Abstract
Despite existing reports on differential DNA methylation in type 2 diabetes (T2D) and obesity, our understanding of its functional relevance remains limited. Here we show the effect of differential methylation in the early phases of T2D pathology by a blood-based epigenome-wide association study of 4808 non-diabetic Europeans in the discovery phase and 11,750 individuals in the replication. We identify CpGs in LETM1, RBM20, IRS2, MAN2A2 and the 1q25.3 region associated with fasting insulin, and in FCRL6, SLAMF1, APOBEC3H and the 15q26.1 region with fasting glucose. In silico cross-omics analyses highlight the role of differential methylation in the crosstalk between the adaptive immune system and glucose homeostasis. The differential methylation explains at least 16.9% of the association between obesity and insulin. Our study sheds light on the biological interactions between genetic variants driving differential methylation and gene expression in the early pathogenesis of T2D.
Subject terms: DNA methylation, Epigenomics, Transcriptomics, Epigenetics, Type 2 diabetes
Our understanding of the functional link between differential DNA methylation and type 2 diabetes and obesity remains limited. Here the authors present a blood-based EWAS of fasting glucose and insulin among 4808 non-diabetic Europeans and identify nine CpGs not previously implicated in glucose, insulin homeostasis and diabetes.
Introduction
Type 2 diabetes (T2D) is a common metabolic disease, characterized by disturbances in glucose and insulin metabolism. The pathogenesis of T2D is driven by inherited and environmental factors1. There is increasing interest in differential DNA methylation in the development of T2D as well as with glucose and insulin metabolism2–6. Depending on the region, DNA methylation may result in gene silencing and thus regulate gene expression and subsequent cellular functions7. Differential methylation in the circulation may predict the development of future T2D beyond traditional risk factors such as age and obesity3,8, but it may also be part of the biological mechanism that links age and/or obesity to glucose, insulin metabolism and/or T2D. A recent longitudinal study with multiple visits reported that most DNA methylation changes occur 80–90 days before detectable glucose elevation9, suggesting that differential DNA methylation evokes changes in glucose and is involved in the early stage(s) of diabetes. Differential DNA methylation is further associated with obesity, which is an important driver of the T2D risk and also precedes the increase in glucose and insulin level in persons developing T2D8. A key question to answer is whether the differential methylation associated with glucose and insulin metabolism is an irrelevant epiphenomenon that is related to obesity acting as a statistical confounder or whether there are functional effects of the differential methylation relevant of obesity that is associated to metabolic pathology.
Here, we aim to determine the relation of differential DNA methylation and fasting glucose and insulin metabolism as markers of early stages of diabetes pathology in non-diabetic subjects, accounting for obesity measured as body mass index (BMI). We identify and replicate nine CpG sites associated with fasting glucose (in FCRL6, SLAMF1, APOBEC3H and the 15q26.1 region) and insulin (in LETM1, RBM20, IRS2, MAN2A2 and the 1q25.3 region). Using cross-omics analyses, we present in silico evidence supporting the functional relevance of the CpG sites on the development and progression of diabetes, in terms of their effect on expression paths and elucidate the genetic networks involved.
Results
Epigenome-wide association analysis and replication
In the discovery phase, we performed a blood-based epigenome-wide association study (EWAS) meta-analysis of four cohorts including 4,808 non-diabetic individuals of European ancestry (Supplementary Data 1), which revealed differential DNA methylation at 28 unique CpG sites in either the baseline model without BMI adjustment or in the second model with BMI adjustment (Table 1 and Supplementary Table 1). The summary statistic results of the EWAS are provided as a Data file [https://figshare.com/s/1a1e8ac0fd9a49e2be30]. These include three CpG sites associated with both insulin and glucose, eight CpG sites associated with fasting glucose only and 17 with fasting insulin (P value < 1.3 × 10−7 in meta-analysis). Of these 28 CpG sites, 13 were identified by earlier EWAS studies of either T2D or related traits, including glucose, insulin, hemoglobin A1c (HbA1c), and homeostatic model assessment-insulin resistance (HOMA-IR)2–5,8,10,11 (Supplementary Table 1). The known CpG sites include three sites located in SLC7A11, CPT1A and SREBF1 that are associated with both glucose and insulin. The remaining ten CpG sites, located in DHCR24, CPT1A, RNF145, ASAM, KDM2B, MYO5C, TMEM49, ABCG1 (harboring two CpG sites) and the 4p15.33 region, are associated with insulin only. All of the previously reported CpG sites with glycemic traits are also associated with BMI in previous EWAS8,10,12–15 (Supplementary Table 1).
Table1.
CpG sites associated with glycemic traits in discovery phase
| Locus | CpG | Chr: Pos | Trait | BetaM1 | P valueM1 | BetaM2 | P valueM2 |
|---|---|---|---|---|---|---|---|
| FCRL6 | cg00936728 | 1: 159772194 | Glucose | −1.79 | 9.1 × 10−8ǂ | −1.60 | 1.9 × 10−7 |
| SLAMF1 | cg18881723 | 1: 160616870 | Glucose | 1.16 | 7.5 × 10−8ǂ | 1.25 | 3.4 × 10−10ǂ |
| 1q25.3 | cg13222915 | 1: 184598594 | Insulin | −1.69 | 2.6 × 10−9ǂ | −1.06 | 4.1 × 10−6 |
| BRE | cg20657709 | 2: 28509570 | Glucose | −1.42 | 2.7 × 10−6 | −1.53 | 4.1 × 10−8ǂ |
| LRPPRC | cg01913188 | 2: 44223249 | Glucose | 1.18 | 9.4 × 10−6 | 1.38 | 5.7 × 10−9ǂ |
| IRAK2 | cg14527942 | 3: 10276383 | Insulin | 2.44 | 3.4 × 10−10ǂ | 2.14 | 2.9 × 10−11ǂ |
| LETM1 | cg13729116 | 4: 1859262 | Insulin | 2.38 | 4.3 × 10−8ǂ | 1.64 | 4.5 × 10−6 |
| RBM20 | cg15880704 | 10: 112546110 | Insulin | 2.50 | 3.8 × 10−9ǂ | 1.38 | 6.7 × 10−5 |
| IRS2 | cg25924746 | 13: 110432935 | Insulin | 2.11 | 3.0 × 10−9ǂ | 1.32 | 4.9 × 10−6 |
| SPTB | cg07119168 | 14: 65225253 | Glucose | −1.64 | 4.4 × 10−7 | −1.63 | 4.9 × 10−8ǂ |
| 15q26.1 | cg18247172 | 15: 91370233 | Glucose | −1.05 | 4.9 × 10−6 | −1.18 | 2.8 × 10−8ǂ |
| MAN2A2 | cg20507228 | 15: 91460071 | Insulin | 1.18 | 5.5 × 10−8ǂ | 0.87 | 9.0 × 10−7 |
| FAM92B | cg06709610 | 16: 85143924 | Insulin | 6.22 | 6.5 × 10−9ǂ | 6.30 | 5.8 × 10−13ǂ |
| CD300A | cg08087047 | 17: 72461209 | Glucose | −1.35 | 5.9 × 10−6 | −1.45 | 1.1 × 10−7ǂ |
| APOBEC3H | cg06229674 | 22: 39492189 | Glucose | −1.62 | 1.8 × 10−6 | −1.70 | 4.7 × 10−8ǂ |
Novel epigenome-wide significant results in the discovery phase (n = 4808) are shown. Model 1 (M1) indicates inverse variance-weighted fixed effect meta-analysis of effect estimates in four cohorts. Each cohort performed a regression model adjusting for age, sex, technical covariates, white blood cell, and smoking status, and accounting for family structure in family-based cohorts. Model 2 (M2) indicates the meta-analysis of the same studies, adjusting for body mass index (BMI) additionally. Locus: the cytogenetic location or the gene symbol of the CpG sites from Illumina annotation. Beta: effect estimate of the meta-analysis. P value shown is genomic controlled after meta-analysis. The effect refers to the increase/ decrease in fasting glucose/ insulin as the outcome in the model
ǂSignificant results (P value < 1.3 × 10−7)
The 15 novel CpG sites were tested using the same statistical models in 11 independent cohorts, including 11,750 non-diabetic participants from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium (Supplementary Data 1). Nine unique CpG-trait associations were replicated when correcting for multiple testing using Bonferroni (15 CpGs, P value threshold for significance < 3.3 × 10−3) and were investigated in the further analyses (Table 2). These include five sites (in LETM1, RBM20, IRS2, MAN2A2 and the 1q25.3 region) associated with fasting insulin and one site (in FCRL6) associated with fasting glucose in the baseline model without adjusting for BMI, and three (in SLAMF1, APOBEC3H and the 15q26.1 region, all associated with fasting glucose) emerging in the BMI-adjusted model. Of note, no locus was found to be associated with fasting insulin in the BMI-adjusted model.
Table2.
CpG sites associated with glycemic traits in replication
| Locus | CpG | Chr: Pos | Trait | BetaM1 | P valueM1 | BetaM2 | P valueM2 |
|---|---|---|---|---|---|---|---|
| FCRL6 | cg00936728 | 1: 159772194 | Glucose | −1.55 × 10−3 | 9.6 × 10−5ǂ | NP | NP |
| SLAMF1 | cg18881723 | 1: 160616870 | Glucose | 1.17 × 10−3 | 7.7 × 10−3 | 1.48 × 10−3 | 1.2 × 10−3ǂ |
| 1q25.3 | cg13222915 | 1: 184598594 | Insulin | −3.77 × 10−3 | 3.3 × 10−16ǂ | NP | NP |
| BRE | cg20657709 | 2: 28509570 | Glucose | NP | NP | −9.40 × 10−4 | 0.036 |
| LRPPRC | cg01913188 | 2: 44223249 | Glucose | NP | NP | 1.64 × 10−5 | 0.90 |
| IRAK2 | cg14527942 | 3: 10276383 | Insulin | −6.49 × 10−5 | 0.48 | −7.72 × 10−5 | 0.45 |
| LETM1 | cg13729116 | 4: 1859262 | Insulin | 1.92 × 10−3 | 7.0 × 10−7ǂ | NP | NP |
| RBM20 | cg15880704 | 10: 112546110 | Insulin | 3.05 × 10−3 | 8.6 × 10−12ǂ | NP | NP |
| IRS2 | cg25924746 | 13: 110432935 | Insulin | 3.38 × 10−3 | 3.0 × 10−11ǂ | NP | NP |
| SPTB | cg07119168 | 14: 65225253 | Glucose | NP | NP | −7.18 × 10−4 | 0.070 |
| 15q26.1 | cg18247172 | 15: 91370233 | Glucose | NP | NP | −1.77 × 10−3 | 5.1 × 10−4ǂ |
| MAN2A2 | cg20507228 | 15: 91460071 | Insulin | 6.11 × 10−3 | 2.3 × 10−15ǂ | NP | NP |
| FAM92B | cg06709610 | 16: 85143924 | Insulin | 2.08 × 10−5 | 0.81 | 5.37 × 10−5 | 0.59 |
| CD300A | cg08087047 | 17: 72461209 | Glucose | NP | NP | −4.92 × 10−4 | 0.28 |
| APOBEC3H | cg06229674 | 22: 39492189 | Glucose | NP | NP | −2.09 × 10−3 | 1.4 × 10−6ǂ |
Novel epigenome-wide significant results in the replication (n = 11,750) are shown. Replication was not performed in the non-significant associated model or trait (NP). Model 1 (M1) indicates inverse variance-weighted fixed effect meta-analysis of effect estimates in the 11 cohorts. Each study performed a regression model adjusting for age, sex, technical covariates, white blood cell, and smoking status, and accounting for family structure in family-based cohorts. Model 2 (M2) indicates the meta-analysis of the same studies, adjusting for body mass index (BMI) additionally. Locus: the cytogenetic location or the gene symbol of the CpG sites from Illumina annotation. Beta: effect estimate of the meta-analysis. The effect refers to the increase/ decrease in methylation as the outcome in the model
ǂSignificant results (P value < 3.3 × 10−3)
Because the replication cohorts also included individuals of African ancestry (AA, n = 4355) and Hispanic ancestry (HA, n = 577), we also performed the replication stratified by ancestry (Supplementary Data 2). Two CpG sites (cg13222915 and cg18247172) were replicated in the AA population when corrected by the number of tests and two (cg00936728 and cg06229674) replicated with nominal significance. In the HA population, cg20507228 was replicated at nominal significance. Two CpG sites (cg18881723 and cg13222915) show the opposite direction for the effect estimate in HA ancestry population as compared to the other two populations. However, the estimates of effect size are not significantly different from zero (P value = 0.63 in cg18881723 and P value = 0.092 in cg13222915).
Glycemic differential DNA methylation and transcriptomics
To determine whether the differential DNA methylation has functional effects on gene expression and subsequent cellular functions, we conducted three series of analyses. Figure 1 shows the overview of the cross-omics analyses. First, we explored the Genotype-Tissue Expression (GTEx)16 database for the expression levels of the genes which annotated to the novel CpG sites. We found that the genes are expressed in a wide range of tissues, including whole blood and spleen (in particular MAN2A2 and RBM20), but also other tissues relevant for glucose and insulin metabolism such as adipose subcutaneous, adipose visceral omentum, liver (in particular, SLAMF1, APOBEC3H, FCRL6 and RBM20), pancreas and skeletal muscle (in particular, SLAMF1, APOBEC3H, FCRL6 and MAN2A2) and small intestine terminal ileum (in particular, MAN2A2, RBM20, FCRL6 and APOBEC3H; Supplementary Figure 1).
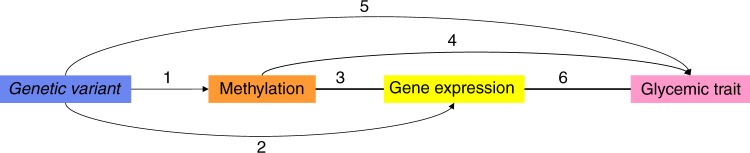
Fig. 1.
Overview of the cross-omics analysis. (1) Methylation quantitative trait loci (meQTL). (2) Expression quantitative trait loci (eQTL). (3) Expression quantitative trait methylation (eQTM). (4) Epigenome-wide association study (EWAS) and Mendelian randomization (MR). (5) Genome-wide association study (GWAS). (6) The association of gene expression expressed in the glucose or insulin metabolism-related tissues and glycemic traits. Results in 1, 2, 3 were extracted from the summary statistics from Biobank-based Integrative Omics Study (BIOS) database (n = 3814). Results in 4 was the results in the current EWAS (discovery phase, n = 4808, replication phase, n = 11,750) and the two-sample Mendelian randomization based on the BIOS database (n = 3814) and GWAS results of Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC). Results in 5 was from the GWAS results of MAGIC or the DIAbetes Genetics Replication And Meta-analysis consortium (DIAGRAM, n = 96,496–452,244). Results in 6 was based on the summary statistics of Genotype-Tissue Expression project (GTEx) and MAGIC or DIAGRAM (n = 153–491)
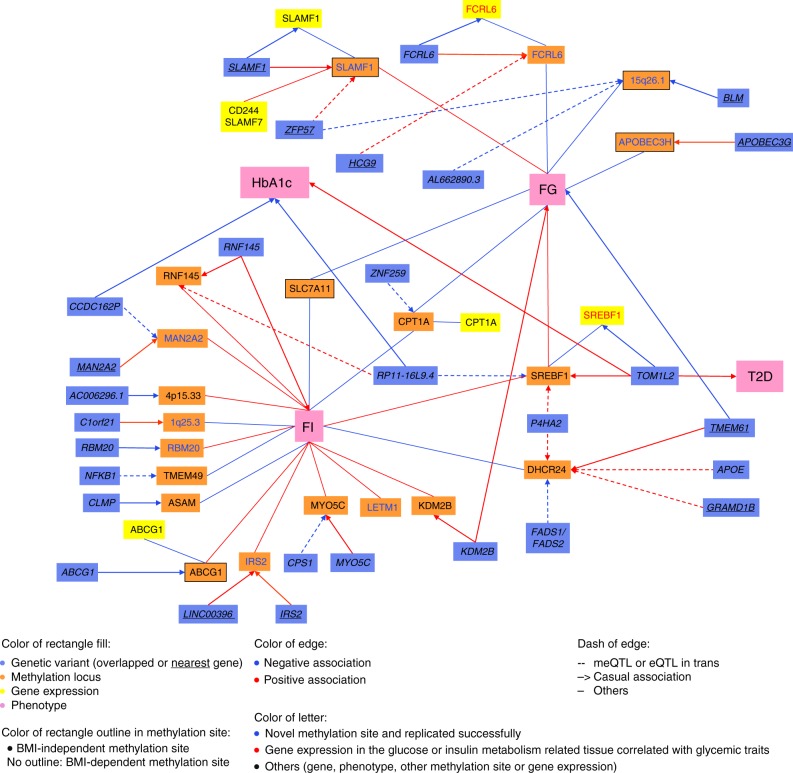
Second, the effect on gene expression in blood of the previously identified 11 independent CpG sites (cg00574958 in CPT1A and cg06500161 in ABCG1 were used) and the nine novel sites from our current study was examined in the Biobank-based Integrative Omics Study (BIOS) database that is part of the Biobanking and BioMolecular Infrastructure of the Netherlands (BBMRI-NL)17 (indicated in Fig. 2 in the orange boxes). We found that five CpG sites, i.e. cg00936728 (FCRL6), cg18881723 (SLAMF1), cg00574958 (CPT1A), cg11024682 (SREBF1) and cg06500161 (ABCG1), are expression quantitative trait methylations (eQTMs), i.e. there is correlation between gene expression and methylation18. In most cases, the differential methylation levels are associated with the expression (indicated in Fig. 2 in the yellow boxes) of their respective genes. Cg18881723 (SLAMF1) is also associated with the expression of two other genes near SLAMF1, i.e. SLAMF7 and CD244 (Supplementary Table 2).
Fig. 2.
Significant associations of the cross-omics integration. The effect allele is standardized across all associations. Only the significant associations which passed the specific P value threshold in each association step and the direction of effects consistent were shown in the figure. FG fasting glucose. FI fasting insulin, T2D type 2 diabetes, HbA1c hemoglobin A1c
Third, we investigated whether the genetically regulated expression of the annotated genes in specific tissues is altered in T2D or related traits, such as glucose, insulin and HbA1c. To answer this question, we mined in the MetaXcan database for genome-wide association studies (GWAS) of T2D, fasting glucose, HbA1c, insulin and HOMA-IR19–23 as a genetic proxy for the traits24. No association was found between glycemic traits and the DNA expression in adipose subcutaneous, adipose visceral omentum and small intestine terminal ileum. Supplementary Table 3 gives the significant findings for tissues known to be implicated in glucose and insulin metabolism including blood, liver, pancreas and skeletal muscle (P value < 0.05 for MetaXcan). As described earlier, we associated the increased expression of SREBF1 with decreased risk of T2D and decreased HbA1c levels in the whole blood25. The increased expression in the whole blood of ABOBEC3H, a methylation locus we identified in the present study, is associated with increased HOMA-IR level, a measure of insulin resistance. In skeletal muscle, the increased fasting glucose is associated with the increased expression of KDM2B and decreased expression of MAN2A2. Moreover, we discovered that increased hepatic expression of FCRL6, which was annotated to the methylation locus associated with fasting glucose in the present study, is associated with the risk of T2D. In the pancreas, the increased expression of the methylation loci MYO5C and RBM20 are associated with increased fasting glucose levels.
Glycemic differential DNA methylation and genomics
Although differential DNA methylation may be the result of environmental exposures, the process is often (partly) heritable with genetic variants (co-)determining the process26. Therefore, we next set out to find whether the differential methylation associated with fasting glucose and insulin levels is driven by genetic variants which referred to as methylation quantitative trait loci (meQTLs). Using the BIOS database (blood-based data)17, we were able to study 18 out of the 20 unique CpG sites in this respect. We associated 2,991 single-nucleotide polymorphisms (SNPs) in 29 unique meQTLs (indicated in Fig. 2 in the blue boxes) with differential methylation either in cis or trans (for details see Supplementary Data 3). Six of these meQTLs (4 cis and 2 trans-acting) are also associated with T2D, fasting glucose, fasting Insulin, or HbA1c in earlier studies21,22,27–29 and the directions of the effect between the SNP, methylation and glycemic traits are consistent (shown in Fig. 2 in the pink boxes, for details see Supplementary Table 4). A genetic locus near TMEM61 is a common genetic driver affecting the differential methylation at nearby CpG cg17901584 (DHCR24) in our study and fasting glucose levels in an earlier study22. Further, the RNF145 locus was found to be a common driver affecting the differential methylation at cg26403843 (RNF145) and fasting insulin levels21. The KDM2B locus affects differential methylation at cg13708645 (KDM2B) and fasting glucose levels22, and the TOM1L2/RAI1 locus affects the differential methylation at cg11024682 (SREBF1) as well as HbA1c and T2D27,28. Two trans-acting loci involve a genetic locus in CCDC162P that is affecting differential methylation at cg20507228 (MAN2A2) and HbA1c27 and the genetic locus in RP11-16L9.4 affecting the differential methylation at cg11024682 (SREBF1) and HbA1c27.
We next explored if these genetic variants associated with differential methylation (meQTLs) are also associated with gene expression, i.e. quantitative trait loci (eQTLs; see the integrated outline of analyses in Fig. 2 and detailed in Supplementary Table 5). We searched specifically for expression profiles earlier associated with glycemic CpG sites in blood (listed in Supplementary Table 2). We associated three genetic variants with both differential methylation and gene expression in blood. These include that: 1) rs11265282 in FCRL6 is positively associated with the differential methylation at cg00936728 (FCRL6) and decreased the expression of FCRL6 in blood, 2) rs1577544 near SLAMF1 is associated with decreased differential methylation at cg18881723 (SLAMF1) and decreased SLAMF1 expression in blood, and 3) rs6502629 in TOM1L2 is associated with increased differential methylation at cg11024682 (SREBF1) and decreased SREBF1 expression in blood.
As we observed that the genes driving glycemic CpG sites overlapped with genetic determinants of T2D or related traits, we studied the causal effect of differential methylation on glucose and insulin metabolism with a generalized summary statistic-based Mendelian randomization (MR) test30. Up to eight independent genetic variants include in the genetic risk score were used as the instrumental variable for each CpG. Thirteen CpG sites out of the initial 20 met the present MR criteria and were tested by MR (Supplementary Data 4). No significant association was detected when adjusting for multiple testing accounting for 13 independent tests (P value threshold for significance < 3.8 × 10−3). The genetic risk score for cg15880704 (RBM20) methylation levels is nominally significantly associated with fasting insulin levels (P value = 0.04), and the genetic risk score for cg18881723 (SLAMF1) levels is nominally associated with fasting glucose levels (P value = 0.05) in the MR tests.
Multi-omics integration and functional annotation
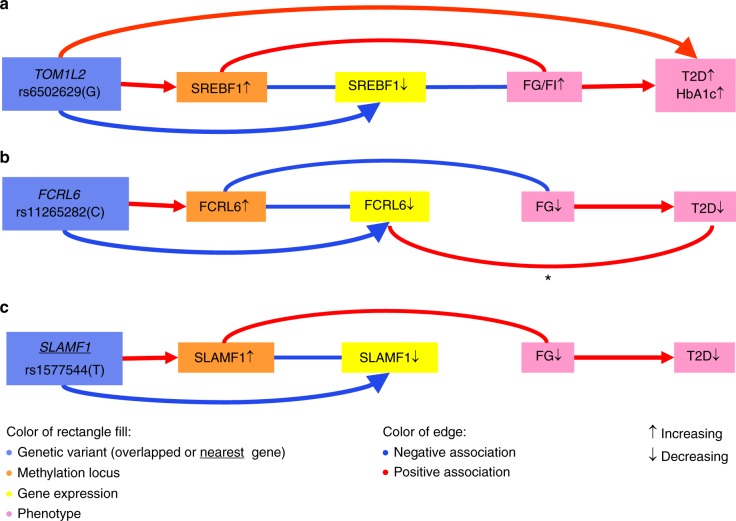
To understand the biological relevance of our findings, we first integrated the cascade of associations into genomics, epigenomics, transcriptomics and glycemic traits through EWAS, eQTM, meQTL and eQTL. There are three pathways emerging when considering the consistency of the direction of the effects between the associations. One pathway involves SREBF1, which in part, was reported earlier3,25,31 but substantially extended in the current report. The other two involve differential methylation of FCRL6 and SLAMF1 (Fig. 3). The C allele of rs11265282 in FCRL6 is associated with increased methylation, which turns down the FCRL6 expression in blood. In addition, the genetically decreased FCRL6 expression in the liver is also associated with a decreased risk of T2D. The T allele of rs1577544 near SLAMF1 increases the differential methylation levels in the blood, which decreases SLAMF1 expression in the circulation, which is consistent with the negative association between the genetic variant and gene expression levels.
Fig. 3.
The cross-omics integration of CpGs in SREBF1 (a), FCRL6 (b) and SLAMF1 (c). Cascading associations cross multi-omics were integrated in the network. * The association happens in the FCRL6 expression in liver. All other differential methylation or gene expression was measured in blood. FG fasting glucose, FI fasting insulin, T2D type 2 diabetes, HbA1c hemoglobin A1c
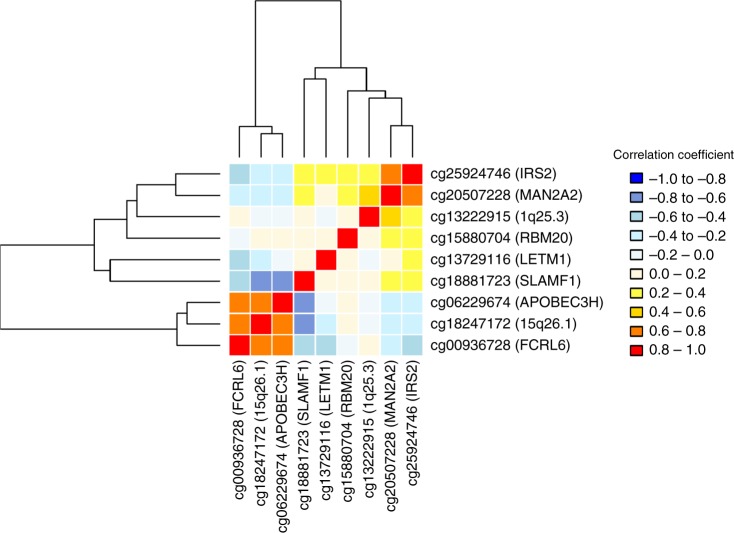
To understand the correlation of the findings, we clustered the normalized differential methylation values of the nine novel CpG sites including those not annotated to a gene. Two clusters emerge, one including IRS2, MAN2A2, 1q25.3 locus (intergenic), RBM20, LETM1 and SLAMF1 and the second one including FCRL6, 15q26.1 (intergenic) and APOBEC3H (Fig. 4 and Supplementary Table 6). Four CpGs in FCRL6, 15q26.1 (intergenic), APOBEC3H and SLAMF1 are highly correlated with each other, in which the absolute correlation coefficients are bigger than 0.6, while they are located in different chromosomes, suggesting a common biological mechanism: SLAMF1 and FCRL6 from chromosome 1, APOBEC3H from chromosome 22 and 15q26.1 from chromosome 15. We next performed gene set enrichment analysis in different pathway databases, including KEGG pathways32, Reactome Pathway Knowledgebase33 and Gene Ontology (GO) biological process classification34. We found that the genes in the first cluster are highly enriched together in multiple pathways, including regulation of leukocyte proliferation, protein secretion and cell activation (SLAMF1 and IRS2), hexose, monosaccharide and carbohydrate metabolism (IRS2 and MAN2A2). Further, SLAMF1 (cluster 1) and APOBEC3H (cluster 2) are both enriched in immune effector processes and innate immune response (Supplementary Table 7).
Fig. 4.
Clustered correlation of the nine novel glycemic CpGs. The correlation of the novel CpG sites was checked by Pearson’s correlation test (n = 1544). The hierarchical cluster analysis was used in the clustering
BMI in the association of methylation and glycemic traits
Of note, among the 20 methylation loci associated with glycemic metabolism in the present analyses, 11 are associated with BMI in the previous EWAS8,10,12–15. These 11 loci are all associated with insulin metabolism (Supplementary Table 1). Based on the bi-direction MR findings performed as part of the previous EWAS of BMI8, we found that BMI appears to drive methylation for cg06500161 (ABCG1, P value = 6.4 × 10−5), a CpG that we associated with insulin levels. Using a marginal P value of 0.05 in their MR results, the differential methylation appears to be a consequence of obesity rather than a cause for three other CpG sites: cg110244682 (SREBF1; P value = 4.1 × 10−3), cg17901584 (DHCR24; P value = 4.1 × 10−3) and cg26403843 (RNF145; P value = 0.011)8. Taken together (Supplementary Table 1), our results raise the question whether BMI is driving differential methylation, which subsequently raises insulin level in the circulation. Such a pathway would predict that the association between BMI and insulin changes when adjusting for differential methylation at ABCG1, SREBF1, DHCR24 and RNF145. We tested this hypothesis in the non-diabetic individuals of the Rotterdam Study by comparing the relationship between BMI and fasting insulin with and without adjusting for the methylation levels at the four CpG sites. The variance explained (R2) by the linear regression model improves significantly from 0.40 to 0.43 (P value = 1.2 × 10−13 by analysis of variance (ANOVA) testing) when adjusting for the CpG effect, while the effect estimates for BMI decrease by 9.2% (beta: 0.065, standard error (SE): 0.003, P value = 1.2 × 10−82 for the model without CpG adjustment compared to beta: 0.059, SE: 0.003, P value = 2.9 × 10−70 adjusting for the four CpGs). When we extended the adjustment to the 16 CpG sites associated with circulating insulin levels, the variance explained by the model improves further (R2 = 0.46, P value = 2.1 × 10−18) and the beta for BMI reduces further by 16.9% (beta: 0.054, SE: 0.003, P value = 4.6 × 10−58 for the model adjusting for 16 CpG sites).
Discussion
The current large-scale EWAS identify and replicate nine CpG sites associated with fasting glucose (in FCRL6, SLAMF1, APOBEC3H and the 15q26.1 region) or insulin (in LETM1, RBM20, IRS2, MAN2A2 and the 1q25.3 region). When we adjust for BMI as a potential confounder, three CpG sites (in SLAMF1, APOBEC3H and the 15q26.1 region) are associated with fasting glucose only after adjustment for BMI. We validate 13 previously reported CpG sites from 11 independent genetic loci2–6,8,10,12–15 and complement the understanding on why these CpG sites are associated with T2D and/or glycemic traits based on comprehensive cross-omics analyses. We present in silico evidence supporting the functional relevance of the CpG sites, in terms of their effect on expression paths and elucidate the genetic networks involved.
Our data show that differential methylation plays a key role in understanding the immunological changes observed in glucose metabolism35. SLAMF1 and APOBC3H are both enriched in immune function and the innate immune response. The differential methylation level at FCRL6, 15q26.1 (intergenic), APOBEC3H and SLAMF1 were highly correlated though they were on three different chromosomes. This finding suggests a common pathway. SLAMF1 belongs to the immunoglobulin gene superfamily and is involved in T-cell stimulation36. APOBEC3H proteins are part of an intrinsic immune defense that has potent activity against a variety of retroelements36 and its expression in whole blood is positively associated with HOMA-IR from the current study. FCRL6 is a distinct indicator of cytotoxic effector T-lymphocytes that is upregulated in diseases characterized by chronic immune stimulation36. Meanwhile, we show that decreased FCRL6 differential methylation increased expression of FCRL6 and fasting glucose in the blood. A key finding that links FCRL6 to glucose metabolism is that the genetically determined FCRL6 expression in the liver is also associated with decreased risk of T2D. In line with a role in immune relation and pathology37,38, the HLA region (6p22.1 region) is a key meQTLs of FCRL6 (rs2523946), 15q26.1 (rs3129055 and rs4324798) and SLAMF1 (rs3129055). Of interest is that in the population of non-diabetic individuals, we found strong signals of the immune system particularly when we adjust the effects attributed to BMI. Remarkably, three out of the four methylation loci at SLAMF1, APOBEC3H and the 15q26.1 region emerged in the BMI-adjusted model, suggesting that these associations were masked by confounding noise of BMI on methylation in opposite effects to that of insulin.
We studied the interplay between BMI, fasting glucose and insulin levels, and differential methylation in the circulation. On the one hand, we find evidence that the differential methylation of the insulin-related CpG sites together explained up to 16.9% of the association between obesity and insulin levels. These findings are in line with the Nature paper on the EWAS of BMI that found that the methylation patterns in blood predict future diabetes8. Our study reveals that insulin is a key player underlying the association reported earlier8. On the other hand, we find evidence that the association between differential methylation and insulin metabolism is attenuated up to 62%, e.g. CpG sites in SREBF1 (62%), ASAM (56%), CPT1A (54%) and TMEM49 (52%), when BMI is accounted for in the model, suggesting that the interplay between BMI, differential methylation and insulin metabolism is extremely complex and differs across CpG sites. BMI may be a confounder of associations for some CpGs but may be in the causal pathway for others.
To our knowledge, we report for the first time that, in blood, differential methylation of IRS2 was associated with fasting insulin level. Expression level of IRS2 (insulin receptor substrate 2) in β-cells in the pancreas are associated with the onset of diabetes39–41. Though the expression level of IRS2 is low in blood, we find its blood-based differential methylation was associated with fasting insulin. We also find an insulin-related genetic locus, MAN2A2 (mannosidase alpha class 2 A member 2) in our EWAS. MAN2A2 encodes an enzyme that forms intermediate asparagine-linked carbohydrates (N-glycans)42. It is related to the hexose/monosaccharide metabolism. In addition, the expression of MAN2A2 in skeletal muscle is negatively associated with fasting glucose level and the meQTL (rs9374080) of MAN2A2 associates with HbA1c27. Together, these findings suggest that regulating the differential methylation level or expression level of MAN2A2 may be relevant to the development of insulin resistance. Another interesting gene that emerged is the familial cardiomyopathy related gene RBM20, which may play a role in cardiovascular complications of diabetes via mediating insulin damage in cardiac tissues43. The expression of RBM20 in the pancreas is also associated with fasting glucose. The meQTL for RBM20 is associated with pulse rate (P value = 4.6 × 10−5) in UKBIOBANK GWAS44, and its mRNA is highly expressed in cardiac tissues45.
One limitation of our study is that the main findings are based on data from blood which was the only accessible tissue in our epidemiological studies and may not be representative of more disease-relevant tissues. However, the concordance of differential methylation between blood and adipose is high for certain pathways46. DNA methylation globally is considered a relatively stable epigenetic mark that can be inherited through multiple cell divisions47,48. However, some changes can be dynamic reflected by recent environmental exposures. This phenomenon could be site-specific. While our study provides a snapshot of associations specific to the fasting state, instant methylation of different CpG sites in the vicinity of IRS2 and KDM2B have been reported earlier49. Such effects may also occur at the loci presented in the present study. Our present MR analyses yield no evidence for the causal effects of CpG sites on fasting glucose or insulin. One limitation in the interpretation of the findings is that low power of the MR due to the fact we lack insight in the genes driving differential methylation. For instance, seven of the 13 performed CpG sites have instrumental variables which explain less than 5% of the exposure. Further studies are needed to include additional biologically relevant tissues and perform MR based on the tissue-specific meQLTs. Last but not least, cg19693031 in TXNIP has been repeatedly associated with type 2 diabetes case-control status earlier3,50,51. Although it did not pass our pre-defined EWAS significance threshold, TXNIP is associated with fasting glucose in the non-diabetic population (P value = 7.6 × 10−7 in the BMI adjustment model) if we take the current study aiming to replicate earlier findings. Of note is that cg19693031 is not associated with fasting insulin (in BMI-unadjusted model, p value = 0.30; in the BMI-adjusted model, p value = 0.37).
In conclusion, our large-scale EWAS and replication identifies nine differentially methylated sites associated with fasting glucose or insulin, and shows that differential methylation explains part of the association between obesity and insulin metabolism. The integrative in silico cross-omics analyses provide insights of glycemic loci into the genetics, epigenetics and transcriptomics pathways. We also highlight that differential methylation is a key point in the involvement of the adaptive immune system in glucose homeostasis. Further studies in the future will benefit from tissue-specific methylation and meQTL databases which are currently the missing piece of the in silico data integration framework.
Methods
Study population
The discovery samples consisted of 4808 European individuals without diabetes from four non-overlapped cohorts, recruited by Rotterdam Study III-1 (RS III-1, n = 626), Rotterdam Study II-3 and Rotterdam Study III-2 (called as RS-BIOS, n = 705), Netherlands Twin Register (NTR, n = 2753) and UK adult Twin registry (TwinsUK, n = 724). The replication sets contained up to 11,750 individuals from 11 independent cohorts from CHARGE, including up to 6818 individuals from European ancestry, 4355 from African ancestry and 577 from Hispanic ancestry (Supplementary Data 1). They are from Atherosclerosis Risk in Communities (ARIC) Study, Baltimore Longitudinal Study of Aging (BLSA), Cardiovascular Health Study (CHS), Framingham Heart Study Cohort (FHS), The Genetic Epidemiology Network of Arteriopathy (GENOA), Genetics of Lipid Lowering Drugs and Diet Network (GOLDN), Hypertension Genetic Epidemiology Network (HyperGEN), Invecchiare in Chianti Study (InCHIANTI), Kooperative Gesundheitsforschung in der Region Augsburg (KORA), Women’s Health Initiative - Broad Agency Award 23 (WHI-BAA23) and Women’s Health Initiative - Epigenetic Mechanisms of PM-Mediated CVD (WHI-EMPC). We excluded individuals with known diabetes and/or fasting glucose ≥ 7 mmol/l and/or those on anti-diabetic treatment. All studies were approved by their respective Institutional Review Boards, and all participants provided written informed consent. Details about the studies have been reported previously, and the key references as well as the summary of the design of each study are reported in Supplementary Note 1.
Glycemic traits and covariates
Venous blood samples were obtained after an overnight fast in all discovery and replication cohorts. BMI was calculated as weight over height squared (kg m−2) based on clinical examinations. Smoking status was divided into current, former and never, based on questionnaires. White blood cell counts were quantified using standard laboratory techniques or predicted from methylation data using the Houseman method52. The cohort-specific measurement of glycemic traits and covariates are shown in Supplementary Note 1.
DNA methylation quantification
The Illumina© Human Methylation450 array was used in all discovery and replication cohorts to quantify genome-wide DNA methylation in blood samples. We obtained DNA methylation levels reported as β values, which represents the cellular average methylation level ranging from 0 (fully unmethylated) to 1 (fully methylated). Study-specific details regarding DNA methylation quantification, normalization and quality control procedures are provided in the Supplementary Note 1.
Epigenome-wide association analysis and replication
All statistical analyses were performed using R statistical software and the two-tailed test was considered. Insulin was natural log transformed. In the discovery analysis, we first performed EWAS in each cohort separately. Linear regression analysis was used to test the association between glucose and insulin with each CpG site in the Rotterdam Study samples. Linear mixed models were used in NTR and TwinsUK, accounting for the family structure. We fitted the following two models for each cohort: (1) the baseline model adjusting for age, sex, technical covariates (chip array number and position on the array), white blood cell counts (lymphocytes, monocytes, and granulocytes) and smoking status, and (2) a second model additionally adjusting for BMI. We removed probes that have evidence of multiple mapping or contain a genetic variant in the CpG site53. All cohort-specific EWAS results for each model were then meta-analysed using inverse variance-weighted fixed effect meta-analysis as implemented in the metafor R package54. In total, we meta-analysed 393,183 CpG sites that passed quality control in all four discovery cohorts. The details of the quality control for each cohort could be found in the Supplementary Note 1. The association was later corrected by the genomic control factor (λ) in each meta-EWAS55. We produced quantile-quantile (QQ) plots of the -log10 (P) to evaluate inflation in the test statistic (Supplementary Figure 2). A Bonferroni correction was used to correct for multiple testing and identify epigenome-wide significant results (P < 1.3 × 10−7). We did not correct the number of glycemic traits and models, as they are highly correlated and not independent. The genome coordinates were provided by Illumina (GRCh37/hg19). The CpG sites were annotated to genes using Infinium HumanMethylation 450 BeadChip annotation resources. The correlation of the CpG sites located in the same gene was further checked in the overall RS III-1 and RS-BIOS samples by Pearson’s correlation test (n = 1544) to find the independent top CpG sites.
For the associations discovered in the meta-EWAS that have not been reported previously, we attempted replication in independent samples using the same traits and regression models as in the discovery analyses. Study-specific details of replication cohorts are provided in Supplementary Data 1 and Supplementary Note 1. Results from each replication cohort were meta-analysed using the same methods as in the discovery analyses. Bonferroni P value < 3.3 × 10−3 (0.05 corrected by 15 CpGs tested for associations) was considered significant.
Glycemic differential DNA methylation and transcriptomics
To explore whether the differential CpG sites were associated with gene expression level in blood, we explored eQTMs17 from the European blood-based BIOS database17 from BBMRI-NL which captured meQTLs, eQTLs and eQTMs from genome-wide database of 3841 Dutch blood samples (See resources of the database in URLs). The associated gene expression probes of the known and replicated CpG sites were searched. We then tested whether the expression of the genes that harbor the identified methylation sites was associated with T2D and related traits in glucose metabolism-related tissues (adipose subcutaneous, adipose visceral omentum, liver, whole blood, pancreas, skeletal muscle and small intestine terminal ileum) using MetaXcan package24,56. MetaXcan associates the expression of the genes with the phenotype by integrating functional data generated by large-scale efforts, e.g. GTEx project16 with that of the GWAS of the trait. MetaXcan is trained on transcriptome models in 44 human tissues from GTEx and is able to estimate their tissue-specific effect on phenotypes from GWAS. For this study, we used the GWAS studies of T2D19, fasting glucose traits21,22, fasting insulin22, HbA1c23 and HOMA-IR20. We used the nominal P value threshold (P value threshold for significance < 0.05) as we had separate assumptions for each terminal pathway between gene expressions and phenotype. The associations with genes in low prediction performance were excluded, i.e. the association of the tissue model’s correlation to the gene’s measured transcriptome is not significant (P value > 0.05).
Glycemic differential DNA methylation and genomics
We identified the genetic determinants of the significant CpG sites known or replicated through the current EWAS using the results of the cis and trans meQTLs from the European blood-based BIOS database17 (See resources of the database in URLs). All the reported SNPs with P value adjusted for false discovery rate (FDR) less than 0.05 in the database were treated as the target genetic variants in the present study. The SNPs were annotated based on the information in the BIOS study17 or the nearest protein-coding gene list from SNPnexus57 on GRCh37/hg19. We also explored the associations of these DNA methylation-related genetic variants with T2D or related traits, i.e., fasting glucose, insulin, HbA1c and HOMA-IR, based on public GWAS data sets in European ancestry20–22,27–29. Meanwhile, we checked the effect direction consistency of the association between the SNPs, CpG sites and T2D or related traits. That is the direction of the association between SNP and T2D or related traits should be a combination of the direction of SNP with CpG sites and CpG sites with T2D or related traits. A multiple-testing correction was performed by Bonferroni adjustment (P value significant threshold < 1.8 × 10−3, 0.05 corrected by the 29 genetic loci shown in Supplementary Data 3). The associations of the DNA methylation-related genetic variants and the gene expression were also looked up in the BIOS database17. This is limited to the expression profiles earlier associated with glycemic CpG sites in blood.
For the significant CpG sites known or replicated through EWAS, we attempted to evaluate the causality effect of CpG sites on their significant traits, either fasting glucose or fasting insulin, using two-sample MR approach as described in detail before by Dastani et al.30,58 based on the summary statistic GWAS results from the BIOS database and the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) database17,21 (Supplementary Figure 3). Briefly, we constructed a weighted genetic risk score for individual CpG on phenotype using independent SNPs as the instrument variables of the CpG, implemented in the R-package gtx. The effect of each score on phenotype was calculated as
where βi is the effect of the CpG-increasing alleles on phenotype, si its corresponding standard error and ωi the SNP effect on the respective CpG. Because the genetic variants might be close (cis) or far (trans) from the methylated site, we also performed MR test in the cis only SNPs if the CpG has both cis and trans genetic markers. All SNPs were mapped to the human genome build hg19. For each test (one CpG with one trait), we extracted all the genetic markers of the CpG in the fasting glucose or insulin GWAS from the MAGIC data set (n = 96,496)21 with their effect estimate and standard error on fasting glucose or insulin. Within the overlapped SNPs, we removed SNPs in potential linkage disequilibrium (LD, pairwise R2 ≥ 0.05) in 1-Mbp window based on the 1000 Genome imputed genotype data set from the general population: Rotterdam Study I (RS I, n = 6291)59. We managed to exclude the genetic loci which were genome-wide associated with glycemic traits, but none of the genetic loci meet this exclusion criterion. The instrumental variables that explain more than 1% of the variance in exposure (DNA methylation) were taken forward for MR test. The Bonferroni P value threshold was used to correct for the 13 CpG sites available for MR (P value < 3.8 × 10−3).
Functional annotation
Further, we integrated the cascade of associations as above among the results of EWAS, eQTM, meQTL and eQTL and showed in Fig. 3. We checked the effect direction consistency of the association between the SNPs, CpG sites, gene expression in blood and glycemic traits. The correlation of the novel CpG sites was checked in the overall RS III-1 and RS-BIOS samples by Pearson’s correlation test (n = 1544). The hierarchical cluster analysis was used in the clustering. Gene set enrichment analyses were performed in the genes of new CpG sites60. We tested if genes of interest were over-represented in any of the pre-defined gene sets from KEGG pathway database32, Reactome Pathway Knowledgebase33 and GO biological process34. Multiple test correction was performed in the tests. Gene sets of KEGG pathway database, Reactome Pathway Knowledgebase were obtained from Molecular Signatures Database (MsigDB) c2 and GO biological process was obtained from MsigDB c560. We used the platform of Functional Mapping and Annotation of Genome-wide Association Studies (FUMA GWAS)61 and GENE2FUNC function to perform the gene set enrichment analysis and the tissue-specific gene expression patterns based on GTEx v616. Besides, the tools Ensembl Human Genes62 (see URLs) and UCSC GRCh37/hg1963 (see URLs) were also used in interpreting genetic determinants, CpG sites and genes.
BMI in the association of methylation and glycemic traits
We used linear regression to check the effect of CpGs on the relationship between BMI and fasting insulin in the non-diabetic individuals in Rotterdam study. The initial model used BMI as the independent variable and the natural log transformed insulin as the dependent variable. The covariates included age, sex, technical covariates (chip array number and position on the array), white blood cell counts, smoking status and data set (RS III-1 and RS-BIOS). The normalized differential methylation values of CpG sites were added as covariates in the advanced model. The differences of the models were compared by ANOVA testing using anova function in R (P value < 0.05).
URLs
BIOS database, https://genenetwork.nl/biosqtlbrowser/ [https://genenetwork.nl/biosqtlbrowser/]; SNPnexus, http://snp-nexus.org/index.html [http://snp-nexus.org/index.html]; GWAS database of glycemic traits, https://www.magicinvestigators.org/ [https://www.magicinvestigators.org/]; GWAS database of T2D, http://diagram-consortium.org/ [http://diagram-consortium.org/]; MetaXcan, https://s3.amazonaws.com/imlab-open/Data/MetaXcan/results/metaxcan_results_database_v0.1.tar.gz [https://s3.amazonaws.com/imlab-open/Data/MetaXcan/results/metaxcan_results_database_v0.1.tar.gz]; NHGRI-EBI Catalog, https://www.ebi.ac.uk/gwas/ [https://www.ebi.ac.uk/gwas/]; Ensembl, https://www.ensembl.org/Homo_sapiens/Info/Index [https://www.ensembl.org/Homo_sapiens/Info/Index]; FUMA, http://fuma.ctglab.nl [http://fuma.ctglab.nl]; UCSC, https://genome.ucsc.edu/cgi-bin/hgGateway [https://genome.ucsc.edu/cgi-bin/hgGateway] (available: 1st Jan 2019)
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We gratefully acknowledge the BIOS consortium (https://www.bbmri.nl/?p=259) of Biobanking and BioMolecular resources Research Infrastructure of the Netherlands (BBMRI-NL) and Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. This work is part of the CardioVasculair Onderzoek Nederland (CVON 2012-03), the Common mechanisms and pathways in Stroke and Alzheimer's disease (CoSTREAM) project (https://www.costream.eu, grant agreement No 667375), Memorabel program (project number 733050814), Netherlands X-omics Research Infrastructure and U01-AG061359 NIA. The full list of funding information of each cohort is found in Supplementary Note 2. J.L., C.M.v.D. and A.Demirkan have used exchange grants from the Personalized pREvention of Chronic DIseases consortium (PRECeDI) (H2020-MSCA-RISE-2014). A.Demirkan is supported by a Veni grant (2015) from ZonMw (VENI 91616165). C.M.v.D. received funding of CardioVasculair Onderzoek Nederland (CVON2012-03) of the Netherlands Heart Foundation. B.A.H. was supported by NHLBI K01 award (K01 HL130609-02). V.W.V.J. received a grant from the Netherlands Organization for Health Research and Development (VIDI 016.136.361) and a Consolidator Grant from the European Research Council (ERC-2014-CoG-648916). J.F.F. has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 633595 (DynaHEALTH). J.B.M. is supported by K24 DK080140. J.T.B. received funding support from the JPI ERA-HDHL DIMENSION project (BBSRC BB/S020845/1) and from the ESRC (ES/N000404/1). The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
Author contributions
J.L., E.C.-M., D.L., A.Dehghan, J.T.B., J.B.J.v.M., A.I., A. Demirkan and C.M.v.D. contributed to study design. J.L., S. Ligthart, P.R.M., M.J.P., L.D., V.W.V.J., J.F.F., G.W., E.J.C.d.G, T.L.A., L.H., E.A.W., N.A., T.D.S., M.-F.H., J.I.R., J.B.J.v.M., A.I., D.I.B., J.T.B. and C.M.v.D. contributed to data collection. V.W.V.J., H.T., G.W., E.J.C.d.G., D.L., J.A.S., N.S., S.B., L.F., D.K.A., H.G., T.L.A., L.H., A.B., E.A.W., A.G.U., E.J.G.S., O.H.F., J.D., J.I.R., J.B.M., J.P., D.I.B., T.D.S. and C.M.v.D. contributed to cohort design and management. J.L., E.C-M., J.v.D., S.Lent, I.N., P.-C.T., T.C.M., R.J., J.F.F., A.Y.C., S.-J.H., R.G., E.S., C.H., B.A.H., T.T., A.Z.M., R.N.L., M.A.J. and A.I. contributed to data analysis. J.L., E.C.-M., S.Ligthart, K.W.v.D., N.A., A.Dehghan, A.I., A. Demirkan and C.M.v.D. contributed to writing of manuscript. J.L., E.C.-M., J.v.D., S. Lent, S. Ligthart, T.C.M., L.D., V.W.V.J., H.T., J.F.F., D.L., J.B., R.G., C.H., B.A.H., N.S., D.K.A., H.G., E.A.W., N.A., E.J.G.S., J.D., M.-F.H., J.I.R., J.B.M., J.P., A.I., J.T.B., A. Demirkan and C.M.v.D. contributed to critical review of manuscript.
Data Availability
All relevant data supporting the key findings of this study are available within the article and its Supplementary Information files; the cohort data sets generated and analyzed during the current study are available from the authors from each cohort upon reasonable request. The summary statistics of each cohort and meta-analysis in the discovery phase and the source data underlying Supplementary Figure 2 are provided as a Data file [https://figshare.com/s/1a1e8ac0fd9a49e2be30]. The web links for the publicly available data sets used in the paper are listed in URLs. In detail, for the BIOS data, the cis-meQTL look-up files were mainly from “Full list of primary cis-meQTLs” and the results in “Cis-meQTLs independent top effects” were also checked. The trans-meQTL look-up file was from “Trans-meQTLs top effects”. The “eQTM” look-up file was from “Cis-eQTMs independent top effects”. The eQTL look-up file was from “Cis-eQTLs Gene-level all primary effects”. Fasting glucose GWAS was from both ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Metabochip_Public_data_release_25Jan.zip [ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Metabochip_Public_data_release_25Jan.zip] and ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Manning_et_al_FastingGlucose_MainEffect.txt.gz [ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Manning_et_al_FastingGlucose_MainEffect.txt.gz]; fasting insulin GWAS was from ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Manning_et_al_lnFastingInsulin_MainEffect.txt.gz [ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Manning_et_al_lnFastingInsulin_MainEffect.txt.gz]; HbA1c was from ftp://ftp.sanger.ac.uk/pub/magic/HbA1c_METAL_European.txt.gz [ftp://ftp.sanger.ac.uk/pub/magic/HbA1c_METAL_European.txt.gz]. The type 2 diabetes GWAS was downloaded from http://diagram-consortium.org/ [http://diagram-consortium.org/] “T2D GWAS meta-analysis - Trans-Ethnic Summary Statistics Published in in Mahajan et al. (2018)”. The file “T2D_TranEthnic.BMIunadjusted.txt” was used. A reporting summary for this article is available as a supplementary information file.
Competing interests
The authors declare no competing interests.
Footnotes
Journal peer review information: Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jun Liu, Elena Carnero-Montoro.
These authors jointly supervised this work: Ayşe Demirkan, Cornelia M. van Duijn
Contributor Information
Jun Liu, Email: jun.liu@ndph.ox.ac.uk.
Ayşe Demirkan, Email: a.demirkan@surrey.ac.uk.
Cornelia M. van Duijn, Email: cornelia.vanduijn@ndph.ox.ac.uk
Supplementary information
Supplementary Information accompanies this paper at 10.1038/s41467-019-10487-4.
References
- 1.American Diabetes A. 2. Classification and diagnosis of diabetes: standards of medical care in diabetes—2018. Diabetes Care. 2018;41:S13–S27. doi: 10.2337/dc18-S002. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo B, et al. Epigenome-wide association study of fasting measures of glucose, insulin, and HOMA-IR in the Genetics of Lipid Lowering Drugs and Diet Network study. Diabetes. 2014;63:801–807. doi: 10.2337/db13-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers JC, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3:526–534. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kriebel J, et al. Association between DNA methylation in whole blood and measures of glucose metabolism: KORA F4 study. PLoS ONE. 2016;11:e0152314. doi: 10.1371/journal.pone.0152314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulkarni H, et al. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum. Mol. Genet. 2015;24:5330–5344. doi: 10.1093/hmg/ddv232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding J, et al. Alterations of a cellular cholesterol metabolism network are a molecular feature of obesity-related type 2 diabetes and cardiovascular disease. Diabetes. 2015;64:3464–3474. doi: 10.2337/db14-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. 2013;5:553–568. doi: 10.2217/epi.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahl S, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, R. etal. Longitudinal personal DNA methylome dynamics in a human with a chronic condition. Nat Med (2018). [DOI] [PMC free article] [PubMed]
- 10.Al Muftah WA, et al. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clin. Epigenetics. 2016;8:13. doi: 10.1186/s13148-016-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walaszczyk E, et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case-control sample of the Lifelines study. Diabetologia. 2018;61:354–368. doi: 10.1007/s00125-017-4497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demerath EW, et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum. Mol. Genet. 2015;24:4464–4479. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aslibekyan S, et al. Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity. 2015;23:1493–1501. doi: 10.1002/oby.21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B, et al. Methylation loci associated with body mass index, waist circumference, and waist-to-hip ratio in Chinese adults: an epigenome-wide analysis. Lancet. 2016;388(Suppl 1):S21. doi: 10.1016/S0140-6736(16)31948-1. [DOI] [Google Scholar]
- 15.Wilson LE, Harlid S, Xu Z, Sandler DP, Taylor JA. An epigenome-wide study of body mass index and DNA methylation in blood using participants from the Sister Study cohort. Int J. Obes. 2017;41:194–199. doi: 10.1038/ijo.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consortium GT, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonder MJ, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 2017;49:131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 18.Jones MJ, Fejes AP, Kobor MS. DNA methylation, genotype and gene expression: who is driving and who is along for the ride? Genome Biol. 2013;14:126. doi: 10.1186/gb-2013-14-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Replication DIG, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manning AK, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott RA, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soranzo N, et al. Common variants at 10 genomic loci influence hemoglobin A1C levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbeira AN, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grarup N, et al. Association of variants in the sterol regulatory element-binding factor 1 (SREBF1) gene with type 2 diabetes, glycemia, and insulin resistance: a study of 15,734 Danish subjects. Diabetes. 2008;57:1136–1142. doi: 10.2337/db07-1534. [DOI] [PubMed] [Google Scholar]
- 26.Jin B, Li Y, Robertson KD. Genes Cancer. 2011. DNA methylation: superior or subordinate in the epigenetic hierarchy? pp. 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheeler E, et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 2017;14:e1002383. doi: 10.1371/journal.pmed.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahajan A, et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat. Genet. 2018;50:559–571. doi: 10.1038/s41588-018-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott RA, et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dastani Z, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8:e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendelson MM, et al. Association of Body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a mendelian randomization approach. PLoS Med. 2017;14:e1002215. doi: 10.1371/journal.pmed.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The Gene Ontology C. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickup JC, Crook MA. Is type II diabetes mellitus a disease of the innate immune system? Diabetologia. 1998;41:1241–1248. doi: 10.1007/s001250051058. [DOI] [PubMed] [Google Scholar]
- 36.GIB, M. D. National Library of Medicine (US), National Center for Biotechnology Information (2004). https://www.ncbi.nlm.nih.gov/
- 37.Di Bernardo MC, et al. Risk of developing chronic lymphocytic leukemia is influenced by HLA-A class I variation. Leukemia. 2013;27:255–258. doi: 10.1038/leu.2012.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomson, G. & Bodmer, W.F. The genetics of HLA and disease associations. Measuring selection in natural populations. Springer, Berlin, Heidelberg, 545–564 (1977).
- 39.Withers DJ, et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 40.Lingohr MK, et al. Decreasing IRS-2 expression in pancreatic beta-cells (INS-1) promotes apoptosis, which can be compensated for by introduction of IRS-4 expression. Mol. Cell Endocrinol. 2003;209:17–31. doi: 10.1016/j.mce.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Hennige AM, et al. Upregulation of insulin receptor substrate-2 in pancreatic beta cells prevents diabetes. J. Clin. Invest. 2003;112:1521–1532. doi: 10.1172/JCI18581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akama TO, et al. Germ cell survival through carbohydrate-mediated interaction with Sertoli cells. Science. 2002;295:124–127. doi: 10.1126/science.1065570. [DOI] [PubMed] [Google Scholar]
- 43.Guo W, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 2012;18:766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Consortium GT. the genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang YT, et al. Epigenome-wide profiling of DNA methylation in paired samples of adipose tissue and blood. Epigenetics. 2016;11:227–236. doi: 10.1080/15592294.2016.1146853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 48.Kim M, Costello J. DNA methylation: an epigenetic mark of cellular memory. Exp. Mol. Med. 2017;49:e322. doi: 10.1038/emm.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rask-Andersen M, et al. Postprandial alterations in whole-blood DNA methylation are mediated by changes in white blood cell composition. Am. J. Clin. Nutr. 2016;104:518–525. doi: 10.3945/ajcn.115.122366. [DOI] [PubMed] [Google Scholar]
- 50.Soriano-Tarraga C, et al. Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum. Mol. Genet. 2016;25:609–619. doi: 10.1093/hmg/ddv493. [DOI] [PubMed] [Google Scholar]
- 51.Florath I, et al. Type 2 diabetes and leucocyte DNA methylation: an epigenome-wide association study in over 1,500 older adults. Diabetologia. 2016;59:130–138. doi: 10.1007/s00125-015-3773-7. [DOI] [PubMed] [Google Scholar]
- 52.Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinforma. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen YA, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010;36:1–48. doi: 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 55.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 56.Gamazon ER, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dayem Ullah AZ, et al. SNPnexus: assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Res. 2018;46:W109–W113. doi: 10.1093/nar/gky399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J, et al. A mendelian randomization study of metabolite profiles, fasting glucose, and type 2. Diabetes Diabetes. 2017;66:2915–2926. doi: 10.2337/db17-0199. [DOI] [PubMed] [Google Scholar]
- 59.Ikram MA, et al. The Rotterdam Study: 2018 update on objectives, design and main results. Eur. J. Epidemiol. 2017;32:807–850. doi: 10.1007/s10654-017-0321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zerbino DR, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haeussler M, et al. The UCSC genome browser database: 2019 update. Nucleic Acids Res. 2019;47:D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All relevant data supporting the key findings of this study are available within the article and its Supplementary Information files; the cohort data sets generated and analyzed during the current study are available from the authors from each cohort upon reasonable request. The summary statistics of each cohort and meta-analysis in the discovery phase and the source data underlying Supplementary Figure 2 are provided as a Data file [https://figshare.com/s/1a1e8ac0fd9a49e2be30]. The web links for the publicly available data sets used in the paper are listed in URLs. In detail, for the BIOS data, the cis-meQTL look-up files were mainly from “Full list of primary cis-meQTLs” and the results in “Cis-meQTLs independent top effects” were also checked. The trans-meQTL look-up file was from “Trans-meQTLs top effects”. The “eQTM” look-up file was from “Cis-eQTMs independent top effects”. The eQTL look-up file was from “Cis-eQTLs Gene-level all primary effects”. Fasting glucose GWAS was from both ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Metabochip_Public_data_release_25Jan.zip [ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Metabochip_Public_data_release_25Jan.zip] and ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Manning_et_al_FastingGlucose_MainEffect.txt.gz [ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Manning_et_al_FastingGlucose_MainEffect.txt.gz]; fasting insulin GWAS was from ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Manning_et_al_lnFastingInsulin_MainEffect.txt.gz [ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Manning_et_al_lnFastingInsulin_MainEffect.txt.gz]; HbA1c was from ftp://ftp.sanger.ac.uk/pub/magic/HbA1c_METAL_European.txt.gz [ftp://ftp.sanger.ac.uk/pub/magic/HbA1c_METAL_European.txt.gz]. The type 2 diabetes GWAS was downloaded from http://diagram-consortium.org/ [http://diagram-consortium.org/] “T2D GWAS meta-analysis - Trans-Ethnic Summary Statistics Published in in Mahajan et al. (2018)”. The file “T2D_TranEthnic.BMIunadjusted.txt” was used. A reporting summary for this article is available as a supplementary information file.