Abstract
The COP9 signalosome (CSN) is a multifunctional protein complex essential for arabidopsis development. One of its functions is to promote Rub1/Nedd8 deconjugation from the cullin subunit of the Skp1-cullin-F-box ubiquitin ligase. Little is known about the specific role of its eight subunits in deneddylation or any of the physiological functions of CSN. In the absence of CSN1 (the fus6 mutant), arabidopsis CSN complex cannot assemble, which destabilizes multiple CSN subunits and contributes, together with the loss of CSN1, to the phenotype of fus6. To distinguish CSN1-specific functions, we attempted to rescue the complex formation with deletion or point-mutation forms of CSN1 expressed as transgenes in fus6. We show that the central domain of CSN1 is critical for complex assembly, whereas the C-terminal domain has a supporting role. By expressing the C231 fragment, which contains the structural information but lacks the presumed functional domain located at the N terminus, we have rescued the complex formation and restored the Rub1/Nedd8 deconjugation activity on cullins (fus6/C231). Nonetheless, fus6/C231 exhibits pleiotropic phenotype, including photomorphogenic defects and growth arrest at seedling stage. We conclude that CSN1 N-terminal domain is not required for the Rub1/Nedd8 deconjugation activity of cullins, but contributes to a significant aspect of CSN functions that are essential for plant development.
INTRODUCTION
The COP9 signalosome (CSN), previously known as the COP9 complex or JAB1-containing signalosome, is a highly conserved protein complex consisting of eight subunits (Wei and Deng, 1999; Deng et al., 2000). In arabidopsis, CSN mutants were isolated by virtue of their inability to undergo etiolation in the dark (constitutively photomorphogenic) and their dark seed color (fusca) (Wei and Deng, 1996; Frankhauser and Chory, 1997). The normal dark-grown arabidopsis seedlings undergo etiolation characterized by elongated hypocotyl, and closed and undeveloped cotyledons with apical hook. The light-grown seedlings undergo photomorphogenesis resulting in short hypocotyl, open and expanded cotyledons, and activated shoot apex to produce true leaves. Accompanying this morphological change is the expression of an array of genes involved in growth and photosynthesis (von Arnim and Deng, 1996; Deng and Quail, 1999). The CSN mutants exhibit shortened hypocotyls, open cotyledons, and expression of the light-induced genes in a light-independent manner, indicating that CSN negatively regulates photomorphogenesis. Beside light responses, the mutants display additional abnormalities that lead to early lethality at late seedling stage (Castle and Meinke, 1994; Misera et al., 1994; Mayer et al., 1996). CSN is also implicated in the plant auxin response (Schwechheimer et al., 2001).
CSN has recently been shown to interact with Skp1-cullin-F-box (SCF) E3 ubiquitin ligases and to promote Rub1/Nedd8 deconjugation of SCF cullin subunit (Lyapina et al., 2001; Schwechheimer et al., 2001). SCF represents a conserved class of E3 enzymes that contains, as core components, Skp1, Cdc53/cullin, and Hrt1/Roc1/Rbx1 (Deshaies, 1999). Rub1/Nedd8 is an ubiquitin-related polypeptide that can be conjugated to members of the cullin family through an enzymatic cascade similar to that of ubiquitin conjugation (Hochstrasser 2000; Jentsch and Pyrowolakis, 2000). Cullin modification appears to be necessary for proper SCF function in higher eukaryotes (Kawakami et al., 2001). In arabidopsis, the Rub1 conjugation pathway is required for the auxin response (del Pozo et al., 1998). The modulation of the Rub1 conjugation by CSN may account, in part, for the involvement of CSN in auxin response (Schwechheimer et al., 2001). It remains unclear, however, whether the lack of Rub1/Nedd8 deconjugation on cullins is solely responsible for the pleiotropic phenotype of the arabidopsis CSN mutant and whether CSN has physiologically relevant functions independent of this activity.
The arabidopsis mutant of the CSN1 subunit is known as fus6 (Castle and Meinke, 1994) or cop11 (Wei et al., 1994b; Staub et al., 1996). This mutant also exhibits defects in CSN8 accumulation and in association of CSN5 with the complex (Wei et al., 1994a; Staub et al., 1996; Kwok et al., 1998). On the other hand, the CSN1 protein cannot be detected in other CSN mutants (Kwok et al., 1998; Serino et al., 1999), a phenomenon that is best explained by the instability of CSN1 in the absence of the complex assembly. CSN1 has been reported to interact with five other subunits, CSN2, 3, 4, 5, and 7 (Kwok et al., 1998; Karniol et al., 1999; Mundt et al., 1999; Serino et al., 1999; Kapelari et al., 2000; Tsuge et al., 2001). Some of these interactions are conserved in multiple organisms such as plants, fission yeast, and mammals. The cross-species conservation in complex assembly is highlighted by the observation that human CSN1 (GPS1 or hCSN1) can replace arabidopsis CSN1 to integrate into the arabidopsis CSN complex, although at a reduced efficiency (Kang et al., 2000). The regions on hCSN1 that mediate interactions with other CSN subunits have been located to the central and the C terminus, encompassing the PCI domain (Freilich et al., 1999; Tsuge et al., 2001; reviewed by Kim et al., 2001). The N terminus (1–196 amino acid [aa] residues) of hCSN1 (CSN1N), on the other hand, was shown to be dispensable for complex assembly but required for the activity of CSN1 in repression of gene expression and c-Jun–dependent transcription in mammalian cells. It was, therefore, proposed that CSN1N domain confers most of the activities of CSN1 (Tsuge et al., 2001).
We are interested to understand the functional contribution of CSN1 to the COP9 signalosome in plant development. We found that the CSN1-specific function cannot be directly inferred from the phenotype of fus6 due to the pleiotropic defects of the entire complex. To produce a mutant specifically deficient in CSN1-mediated functions, we attempted to rescue, in fus6, the formation of a CSN complex that contains a complete set of the subunits except the activity domain of CSN1. Our initial structure-function study of CSN1 in arabidopsis allowed us to generate and characterize a transgenic fus6 variant that we believe has fulfilled this intent.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The fus6-1 mutation (Wassilewskija ecotype) was caused by a T-DNA insert that carries a kanamycin-resistant marker (Castle and Meinke, 1994). This mutant allele is a null fus6 mutant and has been used in combination with all of the transgenes in this study. When not specified, we use fus6 to indicate fus6-1 allele throughout the text. Correspondingly, the wild-type plants used in this study were of the Wassilewskija ecotype. Arabidopsis seeds were surface-sterilized, washed, and plated on a dish containing growth medium (Sigma, St. Louis, MO) with 1% sucrose. The seeds were vernalized by incubating at 4°C for 3–8 d and placed in a standard long-day growth chamber at 22°C (Staub et al., 1996) for periods as specified.
Molecular Analysis of Ethyl Methanesulfonate (EMS)-mutagenized fus6 Alleles
Total RNA from seedlings of the fus6-T236, fus6-T379, fus6-T414, fus6-G236 mutants (Misera et al., 1994) were extracted using TRIzol RNA Purification kit (Invitrogen, Carlsbad, CA). The mutant CSN1 cDNAs were generated by reverse transcription-polymerase chain reaction with 1–2 μg of mutant total RNA, 1 μg/ml specific primer sets: the reverse primer COP11R (5′-GCAGGTCGAC AGTTTAGCAA ACGAGATGTC ATCAAAC-3′) and the forward primer COP11US (5′-TCGACTCGAG TCTAGACCCG GGGGATCCGG ATCCATGGAG CGAGACGAAG AAGCGGGTGG ACC-3′). The Omniscript RT kit and HotStarTaq DNA Polymerase from QIAGEN (Valencia, CA) were used. The BamHI-SalI fragment of the polymerase chain reaction (PCR) product was subcloned into Bluescript KS vector. Multiple Bluescript clones of each mutant allele were sequenced, and the region that contains the mutation was further confirmed by direct sequencing of the PCR products.
Arabidopsis Transformation and Plasmid Construction
The CSN1 constructs were subcloned into the pPZPY122 transformation that has a gentamicin-resistant marker and contains a cauliflower mosaic virus 35S promoter to drive the expression of the transgene (Yamamoto et al., 1998). The FS1 (full-length CSN1) construct was generated by PCR amplification from pCRII-FUS6 (Staub et al., 1996) by using the forward primer COP11–5NFL (5′-CATGCCATGG ATCCACATAT GGACTACAAA GACGATGACG ATAAAGCAGA GCTCCATATG GAGCGAGACG AAG-3′), which contains the sequence for one copy of flag epitope tag, and the reverse primer COP11R (5′-GCAGGTCGAC AGTTTAGCAA ACGAGAT-CTC ATCAAAG-3′). The C231 fragment was PCR amplified by using the forward primer COP11-A2 (5′-CCTGGATCCT ATGGTTAATG CAAAACTGCG-3′) and the T7 primer. The N210 fragment was amplified using the forward primer COP11–5NFL and the reverse primer COP11-A3DS (5′-CATGCCATGG TACCGTCGAC AGGTTCAAGG GTTTCAGG-3′). The Expand High Fidelity PCR System (Roche Molecular Biochemicals, Indianapolis, IN) was used, and the PCR products for FS1 and N210 were first cloned into pCR2.1-TOPO vector (Invitrogen). After confirmation of the sequences, the BamHI-SalI fragments were subcloned into pPZPY122. For N270 construct, the 5′ fragment between EcoRI (5′-cloning site) and the HindIII sites of pCRII-FUS6 was first subcloned into a Bluescript vector. Then BamHI-SalI fragment from this plasmid was subcloned into pPZPY122. The G222R construct was made by replacing the FS1 SacI-HindII fragment with the corresponding fragment from pBS-T236, which contains and CSN1 mutant cDNA from fus6-T236 as described in above. FS1, N210, and G222R, but not N270 and C231, constructs contain a Flag epitope tag translationally fused at the amino terminus.
For stable transformation, the DNA constructs were electroporated into the Agrobacterium strain GV3101 (pMP90). Arabidopsis transformation was performed by floral dip method (Clough and Bent, 1998). The FS1, N210, and G222R constructs were directly introduced into FUS6 +/− heterozygous plants that were selected from plates containing kanamycin (100 μg/ml; Sigma). The N270 and C231 constructs were first introduced into wild-type plants and then crossed into fus6. Transgenic plants were selected with gentamicin (100 μg/ml; Sigma). The presence and the number of transgenic loci were determined by segregation of the resistant markers: gentamicin for the transgenes and kanamycin for the fus6-1 allele.
Yeast Two-Hybrid Assay
CSN1, CSN2, CSN4, and G222R were fused at the amino terminus to Lex A-DNA binding domain in pEG202 and to a transcription activation domain in pJG4-5 (Golemis et al., 1994). The pEG202 constructs were transformed into the yeast strain EGY48, whereas the pJG4-5 constructs were transformed in the yeast strain L40. The resulting transformants were mated and the diploid strains carrying the different construct combinations as indicated were tested for β-galactosidase activity in the plate assay as shown. The liquid assay measuring the β-galactosidase activity (Golemis et al., 1994) was performed to confirm the results.
Northern Blot Analysis
Seedlings were ground in liquid N2 and extracted for RNA by using the RNeasy kit (QIAGEN). The RNA amount of 20 μg/lane was loaded and then transferred to Hybond N+ membrane (Amersham Biosciences, Piscataway, NJ) for standard Northern hybridization in a buffer containing 5× Denhart's solution, 6 × SSC, and 0.5% SDS. The probes used for detecting chalcone synthase (CHS), chlorophyll a/b binding protein (CAB), and 18S rRNA (Deng et al., 1991) were prepared by random priming (Roche Molecular Biochemicals). The RNA probes used for detecting transgene expression were prepared using MAXIscript T7/T3 kit (Ambion, Austin, TX). The probes corresponding to the N210 fragment (5′ probe) and to the C231 fragment (3′ probe) were mixed in the hybridization.
Protein Extraction, Gel Filtration Chromatography, and Immunoblot Analysis
Seedlings were collected and ground in liquid N2. The procedure and the buffers for homogenization, gel filtration, and later analysis have been described (Kang et al., 2000). A Superdex 200 HR 10/30 column (Amersham Biosciences) was used for gel filtration, and fractions of 0.5 ml were collected according to Kang et al. (2000). The fractions were separated by 12% SDS-PAGE and transferred to a polyvinylidene difluoride Immobilon membrane (Millipore, Bedford, MA). The antibodies against CSN1 (Staub et al., 1996), CSN8 (Wei et al., 1994a), and HY5 (Osterlund et al., 2000) were affinity purified. The antibodies against CSN4 (Serino et al., 1999), CSN5 (Kwok et al., 1998), CSN7 (Karniol et al., 1999), Rpn6 (Kwok et al., 1999), and PhyA (Parks and Quail, 1993) have been described. To produce antibodies against AtCul1, a fragment corresponding to amino acid 1–320 of AtCUL1 was PCR amplified by using the oligonucleotides CULATG 5′-GAATTCGAGC GCAAGACTAT TGAC-3′ and CULAA320 5′-CTCGAGACTT CCAGCAACCG TTTTG-3′ and expressed as a glutathione S-transferase fusion in the vector pGEX-4T-1. Gel-eluted recombinant protein was used for production of polyclonal antibodies in rabbits.
For direct immunoblot analysis of plant extract, tissues were homogenized in the extraction buffer containing 50 mM Tris-HCl, pH 7.2, 150 mM NaCl, 10 mM MgCl2, 10% glycerol, freshly added 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and Complete Protease-Inhibitor Cocktail (Roche Molecular Biochemicals). The extract was centrifuged at 4°C for 10 min and the protein concentration in the supernatant was measured by Bradford assay (Bio-Rad, Hercules, CA). In all cases where equal loading of the samples was required, the same samples or the blot was also probed with anti-Rpn6 to confirm the equal loading. Light-grown seedlings were used in all of the analyses when not specified.
RESULTS
CSN1 Subunit Is Necessary for Proper Assembly of CSN Complex
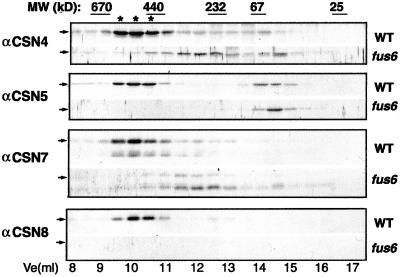
We first reevaluated the structural role of CSN1 in more detail by examining the size fractionation profiles of additional CSN subunits in fus6 (fus6-1, when not specified). The fus6 mutant still contains CSN5, CSN7, and a few other CSNs (Kwok et al., 1998; Karniol et al., 1999; our unpublished data), although, with the exception of CSN5 (Kwok et al., 1998), it is not known whether these subunits exist as complex-associated form or as monomers. We examined the gel filtration profiles of CSN4, CSN5, CSN7, and CSN8 in the crude extract of fus6 by immunoblots. Whereas the wild-type extract contains all four subunits in the fractions corresponding to CSN complex of ∼500 kDa (Figure 1, marked by asterisks), the fus6 mutant lacks this 500-kDa peak for all of the subunits examined. Instead, most of CSN4 and CSN7 are in the fractions of 250–300 kDa, and a small portion is in the fraction corresponding to the monomeric size (Figure 1). CSN7 shows as two bands that may correspond to the 33- and 30-kDa forms (Karniol et al., 1999), with the 30-kDa form being stronger in fus6. We also noted that the amounts of total CSN4 and CSN7 are markedly reduced compared with their levels in wild-type extract (our unpublished data). As previously reported for fus6, CSN5 accumulates only in the free forms and the intact CSN8 is missing (Figure 1; Wei et al., 1994a; Kwok et al., 1998; Kang et al., 2000). This result further illustrates the fact that CSN1 has an indispensable role in the formation of intact CSN complex.
Figure 1.
CSN1 is essential for the structural integrity of the COP9 signalosome. Extracts from the light-grown seedlings of wild-type (WT) or fus6-1 (fus6) were fractionated over a Superdex-200 gel filtration column. Fractions of 0.5 ml were collected and blotted using the antibodies against arabidopsis CSN4 (COP8), CSN5 (AJH), CSN7 (FUS5), and CSN8 (COP9), respectively, as indicated on the left. Ve indicates the elution volume of the column. The positions of the molecular weight standards are labeled at the top. Asterisks indicate the peak fractions containing the COP9 signalosome.
Point Mutation in fus6-T236 Disrupts the CSN1 Binding to CSN2 and CSN4
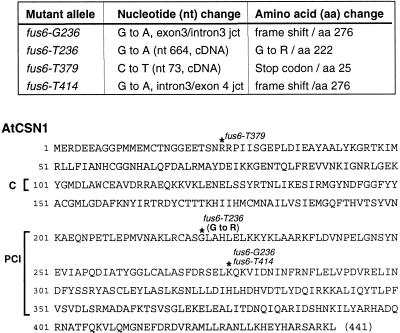
Multiple EMS-mutagenized alleles of fus6 mutants have been isolated (Misera et al., 1994). Similar to the fus6-1 null mutant, all of these mutants are lethal and lack the intact CSN (our unpublished data). We set out to determine the molecular lesions in these mutants because it may provide us information about the structural and functional constrains on CSN1. The result is summarized in Figure 2. The fus6-T379 mutation results in a stop-codon upstream in the coding region, which likely knock outs the production of CSN1. Mutations in fus6-G236 and fus6-T414 both caused frame shifts in the PCI domain, which would delete the C-terminal 165 aa residues.
Figure 2.
Identification of the mutations in four EMS mutant alleles of fus6. Top, nucleotide mutations and the corresponding changes in amino acid sequences are summarized. Bottom, diagram indicating the positions of the mutation along the amino acid sequence of arabidopsis CSN1 is shown.
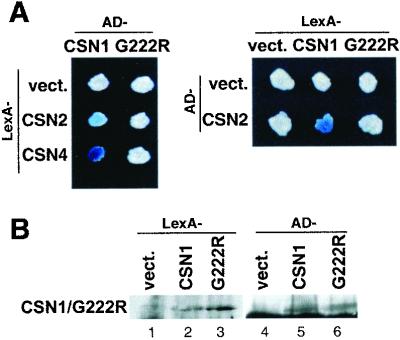
Interestingly, the fus6-T236 mutant results in a missense mutation from glycine to arginine at aa 222. This glycine residue is conserved among CSN1 orthologs from plant, human, and fission yeast (Kang et al., 2000). In addition, the G222R mutation is near the start of the PCI domain and falls into the corresponding region required for interaction with CSN2 and CSN4 in an hCSN1 study (Tsuge et al., 2001). We suspected that the fus6-T236 mutation (G222R) might affect its binding to CSN2 and CSN4, and jeopardize its capacity to support complex assembly. To test this hypothesis, we made the identical mutation in the CSN1 yeast two-hybrid constructs (G222R), and assayed its interaction with CSN2 and CSN4 in yeast. Like hCSN1, arabidopsis CSN1 was also found to interact with CSN2 and CSN4 (Figure 3), although the interaction with CSN4 was detected only with AD-CSN4 but not with lexA-CSN4 (Serino et al., 1999; our unpublished data). In all cases, G222R failed to interact with these two subunits (Figure 3A). As shown in Figure 3B, the G222R fusion proteins were expressed in yeast cells. These results showed that the G222R mutation abolished the binding of CSN1 with CSN2 and CSN4 in the yeast two-hybrid assay.
Figure 3.
Point mutation in fus6-T236 disrupts CSN1 interactions with CSN2 and CSN4 in a yeast-two-hybrid assay. (A) CSN1 and its corresponding point mutation G222R as well as CSN2 and CSN4 were expressed as fusion proteins to the LexA DNA binding domain (LexA) and to the transcription activation domain (AD). Plate assays of lac Z staining are shown with both fusion constructs. A positive interaction is indicated by a blue colony. Note that the colony containing AD-CSN1 and LexA-CSN2 showed a light blue color. In all cases, colonies containing G222R fusion proteins were white. (B) Immunoblot of the yeast extracts shows that the G222R protein was expressed.
Transgenic Approach to Dissect Arabidopsis CSN1
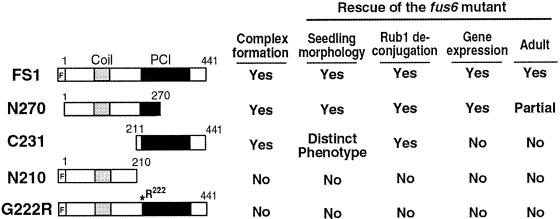
To confirm the consequence of G222R mutation on complex assembly in vivo and to further dissect the structure and function of CSN1 in the context of plant development, we made the following CSN1 mutant constructs for plant transformation. As depicted in Figure 4, the deletion mutant N270 (aa 1–270) contains the N-terminal and the central domain, including that (aa 161–268) corresponding to a complete CSN4 binding site for hCSN1 (Tsuge et al., 2001). This fragment is largely equivalent to the presumed CSN1 product in fus6-G236/T414. The C231 mutant (aa 211–441) contains the central and the C-terminal regions but lacks the N terminus. The N210 construct (aa 1–210) contains just the N terminus of CSN1. The G222R construct contains the identical point mutation as in fus6-T236. As a control, full-length CSN1 (FS1) was subcloned into the same vector. All of the constructs are under the 35S promoter (Yamamoto et al., 1998). We use the TRANSGENE-NAMES to represent the transgenic lines that are in wild-type (FUS6+/+) or FUS6+/− backgrounds, and use fus6/TRANSGENE-NAME to refer to those transgenic plants that are in fus6 −/− background.
Figure 4.
Diagram showing the CSN1 constructs used to generate stable transgenic plants. The full-length CSN1 (FS1) construct contains the complete open reading frame of 1–441 aa residues. A Flag epitope tag is translationally fused at the amino termini (as represented by a blank bar labeled with F). N270 construct contains N-terminal 1–270 aa of CSN1, and C231 contains the C-terminal 231 aa residues from 211–441 aa. The N210 construct contains the N-terminal 1–211 aa. The G222R construct is otherwise identical to CSN1 except that it contains a glycine-to-arginine mutation at the aa 222, as marked by the asterisk. Filled bars indi-cate the PCI domain. A summary of these transgenes with regard to their abilities to complement the fus6 phenotype and to form the complex is shown to the right of each corresponding construct.
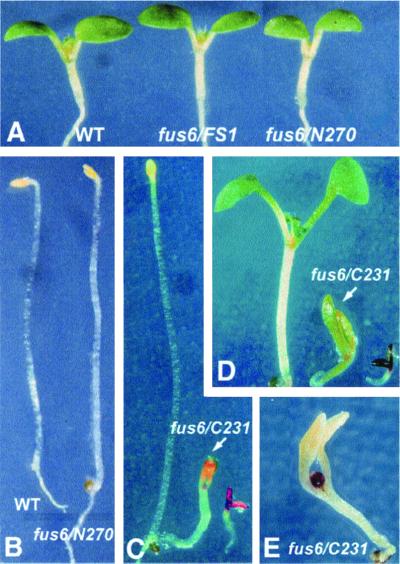
All of the 16 transgenic lines of N210 as well as 30 lines of G222R examined failed to rescue the fus6 mutant phenotype (Figure 4, right), nor did they display any new phenotypes in wild-type or mutant backgrounds. The FS1, N270, and C231 transgenes, on the other hand, completely or partially rescued the mutant (Figure 5). Under the light conditions (Figure 5A) or in complete darkness (Figure 5B), fus6/N270 and fus6/FS1 seedlings look indistinguishable from the wild-type seedlings.
Figure 5.
Characterization of fus6/N270 and fus6/C231 seedling phenotypes. (A) Light-grown fus6/FS1 and fus6/N270 seedlings appear indistinguishable from the wild-type (WT) seedling. (B) Dark-grown fus6/N270 shows etiolated phenotype identical to the etiolated WT seedling. (C) Morphology of a 9-d-old dark-grown fus6/C231 mutant (indicated by the arrow) in comparison with wild-type (right) and the fus6 seedlings (left). (D) Morphology of a 7-d-old light-grown fus6/C231 seedling (indicated by the arrow) in comparison with wild-type (left) and the fus6 seedling (right). (E) Typical morphology of a senesced fus6/C231 seedling. The mutant was grown under the light condition for 22 d. Note the pigmented shoot apex and the absence of true leaves.
Unlike FS1 and N270, C231 is unable to fully rescue fus6, and it confers, instead, a phenotype that is distinct from regular fus6. A characteristic feature of the fus6/C231 plants is closed cotyledons under both dark (Figure 5C) and light (Figure 5D) conditions, in contrast to the constitutively open-cotyledon phenotype of fus6 (Wei et al., 1994b; Figure 5, C and D). Additionally, fus6/C231 displays light-dependent cotyledon enlargement, resulting in a larger seedling size compared with fus6 when grown under the light (Figure 5D). In this sense, C231 partially rescued the fus6 seedling phenotype. However, fus6/C231 resembles fus6 in hypocotyl shortening (only slightly longer than fus6) and in developmental arrest at seedling stage. After prolonged growth period, fus6/C231 eventually senesces with a terminal morphology of enlarged but closed cotyledons and, intriguingly, a highly pigmented shoot apex that is unable to produce true leaves (Figure 5E). It is important to point out that the fus6/C231 phenotype is exclusively associated with the fus6 mutant background. The C231 transgenic plants in wild-type or FUS6 heterozygous backgrounds do not display obvious abnormalities.
Complex Assembly Is a Prerequisite for CSN1 Protein Accumulation and Phenotype Rescue
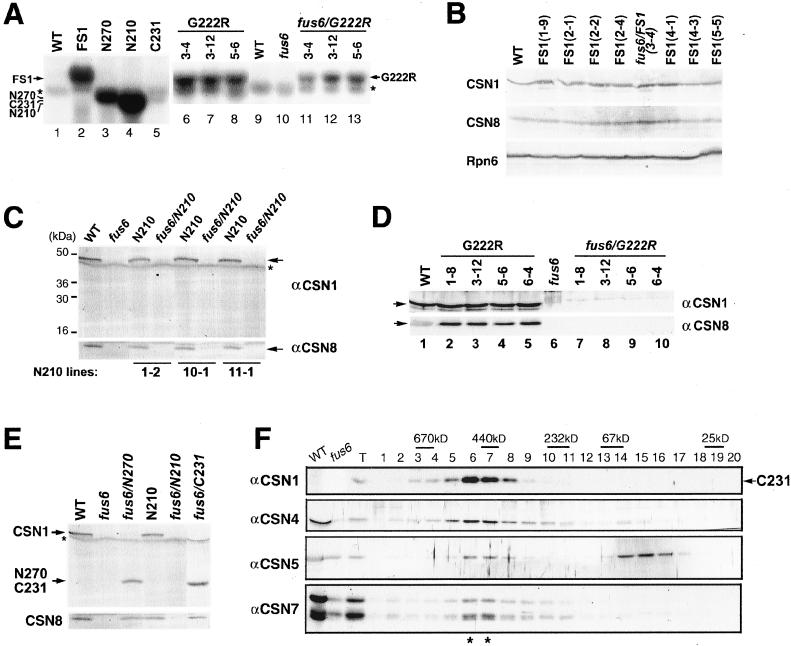
The failure of N210 and G222R transgenic lines in the complementation test prompted us to examine the expression of the transgenes. Northern blot analysis (Figure 6A) from representative transgenic lines showed that all of the transgenes, including C231, are overexpressed to different extents at the mRNA level compared with the endogenous CSN1 mRNA, the amount of which was too low to be detected in this experiment. Nonetheless, as shown in the immunoblot of representative FS1 lines (Figure 6B), the protein products were not overproduced compared with the endogenous CSN1 in wild-type seedlings.
Figure 6.
Expression of the transgenes. (A) Northern blot analysis to show the expression of all of the transgenes at mRNA level. The weak band indicated by the asterisk is the background band of rRNA. (B) Equal amounts of protein from 6-d-old FS1 transgenic or wild-type seedlings were analyzed for CSN1 accumulation. The same samples were probed with antibodies against CSN8 (middle), and a proteasome subunit Rpn6 (bottom). The Rpn6 blot indicates that approximately equal amounts of protein were loaded for each sample. (C) Absence of N210 protein accumulation in the transgenic seedlings. The mutants (fus6/N210) and the wild-type-like seedlings (N210) segregated from N210 lines (FUS6+/−) were examined. The asterisk denotes the nonspecific band for the batch of CSN1 antibody. Note that endogenous CSN1 and CSN8 were not found in the fus6/N210 mutants. (D) Immunoblot analysis of G222R transgenic seedlings. The mutants and the wild-type-like seedlings segregated from G222R lines (FUS6+/−) were collected and probed with anti-CSN1 antibody. Note that the G222R band is hardly detectable in fus6/T236 seedlings (lanes 7–10), compared with the strong band of endogenous CSN1 in G222R (wild-type-like) seedlings (lanes 2–5). (E) Immunoblot showing the expression of N270 and C231 proteins in the transgenic mutants in comparison with the N210 seedlings. The asterisk denotes the nonspecific band. (F) Gel filtration analysis of fus6/C231. The fus6/C231 seedling extract was size fractionated through a Superdex-200 column. Fractions were immunoblotted using antibodies against CSN1 (C231), CSN4, 5, and 7. Lanes WT and fus6 indicates wild-type and the fus6 mutant extracts, respectively. Note loadings were not calibrated and the wild-type sample was overloaded. Lane T was loaded with fus6/C231 extract before fractionation. Note restoration of the CSN peak (marked by asterisks).
Despite of the mRNA overexpression, the N210 protein product could not be found in any of the transgenic seedlings of either wild-type or mutant backgrounds (Figure 6C). As expected, the accumulation of CSN8, which is an indicator for the formation of the complex, was not rescued in the fus6/N210 mutants (Figure 6C). Clearly, either N210 was not translated and/or it was rapidly degraded. This result is consistent with a CSN1 model (Tsuge et al., 2001), according to which N210 lacks the domain necessary for integration of the subunit into the complex.
The G222R transgene product was barely detected (Figure 6D). Moreover, intact CSN8 was not found (Figure 6D), suggesting that the complex could not form. Thus, the G222R point mutation, which disrupts the interaction of CSN1 with other subunits, simultaneously abolished complex formation, protein accumulation, and therefore ability to rescue the mutant.
Central Domain of CSN1 Is Critical for Complex Assembly
The transgene products of the N270 and C231were readily detected (Figure 6E). More significantly, in the absence of endogenous CSN1, accumulation of CSN8 was rescued in fus6/N270, fus6/C231, fus6/FS1 seedlings (Figure 6, B and E), hinting at the formation of the CSN complex in these mutants. As expected from complete rescue of the mutant phenotype, the gel filtration profile of CSN in FS1, fus6/FS1, N270, and fus6/N270 seedlings (our unpublished data) are much alike that of the wild-type seedlings (Figure 1).
In light of the lethal phenotype of fus6/C231, we were particularly interested to know whether C231 can rescue the complex formation. Size fractionation of fus6/C231 seedlings showed clear restoration of the CSN complex, because all subunits examined cofractionated in the peak of ∼500 kDa (Figure 6F). We refer to the CSN complexes formed in the fus6/N270 and fus6/C231 mutants as CSNS1-N270 and CSNS1-C231 complexes, respectively, to recognize the fact that these complexes contain the truncated CSN1 instead of wild-type CSN1. We noted that N270 and C231 proteins are not overproduced compared with the level of wild-type CSN1 (Figure 6E). This correlates with the observation that both proteins do not appear to accumulate outside of the CSN complex (Figure 6F; our unpublished data). Together, these results argue that the phenotypes of fus6/C231 most likely reflect the loss-of-function defects of CSNS1-C231 complexes as opposed to a dominant negative or dominant gain-of-function phenotype. Because N270 and C231 fragments overlap at the central region of CSN1, the findings that both of these transgenes are capable, and that G222R is incapable, of supporting complex assembly underscore the importance of the central region of CSN1 in complex assembly.
Both N270 and C231 Can Rescue Rub1 Deconjugation on Cullins, but Only N270 Can Completely Rescue the Gene Expression Defects of fus6
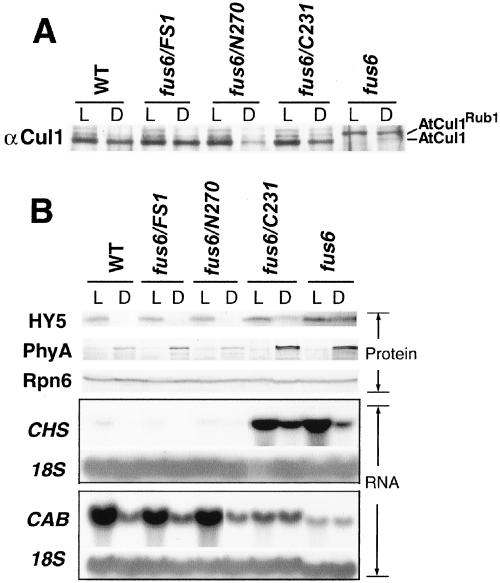
We further characterized the transgenic seedlings with regard to the newly discovered Rub1/Nedd8 deconjugation activity and the gene expression pattern related to photomorphogenesis. A small fraction of arabidopsis Cul1 (AtCul1) can be found as Rub1-modified form in seedlings (del Pozo and Estelle, 1999). In CSN mutants such as cop9 (CSN8 mutant), however, AtCul1 accumulates nearly completely in the Rub1-modified form (Schwechheimer et al., 2001). This is because the CSN-mediated Rub1/Nedd8 deconjugation activity is missing in the CSN mutants (Lyapina et al., 2001). To determine whether this activity can be rescued by the transgenes, we examined the AtCul1 modification pattern by immunoblot (Figure 7A). Like the cop9 mutant, AtCul1 accumulates only as Rub1-modified form in fus6. Notably, all of the transgenes, including C231 completely rescued this defect, because AtCul1 predominantly accumulates in the unmodified form in fus6/FS1, fus6/N270, and fus6/C231, similar to the wild type (Figure 7A). This result shows that CSNS1-N270 and CSNS1-C231 complexes are functional with regard to the Rub1/Nedd8 deconjugation activity on AtCul1.
Figure 7.
Molecular characterization of the fus6/N270 and fus6/C231 phenotype. (A) Total extracts from transgenic mutant seedling were probed with anti-AtCul1 antibody. The established pattern of Rub1-modified and unmodified forms can be readily distinguished as labeled. Note all of the transgenic seedlings showed similar AtCul1 modification pattern as in wild type. (B) Light- (L) and dark (D)-grown seedlings of 4 d old were collected. Equal amounts of the protein extract (40 μg) were loaded in each lane. The anti-HY5 and anti-PhyA blots were done with the same filter. The blot for Rpn6 was to confirm equal loading of the samples. The Northern analysis is shown in the lower part. L- and D-grown seedlings of 7 d old were collected for RNA extraction. Total RNA of 20 μg was loaded in each lane. The blot was hybridized with a CHS probe and a CAB probe (Deng et al., 1991). The 18S ribosomal blot serves as a loading control.
Because CSN has been implicated in the ubiquitin-proteasome pathway, we examined the cellular level of two well-established molecules, HY5 and phytochrome A (PhyA), both of which undergo proteolysis through the 26S proteasome in a light signal-dependent manner. Degradation of the transcription factor HY5 occurs in the dark of wild-type seedlings but not in the cop/det/fus mutants, including a CSN mutant, cop8-1 (mutant of CSN4 subunit) (Osterlund et al., 2000). As predicted, fus6 also exhibit HY5 accumulation in the dark (Figure 7B). fus6/FS1 and fus6/N270 completely and fus6/C231 partially rescued the light regulation of HY5 (Figure 7B, top). In contrast to HY5, photoreceptor PhyA is stabilized in the dark and is rapidly degraded once exposed to light (Clough and Vierstra, 1997). We found that degradation of PhyA is not defective in fus6 or any of the transgenic mutant seedlings. In fact, PhyA protein in the dark appears more abundant in the fus6 and fus6/C231 mutants (Figure 7B). Thus, our data show that fus6 is unable to degrade HY5 in the dark but is capable to degrade PhyA in the light, suggesting that modulation of the proteasome-mediated degradation by CSN is substrate specific and/or light condition dependent.
We next examined whether N270 and C231 can rescue the gene expression defects of fus6. The expression of CHS gene can be induced by light, especially UV light, and by environmental stresses (Fuglevand et al., 1996; Richard et al., 2000). Clearly, N270 and FS1 but not C231 transgenes can rescue the CHS hyperexpression of fus6 (Figure 7B). In addition, CSN mutants are characterized by light-independent expression of normally light-induced genes such as CAB (Wei et al., 1994b; Kwok et al., 1996). Again, fus6/FS1 and fus6/N270 resembles wild type, whereas fus6/C231 resembles fus6, in the pattern of CAB gene expression (Figure 7). We conclude from these results that N270, but not C231, can rescue the gene expression defect of fus6.
DISCUSSION
In this report, we demonstrated that the structural role of CSN1 can be separated from its role as a functional module of the COP9 signalosome as manifested in fus6/C231. This mutant variant allowed us to distinguish the CSN1N-dependent functions apart from other aspects of the CSN functions such as the Rub/Nedd8 deconjugation activity. Clearly, the CSN1N-mediated activities are essential for proper photomorphogenic response and seedling development.
fus6/C231 Represents a CSN1-specific Loss-of-Function Mutant
In the fus6 mutant, not only CSN1 is missing but also multiple CSN subunits display defects as a secondary result of the mutation (Figure 1; Kwok et al., 1998). Clearly, the activities mediated by these subunits are simultaneously lost or altered in fus6, resulting in a phenotype that probably reflects the loss of the entire complex rather than the single CSN1 subunit. This is further supported by the fact that the CSN mutants of arabidopsis display similar phenotypes, even although the mutations target to distinct subunits, such as fus6(CSN1), fus4(CSN4), fus5(CSN7), and fus7(CSN8). To generate a CSN1-specific mutant, the integrity of the complex needs to be preserved.
In the case of hCSN1, the functional domain has been located to the N-terminal 196 aa residues (CSN1N), whereas the structural domain assigned to the central and C-terminal region through a cell-based transient expression study (Tsuge et al., 2001). Given the ability of hCSN1 to form the complex with the arabidopsis CSN subunits (Kang et al., 2000), it is not surprising that our data agree with the domain assignment of hCSN1. We show that the C231 fragment is able to rescue the complex (CSNS1-C231) formation (Figure 6, E and F). However, as a result of CSN1N deletion, many aspects of the CSN functions are severely crippled, which is manifested by the seedling lethality and the photomorphogenic defects of fus6/C231. We cannot exclude the possibility that the amount of CSNS1-C231 complex may be slightly lower than the wild-type complex, perhaps as a result of its compromised stability for having an incomplete CSN4 binding site. Although the slight reduction in the level of the CSNS1-C231 complex may enhance mutant severity, we believe that it cannot account for the fus6/C231 phenotypes for the following reasons. First, mutants with significantly reduced CSN levels exhibit much healthier phenotypes compared with fus6/C231 (Peng et al., 2001; Schwechheimer et al., 2001). Second, although Rub1/Nedd8 deconjugation from cullins is defective in a CSN low-level mutant (Schwechheimer et al., 2001), this activity is not at all compromised in fus6/C231, demonstrating the full competence of the CSNS1-C231 complex in certain aspects of the CSN functions. In addition, compared with CSN low-level mutants (Peng et al., 2001) and fus6/N270 (our unpublished data), which display adult plant phenotypes that are variable and progressive, the phenotype of fus6/C231 is highly uniform and stable. This is consistent with the idea that an activity (or activities) is absent from fus6/C231 rather than quantitatively reducing. Taken together, it is safe to conclude that the phenotype of fus6/C231 reflects the loss of CSN1-specific function more accurately than regular fus6.
The phenotype of fus6/C231 suggests that the CSN1N-dependent function(s) is required for hypocotyl elongation, down-regulation of CHS, and light dependency of CAB expression. In agreement, overexpression of rice CSN1 (ozCSN1) confers the light-grown seedling with longer hypocotyl and reduced anthocyanin pigmentation (Kang et al., 2000). In addition, fus6/C231 shows a constitutively closed cotyledon phenotype, which suggests that the CSN1N domain is required for light-dependent cotyledon opening. More importantly, CSN1N is required for the shoot apex to produce true leaves, an event that usually symbolizes the transition from seedling to vegetative development. Further characterization of the fus6/C231 mutant will be likely to reveal additional knowledge about the functions of CSN1N.
CSN Functions and Rub1/Nedd8 Deconjugation Activity
The CSN mutants in fission yeast and arabidopsis lack Rub1/Nedd8 deconjugation activity on cullins (Lyapina et al. 2001; Schwechheimer et al., 2001). We found that Rub1 deconjugation activity was fully restored in multiple cullins in fus6/C231 (Figure 7; our unpublished data), indicating that CSN1N is not required for this activity. Considering the severe phenotype of fus6/C231, our data imply that restoration of Rub1 deconjugation activity on cullins is insufficient to overcome the seedling lethality or to rescue the gene expression defect of fus6 (Figure 7). Accordingly, the loss of Rub1 deconjugation activity on cullins alone is insufficient to account for the altered photomorphogenic response and seedling lethality. This is consistent with a recent report suggesting that the neddylation defect on cullins cannot account for the slow-growing phenotype of the fission yeast CSN1 mutant (Zhou et al., 2001). However, this study does not reveal the physiological function of CSN-mediated Rub1/Nedd8 deconjugation, nor does it exclude a role of this activity in photomorphogenesis. Ultimately, a CSN mutant specifically defective in Rub1/Nedd8 deconjugation activity would be necessary to properly address the physiological consequence of the loss of such activity. In addition, because cullins are the only known targets of Rub1/Nedd8 modification (Hochstrasser 2000; Jentsch and Pyrowolakis, 2000), we cannot rule out that Rub1/Nedd8 deconjugation from other targets, if any, might be defective in fus6/C231.
CSN1 and CSN1N domain have been shown to interact with the RING-finger subunit (Hrt1/Roc1/Rbx1) of SCF in yeast two-hybrid assays (Lyapina et al., 2001; Schwechheimer et al., 2001). Hence, it is somewhat surprising to find that the CSN1N domain is dispensable for CSN′s Rub1 deconjugation activity on cullin. Hrt1/Roc1/Rbx1 has been shown to bind to human Cul1 and facilitate its Nedd8/Rub1 modification (Furukawa et al., 2000). It is possible that the interaction of CSN1with this RING-finger protein may influence other aspects of SCF function.
CSN1 Is Essential for Structural Integrity of COP9 Signalosome
The dramatic effect of G222R mutation as in fus6-T236 highlights the predominant role of the central region of CSN1 in complex assembly. When this region is preserved as in N270 and C231 transgenes, a full-size complex is formed. Interestingly, the N270 fragment is largely equivalent to the lethal mutants fus6-T414 and fus6-G236 mutants in that the C-terminal 170 aa residues are missing. Yet, although fus6/N270 appears normal as seedlings and contains the complex (Figures 5 and 6), fus6-T414 and fus6-G236 exhibit a typical fusca phenotype (Misera et al., 1994) and lack the complex (our unpublished data). The CSN1 accumulation in fus6-G236 reported by Staub et al. (1996) might possibly be due to wild-type contamination. We suggest that the 35S-promoter–mediated overexpression of N270, at least at mRNA level, may partially compensate for the inferior ability of N270 in maintaining a stable complex. The fus6/N270 adult plants display signs of complex deficiency and instability at later developmental stages (our unpublished data), agreeing with the hypothesis that the C-terminal domain of CSN1 assists in maintaining structural stability of the complex. In the absence of CSN1, CSN4 and CSN7 cofractionate primarily in fractions corresponding to 250–300 kDa (Figure 1). This small complex may represent an assembly intermediate, or part of another complex. We have been unsuccessful in further characterizing this small complex, probably due to its low abundance and instability.
Among all of the CSN1 transgenes generated, we observed a strict correlation between CSN1 protein accumulation and complex assembly. Thus, the described CSN1 transgenic mutant series nicely illustrated a point, which has been suggested from the characterization of CSN mutants (Wei and Deng et al., 1999), that the CSN1 protein accumulation in planta depends on the complex formation. This notion is further reinforced by the findings that CSN1 proteins, either wild-type or mutated forms, have not been found to accumulate outside of the CSN complex in arabidopsis. Curiously, the correlation of integration and accumulation does not apply to transient expression systems, because the CSN1N fragment can be transiently expressed to high levels in mammalian cells (Tsuge et al., 2001) or in onion cells (our unpublished data).
The nature of CSN1N-dependent activity remains to be revealed. As an essential aspect of the CSN functions, CSN1N could confer proper subcellular localization, act as a docking site for critical molecules or complexes, or provide essential enzymatic activities to the complex. Elucidation of such activities will help to understand the molecular mechanisms underlying the functions of the COP9 signalosome.
ACKNOWLEDGMENTS
We thank Dr. Xingwang Deng for plant growth facility and the genetic materials and antibodies, Dr. D. Chamovitz for CSN7 antibody, and Dr. P. Quail for PhyA antibody. We thank Dr. Tomohiko Tsuge for critical reading of the manuscript. This research is supported by a U.S. Department of Agriculture grant (99-35304-8641), and a national Institutes of Health grant (1 R01 GM-61812-01) to N.W.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–08-0427. Article and publication date are at www.molbiolcell.org/cgi/10.1091/mbc.01–08-0427.
REFERENCES
- Castle L, Meinke D. A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell. 1994;6:25–41. doi: 10.1105/tpc.6.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Clough RC, Vierstra RD. Phytochrome degradation. Plant Cell Environ. 1997;20:713–721. [Google Scholar]
- del Pozo JC, Estelle M. The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc Natl Acad Sci USA. 1999;96:153. doi: 10.1073/pnas.96.26.15342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Timpte C, Tan S, Callis J, Estelle M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- Deng XW, Caspar T, Quail PH. COP1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- Deng XW, et al. Unified nomenclature for the COP9 signalosome: an essential regulator of development. Trends Genet. 2000;16:289. doi: 10.1016/s0168-9525(00)02071-0. [DOI] [PubMed] [Google Scholar]
- Deng XW, Quail PH. Signaling in light-controlled development. Semin Cell Dev Biol. 1999;10:121–129. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Frankhauser C, Chory J. Light control of plant development. Annu Rev Plant Physiol Plant Mol Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Freilich S, Oron E, Kapp Y, Nevo-Caspi Y, Orgad S, Segal D, Chamovitz DA. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr Biol. 1999;9:1187–1190. doi: 10.1016/S0960-9822(00)80023-8. [DOI] [PubMed] [Google Scholar]
- Fuglevand G, Jackson JA, Jenkins GI. UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell. 1996;8:2347–2357. doi: 10.1105/tpc.8.12.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M, Zhang Y, McCarville J, Ohta T, Xiong Y. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol Cell Biol. 2000;20:8185–8197. doi: 10.1128/mcb.20.21.8185-8197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis EA, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins. In: Ausbel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology, 27 (suppl) New York: John Wiley & Sons; 1994. pp. 13.14.1–13.14.17. [Google Scholar]
- Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol. 2000;2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Pyrowolakis G. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 2000;10:335–342. doi: 10.1016/s0962-8924(00)01785-2. [DOI] [PubMed] [Google Scholar]
- Kang D, Wang X, Cao K, Sun C, Deng XW, Wei N. A gain-of-function phenotype conferred by over-expression of functional subunits of the COP9 signalosome in Arabidopsis. Plant J. 2000;23:597–608. doi: 10.1046/j.1365-313x.2000.00824.x. [DOI] [PubMed] [Google Scholar]
- Kapelari B, Bech-Otschir D, Hegerl R, Schade R, Dumdey R, Dubiel W. Electron microscopy and subunit-subunit interaction studies reveal a first architecture of COP9 signalosome. J Mol Biol. 2000;300:1169–1178. doi: 10.1006/jmbi.2000.3912. [DOI] [PubMed] [Google Scholar]
- Karniol B, Malec P, Chamovitz DA. Arabidopsis FUSCA5 encodes a novel phosphoprotein that is a component of the COP9 complex. Plant Cell. 1999;11:839–848. doi: 10.1105/tpc.11.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Hofmann K, von Arnim AG, Chamovitz DA. PCI complexes: pretty complex interactions in diverse signaling pathways. Trends Plant Sci. 2001;6:379–386. doi: 10.1016/s1360-1385(01)02015-5. [DOI] [PubMed] [Google Scholar]
- Kwok SF, Piekos B, Misera S, Deng XW. A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 1996;110:732–742. doi: 10.1104/pp.110.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok FS, Solano R, Tsuge T, Chamovitz DA, Matsui M, Ecker JR, Deng XW. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell. 1998;10:1779–1790. doi: 10.1105/tpc.10.11.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok SF, Staub JM, Deng XW. Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J Mol Biol. 1999;285:85–95. doi: 10.1006/jmbi.1998.2315. [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Mayer R, Raventos D, Chua NH. det1, cop1, and cop9 mutations cause inappropriate expression of several gene sets. Plant Cell. 1996;8:1951–1959. doi: 10.1105/tpc.8.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misera S, Muller AJ, Weiland-Heidecker U, Jurgens G. The FUSCA genes of arabidopsis: negative regulators of light responses. Mol Gen Genet. 1994;244:242–252. doi: 10.1007/BF00285451. [DOI] [PubMed] [Google Scholar]
- Mundt KE, et al. The C.O.P9 signalosome complex is conserved in fission yeast and has a role in S. phase. Curr Biol. 1999;9:1427–1430. doi: 10.1016/s0960-9822(00)80091-3. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CH, Deng XW. Targeted destabilization of HY5 during light-regulated Arabidopsis development. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Parks BM, Quail PH. hy8, a new class of Arabidopsis mutants deficient in functional phytochrome A. Plant Cell. 1993;5:39–48. doi: 10.1105/tpc.5.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Serino G, Deng X-W. Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted development processes in arabidopsis. Plant Cell. 2001;13:2393–2407. doi: 10.1105/tpc.010248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S, Lapointe G, Rutledge RG, Seguin A. Induction of chalcone synthase expression in white spruce by wounding and jasmonate. Plant Cell Physiol. 2000;41:982–987. doi: 10.1093/pcp/pcd017. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292:1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- Serino G, Tsuge T, Kwok SF, Matsui M, Wei N, Deng XW. Arabidopsis cop8 and fus4 mutations define the same locus that encodes subunit 4 of the COP9 signalosome. Plant Cell. 1999;11:1967–1979. doi: 10.1105/tpc.11.10.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Wei N, Deng XW. Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell. 1996;8:2047–2056. doi: 10.1105/tpc.8.11.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T, Matsui M, Wei N. The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J Mol Biol. 2001;305:1–9. doi: 10.1006/jmbi.2000.4288. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. Light control of seedling development. Annu Rev Plant Physiol. 1996;47:215–243. doi: 10.1146/annurev.arplant.47.1.215. [DOI] [PubMed] [Google Scholar]
- Wei N, Chamovitz DA, Deng XW. Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell. 1994a;78:117–124. doi: 10.1016/0092-8674(94)90578-9. [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW. The role of pleiotropic COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 1996;112:871–878. doi: 10.1104/pp.112.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. Making sense of the COP9 signalosome. Trends Genet. 1999;15:98–103. doi: 10.1016/s0168-9525(98)01670-9. [DOI] [PubMed] [Google Scholar]
- Wei N, Kwok SF, von Arnim AG, Lee A, McNellis T, Piekos B, Deng XW. Arabidopsis COP8, COP10, COP11 genes are involved in repression of photomorphogenic developmental pathway in darkness. Plant Cell. 1994b;6:629–643. doi: 10.1105/tpc.6.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Matsui M, Ang LH, Deng XW. Role of COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell. 1998;10:1083–1094. doi: 10.1105/tpc.10.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Seibert V, Geyer R, Rhee E, Lyapina S, Cope G, Deshaies RJ, Wolf DA. The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem. 2001;2:7. doi: 10.1186/1471-2091-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]