Summary
Hemophilia A (HA) is caused by genetic mutations in the blood coagulation factor VIII (FVIII) gene. Genome-editing approaches can be used to target the mutated site itself in patient-derived induced pluripotent stem cells (iPSCs). However, these approaches can be hampered by difficulty in preparing thousands of editing platforms for each corresponding variant found in HA patients. Here, we report a universal approach to correct the various mutations in HA patient iPSCs by the targeted insertion of the FVIII gene into the human H11 site via CRISPR/Cas9. We derived corrected clones from two types of patient iPSCs with frequencies of up to 64% and 66%, respectively, without detectable unwanted off-target mutations. Moreover, we demonstrated that endothelial cells differentiated from the corrected iPSCs successfully secreted functional protein. This strategy may provide a universal therapeutic method for correcting all genetic variants found in HA patients.
Keywords: genome editing, H11 locus, CRISPR/Cas9, hemophilia, iPSCs
Highlights
-
•
Two types of FVIII mutations were corrected using Cas9-mediated KI in patient iPSCs
-
•
Targeted KI of the FVIII into the H11 site induced the production of functional protein
-
•
Whole-genome sequencing analyses revealed no off-target mutations in the corrected iPSCs
Hemophilia A (HA) is caused by various genetic mutations within the blood coagulation factor VIII (FVIII) gene. In this article, Kim and colleagues attempt the targeted insertion of the FVIII gene into the human H11 site in two types of HA patient iPSCs. This approach may offer a universal therapeutic method for correcting all genetic variants found in HA patients.
Introduction
Hemophilia A (HA) is one of the most common inherited bleeding disorders, with an incidence of 1 in 5,000 males worldwide (Berntorp and Shapiro, 2012). HA is caused by various genetic mutations (2,015 unique variants) within the X-linked coagulation factor VIII (FVIII) gene, including large deletions, insertions, inversions, and point mutations (FVIII variant database; www.factorviii-db.org/). At present, the intravenous infusion of recombinant FVIII protein is an available treatment option; however, this therapy is not curative and is associated with high costs, lifelong treatment, and the formation of FVIII-inactivating antibodies. Thus, the development of a fundamental method for treating HA is required.
Human induced pluripotent stem cells (iPSCs) are a versatile cell source for transplanting autologous cells with restored genes to compensate for mutated genes, for understanding cellular and molecular disease mechanisms and disease modeling (Cherry and Daley, 2013), and for therapeutic applications including drug discovery (Shi et al., 2017). Indeed, autologous retinal pigment epithelial cells differentiated from iPSCs were transplanted into a patient with neovascular age-related macular degeneration (Mandai et al., 2017). In addition, patient iPSCs were used for genome editing to correct genetic mutations (Li et al., 2015, Park et al., 2015a, Xu et al., 2017).
The type II clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system is a versatile tool for genome editing (Jinek et al., 2012). Recently, nuclease-mediated genome editing was performed in HA patient iPSCs to correct the endogenous FVIII locus to the normal gene orientation without using ectopic donor DNA or with the targeted insertion of donor DNA harboring partial FVIII exons (Park et al., 2015b, Wu et al., 2016), strategies that are typically applied to inversion mutations. Interestingly, more than 60% of HA patients exhibit other genetic variations including point mutations, large deletions, insertions, or duplications (Graw et al., 2005), which suggests that the application of a universal correction strategy is required for all types of genetic variations that occur in HA. In addition, protocols to differentiate iPSCs into liver sinusoidal endothelial cells (LSECs), which are the primary producers of the FVIII protein, have also not been available to date (Park et al., 2016a). Thus, to overcome these limitations we have developed a universal correction approach by accessing a genomic safe harbor site and expressing functional FVIII without any restrictions in the type of variation or the cell type used for transplantation.
In this study, we chose the human H11 locus as a safe harbor site and inserted the functional FVIII gene into this site using the CRISPR/Cas9 system in a targeted manner in both deleted- and inverted-patient iPSCs. Importantly, we found that the mRNA expression induced FVIII activity in the cultured supernatants. Moreover, we demonstrated that no off-target mutations were found in the corrected clones. To our knowledge, this is the first report to demonstrate a targeted FVIII insertion in a safe harbor locus that resulted in the phenotypic correction of the FVIII deficiency in HA patient iPSCs. This approach may provide a universal correction method for application to all types of genetic variations found in HA patients.
Results
Generation of FVIII Deleted Patient iPSCs
First, we derived FVIII deleted iPSC clones from adipose tissue-derived mesenchymal stem cells obtained from a patient diagnosed with several exon deletions (exons 8–22) using episomal vectors. We selected a total of nine embryonic stem cell-like colonies (termed Epi1 to Epi9), which were maintained onto a feeder layer followed by adaptation in feeder-free culture conditions (Figure S1A). After seven passages, we confirmed the absence of the episomal vectors in all the clones except one by PCR (Figure S1B). For the remaining experiments we chose the Epi6 line, which does not contain the EBNA-1 sequence encoded in the vectors and expresses pluripotency markers including SSEA4, TRA-1-60, OCT4, NANOG (Figure S1C), SOX2, and Lin28 (Figure S1D). The ability to differentiate in vitro was further confirmed in the Epi6 line (Figure S1E), which also exhibited a normal karyotype (Figure S1F).
Synthetic Single Guide RNA Design and Validation of Nuclease Activity
To insert FVIII into the H11 locus using a targeted approach, we designed the synthetic single guide RNA (sgRNA) that recognized the target site using web-based in silico tools (crispr.mit.edu) (Figure 1A). We co-transfected either HEK-293T cells or each iPSC clone with both Cas9 and sgRNA vectors. To test the nuclease activity, we performed a T7 endonuclease I (T7E1) assay. The nuclease activity was relatively high, inducing small insertions and deletions (indels) mutations with a frequency of 15% at the H11 locus based on the T7E1 assay results (Figure S2A). However, the nuclease activity in each iPSC clone was relatively low due to low transfection efficiency, inducing indels with a frequency of 4% and 4.3%, respectively (Figure S2A). In addition, deep sequencing analyses revealed that the frequency of indels at the target site was 27.9% (Figure S2B), with various indels found at the target site (Figure S2C). Moreover, no off-target mutations were detectably induced at eight homologous sites that differed from the on-target site by up to four nucleotides (Figure S2B).
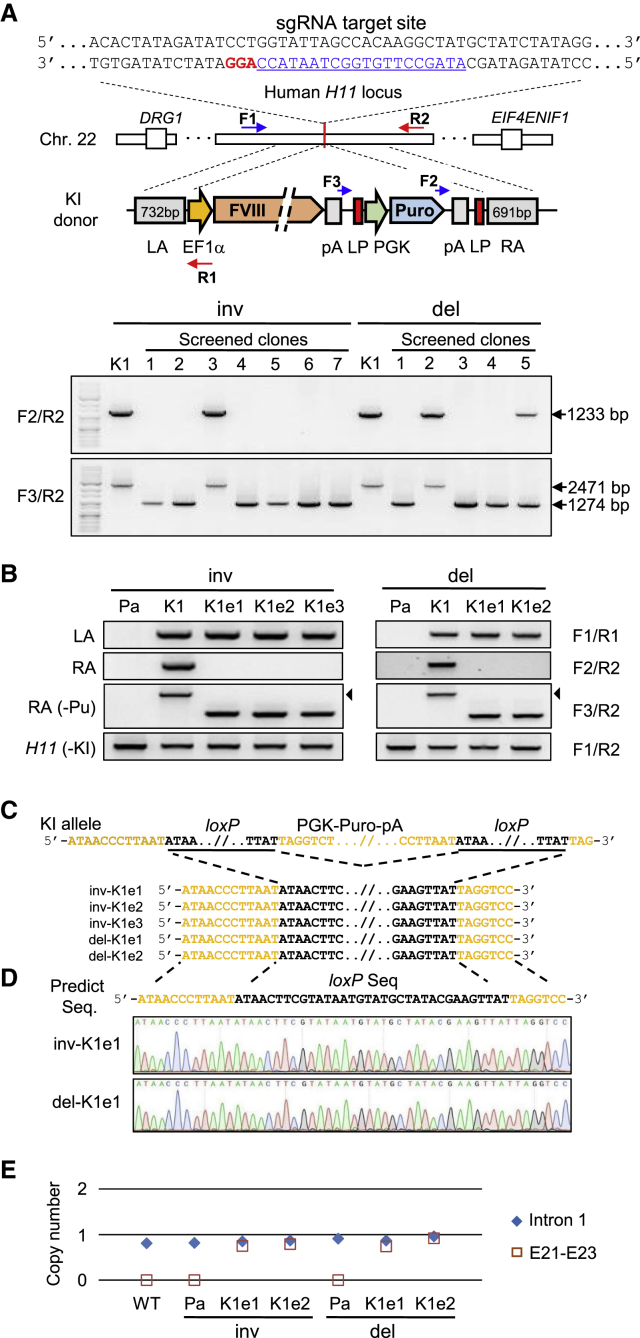
Figure 1.
Targeted Insertions of the FVIII Gene into the Human H11 Locus of HA Patient-Derived iPSCs
(A) Schematic overview depicting the FVIII gene knockin strategy into the H11 locus located in human chromosome 22. The bases underlined in blue indicate the sgRNA target site. The protospacer adjacent motif sequence is shown in red. The five specific primers used for genotyping are shown. Lower panel shows PCR-based screening for excision of the selection cassette.
(B) PCR-based genotype analysis to confirm the removal of the puromycin expression cassette in the targeted iPSC clones. Black arrowheads indicate the DNA bands containing the puromycin cassette in the inv-K1 and del-K1 clones.
(C) Post-excision DNA sequence analysis between the two underlined loxP sites.
(D) Chromatograms showing the targeted excision of the puromycin cassette from the knockin allele.
(E) Droplet digital PCR (ddPCR) analysis to determine the copy number of the inserted fragment in each clone. The RPP30 gene served as a reference. The primer sequences for the intron 1 locus (Intron 1) of the endogenous FVIII gene and for exon 21 to exon 23 (E21–E23) of the knockin donor template are described in the ddPCR analysis section of Experimental Procedures.
See also Figures S2 and S3; Tables S1 and S2.
Targeted Knockin of the FVIII Gene into the Human H11 Locus
In parallel, we cloned the FVIII expression cassette under the control of the human elongation factor 1α (EF1α) promoter and a puromycin expression cassette into the backbone vector to construct donor DNA, followed by cloning flanked by both homology arms (Figures 1A and S3A). Thereafter, we electroporated Cas9 ribonucleoproteins (RNPs) and donor DNA into both FVIII inverted iPSCs (inv-Pa) and FVIII deleted iPSCs (del-Pa). Following additional culturing and puromycin selection, we screened drug-resistant colonies by PCR-based genotyping to find colonies harboring the targeted knockin using the specific primer sets listed in Table S1 (Figure S3A). Nine out of 14 colonies (64.3%, in case of inv-Pa) and 18 out of 27 colonies (66.7%) exhibited positive PCR bands for the inv-Pa and del-Pa knockin junctions on an agarose gel, respectively (Table S2). Following three passages, we derived two clones (inv-K1 and inv-K2) from inv-Pa iPSCs and three clones (del-K1 to del-K3) from del-Pa iPSCs, and demonstrated that all of the clones were single-allele knockin clones (Figure S3B). Targeted knockins from all the clones were further verified by Sanger sequencing of PCR amplicons (Figures S3C and S3D). Next, to excise the puromycin expression cassette from the clones, we chose the inv-K1 and del-K1 clones because they had no indels, even in the untargeted allele (−KI allele) (Figure S3C). After the transient expression of Cre recombinase in these clones, we screened seven colonies by PCR-based genotyping using the F3/R2 primers (Figure 1A). The successful excision of the selection cassette was confirmed by PCR-based genotyping using specific primers (Figure 1B and Table S1). Following the verifications of recombination between the two loxP sites by Sanger sequencing (Figures 1C and 1D), we derived the three puromycin-excised clones (inv-K1e1 to inv-K1e3) from the inv-K1 clone, and the two clones (del-K1e1 and del-K1e2) from the del-K1 clone (Figure 1B). Next, we confirmed that four knockin clones (inv-K1e1, inv-K1e2, del-K1e1, and del-K1e2) contained only one copy of the FVIII knockin fragment using droplet digital PCR analysis (Figure 1E). These results revealed targeted integration of the FVIII gene at the H11 locus.
Analyses of Pluripotency and Off-Target Mutations in Genetically Corrected Clones
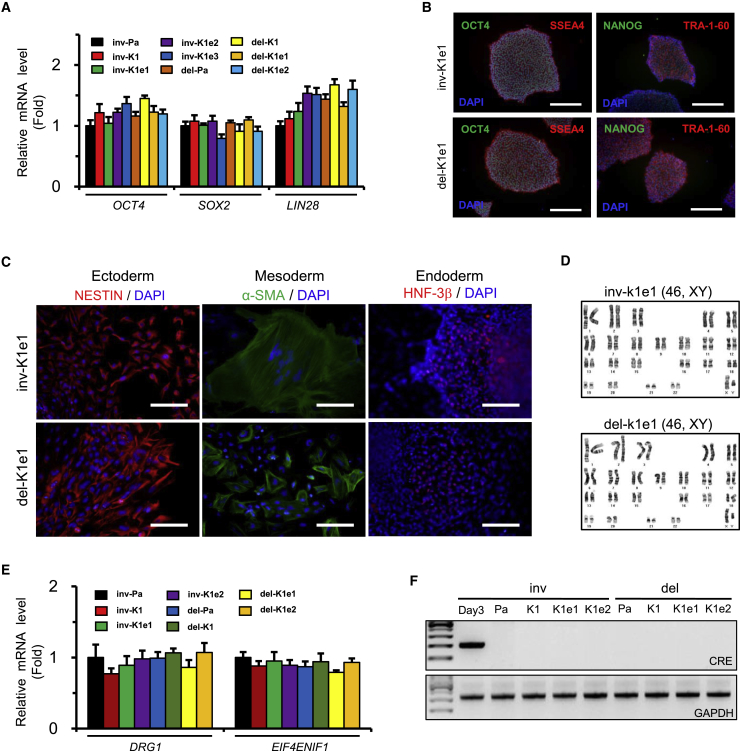
Next, we investigated whether the corrected clones maintained their pluripotent characteristics compared with the parental clones. Indeed, all the corrected clones actively transcribed pluripotency genes, including OCT4, SOX2, and LIN28 (Figure 2A), and maintained similar levels of pluripotency marker proteins such as SSEA4, TRA-1-60, OCT4, and NANOG compared with their parental iPSC clones (Figure 2B). In addition, the corrected clones successfully differentiated into the three germ layers, as shown by positive immunostaining for NESTIN (ectoderm), α-smooth muscle actin (mesoderm), and hepatocyte nuclear factor-3β (endoderm) (Figure 2C). Furthermore, the clones exhibited a normal 46, XY karyotype by G-banding (Figure 2D). We then confirmed by qPCR analysis that no significant differences were found in the expression levels of DRG1 and EIF4ENIF1 genes between the parental and knockin iPSC clones (Figure 2E). No PCR bands corresponding to the Cre sequence were amplified from each knockin clone (Figure 2F). We then investigated whether off-target mutations were induced in the corrected clones by the nuclease used in this study. Ten potential off-target sites were examined by targeted deep sequencing in the four corrected clones and the two types of parental clones. We verified that no significant off-target mutations were induced at the sites listed in Table S3 (Figure S4). To further verify off-target mutations, we performed whole-genome sequencing analysis for three iPSC clones (del-Pa, del-K1e1, and del-K1e2) using Illumina NovaSeq6000 and yielding 40× coverage. We demonstrated that no off-target mutations were found (Table S4). These results revealed that the nuclease used in this study did not induce off-target mutations in two of the knockin clones, which is in line with a recent study demonstrating the high specificity of Cas9-mediated nuclease in the clonal populations of pluripotent stem cells (Park et al., 2015b, Veres et al., 2014).
Figure 2.
Analyses of Pluripotency from the Corrected iPSC Clones
(A) Expression of endogenous OCT4, SOX2, and LIN28 in parental patient and corrected iPSC clones. The expression level of each gene was normalized to that of GAPDH. Data are means ± SEM of three independent experiments.
(B) Expression of the pluripotency markers OCT4, NANOG, SSEA-4, and TRA-1-60 detected by immunocytochemistry. The DAPI signal indicates the total cell content in the image. Scale bars, 200 μm.
(C) Expression of marker proteins representing the ectoderm (NESTIN), mesoderm (α-smooth muscle actin [α-SMA]), and endoderm (hepatocyte nuclear factor-3β [HNF-3β]). The DAPI signal indicates the total cell content in the image. Scale bars, 100 μm.
(D) Karyotype analyses were performed in the corrected iPSC clones.
(E) Expression of DRG1 and EIF4ENIF1 in parental and corrected patient iPSC clones. The expression level of each gene was normalized to that of GAPDH. Data are means ± SEM of three independent experiments.
(F) Detection of the Cre expression vector sequence remaining in each clone by PCR. The GAPDH gene was used as a quality control for the total isolated DNA. Total DNA isolated from the cells 3 days after electroporation was used as the positive control for the Cre expression vector.
Phenotypic Correction of the FVIII Deficiency In Vitro
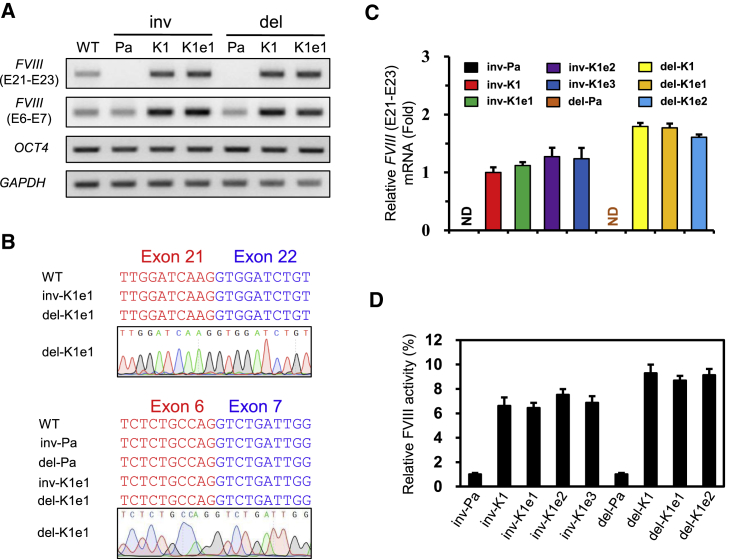
After the successful targeted insertion of the FVIII gene in the H11 locus, we examined the phenotype of the corrected clones using an in vitro culture system. Using semiquantitative RT-PCR, the FVIII mRNA expression levels in the H11 locus were measured at the undifferentiated stage. As expected, no PCR bands corresponding to FVIII exons 21 and 23 were amplified from the two types of patient iPSCs due to either incorrect splicing (inv-Pa) or deletion of the corresponding region (del-Pa); however, corresponding bands to FVIII exons 6 and 7 were amplified from both inv-Pa and del-Pa iPSC clones (Figure 3A). In contrast, PCR bands corresponding to exons 21 and 23 were amplified in the corrected iPSCs, and were detected regardless of the presence or absence of the selection cassette (Figure 3A). We also performed Sanger sequencing to verify the sequences of the amplified DNA bands (Figure 3B). In additional qPCR analyses, we demonstrated that there were no differences in the expression of the FVIII gene between each post-excision clone (inv-K1e1 to inv-K1e3, or del-K1e1 and del-K1e2) (Figure 3C). We then investigated whether the induction of FVIII expression corresponded to increased FVIII activity in the corrected clones. As shown in Figure 3D, all of the corrected clones, regardless of whether they had the selection cassette, exhibited highly elevated FVIII activity levels compared with levels in the parental patient iPSCs.
Figure 3.
Phenotypic Rescue of the Expression of FVIII Gene from the Corrected iPSC Clones
(A) RT-PCR analysis to detect the expression of FVIII and OCT4 in undifferentiated, corrected iPSC clones. GAPDH expression was used for normalization.
(B) Chromatograms showing the sequences of amplified DNA bands from each clone (related to A).
(C) Results of the qPCR analysis showing the FVIII expression levels in undifferentiated patient iPSC clones and corrected clones. GAPDH expression was used for normalization. Data are means ± SEM of three independent experiments. ND, not detected.
(D) The FVIII activity was determined after 20-fold concentration in supernatants obtained from either patient or corrected clones. Data indicate activity detected per 1 × 106 iPSCs. Data are means ± SEM of three independent experiments.
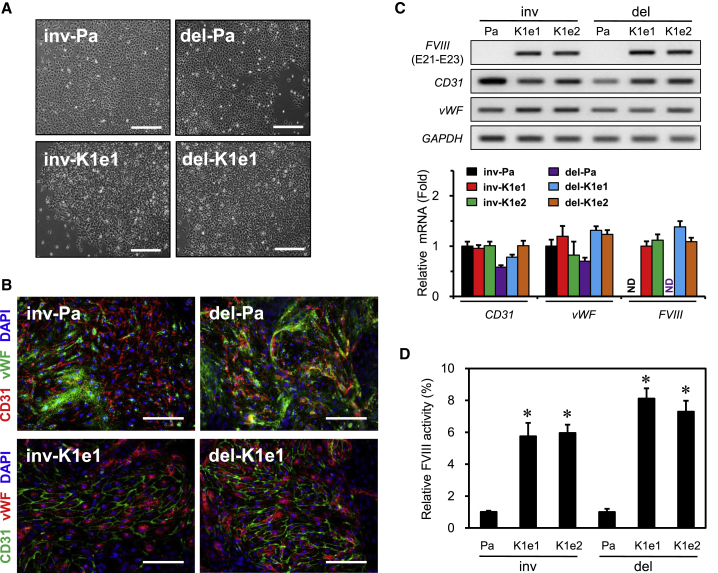
Next, the four corrected iPSC clones, including the two types of patient iPSCs, were differentiated into endothelial cells, a source of FVIII production (Shahani et al., 2010), as previously reported (Harding et al., 2017). We did not detect any impairment in the differentiation of the clones into endothelial cells, which exhibited cobblestone-like morphologies at the time of differentiation on day 4 (Figure 4A) and positive staining for endothelial cell markers such as CD31 and von Willebrand factor (vWF) at the end of differentiation (Figure 4B). We further evaluated the levels of FVIII mRNA using RT-PCR and qPCR. No PCR bands corresponding to FVIII exons 21 and 23 were detected in the patient endothelial cells that differentiated from inv-Pa and del-Pa clones (Figure 4C). However, as expected, the FVIII mRNA was detected in endothelial cells differentiated from the corrected iPSC clones (Figure 4C). In addition, all cells that differentiated from the corrected clones showed significantly elevated FVIII activity levels compared with the patient iPSCs (Figure 4D). These results indicated that the phenotype of the patient iPSCs could be corrected by the expression of the FVIII gene via targeted knockin at the H11 locus.
Figure 4.
Functional Correction of FVIII Deficiency in Endothelial Cells Differentiated from Corrected iPSC Clones
(A) Phase image of cobblestone-like morphologies at the time of differentiation on day 4 in the indicated clones. Scale bars, 200 μm.
(B) Expression of marker proteins (CD31 and vWF) representing endothelial cells (ECs) derived from parental patient and corrected clones. The DAPI signal indicates the total cell content in the image. Scale bars, 100 μm.
(C) Expression of FVIII mRNA including EC marker genes were verified in cells differentiated from patients and corrected clones using RT-PCR (upper panel) and qPCR (lower panel). GAPDH expression was used for normalization. Data are means ± SEM of three independent experiments. ND, not detected.
(D) The FVIII activity was determined after 20-fold concentration in supernatants obtained from patient or corrected clones after differentiation into ECs. Data indicate activity detected per 1 × 106 ECs. Data are means ± SEM of three independent experiments. ∗p < 0.001 compared with the parental cells (Student's t test).
Discussion
In typical genome editing, correcting the endogenous locus itself is an ideal strategy (Park et al., 2016b). However, there are some current limitations to targeting the endogenous site in HA: (1) there are no protocols for differentiating iPSCs into LSECs, which are the primary producers of the FVIII protein; (2) microvascular endothelial cells cannot fully correct the phenotype because they produce only a small amount of FVIII protein; and (3) it is not feasible to prepare sgRNA, including donor DNA, for the 2,015 unique mutations found in HA. To overcome these limitations, we attempted the targeted knockin of the functional FVIII gene into a safe harbor site for a universal correction regardless of mutation types. Although there was an attempt to target FVIII gene at ribosomal DNA locus in HA patient iPSCs using TALENickases, this attempt has not demonstrated successful functional restoration in an in vivo system (Pang et al., 2016).
The H11 locus was identified as a safe harbor site and used to express transgene in iPSCs (Turan et al., 2016, Zhu et al., 2014) or animal model (Ruan et al., 2015). Interestingly, this locus is an intergenic sequence; thus, knockin of a transgene into the site does not induce the gene disruption observed with other safe harbor sites such as AAVS1 and the albumin locus. Using this advantage, we chose the H11 locus to develop our universal correction platform via targeted integration of the FVIII gene. In addition, we used two types of patient iPSCs, FVIII inverted and large deleted patient iPSCs, for a proof of principle. In our experiments, we generated corrected clones with high efficiency in the two different types of iPSCs and demonstrated the functional recovery of mRNA expression as well as the secretion of the FVIII protein from the corrected cells in vitro. Nevertheless, the functional effects following transplantation in an in vivo system still need to be confirmed.
Off-target mutations are a concern regarding the therapeutic use of engineered nucleases. To avoid off-target effects, we chose unique target sequences that differed from any other site in the human genome by at least three nucleotides. We also used 5′-GGX20 sgRNAs transcribed in vitro, which reduced off-target mutations without reducing on-target activity as previously reported (Cho et al., 2014), and electroporated Cas9 RNPs into the patient iPSCs (Kim et al., 2014). In addition, targeted deep sequencing was performed to validate the absence of unwanted mutations in the corrected clones following the removal of the puromycin expression cassette, resulting in no off-target mutations. These results are consistent with previous reports of the reduced induction of off-target mutations in individual clones of edited cells (Park et al., 2015b, Veres et al., 2014).
In summary, we used the H11 locus to develop a universal correction platform, and precisely targeted the FVIII gene via error-free knockin with high efficiency. Finally, we verified that endothelial cells (ECs) successfully differentiated from the corrected iPSCs containing the FVIII gene and secreted functional protein in vitro system. This approach is not only simple but may also provide a universal platform to correct for the various mutations found in HA using the same sgRNA and donor DNA for one target site, the human H11 locus.
Experimental Procedures
Ethical Statement
The generation and analyses of iPSCs from the HA patients were approved by Yonsei University Institutional Review Board (IRB #4-2012-0028). All volunteers who participated in this study signed written informed consent forms before donating cells for the generation of iPSCs.
Preparations of Donor Plasmid and Guide RNA for SpCas9
To construct the donor plasmid, we used the pCDNA4/BDD-FVIII plasmid (www.addgene.org, no. 41035) (Peters et al., 2013) as a backbone. See Supplemental Experimental Procedures for details of the protocol.
Off-Target Analysis and Targeted Deep Sequencing
Ten potential off-target sites differing by up to four nucleotides from the on-target site were searched using a web-based in silico tool (crispr.mit.edu) (Ran et al., 2013). For deep sequencing analysis, PCR amplicons for each off-target site and the on-target site were prepared from genomic DNA using high-fidelity PrimeSTAR Max DNA polymerase (Takara Bio) and the specific primer sets listed in Table S3. Following PCR purification of the resulting PCR products using an AccuPrep PCR Purification Kit (Bioneer, Korea), the PCR amplicons were subjected to paired-end sequencing using a MiSeq system (Illumina) at LAS (Korea).
CRISPR/Cas9-Mediated Correction by FVIII Knockin
Cas9 RNPs and FVIII knockin donor DNA were electroporated into the patient iPSCs as previously described (Park et al., 2015b) with slight modifications. See Supplemental Experimental Procedures for details of the protocol.
Excision of the Puromycin Selection Cassette
To excise the puromycin expression cassette from the FVIII knockin clones, we electroporated 2 × 105 knockin iPSCs with 1 μg of the Cre expression plasmid (pCAG-Cre:GFP; no. 13776, www.addgene.org). The isolation of clones was performed by single cell passaging as previously described (Park et al., 2016c). The excision of the puromycin cassette was confirmed by PCR-based genotyping using the F3 and R2 primers. After removal of the puromycin expression cassette, we confirmed the absence of the Cre expression vector in each clone by PCR using the CRE-F and CRE-R primers listed in Table S1.
Differentiation into Endothelial Cells
To induce the differentiation of the iPSCs into ECs, we used a previously described protocol (Harding et al., 2017) with slight modifications. See Supplemental Experimental Procedures for details of the protocol.
Measurement of FVIII Activity
To measure FVIII activity, we concentrated culture supernatants 20-fold using an Amicon Ultra-4 centrifugal filter (Millipore). The FVIII activities were measured in the culture supernatants using the commercially available Coamatic Factor VIII chromogenic assay kit (Instrumentation Laboratory) according to the manufacturer's instructions. See Supplemental Experimental Procedures for details of the protocol.
Statistical Analysis
All data are expressed as means ± SEM of at least three independent experiments. Statistically significant differences were estimated using a Student's t test. A resulting p value of <0.05 was considered statistically significant.
Author Contributions
C.-Y.P. designed and carried out the experiments. S.-R.C. and J.J.S. helped with iPSC clone derivation and validation. C.-Y.P. and D.-W.K. interpreted the results. C.-Y.P., J.K., and D.-W.K. were in charge of critical revision. C.-Y.P. and D.-W.K. wrote the manuscript.
Acknowledgments
C.-Y.P. was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (2016R1C1B1008742). D.-W.K. was supported by the Bio & Medical Technology Development Program of the NRF (2017M3A9B4042580), the Korea Health Technology R&D Project from the Ministry of Health and Welfare (HI18C0829), and the Faculty Research Grant of Yonsei University College of Medicine (6-2017-0190).
Published: May 16, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.04.016.
Accession Numbers
The deep sequencing data files reported in this study have been deposited in the GEO database repository (www.ncbi.nlm.nih.gov/geo/) under the accession number GEO: GSE124663. The whole-genome sequencing data files have also been deposited in the SRA database (www.ncbi.nlm.gov/sra/) under the accession number PRJNA515982.
Supplemental Information
References
- Berntorp E., Shapiro A.D. Modern haemophilia care. Lancet. 2012;379:1447–1456. doi: 10.1016/S0140-6736(11)61139-2. [DOI] [PubMed] [Google Scholar]; Berntorp, E., and Shapiro, A.D. (2012). Modern haemophilia care. Lancet 379, 1447-1456. [DOI] [PubMed]
- Cherry A.B., Daley G.Q. Reprogrammed cells for disease modeling and regenerative medicine. Annu. Rev. Med. 2013;64:277–290. doi: 10.1146/annurev-med-050311-163324. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cherry, A.B., and Daley, G.Q. (2013). Reprogrammed cells for disease modeling and regenerative medicine. Annu. Rev. Med. 64, 277-290. [DOI] [PMC free article] [PubMed]
- Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cho, S.W., Kim, S., Kim, Y., Kweon, J., Kim, H.S., Bae, S., and Kim, J.S. (2014). Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24, 132-141. [DOI] [PMC free article] [PubMed]
- Graw J., Brackmann H.H., Oldenburg J., Schneppenheim R., Spannagl M., Schwaab R. Haemophilia A: from mutation analysis to new therapies. Nat. Rev. Genet. 2005;6:488–501. doi: 10.1038/nrg1617. [DOI] [PubMed] [Google Scholar]; Graw, J., Brackmann, H.H., Oldenburg, J., Schneppenheim, R., Spannagl, M., and Schwaab, R. (2005). Haemophilia A: from mutation analysis to new therapies. Nat. Rev. Genet. 6, 488-501. [DOI] [PubMed]
- Harding A., Cortez-Toledo E., Magner N.L., Beegle J.R., Coleal-Bergum D.P., Hao D., Wang A., Nolta J.A., Zhou P. Highly efficient differentiation of endothelial cells from pluripotent stem cells requires the MAPK and the PI3K pathways. Stem Cells. 2017;35:909–919. doi: 10.1002/stem.2577. [DOI] [PubMed] [Google Scholar]; Harding, A., Cortez-Toledo, E., Magner, N.L., Beegle, J.R., Coleal-Bergum, D.P., Hao, D., Wang, A., Nolta, J.A., and Zhou, P. (2017). Highly efficient differentiation of endothelial cells from pluripotent stem cells requires the MAPK and the PI3K pathways. Stem Cells 35, 909-919. [DOI] [PubMed]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821. [DOI] [PMC free article] [PubMed]
- Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kim, S., Kim, D., Cho, S.W., Kim, J., and Kim, J.S. (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012-1019. [DOI] [PMC free article] [PubMed]
- Li H.L., Fujimoto N., Sasakawa N., Shirai S., Ohkame T., Sakuma T., Tanaka M., Amano N., Watanabe A., Sakurai H. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4:143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, H.L., Fujimoto, N., Sasakawa, N., Shirai, S., Ohkame, T., Sakuma, T., Tanaka, M., Amano, N., Watanabe, A., Sakurai, H., et al. (2015). Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 4, 143-154. [DOI] [PMC free article] [PubMed]
- Mandai M., Watanabe A., Kurimoto Y., Hirami Y., Morinaga C., Daimon T., Fujihara M., Akimaru H., Sakai N., Shibata Y. Autologous induced stem-cell-derived retinal cells for macular degeneration. N. Engl. J. Med. 2017;376:1038–1046. doi: 10.1056/NEJMoa1608368. [DOI] [PubMed] [Google Scholar]; Mandai, M., Watanabe, A., Kurimoto, Y., Hirami, Y., Morinaga, C., Daimon, T., Fujihara, M., Akimaru, H., Sakai, N., Shibata, Y., et al. (2017). Autologous induced stem-cell-derived retinal cells for macular degeneration. N. Engl. J. Med. 376, 1038-1046. [DOI] [PubMed]
- Pang J., Wu Y., Li Z., Hu Z., Wang X., Hu X., Liu X., Zhou M., Liu B., Wang Y. Targeting of the human F8 at the multicopy rDNA locus in Hemophilia A patient-derived iPSCs using TALENickases. Biochem. Biophys. Res. Commun. 2016;472:144–149. doi: 10.1016/j.bbrc.2016.02.083. [DOI] [PubMed] [Google Scholar]; Pang, J., Wu, Y., Li, Z., Hu, Z., Wang, X., Hu, X., Liu, X., Zhou, M., Liu, B., Wang, Y., et al. (2016). Targeting of the human F8 at the multicopy rDNA locus in Hemophilia A patient-derived iPSCs using TALENickases. Biochem. Biophys. Res. Commun. 472, 144-149. [DOI] [PubMed]
- Park C.Y., Halevy T., Lee D.R., Sung J.J., Lee J.S., Yanuka O., Benvenisty N., Kim D.W. Reversion of FMR1 methylation and silencing by editing the triplet repeats in fragile X iPSC-derived neurons. Cell Rep. 2015;13:234–241. doi: 10.1016/j.celrep.2015.08.084. [DOI] [PubMed] [Google Scholar]; Park, C.Y., Halevy, T., Lee, D.R., Sung, J.J., Lee, J.S., Yanuka, O., Benvenisty, N., and Kim, D.W. (2015a). Reversion of FMR1 methylation and silencing by editing the triplet repeats in fragile X iPSC-derived neurons. Cell Rep. 13, 234-241. [DOI] [PubMed]
- Park C.Y., Kim D.H., Son J.S., Sung J.J., Lee J., Bae S., Kim J.H., Kim D.W., Kim J.S. Functional correction of large factor VIII gene chromosomal inversions in hemophilia a patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 2015;17:213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]; Park, C.Y., Kim, D.H., Son, J.S., Sung, J.J., Lee, J., Bae, S., Kim, J.H., Kim, D.W., and Kim, J.S. (2015b). Functional correction of large factor VIII gene chromosomal inversions in hemophilia a patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell 17, 213-220. [DOI] [PubMed]
- Park C.Y., Lee D.R., Sung J.J., Kim D.W. Genome-editing technologies for gene correction of hemophilia. Hum. Genet. 2016;135:977–981. doi: 10.1007/s00439-016-1699-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Park, C.Y., Lee, D.R., Sung, J.J., and Kim, D.W. (2016a). Genome-editing technologies for gene correction of hemophilia. Hum. Genet. 135, 977-981. [DOI] [PMC free article] [PubMed]
- Park C.Y., Sung J.J., Kim D.W. Genome editing of structural variations: modeling and gene correction. Trends Biotechnol. 2016;34:548–561. doi: 10.1016/j.tibtech.2016.02.011. [DOI] [PubMed] [Google Scholar]; Park, C.Y., Sung, J.J., and Kim, D.W. (2016b). Genome editing of structural variations: modeling and gene correction. Trends Biotechnol. 34, 548-561. [DOI] [PubMed]
- Park C.Y., Sung J.J., Choi S.H., Lee D.R., Park I.H., Kim D.W. Modeling and correction of structural variations in patient-derived iPSCs using CRISPR/Cas9. Nat. Protoc. 2016;11:2154–2169. doi: 10.1038/nprot.2016.129. [DOI] [PubMed] [Google Scholar]; Park, C.Y., Sung, J.J., Choi, S.H., Lee, D.R., Park, I.H., and Kim, D.W. (2016c). Modeling and correction of structural variations in patient-derived iPSCs using CRISPR/Cas9. Nat. Protoc. 11, 2154-2169. [DOI] [PubMed]
- Peters R.T., Toby G., Lu Q., Liu T., Kulman J.D., Low S.C., Bitonti A.J., Pierce G.F. Biochemical and functional characterization of a recombinant monomeric factor VIII-Fc fusion protein. J. Thromb. Haemost. 2013;11:132–141. doi: 10.1111/jth.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peters, R.T., Toby, G., Lu, Q., Liu, T., Kulman, J.D., Low, S.C., Bitonti, A.J., and Pierce, G.F. (2013). Biochemical and functional characterization of a recombinant monomeric factor VIII-Fc fusion protein. J. Thromb. Haemost. 11, 132-141. [DOI] [PMC free article] [PubMed]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ran, F.A., Hsu, P.D., Wright, J., Agarwala, V., Scott, D.A., and Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281-2308. [DOI] [PMC free article] [PubMed]
- Ruan J., Li H., Xu K., Wu T., Wei J., Zhou R., Liu Z., Mu Y., Yang S., Ouyang H. Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Sci. Rep. 2015;5:14253. doi: 10.1038/srep14253. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ruan, J., Li, H., Xu, K., Wu, T., Wei, J., Zhou, R., Liu, Z., Mu, Y., Yang, S., Ouyang, H., et al. (2015). Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Sci. Rep. 5, 14253. [DOI] [PMC free article] [PubMed]
- Shahani T., Lavend'homme R., Luttun A., Saint-Remy J.M., Peerlinck K., Jacquemin M. Activation of human endothelial cells from specific vascular beds induces the release of a FVIII storage pool. Blood. 2010;115:4902–4909. doi: 10.1182/blood-2009-07-232546. [DOI] [PubMed] [Google Scholar]; Shahani, T., Lavend'homme, R., Luttun, A., Saint-Remy, J.M., Peerlinck, K., and Jacquemin, M. (2010). Activation of human endothelial cells from specific vascular beds induces the release of a FVIII storage pool. Blood 115, 4902-4909. [DOI] [PubMed]
- Shi Y., Inoue H., Wu J.C., Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shi, Y., Inoue, H., Wu, J.C., and Yamanaka, S. (2017). Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 16, 115-130. [DOI] [PMC free article] [PubMed]
- Turan S., Farruggio A.P., Srifa W., Day J.W., Calos M.P. Precise correction of disease mutations in induced pluripotent stem cells derived from patients with limb girdle muscular dystrophy. Mol. Ther. 2016;24:685–696. doi: 10.1038/mt.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]; Turan, S., Farruggio, A.P., Srifa, W., Day, J.W., and Calos, M.P. (2016). Precise correction of disease mutations in induced pluripotent stem cells derived from patients with limb girdle muscular dystrophy. Mol. Ther. 24, 685-696. [DOI] [PMC free article] [PubMed]
- Veres A., Gosis B.S., Ding Q., Collins R., Ragavendran A., Brand H., Erdin S., Talkowski M.E., Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Veres, A., Gosis, B.S., Ding, Q., Collins, R., Ragavendran, A., Brand, H., Erdin, S., Talkowski, M.E., and Musunuru, K. (2014). Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15, 27-30. [DOI] [PMC free article] [PubMed]
- Wu Y., Hu Z., Li Z., Pang J., Feng M., Hu X., Wang X., Lin-Peng S., Liu B., Chen F. In situ genetic correction of F8 intron 22 inversion in hemophilia A patient-specific iPSCs. Sci. Rep. 2016;6:18865. doi: 10.1038/srep18865. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu, Y., Hu, Z., Li, Z., Pang, J., Feng, M., Hu, X., Wang, X., Lin-Peng, S., Liu, B., Chen, F., et al. (2016). In situ genetic correction of F8 intron 22 inversion in hemophilia A patient-specific iPSCs. Sci. Rep. 6, 18865. [DOI] [PMC free article] [PubMed]
- Xu X., Tay Y., Sim B., Yoon S.I., Huang Y., Ooi J., Utami K.H., Ziaei A., Ng B., Radulescu C. Reversal of phenotypic abnormalities by CRISPR/Cas9-mediated gene correction in Huntington disease patient-derived induced pluripotent stem cells. Stem Cell Reports. 2017;8:619–633. doi: 10.1016/j.stemcr.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu, X., Tay, Y., Sim, B., Yoon, S.I., Huang, Y., Ooi, J., Utami, K.H., Ziaei, A., Ng, B., Radulescu, C., et al. (2017). Reversal of phenotypic abnormalities by CRISPR/Cas9-mediated gene correction in huntington disease patient-derived induced pluripotent stem cells. Stem Cell Reports 8, 619-633. [DOI] [PMC free article] [PubMed]
- Zhu F., Gamboa M., Farruggio A.P., Hippenmeyer S., Tasic B., Schule B., Chen-Tsai Y., Calos M.P. DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Res. 2014;42:e34. doi: 10.1093/nar/gkt1290. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhu, F., Gamboa, M., Farruggio, A.P., Hippenmeyer, S., Tasic, B., Schule, B., Chen-Tsai, Y., and Calos, M.P. (2014). DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Res. 42, e34. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.