Abstract
Vitis vinifera is widely grown worldwide for making wine and for use as table grapes. Of the existing cultivars, some have a floral and fruity flavour, referred to as a Muscat flavour. It is well-documented that this flavour originates from a series of terpene compounds, but the mechanism of terpene content differences among the Muscat-type cultivars remains unclear. Transcript and terpene metabolite profiles were integrated to elucidate the molecular mechanism of this phenomenon. In this research, three genotypes with different aromatic strengths were investigated by RNA sequencing. A total of 27 fruit samples from three biological replicates were sequenced on Illumina HiSeq2000 at three stages, corresponding to the veraison; berries had intermediate Brix value and were harvest-ripe. After quality assessment and data clearance, a total of 254.18 Gb of data with more than 97% Q20 bases were obtained, approximately 9.41 Gb data were generated per sample. These results will provide a valuable dataset for the discovery of the mechanism of terpene biosynthesis.
Subject terms: Plant molecular biology, Secondary metabolism, RNA sequencing
| Design Type(s) | transcription profiling design • gene expression analysis objective |
| Measurement Type(s) | transcription profiling assay |
| Technology Type(s) | RNA sequencing |
| Factor Type(s) | cultivar • biological replicate • developmental stage |
| Sample Characteristic(s) | Vitis vinifera • berry |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Background & Summary
The trait of aroma is one of the most important parameters for the quality of grapes and is the main concern when consumers buy grape products. For genetic improvement research and breeding, the biosynthesis mechanism of aromatic compounds and their regulation has attracted much attention. Terpenes are the typical aromatic compounds in Muscat grapes, and they belong to the second metabolites1–4; they have a low olfactory threshold and can be easily precepted by humans. The terpenes mainly exist in the pericarp and in the flesh of some cultivars5, with their content being affected by the genotype6,7, developmental stage8,9, environment and management of the grape10–13. Terpenes have two forms: the free form, which directly leads to the aromatic flavour, and the glycoside bound form, in which the potential aromatic compounds transfer to the free form by hydrolysis14–16.
Biologically, the biosynthesis of terpene compounds in plants are synthesized by two pathways, the methyl-erythritol-4-phosphate pathway (DXP/MEP) in the plastid and the mevalonate pathway (MVA) in the cytoplasm17, with terpenes located in the mesocarp and pericarp18. Starting from pyruvic acid and 3-phosphate glyceraldehyde, by 1-deoxy-D-xylulose-5-phosphate synthase (DXS), which is the entrance enzyme in the MEP pathway, the two compounds were changed into 1-deoxy-D-xyulose-5-phosphate and, then, through six enzymatic reactions, were converted into geranyl-diphosphate (GPP). Geranyl-diphosphate was the substrate for all the terpenes. Then, by a series of terpene synthases, the GPP was synthesized into hemiterpenes (C5), monoterpenes (C10), sesquiterpenes (C15) or diterpenes (C20)19–22.
The genetic mechanism of Muscat flavour in grapevines has been studied through quantitative trait loci analysis (QTL) in different F1 populations23,24, and in selfing populations, it has been shown that VvDXS is a structural candidate gene for geraniol, nerol, and linalool concentrations in wine grapes25. Battilana reported that single nucleotide polymorphism (SNP) mutations in VvDXS are the main cause of the Muscat flavour. The substitution of a lysine with an asparagine at position 284 of the VvDXS amino acid sequence affects the monoterpene content of Muscat flavour and neutral cultivars26.
In Muscat grapes, some cultivars have a very strong flavour, while others have moderate or light flavour. The terpene type and concentration varied among the cultivars. To date, terpene accumulation has been well-documented in some wine grapes. Terpene accumulation in developing Gewurztraminer grapes has been shown to be correlated with an increase in the transcript abundances of early terpenoid pathway enzymes27. Some transcription factors involved in controlling terpene biosynthesis have been predicted in the grapevine cultivar Muscat Blanc à Petits Grains through gene co-expression network analysis28. A Nudix hydrolase was also found to be a component of a terpene synthase-independent pathway, with cytochrome P450 hydroxylases, epoxide hydrolases and glucosyltransferases genes potentially involved in monoterpene metabolism29. However, there are few reports on the table grape30.
In this study, we present the transcriptome analysis of three genotypes of table grapes. During berry development, 27 samples, in total, were sequenced on the Illumina HiSeq Platform. After quality assessment and data clearance, a total of 254.18 Gb high-quality base pairs with more than 97% Q20 bases were obtained, and an approximately 9.41 Gb per sample. In the aggregate, a total of 776 million reads were yielded, with an average of 31.66 million reads per sample. Furthermore, approximately 76.65% of the total reads were uniquely aligned to the grape genome (V2)31. These data will provide useful information for investigating terpene biosynthesis.
Methods
Overview of the experimental design
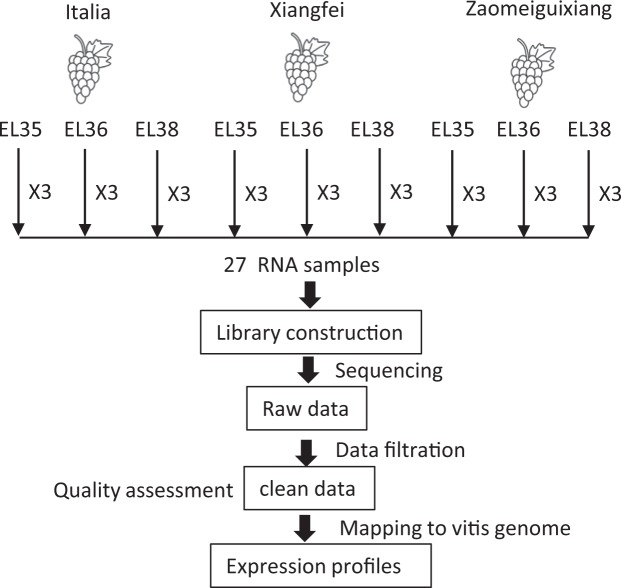
The berries of three genotypes were collected at three developmental stages. Approximately 300 grape berries were randomly collected for each replicate, with three replicates harvested for each stage. The experimental design and analysis pipeline are shown in Fig. 1.
Fig. 1.
Flowchart of the experimental design. Berry samples were collected at three developmental stages, and three biological replicates per sample were used for transcriptome sequencing. All raw reads were quality controlled and assessed. Then, the clean data were mapped to the V. vinifera reference genome (V2) by Hisat2. Gene expression levels were calculated with RSEM.
Materials and methods
Plant materials
Three V. vinifera cultivars were used for transcript study. ‘Xiangfei’ was registered by our team and has a strong Muscat flavour and a green to golden skin colour, while ‘Italia,’ the famous mid-late season table grape cultivar that originated in Italy, has a moderate Muscat flavour. ‘Zaomeiguixiang’ has a purple-reddish colour and a strong Muscat flavour.
Sampling
The vines were grown in the experimental vineyard at the Beijing Academy of Forestry and Pomology Sciences in China (39°58′N and 116°13′E) under a plastic cover and were trained into a two-wire vertical trellis system with a 2.5-m row space and a 0.75 m plant space. In 2017, berry samples from three vines were harvested at the developmental stages corresponding to EL35, EL36, and EL3832. The berry begins to colour and soften at EL 35 (about 5% of the berries started to colour and soften), progresses to the complete veraison with an intermediate Brix of EL 36, and reaches harvest ripeness at EL38. At each stage, three replicates were harvested; approximately 300 grape berries were randomly collected for each replicate.
Physiochemical parameters
Fifty berries of each replicate were pressed and centrifuged to determine total soluble solids (TSS), pH value and titratable acidity. TSS was measured by a digital refractometer (PAL-1, Atago, Tokyo, Japan). The pH value was measured by a pH meter (FiveGo F2-Standard, Mettler Toledo, Switzerland). Titratable acidity was analysed by titration with NaOH (0.1M) to the end point of pH 8.2 and expressed as tartaric acid equivalents in accordance with the National Standard of People’s Republic of China (GB/T15038-2006, 2006). The other berries were then frozen in liquid nitrogen and stored at −80 °C.
RNA extraction and sequencing
The extraction of total RNA from the berries was carried out by a Plant RNA extraction kit (Aidlab Biotechnologies, Beijing, China). The quality of the RNA was verified by agarose gel electrophoresis, and the concentration was determined by the absorbance ratio (A260/A280, 1.8–2.0) on an Implen P330 nanophotometer (Implen GmbH, Munich, Germany).
The RNA-Seq libraries were constructed from 27 samples according to the methods of Wang33. The enriched mRNA was obtained by using oligo (dT) magnetic beads then fragmented by 94 °C for 5 min. cDNA was synthesized by Superscript®III Reverse Transcriptase, followed by purification, end repair and dA-tailing and was then ligated with the sequencing adaptor. Afterwards, PCR amplification was conducted by indexed primers. The constructed library was QC checked by Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-Time PCR System and then sequenced by Illumina HiSeq2000 platform at BGI Life Tech Co., Ltd. (Shenzhen, China). Low quality reads (more than 20% of the base qualities are lower than 10), reads with adaptors and reads with unknown bases (N bases more than 5%) were filtered to get clean reads and were stored in FASTQ format. The clean reads were mapped onto the reference grapevine genome (V2) using Hisat234.
Data Records
The RNA-Seq clean data of the 27 samples were deposited at the NCBI Sequence Read Archive with accessions SRP18415235. The files of gene expression level were deposited in NCBI’s Gene Expression Omnibus (GEO), and are accessible through GEO Series accession number GSE13038636. The information of the differentially expressed genes (DEGs) between samples were deposited in figshare37.
Technical Validation
Quality control
The physiochemical parameter of the samples was shown in Table 1. A total of 27 RNA samples were prepared and sequenced, with the sequencing depth ranging between 22.48 and 33.08 million reads; the Q20 values for the clean reads were above 97%, and the average mapping ratio was 84.72% (Online-only Table 1).
Table 1.
Physiochemical parameters for each sample.
| Sample name | Total soluble solids | Titratable acidity(g/l) | pH |
|---|---|---|---|
| X-EL35-1 | 10.84 | 4.25 | 3.11 |
| X-EL35-2 | 10.80 | 4.20 | 3.15 |
| X-EL35-3 | 10.95 | 4.26 | 3.16 |
| X-EL36-1 | 13.46 | 4.01 | 3.53 |
| X-EL36-2 | 13.30 | 3.98 | 3.50 |
| X-EL36-3 | 13.80 | 4.05 | 3.58 |
| X-EL38-1 | 16.62 | 3.73 | 3.75 |
| X-EL38-2 | 16.40 | 3.70 | 3.71 |
| X-EL38-3 | 16.42 | 3.68 | 3.77 |
| Y-EL35-1 | 5.18 | 5.15 | 3.07 |
| Y-EL35-2 | 5.20 | 5.20 | 3.05 |
| Y-EL35-3 | 5.18 | 5.22 | 3.01 |
| Y-EL36-1 | 7.61 | 4.85 | 3.14 |
| Y-EL36-2 | 7.45 | 4.80 | 3.18 |
| Y-EL36-3 | 7.40 | 4.79 | 3.17 |
| Y-EL38-1 | 14.80 | 4.51 | 3.47 |
| Y-EL38-2 | 14.50 | 4.52 | 3.48 |
| Y-EL38-3 | 14.57 | 4.48 | 3.45 |
| Z-EL35-1 | 9.78 | 3.96 | 3.30 |
| Z-EL35-2 | 9.70 | 3.95 | 3.32 |
| Z-EL35-3 | 9.80 | 3.99 | 3.32 |
| Z-EL36-1 | 12.90 | 3.48 | 3.75 |
| Z-EL36-2 | 12.95 | 3.55 | 3.78 |
| Z-EL36-3 | 12.88 | 3.52 | 3.71 |
| Z-EL38-1 | 17.25 | 3.05 | 3.85 |
| Z-EL38-2 | 17.20 | 2.96 | 3.80 |
| Z-EL38-3 | 17.29 | 3.07 | 3.82 |
X stands for cultivar Xiangfei, Y for cultivar Italia and Z for cultivar Zaomeiguixiang. EL35: the berry begins to colour and soften, EL36: complete of veraison with an intermediate Brix, EL38: berry reaches harvest ripeness.
Online-only Table 1.
Statistics of sequencing data for each sample.
| Sample | Total Raw Reads(Mb) | Total Clean Reads(Mb) | Total Clean Bases(Gb) | Clean Reads Q20(%) | Total Mapping Ratio% | Uniquely MappingRatio% | Accession number (Biosample) |
|---|---|---|---|---|---|---|---|
| X-EL35-1 | 32.79 | 30.31 | 9.78 | 98.75 | 82.79 | 76.97 | SAMN10880015 |
| X-EL35-2 | 32.67 | 30.17 | 9.69 | 98.82 | 80.71 | 72.02 | SAMN10880016 |
| X-EL35-3 | 32.58 | 30.04 | 9.70 | 98.68 | 81.33 | 76.37 | SAMN10880017 |
| X-EL36-1 | 32.28 | 29.26 | 9.59 | 98.34 | 85.30 | 76.21 | SAMN10880018 |
| X-EL36-2 | 31.96 | 28.86 | 9.50 | 98.61 | 94.53 | 79.67 | SAMN10880019 |
| X-EL36-3 | 30.29 | 27.61 | 8.96 | 98.73 | 91.97 | 80.36 | SAMN10880020 |
| X-EL38-1 | 32.05 | 29.53 | 9.50 | 98.79 | 79.21 | 73.29 | SAMN10880021 |
| X-EL38-2 | 31.58 | 29.07 | 9.38 | 98.79 | 85.37 | 77.24 | SAMN10880022 |
| X-EL38-3 | 32.46 | 29.62 | 9.63 | 98.26 | 89.61 | 81.99 | SAMN10880023 |
| Y-EL35-1 | 32.70 | 30.22 | 9.74 | 98.74 | 71.65 | 68.36 | SAMN10880024 |
| Y-EL35-2 | 32.63 | 29.85 | 9.71 | 98.66 | 70.39 | 64.96 | SAMN10880025 |
| Y-EL35-3 | 33.08 | 30.73 | 9.86 | 98.80 | 82.73 | 76.09 | SAMN10880026 |
| Y-EL36-1 | 32.71 | 29.77 | 9.72 | 98.68 | 83.51 | 70.43 | SAMN10880027 |
| Y-EL36-2 | 32.89 | 30.02 | 9.78 | 98.67 | 84.90 | 75.87 | SAMN10880028 |
| Y-EL36-3 | 32.54 | 29.09 | 9.69 | 98.48 | 82.47 | 74.05 | SAMN10880029 |
| Y-EL38-1 | 32.37 | 29.26 | 9.62 | 98.18 | 66.95 | 58.86 | SAMN10880030 |
| Y-EL38-2 | 32.98 | 30.29 | 9.82 | 98.71 | 82.08 | 74.71 | SAMN10880031 |
| Y-EL38-3 | 31.89 | 27.00 | 9.50 | 97.75 | 83.90 | 73.87 | SAMN10880032 |
| Z-EL35-1 | 33.01 | 30.53 | 9.83 | 98.76 | 94.94 | 87.08 | SAMN10880033 |
| Z-EL35-2 | 28.17 | 24.35 | 8.38 | 97.97 | 80.48 | 71.95 | SAMN10880034 |
| Z-EL35-3 | 32.90 | 30.23 | 9.78 | 98.66 | 94.43 | 87.15 | SAMN10880035 |
| Z-EL36-1 | 32.24 | 29.09 | 9.58 | 98.30 | 86.69 | 79.18 | SAMN10880036 |
| Z-EL36-2 | 32.68 | 29.88 | 9.71 | 98.59 | 93.01 | 84.9 | SAMN10880037 |
| Z-EL36-3 | 32.81 | 29.80 | 9.75 | 98.56 | 90.54 | 82.82 | SAMN10880038 |
| Z-EL38-1 | 25.26 | 22.06 | 7.50 | 98.05 | 89.97 | 82.28 | SAMN10880039 |
| Z-EL38-2 | 32.92 | 30.20 | 9.78 | 98.65 | 88.41 | 82.38 | SAMN10880040 |
| Z-EL38-3 | 22.48 | 19.16 | 6.70 | 97.86 | 89.56 | 80.46 | SAMN10880041 |
X stands for Xiangfei, Y for Italia and Z for Zaomeiguixiang, Total Raw Reads (Mb): The reads amount before filtering, Unit: Mb. Total Clean Reads (Mb): The reads amount after filtering, Unit: Mb. Total Clean Bases (Gb): The total base amount after filtering, Unit: Gb. Clean Reads Q20(%): The Q20 value for the clean reads. Total Mapping Ratio: The percentage of mapped reads. Uniquely Mapping Ratio: The percentage of reads that map to only one location of reference. EL35: the berry begins to colour and soften, EL36: complete of veraison with an intermediate Brix, EL38: berry reaches harvest ripeness.
Analysis of RNA-Seq data
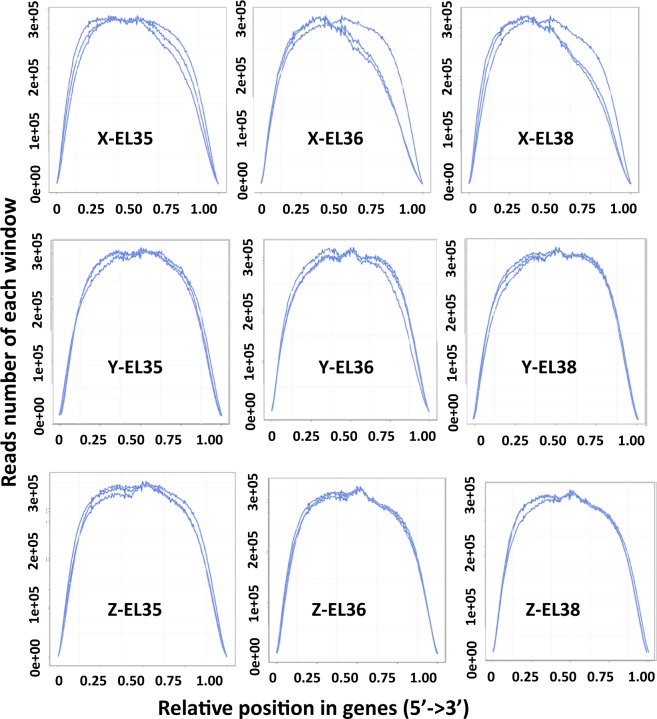
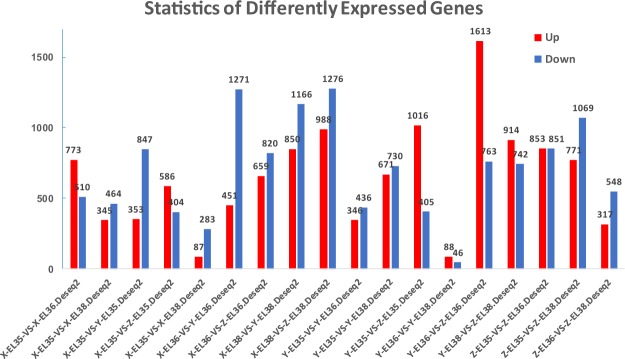
After novel transcript detection, novel coding transcripts were merged with reference transcripts to get a complete reference. Clean reads were mapped to the transcript by using Bowtie238. Gene expression levels were calculated with RSEM39. The distribution of reads based on the detection of read coverage skewness showed good fragmentation randomness (Fig. 2). The differentially expressed genes (DEGs) between samples were identified by the R package, DESeq240. The DEGs with a |log2ratio| ≥ 1 and a false discovery rate probability ≤ 0.001 were considered statistically significant. The statistical analyses of DEG are shown in Fig. 3.
Fig. 2.
Reads distribution on transcripts. The x-axis represents the position along transcripts, and the y-axis represents the number of reads.
Fig. 3.
Statistics of differently expressed genes. The X-axis represents the comparison method between groups and the y-axis represents DEG numbers. The red colour represents upregulated DEGs, and the blue colour represents downregulated DEGs.
Usage Notes
The RNA-Seq fastq.gz files were deposited at Gene Expression Omnibus and can be downloaded using the fastq-dump tool of the SRA Toolkit (https://www.ncbi.nlm.nih.gov). The V2 reference genome of V. vinifera, the annotated file, could be retrieved at (http://genomes.cribi.unipd.it/grape/).
ISA-Tab metadata file
Acknowledgements
This work was supported by the Science and Technology Innovation Ability Construction Projects of Beijing Academy of Agricultural and Forestry Sciences (KJCX20180411), Earmarked Fund for China Agriculture Research System (CARS-29) and Beijing Municipal Natural Science Foundation (6192017).
Online-only Table
Author Contributions
L.S. designed the experiments and wrote the manuscript. B.Q.Z. analysed the data. X.Y.Z. collected the samples and extracted RNA. G.J.Z., A.L.Y., H.L.W. and X.Y.W. reviewed the manuscript. H.Y.X. designed the experiments, reviewed the manuscript and supervised the study.
Code Availability
SOAPnuke: https://github.com/BGI-flexlab/SOAPnuke. Version: v1.5.2. Parameters: -l 5 -q 0.51 -n 0.55 -i -Q 2–seqType 1.
HISAT2: http://www.ccb.jhu.edu/software/hisat. Version:v2.0.4.Parameters:–phred64–sensitive–no-discordant–no-mixed -I 1 -X 1000.
Bowtie2: http://bowtie-bio.sourceforge.net/Bowtie2. Version: v2.2.5. Parameters: -q–phred64–sensitive–dpad 0–gbar 99999999–mp 1,1–np 1–score-min L,0, −0.1 -I 1 -X 1000–no-mixed–no-discordant -p 1 -k 200.
RSEM: http://deweylab.biostat.wisc.edu/RSEM. Version: v1.2.12. Parameters: default.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
ISA-Tab metadata
is available for this paper at 10.1038/s41597-019-0101-y.
References
- 1.Bohlmann J, Keeling CI. Terpenoid biomaterials. Plant J. 2008;54:656–669. doi: 10.1111/j.1365-313X.2008.03449.x. [DOI] [PubMed] [Google Scholar]
- 2.Magnard JL, et al. Plant volatiles. Biosynthesis of monoterpene scent compounds in roses. Science. 2015;349:81–83. doi: 10.1126/science.aab0696. [DOI] [PubMed] [Google Scholar]
- 3.Fenoll J, Martniez MDA, Hellin P, Flores P. Changes of free and glycosidically bound monoterpenes and aromatic alcohols in Muscatel and Ruby Seedless table grapes during development. J. Inter. Des Sciences. De La Vigne Et Du Vin. 2012;46:41–50. [Google Scholar]
- 4.Croteau R. Biosynthesis and catabolism of monoterpenoids. Chem. Rev. 1987;87:929–954. doi: 10.1021/cr00081a004. [DOI] [Google Scholar]
- 5.Luan F, Mosandl A, Munch A, Wust M. Metabolism of geraniol in grape berry mesocarp of Vitis vinifera L. cv. Scheurebe: demonstration of stereoselective reduction, E/Z-isomerization, oxidation and glycosylation. Phytochemistry. 2005;66:295–303. doi: 10.1016/j.phytochem.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 6.Fenoll J, Manso A, Hellin P, Ruiz L, Flores P. Changes in the aromatic composition of the Vitis vinifera grape Muscat Hamburg during ripening. Food Chem. 2009;114:420–428. doi: 10.1016/j.foodchem.2008.09.060. [DOI] [Google Scholar]
- 7.Liu B, et al. The free and enzyme-released volatile compounds of distinctive Vitis amurensis var. Zuoshanyi grapes in China. Eur. Food Res. Technol. 2015;240:985–997. doi: 10.1007/s00217-014-2403-9. [DOI] [Google Scholar]
- 8.Kalua CM, Boss PK. Evolution of volatile compounds during the development of Cabernet Sauvignon grapes (Vitis vinifera L.) J. Agric. Food Chem. 2009;57:3818–3830. doi: 10.1021/jf803471n. [DOI] [PubMed] [Google Scholar]
- 9.Kalua CM, Boss PK. Comparison of major volatile compounds from Reisling and Cabernet Sauvignon grapes (Vitis vinifera L.) from fruitset to harvest. Aus. J. Grape and Wine Res. 2010;16:337–348. doi: 10.1111/j.1755-0238.2010.00096.x. [DOI] [Google Scholar]
- 10.Bureau SM, Razungles AJ, Baumes RL. The aroma of Muscat of Frontignac grapes: effect of the light environment of vine or bunch on volatiles and glycoconjugates. J. Sci. Food Agric. 2000;80:2012–2020. doi: 10.1002/1097-0010(200011)80:14<2012::AID-JSFA738>3.0.CO;2-X. [DOI] [Google Scholar]
- 11.Wang Y, et al. Effects of cluster thinning on vine photosynthesis, berry ripeness and flavonoid composition of Cabernet Sauvignon. Food Chem. 2018;248:101–110. doi: 10.1016/j.foodchem.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 12.Xu XQ, et al. Differences in volatile profiles of Cabernet Sauvignon grapes grown in two distinct regions of China and their responses to weather conditions. Plant Physiol. Biochem. 2015;89:123–133. doi: 10.1016/j.plaphy.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Koundouras S, Marinos V, Gkoulioti A, Kotseridis Y, Van LC. Influence of vineyard location and vine water status on fruit maturation of non-irrigated cv. Agiorgitiko (Vitis vinifera L.). effects on wine phenolic and aroma components. J. Agric. Food Chem. 2006;54:5077–5086. doi: 10.1021/jf0605446. [DOI] [PubMed] [Google Scholar]
- 14.Wilson B, Strauss CR, Williams PJ. The distribution of free and glycosidically-bound monoterpenes among skin, juice, and pulp fractions of some white grape varieties. Amer. J. Enol.Viticult. 1986;37:107–111. [Google Scholar]
- 15.Hjelmeland AK, Ebeler SE. Glycosidically bound volatile aroma compounds in grapes and wine: a review. Amer. J. Enol. Viticult. 2015;66:1–11. doi: 10.5344/ajev.2014.14104. [DOI] [Google Scholar]
- 16.Voirin SG, Baumes RL, Bitteur SM, Gunata ZY, Bayonove CL. Novel monoterpene disaccharide glycosides of vitis vinifera grapes. J. Agric. Food Chem. 1990;38:1373–1378. doi: 10.1021/jf00096a016. [DOI] [Google Scholar]
- 17.Dubey VS, Bhalla R, Luthra R. An overview of the non-mevalonate pathway for terpenoid biosynthesis in plants. J. Biosci. 2003;28:637–646. doi: 10.1007/BF02703339. [DOI] [PubMed] [Google Scholar]
- 18.Luan F, Wust M. Differential incorporation of 1-deoxy-Dxylulose into (3S)-linalool and geraniol in grape berry exocarp and mesocarp. Phytochemistry. 2002;60:451–459. doi: 10.1016/S0031-9422(02)00147-4. [DOI] [PubMed] [Google Scholar]
- 19.Dudareva N, Klempien A, Muhlemann JK, Kaplan I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013;198:16–32. doi: 10.1111/nph.12145. [DOI] [PubMed] [Google Scholar]
- 20.Schwab W, Davidovich-Rikanati R, Lewinsohn E. Biosynthesis of plant-derived flavor compounds. The Plant J. 2008;54:712–732. doi: 10.1111/j.1365-313X.2008.03446.x. [DOI] [PubMed] [Google Scholar]
- 21.Withers ST, Keasling JD. Biosynthesis and engineering of isoprenoid small molecules. Appl. Microbiol. Biotechnol. 2007;73:980–990. doi: 10.1007/s00253-006-0593-1. [DOI] [PubMed] [Google Scholar]
- 22.Degenhardt J, Kollner TG, Gershenzon J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry. 2009;70:1621–1637. doi: 10.1016/j.phytochem.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 23.Doligez A, Audiot E, Baumes R, This P. QTLs for muscat flavour and monoterpenic odorant content in grapevine (Vitis vinifera L.) Mol. Breeding. 2006;18:109–125. doi: 10.1007/s11032-006-9016-3. [DOI] [Google Scholar]
- 24.Battilana J, et al. The 1-deoxy-D: -xylulose 5-phosphate synthase gene co-localizes with a major QTL affecting monoterpene content in grapevine. Theor. Appl. Genet. 2009;118:653–669. doi: 10.1007/s00122-008-0927-8. [DOI] [PubMed] [Google Scholar]
- 25.Duchene E, et al. A grapevine (Vitis vinifera L.) deoxy-d-xylulose synthase gene colocates with a major quantitative trait loci for terpenol content. Theor. Appl. Genet. 2009;118:541–552. doi: 10.1007/s00122-008-0919-8. [DOI] [PubMed] [Google Scholar]
- 26.Battilana J, et al. Functional effect of grapevine 1-deoxy-D-xylulose 5-phosphate synthase substitution K284N on Muscat flavour formation. J. Exp. Bot. 2011;62:5497–5508. doi: 10.1093/jxb/err231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin DM, Chiang A, Lund ST, Bohlmann J. Biosynthesis of wine aroma: transcript profiles of hydroxymethylbutenyl diphosphate reductase, geranyl diphosphate synthase, and linalool/nerolidol synthase parallel monoterpenol glycoside accumulation in Gewürztraminer grapes. Planta. 2012;236:919–929. doi: 10.1007/s00425-012-1704-0. [DOI] [PubMed] [Google Scholar]
- 28.Wen YQ, et al. Using the combined analysis of transcripts and metabolites to propose key genes for differential terpene accumulation across two regions. BMC Plant Biol. 2015;15:240. doi: 10.1186/s12870-015-0631-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costantini L, et al. Drawing Links from Transcriptome to Metabolites: The Evolution of Aroma in the Ripening Berry of Moscato Bianco. (Vitis vinifera L.) Front. Plant. Sci. 2017;8:780. doi: 10.3389/fpls.2017.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y, et al. Aroma characterization based on aromatic series analysis in table grapes. Sci. Rep. 2016;6:31116. doi: 10.1038/srep31116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vitulo N, et al. A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 2014;14:99. doi: 10.1186/1471-2229-14-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coombe BG. Adoption of a system for identifying grapevine growth stages. Aust. J. Grape Wine Res. 1995;1:100–110. doi: 10.1111/j.1755-0238.1995.tb00086.x. [DOI] [Google Scholar]
- 33.Wang L, Si Y, Dedow LK, Shao Y, Liu P, Brutnell TP. A low-cost library construction protocol and data analysis pipeline for Illumina-based strand specific multiplex RNA-seq. PLoS One. 2011;6:e26426. doi: 10.1371/journal.pone.0026426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D, Langmead B, Salzberg SL. HISAT: a fast-spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.NCBI Sequence Read Archive. http://identifiers.org/ncbi/insdc.sra:SRP184152 (2019).
- 36.Sun, L. & Zhu, B. Transcriptome profiles of three Muscat table grape cultivars at three developmental stage. Gene Expression Omnibus, http://identifiers.org/geo:GSE130386 (2019).
- 37.Sun, L. et al. Transcriptome profiles of three Muscat table grape cultivars to dissect the mechanism of terpene biosynthesis. figshare, 10.6084/m9.figshare.c.4378256.v1 (2019). [DOI] [PMC free article] [PubMed]
- 38.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SOAPnuke: https://github.com/BGI-flexlab/SOAPnuke. Version: v1.5.2. Parameters: -l 5 -q 0.51 -n 0.55 -i -Q 2–seqType 1.
HISAT2: http://www.ccb.jhu.edu/software/hisat. Version:v2.0.4.Parameters:–phred64–sensitive–no-discordant–no-mixed -I 1 -X 1000.
Bowtie2: http://bowtie-bio.sourceforge.net/Bowtie2. Version: v2.2.5. Parameters: -q–phred64–sensitive–dpad 0–gbar 99999999–mp 1,1–np 1–score-min L,0, −0.1 -I 1 -X 1000–no-mixed–no-discordant -p 1 -k 200.
RSEM: http://deweylab.biostat.wisc.edu/RSEM. Version: v1.2.12. Parameters: default.