOI] [PubMed] [Google Scholar]
- Wang, N. , Wang, L. i. , Yang, H. , Zhang, H. Q. , Lan, B. , He, X. , … Chen, C. (2015). Multiple genetic variants are associated with colorectal cancer risk in the Han Chinese population. European Journal of Cancer Prevention: the Official Journal of the European Cancer Prevention Organisation, 24(1), 1–5. 10.1097/CEJ.0000000000000012 [DOI] [PubMed] [Google Scholar]
- Wang, R. , Wang, C. F. , Qin, H. M. , Lu, Y. L. , Wei, G. J. , Huang, H. T. , & Wei, Y. S. (2018). Association between polymorphisms in the promoter region of miR‐17‐92 cluster and systemic lupus erythematosus in a Chinese population. Journal of Cellular and Molecular Medicine, 10.1111/jcmm.13672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. e. , Tang, J.‐Q. , Tian, M.‐L. , Li, H. , Wang, X. , Wu, T. , … Wan, Y.‐L. (2012). Prognostic values of the miR‐17‐92 cluster and its paralogs in colon cancer. Journal of Surgical Oncology, 106(3), 232–237. 10.1002/jso.22138 [DOI] [PubMed] [Google Scholar]
- Zhang, G. J. , Li, Y. , Zhou, H. , Xiao, H. X. , & Zhou, T. (2014a). miR20a is an independent prognostic factor in colorectal cancer and is involved in cell metastasis. Molecular Medicine Reports, 10(1), 283–291. 10.3892/mmr.2014.2144 [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Li, X. , Du, Q. , Gong, S. , Wu, M. , Mao, Z. , … Wang, J. (2014b). DUSP10 gene polymorphism and risk of colorectal cancer in the Han Chinese population. European Journal of Cancer Prevention: the Official Journal of the European Cancer Prevention Organisation, 23(3), 173–176. 10.1097/CEJ.0b013e3283647408 [DOI] [PubMed] [Google Scholar]
- Zhou, T. , Zhang, G. , Liu, Z. , Xia, S. , & Tian, H. (2013). Overexpression of miR‐92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. International Journal of Colorectal Disease, 28(1), 19–24. 10.1007/s00384-012-1528-1 [DOI] [PubMed] [Google Scholar]


