Highlights
-
•
FHF2A and B alter Nav1.7 properties in a cell environment-dependent manner.
-
•
FHF2A and B regulate Nav1.7 properties in an isoform-specific manner.
-
•
FHF2A and 2B knockdown in DRG neurons leads to gain-of-function in Nav1.7.
Keywords: Sodium channel, Patch-clamp, HEK293, FGF13, FHF2, Nav1.7
Abstract
Fibroblast Growth Factor Homologous Factors (FHF) constitute a subfamily of FGF proteins with four prototypes (FHF1-4; also known as FGF11-14). FHF proteins have been shown to bind directly to the membrane-proximal segment of the C-terminus in voltage-gated sodium channels (Nav), and regulate current density, availability, and frequency-dependent inhibition of sodium currents. Members of the FHF2 subfamily, FHF2A and FHF2B, differ in the length and sequence of their N-termini, and, importantly, differentially regulate Nav1.6 gating properties. Using immunohistochemistry, we show that FHF2 isoforms are expressed in adult dorsal root ganglion (DRG) neurons where they co-localize with Nav1.6 and Nav1.7. FHF2A and FHF2B show differential localization in neuronal compartments in DRG neurons, and levels of expression of FHF2 factors are down-regulated following sciatic nerve axotomy. Because Nav1.7 in nociceptors plays a critical role in pain, we reasoned that its interaction with FHF2 isoforms might regulate its current properties. Using whole-cell patch clamp in heterologous expression systems, we show that the expression of FHF2A in HEK293 cell line stably expressing Nav1.7 channels causes no change in activation, whereas FHF2B depolarizes activation. Both FHF2 isoforms depolarize fast-inactivation. Additionally, FHF2A causes an accumulation of inactivated channels at all frequencies tested due to a slowing of recovery from inactivation, whereas FHF2B has little effect on these properties of Nav1.7. Measurements of the Nav1.7 current in DRG neurons in which FHF2 levels are knocked down confirmed the effects of FHF2A on repriming, and FHF2B on activation, however FHF2A and B did not have an effect on fast inactivation. Our data demonstrates that FHF2 does indeed regulate the current properties of Nav1.7 and does so in an isoform and cell-specific manner.
1. Introduction
The voltage-gated sodium channel Nav1.7 is preferentially expressed in nociceptive neurons in dorsal root ganglia (DRG), and gain-of-function mutations in this channel have been shown to cause severe human pain disorders, for example Inherited Erythromelalgia or the complete absence of pain (Dib-Hajj et al., 2013, Dib-Hajj et al., 2017). Currents generated by voltage-gated sodium channels from DRG neurons are classified by their sensitivity to tetrodotoxin (TTX), with Nav1.7 contributing 60–70% of the fast-gating TTX-sensitive (TTX-S) component (Shields et al., 2012, Vasylyev et al., 2014), consistent with its robust expression in these neurons (Black et al., 1996, Vasylyev et al., 2014, Usoskin et al., 2015). When comparing properties of the native TTX-S current with recordings from sodium channels that are expressed in heterologous expression systems, for example human embryonic kidney 293 (HEK293) cell line, it is clear that there are distinct biophysical features that are different in the two cell types (Cummins et al., 2001), suggesting that these differences are caused by the association of the sodium channel with other proteins in the neuronal cell environment, such as beta subunits or other cofactors.
Sodium channels interact with a variety of intracellular proteins including intracellular members of the fibroblast growth factors (FGF11-14; also known as fibroblast homologous factors FHF1-4), which can modulate their properties (Dib-Hajj and Waxman, 2010). FGF11-14/FHF1-4 represent a subfamily which lacks a specific signal peptide for secretion, does not bind to or activate the FGF receptors (Olsen et al., 2003) and plays a regulatory role in intracellular processes through its binding to scaffold proteins and recruitment of MAPK (Schoorlemmer and Goldfarb, 2002). FGF11-14/FHF1-4 have been shown to interact with and regulate sodium channels (Pitt and Lee, 2016). Each member of the FHF family has at least two splice isoforms, and FHF2 is known to have at least five isoforms with distinct amino termini (Munoz-Sanjuan et al., 2000). We have previously shown that Nav1.6 interacts with FGF13/FHF2 (Wittmack et al., 2004, Rush et al., 2006). The two main isoforms of FHF2, FHF2A and FHF2B, are present in DRG neurons, but FHF2A is not detected at nodes of Ranvier (Rush et al., 2006), while FHF2B can be found at nodes of Ranvier along the dorsal but not ventral root axons (Wittmack et al., 2004).
The interaction of FHF proteins with sodium channels produce isoform-dependent changes in current properties. The interaction of FHF2B with Nav1.6 in the DRG-derived neuronal cell line ND7/23 leads to an increase in current density and a rightward shift of steady-state inactivation (Wittmack et al., 2004) while FHF2A increases the current density and causes a depolarizing shift in the voltage-dependence of inactivation, and leads to a pronounced attenuation of recovery from inactivation of Nav1.6 (Rush et al., 2006). Recently, FHF2A has been shown to reduce the Nav1.6 resurgent current, while FHF2B appears to enhance it (Barbosa et al., 2017). FHF4 has been shown to have differential effects on Nav1.1, Nav1.5 and sodium currents of hippocampal neurons which presumably include Nav1.1, Nav1.2 and Nav1.6 (Lou et al., 2005). As many of these changes directly influence the availability of voltage-gated sodium channels, they are likely to have an impact on excitability. Thus far, published data suggests that all tested sodium channels may interact with FHFs and that co-expression causes changes in the biophysical properties of these channels. However, effects on Nav1.7, a threshold channel which is important in the development of pain, have yet to be completely explored. FHF2 has been implicated as an important component in the mechanism of heat nociception in DRG cells via its interaction with Nav1.7, in part by stabilizing Nav1.7 at the membrane (Yang et al., 2017). While that study focuses heavily on the involvement of FHF2B, it does not address the effects that FHF2A might have on the electrophysiological behavior of Nav1.7, or on the specific effect of the FHF interaction on Nav1.7 currents studied in isolation in DRG neurons.
In the present study we investigated the interaction of Nav1.7 with FHF2A and FHF2B and examined the biophysical effects caused by co-expression of these proteins in a HEK293 cell system, and by depleting endogenous levels of FHF2 in dorsal root ganglion neurons. Our voltage-clamp recordings of DRG neurons show that FHF2 regulates Nav1.7 current in a cell-type dependent manner, indicating that caution must be exercised in interpreting data from heterologous expression systems.
2. Methods
2.1. Antibodies
The anti-GFP antibody used in this study was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Monoclonal pan sodium channel antibody was purchased from Sigma (St. Louis, MO). The polyclonal FHF2 antibody was a gift from M. Goldfarb (Hunter College, City University, NY). Peptide-specific polyclonal antibodies were raised against a unique peptide (CDKNKLNVFSRVKLFGS) in the N-terminus of the FHF2A protein and has been previously characterized (Rush et al., 2006). The mouse anti-IgG antibody used as a control for immunoprecipitation was purchased from Vector Laboratories (Burlingame, CA) and the secondary anti-rabbit and anti-mouse antibodies for western blot analysis were purchased from Dako (Carpintera, CA).
2.2. Immunohistochemistry
Adult Sprague Dawley rats were anesthetized with ketamine (80 mg/kg, i.p.) and xylazine (10 mg/kg, i.p.) and dorsal root ganglion neurons were excised, and immersion fixed in 4% paraformaldehyde in 0.14 M Sorensen’s phosphate buffer, pH 7.4, for 20 min at 4 °C. Tissue was rinsed with PBS, cyroprotected with 30% sucrose in PBS, and 8 µm cryosections cut and mounted on SuperFrost Plus glass slides (Fisher, Pittsburgh, PA). Sections were incubated sequentially in: 1) blocking solution (PBS with 5% normal goat serum, 1% BSA and 0.1% Triton X-100) 2) primary antibodies [monoclonal Nav1.2, 1:100; NeuroMab, Davis, CA; polyclonal FHF2A, 1:1000 (Rush et al., 2006)] 3) PBS 4) secondary antibodies goat-anti-mouse IgG-Alexa Fluor 488 and goat anti-rabbit 546 (1:1000, Invitrogen, Carlsbad, CA) and 5) PBS. Images were obtained with a Zeiss 510 META confocal microscope and composed with Photoshop CS (Adobe, San Jose, CA).
2.3. Plasmids
FHF2A, FHF2B and FHF1A were cloned into the mammalian expression vector pEGFP-N1 (Clontech, Palo Alto, CA) as described previously (Wittmack et al., 2004, Rush et al., 2006). The translated products are FHF2A/B-GFP and FHF1A-GFP-fusion proteins, which permit detection of the transfected molecule via either fluorescence of the GFP moiety or via an antibody against GFP, ensuring that every fluorescing cell is expressing the FHF subtype. The expressions FHF2A, FHF2B and FHF1A in the context of transfection in this study always refer to the GFP-fusion proteins or their DNA, unless stated otherwise.
The human Nav1.7 construct was previously described (Herzog et al., 2003). Briefly, the cDNA was cloned into the mammalian expression vector pcDNA3.1 (Klugbauer et al., 1995), and Nav1.7R is a TTX resistant derivative modified by changing tyrosine 362 to serine (Y362S), a change that does not alter other characteristics of the channel (Herzog et al., 2003). Throughout the paper, Nav1.7 refers to this TTX resistant Nav1.7R construct. HEK293 cell line stably expressing Nav1.7 was described previously (Lampert et al., 2006).
For experiments in mouse DRG cells, the sequences of the commercially available siRNAs (against FHF2A or against the core region of FHF2, henceforth referred to as FHF2core) were converted into artificial miRNAs using the protocol described in (Boudreau and Davidson, 2012). Briefly, the sense and antisense sequences of each siRNA were cloned to flank a loop sequence derived from the miR-30 pri-miRNA. This hairpin structure was then cloned into a pFBAAV CMV-eGFP vector under the control of a mouse U6 promoter.
2.4. Transfections
For electrophysiology experiments, HEK293-Nav1.7 cells were cultured on 10 cm dishes or 1 cm coverslips for 24 h before being transfected with GFP, FHF2A or FHF2B at 80% confluency using the LipofectAMINE 2000 (Invitrogen, Carlsbad, CA) as described previously (Lampert et al., 2008). Briefly, 20 μg of pEGFP-N1, pEGFP-FHF2A, or pEGFP-FHF2B were added to 1.5 ml of DMEM without serum and antibiotics. LipofectAMINE 2000 (60 μl) was added to a separate tube containing 1.5 ml of DMEM (without serum and antibiotics). The two solutions were mixed together gently and incubated at room temperature for 20 min before being added to the HEK-Nav1.7 cells. The cells were incubated at 37 °C for 24 h before performing biochemical or electrophysiological experiments.
For experiments to determine the suppression efficiency of the siRNAs, HEK-293 cells were cultured in 10 cm dishes until 80% confluency, and then transferred to 24 well plates for 24 h. One the day of transfection, media in each well was exchanged for 0.5 ml of fresh DMEM with no added serum or antibiotics. The cells were then co-transfected using the Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA) according to the standard protocol. Briefly, for each transfection condition, Lipofectamine RNAiMAX (3 μl) was added to 50 μl of Opti-MEM medium. Separately, 1 μg of pEGFP-FHF2A was added to 50 μl of Opti-MEM medium alone (in the case of positive controls) with a scrambled sequence siRNA (in the case of negative controls) or with siRNA against FHF2A. The two solutions were then added together and incubated at room temperature for 5 min before being added to the HEK293 cells. The siRNA was added such that the final concentrations in the 24 well plate was 3, 10 or 30 μM.
Evaluation of the suppression efficiencies of the miRNA constructs was conducted in an analogous way; 1 μg of either pEGFP-FHF2A or pEGFP-FHF2B was co-transfected with 1 μg of either miRNA-FHF2A, miRNA-FHF2B, miRNA-FHF2core or miRNA-SAFE (scrambled sequence).
2.5. Co-immunoprecipitation of FHF2 and Nav1.7 from HEK293-Nav1.7 cells
Each 10 cm dish of transfected HEK 293 (GFP; FHF2A-GFP + Nav1.7; FHF2B + Nav1.7) cells was suspended in 500 μl of IP buffer and immunoprecipitation of the protein complex by anti-GFP antibody (0.1 μg/ml, Santa Cruz) and western blot assay using anti-pan sodium channel antibody was performed. The proteins were separated on a 4–15% SDS-PAGE gel and transferred to an immuno-blot PVDF membrane overnight at 20 V. The membrane was blocked in 10% dry milk for one hour and incubated in primary anti-pan sodium channel antibody (1 μg/ml) for 2 h. The membrane was then washed for 30 min and incubated with secondary antibody (anti-mouse IgG, as described above) for 1 h at room temperature. The membrane was washed, and the signal was detected using the ECL Plus chemiluminescent system. Western blotting of cell lysates with the anti-GFP and anti-pan sodium channel antibodies was used to show expression of GFP, the GFP fusion proteins FHF2A or FHF2A and Nav1.7 in the transfected cells.
2.6. Quantitative real-time PCR
HEK293 cells were harvested 48 h after transfection; they were washed once with PBS, followed by a 5-minute digestion with 0.05% trypsin at 35 °C. The cells were pelleted by centrifugation for 3 min at 500×g and washed with PBS twice more before proceeding. Total RNA was purified from the cells using the Qiagen RNeasy Plus mini kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions. Then, total RNA (500 ng) was reverse transcribed into complementary DNA (cDNA) using iScript Reverse transcription supermix (Bio-Rad, Hercules, CA). Quantitative real-time PCR was then carried out on a Bio-Rad CFX96 Real-Time PCR system using 10 ng of cDNA as template, 2× TaqMan Gene Expression Master Mix (Applied Biosystems, Foster City, CA) and 20× TaqMan Gene Expression Assays (Applied Biosystems) in a total volume of 10 μl. The quantification cycle (Cq) values were acquired with the BioRad CFX Manager software (Bio-Rad) with the following cycle conditions: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Three total RNA extractions and three technical replicates were analyzed for each sample. Positive controls (no siRNA) and no template controls were included on each plate. Glyceraldehyde 3-phosphate dehydrogenase (hGAPDH) was used as a housekeeping gene.
2.7. Isolation and transfection of primary DRG neurons
Animal studies were approved by US Veterans Affairs West Haven Medical Center Animal Care and Use Committee. DRG neurons were isolated, as previously reported (Dib-Hajj et al., 2009), from 4 to 8-week-old Nav1.6 and 1.8 double knock-out mice (homozygous Nav1.6 floxed, homozygous Nav1.8-cre mice and the Cre-dependent tdtomato reporter) that lack functional endogenous TTX-R NaV1.8 current and the TTX-S Nav1.6 current in neurons that are marked by red fluorescence. Briefly, after harvesting the DRGs from the double knock-out mice, they were incubated at 37 °C for 20 min in complete saline solution (CSS) (which is comprised of 137 mM NaCl, 5.3 mM KCl, 1 mM MgCl2, 25 mM sorbitol, 3 mM CaCl2, and 10 mM HEPES, pH 7.2 adjusted with NaOH) and supplemented with 0.5 U/mL Liberase TM (Roche, Basel, Switzerland) and 0.6 mM EDTA. The tissue was subsequently incubated in CSS with 0.5 U/ml Liberase TL (Roche), 0.6 mM EDTA, and 30 U/ml papain (Worthington Biochemical) for 15 min at 37 °C. The DRGs were then centrifuged and triturated in 0.5 ml of DRG culture medium containing 1.5 mg/ml BSA (low endotoxin) and 1.5 mg/ml trypsin inhibitor (Sigma, St. Louis, MO). After pelleting the DRG neurons (100×g for 3 min) the cells were transfected with constructs containing either miRNA-FHF2A, miRNA-FHF2core or miRNA-Scrambled using a Nucleofector IIS (Lonza, Basel, Switzerland) and Amaxa Basic Neuron SCN Nucleofector Kit (VSPI-1003). The cell pellet was resuspended in 20 μl of Nucleofector solution mixed with 1 μg of miRNA construct and transfected using protocol SCN-BNP 6. After electroporation, 100 μl of Ca-free DMEM was added, followed by a 5 min recovery period at 37 °C. The cell mixture was then diluted in DRG media supplemented with BSA and trypsin inhibitor, seeded onto pol-D-lysine/laminin-coated coverslips (BD) and incubated at 37 °C for 45 min in a 95% air/5% CO2 (v/v) incubator to allow the neurons to attach to the coverslips. After 45 min, the volume in each well was brought up to 1 ml with DRG media and the cells were maintained at 37 °C for 24 h prior to voltage-clamp recording.
2.8. Electrophysiology
Whole-cell voltage-clamp recordings of HEK293, which stably expressed Nav1.7 (Lampert et al., 2006) and cultured DRG neurons, were performed with an EPC-9 amplifier (HEKA electronics, Lambrecht/Pfalz, Germany) using fire polished 0.5–2 MΩ glass electrodes, as previously described (Cummins et al., 2009). Briefly, the pipette solution contained (in mM): 140 CsF, 10 NaCl2, 1 EGTA, and 10 HEPES (pH 7.38, CsOH) and the extracellular bath contained (in mM): 140 NaCl, 3 KCl, 1 MgCl2, 1 CaCl2, 10 Hepes, 10 glucose (pH 7.38, NaOH) and 0.25 µM TTX. All recordings were conducted at room temperature (∼21 °C). Only cells with a robust GFP fluorescence signal (GFP, FHF2A-GFP or FHF2B-GFP) and a clearly detectable current (>0.1 nA) were used for recording. The pipette potential was adjusted to zero before seal formation, and the voltages were not corrected for liquid junction potential. Capacity transients were cancelled, and series resistance was compensated by 75–95%. Leakage current was subtracted digitally online using hyperpolarizing potentials applied after the test pulse (P/4 procedure). Currents were acquired using Pulse software (HEKA electronics, Lambrecht/Pfalz, Germany), filtered at 10 kHz and sampled at a rate between 62.5 kHz for current–voltage families and 20 kHz for recovery from inactivation protocols. For current density measurements, the currents were divided by the cell capacitance, revealed by capacitance compensation.
Voltage protocols were implemented at predetermined times from establishing cell access. Standard current-voltage (I-V) families were obtained using 40 ms pulses from a holding potential of −120 mV to a range of potentials (−100 to +70 mV) with ten seconds between pulses. The peak value at each potential was plotted to form I-V curves. Activation curves were obtained by calculating the conductance G at each voltage V:
with Vrev being the reversal potential, which was determined for each cell individually. Activation curves were fitted with the following Boltzmann distribution equation:
where GNa is the voltage-dependent sodium conductance, GNa,max is the maximal sodium conductance, V1/2 is the potential at which activation is half-maximal, Vm is the membrane potential, and k is the slope. Availability protocols consisted of a series of pre-pulses (−130 to −10 mV) lasting 500 ms from the holding potential of −120 mV, followed by a 40 ms depolarization to −10 mV every 10 s. The normalized curves were fitted using a Boltzmann distribution equation:
where INa, max is the peak sodium current elicited after the most hyperpolarized prepulse, Vm is the preconditioning pulse potential, V1/2 is the half-maximal sodium current, and k is the slope factor.
For recovery from inactivation experiments, two 40 and 10 ms depolarization pulses to −10 mV were given from the holding potential of −120 mV, with a variable recovery time period at −120 mV, in the range of 0.1 ms–3276 ms. Curves were fitted with a single rising exponential function
revealing the time constant τ.
Data are expressed as mean ± SEM, and statistical analyses were performed using ANOVA and Tukey post-hoc analysis (significance at least p < 0.05). Data were acquired in two groups, HEK293-Nav1.7 cotransfected with GFP or FHF2A and separately, with GFP or FHF2B. Both batches of data contained appropriate GFP transfected control data that were recorded under the same conditions as the FHF2A or FHF2B cotransfected cells. For all tested parameters, except for current density, no differences between the two control groups were detected and these data were pooled.
2.9. Electrophysiology in mouse DRGs
Whole-cell voltage-clamp recordings of DRG neurons were performed with an EPC-10 amplifier and Patchmaster software (HEKA electronics, Lambrecht/Pfalz, Germany) at room temperature (20 ± 1 °C) as described previously (Cummins et al., 2009). Briefly, the extracellular bath contained (in mM): 140 NaCl, 3 KCl, 1 MgCl2, 1 CaCl2, 10 HEPES, 5 CsCl, 20 tetraethylammonium chloride, 0.1 CdCl2 and 5 4-aminopyridine (pH 7.30, 330 mOsmol/L). The intracellular pipette solution contained (in mM): 140 CsF, 10 NaCl, 1 EGTA, 10 HEPES and 10 dextrose (pH 7.30, 300 mOsmol/L with sucrose). Patch electrodes were fabricated from boroscillicate glass (1.65/1.1, OD/ID; World Precision Instruments) using a Sutter Instruments P-97 puller and had a resistance of 0.85–1.4 MΩ when filled with pipette solutions. DRG neurons were transfected with scrambled miRNA, or miRNAs that targeted both FHF2A and FHF2B (called FHF Core) or FHF2A alone. The electrophysiologist was blinded to the type of miRNA transfected during data acquisition. Small DRG neurons (<30 µm) with both red and green fluorescence, suggesting double knock-outs of Nav1.6 and Nav1.8 channels (red fluorescence) as well as effective transfection of miRNA (green fluorescence), were held at −100 mV to record the total sodium currents which consist primarily of Nav1.7 and Nav1.9 currents. The average diameter of DRG neurons did not differ among the three experimental groups (Fig. S1). Nav1.9 channels activate at a much more hyperpolarized voltage (∼−80 mV) than Nav1.7 channels (∼−45 mV) and produce ultraslow currents that are discernible. Peak current of Nav1.9 channels normally occur at −45 mV, which typically is the voltage threshold for Nav1.7 channels that normally peak at −10 mV. Therefore, we calculated the ratio of peak sodium currents recorded at −45 mV and −10 mV (I−45/I−10) as an indicator of percentage of Nav1.9 currents in the total sodium currents, and data with a ratio >20% were excluded from analysis. Voltage errors were calculated according to Verr = Ipeak * Rs * (1 − Comp%), where Ipeak is the peak sodium currents, Rs is the series resistance and Comp% is the percentage of compensation. In order to minimize voltage errors 70–95% series resistance compensation was applied. Sodium currents with voltage errors >5 mV were excluded from data analysis. Voltage-clamp recordings were acquired 5 min after establishing whole-cell configuration to allow steady-state dialysis between the cytoplasm and pipette solution. Recordings were acquired at 2.9 kHz with low-pass Bessel filter and digitized at 50 kHz. Linear leak and residual capacitance artifacts were removed using P/6 subtraction. Cells with leak currents at the holding potential of −100 mV either >500 pA or >10% of the peak current were excluded from data analysis. Data were analyzed using FitMaster (HEKA Electronik) and Origin (Microcal Software, Northhampton, MA) software. Data were presented as the mean ± SEM. Statistical significance was determined using one-way ANOVA and Tukey’s post hoc tests. A P value of less than 0.05 was considered significant.
3. Results
3.1. Nav1.7 colocalizes with FHF2
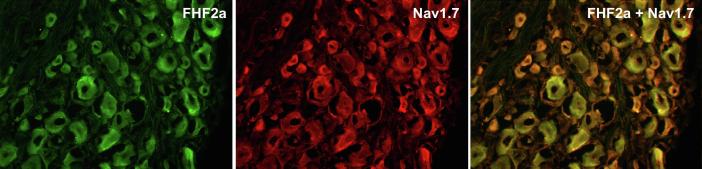
In order to investigate the distribution of endogenous FHF2A, adult rat DRG neurons were harvested and cultured. Anti-FHF2A and anti-Nav1.7 antibodies were used to immunolabel the rat DRG neurons in primary culture. Fig. 1 shows DRG neurons with immunolabelled FHF2A (green, panel A) and with immunolabelled Nav1.7 (red, panel B). In panel C, we show that FHF2A colocalizes with Nav1.7.
Fig. 1.
Adult rat DRG neurons show colocalization of FHF2A with Nav1.7 in small diameter DRG neurons.
3.2. Co-IP shows interaction between FHF2A/B and Nav1.7
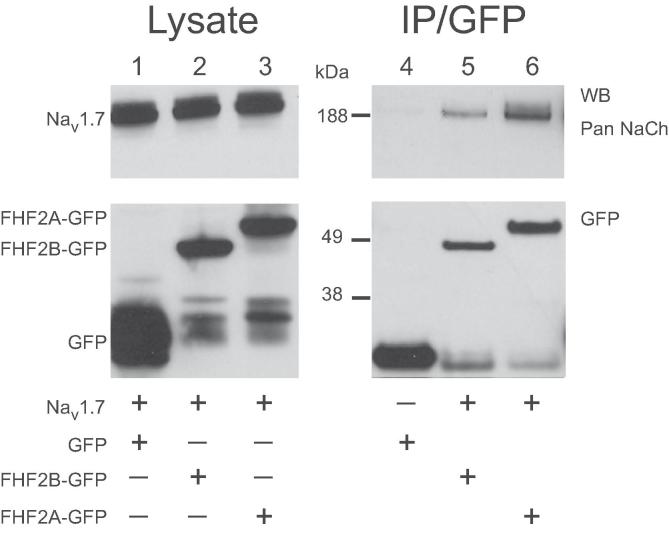
To begin to assess the interaction between FHF2 and Navs, HEK293 cells were co-transfected with Nav1.7 and either GFP, FHF2B-GFP or FHF2A-GFP (Fig. 2, lanes 1, 2 and 3 respectively). Successful and equal expression of Nav1.7 was confirmed in each of the three conditions (Fig. 2, left upper panel). In addition, successful expression of FHF2A-GFP, FHF2B-GFP and GFP was verified (Fig. 2, left lower panel). Each of these proteins migrates at the expected molecular weight when analyzed by SDS-PAGE.
Fig. 2.
FHF2 associates with Nav1.7 in a complex.
To investigate the ability of FHF2 to associate with sodium channels in a complex, FHF2A-GFP and FHF2B-GFP fusion proteins were utilized. HEK293 cells were transfected with either GFP alone (lane 4) or co-transfected with both Nav1.7 and either FHF2B-GFP or FHF2A-GFP (lanes 5 and 6 respectively). Immunoprecipitations were done using anti-GFP antibody, and the resultant precipitates were probed with anti-pan sodium channel antibody (upper right panel) or anti-GFP antibody (lower right panel). Robust immunoreactive signals at expected molecular weights were seen in the anti-GFP blot for all conditions, indicating successful transfection, expression and precipitation of GFP and FHF2A/B-GFP. Probing of the precipitates using anti-pan sodium channel antibody only produced immunoreactive signals when the cells were co-transfected with an isoform of FHF2 (lanes 5 and 6). This data suggests that both FHF2A and FHF2B form a complex with Nav1.7.
3.3. Addition of FHF2A or FHF2B does not change current density of Nav1.7 in HEK293 cells
For current density the values of the two GFP transfected controls were different, and therefore these data are given separately: Batch one: GFP: 315.6 ± 74.6 pA/pF, n = 9; FHF2A: 316.5 ± 43.7 pA/pF, n = 11. Batch two: GFP: 698.0 ± 112.8 pA/pF, n = 7; FHF2A: 523.9 ± 104.4 pA/pF, n = 13. There are no significant differences in current density between GFP and FHF2A/B transfected HEK-Nav1.7 cells in each batch.
3.4. FHF2B causes pronounced shift in activation of Nav1.7 in HEK293 cells
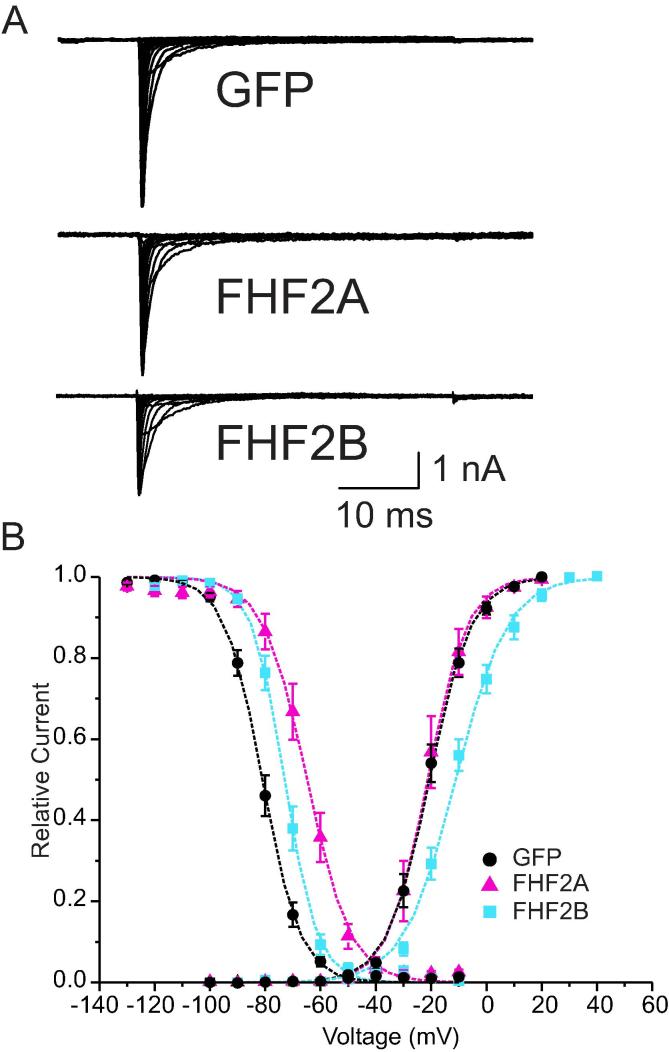
To test for functional implications resulting from the interaction of FHF2 with Nav1.7, we expressed Nav1.7 stably in HEK293 cells and performed whole-cell voltage-clamp recordings. Whole cell patch clamp after transfection of GFP, FHF2A or FHF2B into HEK-Nav1.7 cells showed fast-activating and inactivating currents, with comparable current densities (Fig. 3A). When coexpressed with GFP or FHF2A, the channel activated at potentials close to −50 mV and the current peaked at potentials close to −20 mV. The presence of FHF2B on the other hand shifted the activation voltage-dependence to more positive potentials, causing a shift in the midpoint of activation by 9.7 mV (Fig. 3B, midpoint of activation for GFP −20.6 ± 1.5 mV, n = 16; FHF2A −20.9 ± 2.5 mV, n = 11; FHF2B −10.9 ± 1.7 mV, n = 11, p < 0.005). FHF2B coexpression flattened the activation curve, resulting in a lower voltage-dependency and thus a higher slope factor (slope factors for GFP 6.7 ± 0.4 mV, FHF2B 8.8 ± 0.5 mV). A small steepening of activation curve was observed, when FHF2A was cotransfected (slope 5.2 ± 0.5 mV). FHF2B shifts the voltage-dependence of activation to more positive potentials, which might possibly lead to a decreased excitability of FHF2B expressing neurons.
Fig. 3.
FHF2 alters steady-state kinetics of Nav1.7 in HEK293 cells.
3.5. FHF2 shifts steady-state inactivation of Nav1.7 to more depolarized potentials in HEK293 cells
Nav1.7 expressed in HEK293 cells shows a hyperpolarized voltage-dependence of steady-state fast-inactivation relative to those reported for other sodium channels and for the TTX-sensitive currents in DRG neurons. The midpoint of the Boltzmann fit of steady-state fast-inactivation is −80.9 ± 1.3 mV (n = 15, Fig. 3B) (for Nav1.6: −70.1 ± 0.3 mV (Rush et al., 2006) for TTX-sensitive currents in DRG neurons it was reported to be −69.3 ± 6.6 mV, (Cummins and Waxman, 1997). Both FHF2 subtypes shift the voltage dependence of steady-state fast-inactivation to more depolarized potentials. FHF2A cotransfection leads to a fast-inactivation midpoint of −65.0 ± 2.1 mV (n = 11, p < 0.001), which is a shift of 15.9 mV compared to GFP transfected cells. The 8 mV shift induced by FHF2B cotransfection to −72.9 ± 1.3 mV (n = 11, p < 0.005) is half as much as that of FHF2A. The slope factor of fast-inactivation remained unchanged (GFP 6.0 ± 0.2 mV, FHF2A 6.7 ± 0.6 mV, FHF2B 5.4 ± 0.2 mV).
3.6. FHF2A co-transfection of Nav1.7 leads to enhanced rundown during high frequency stimulation
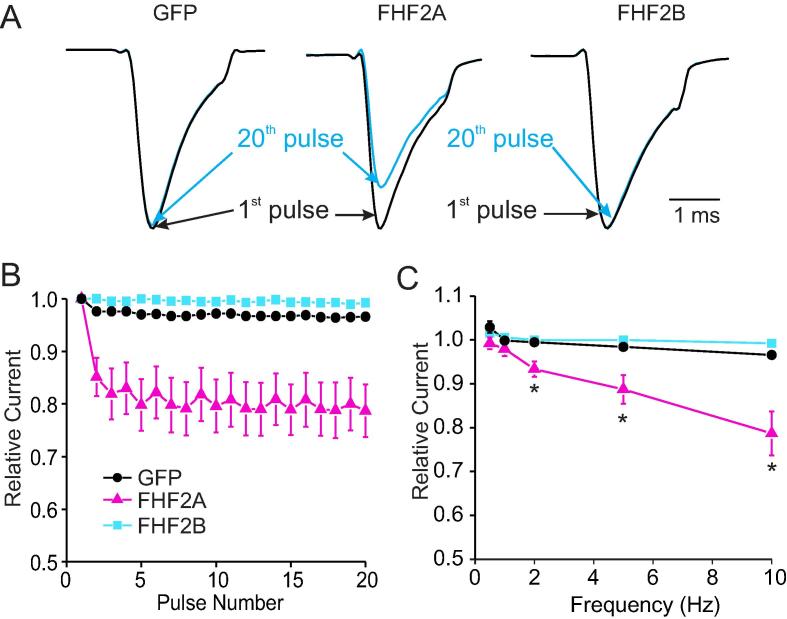
Previous studies had shown that FHF2A coxpression with Nav1.6 leads to a pronounced increase of use-dependent inhibition upon repetitive stimulation. This might have an important impact on firing, bursting or action potential generation after repeated stimulation. To investigate whether Nav1.7, a major contributor to the TTX-S current in small DRG neurons, is influenced by FHF2 coexpression in a similar manner, we applied 20 brief (2 ms) pulses to −10 mV to the cells at various frequencies. At 10 Hz, neither GFP nor FHF2B cotransfected HEK293-Nav1.7 cells showed a reduction of current response at the 20th pulse applied (Fig. 4A and B). By contrast, FHF2A cotransfection clearly reduced the peak current measured at the 20th pulse. This phenomenon was observed at all frequencies tested, reaching 20% inhibition at 10 Hz (Fig. 4C), which could contribute to an increased threshold for firing after repeated stimulation.
Fig. 4.
FHF2A enhances use dependency of Nav1.7.
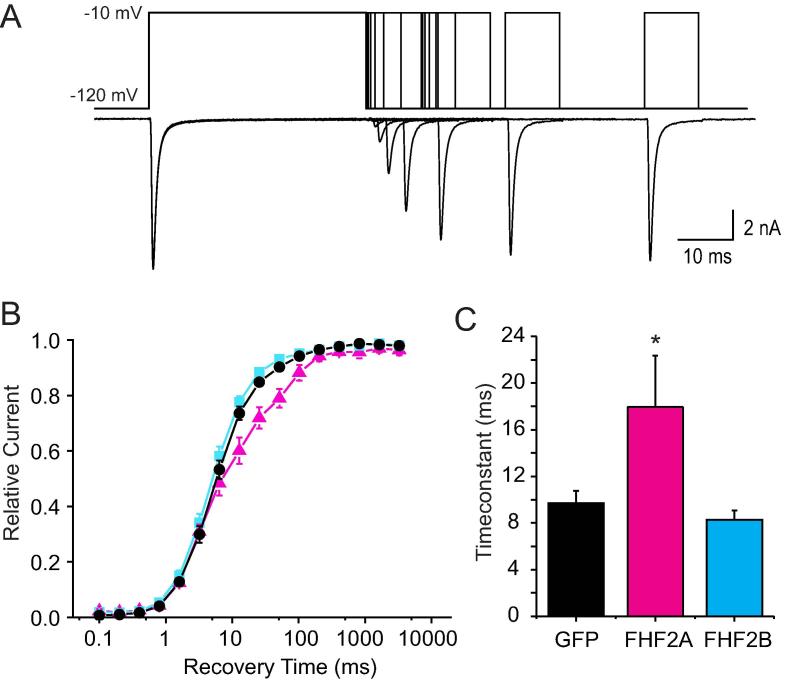
3.7. FHF2A slows recovery from inactivation of Nav1.7
Because inactivated sodium channels are not available for activation upon a depolarizing voltage step, the rate of recovery from inactivation is important for firing properties. Previously, it has been shown that FHF2A markedly slows recovery from inactivation of Nav1.6 (Rush et al., 2006), thus we used similar methods to investigate the influence of FHF2 isoforms on repriming of Nav1.7. With a two-voltage step protocol (Fig. 5A) cells were held at –120 mV and subsequently depolarized to −10 mV for 40 ms to allow complete fast inactivation to occur. After increasing amounts of time at −120 mV (between 0.1 ms and 3276 ms), a second test pulse to −10 mV was applied, and the level of recovery was assessed. FHF2A slowed recovery from fast-inactivation of Nav1.7 (Fig. 5B, red triangles). Multigroup analysis of a single exponential fit of the time courses of Nav1.7 recovery showed an increase of the time constant τ induced by FHF2A co-transfection (GFP: 9.7 ms ± 1.1 ms, n = 10; FHF2A: 18.0 ms ± 4.4 ms, n = 6, p < 0.05) but showed no change induced by FHF2B (8.3 ms ± 0.8 ms, n = 12, p > 0.05) (Fig. 5C).
Fig. 5.
FHF2A slows recovery of Nav1.7 from inactivation.
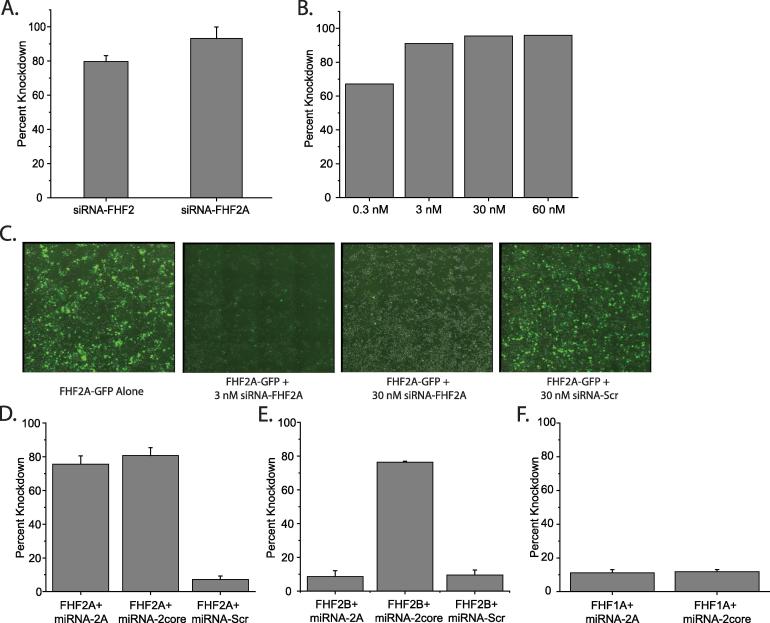
3.8. siRNAs suppress expression of FHF2
Two commercially available siRNAs, one which binds to the common core region of FHF2, and thus suppresses both FHF2A and 2B (miRNA-FHF2core, 79.6% ± 3.4%) and one which suppresses FHF2A only (miRNA-FHF2A, 93% ± 6.6%) were tested in HEK293 cells (Fig. 6A).
Fig. 6.
Knockdown of FHF2A and B using siRNA and miRNA.
To assess what concentration of siRNA was needed to achieve adequate suppression, a concentration series (0.3, 3, 30, 60 nM) was initially tested in HEK293 cells. Concentrations above 3 nM yielded very little further improvement in suppression efficiency (67.1%, 91.1%, 95.5% and 95.9% respectively) (Fig. 6B). This was also apparent on visual inspection of fluorescence levels in transfected HEK293 cells (Fig. 6C).
3.9. miRNAs created from siRNAs suppress expression of FHF2A, FHF2B
Because of the challenges involved in transfecting DRG neurons with single stranded RNAs, the siRNAs were converted to miRNAs against FHF2A and FHF2core and incorporated into a plasmid. The suppression efficiencies of these miRNAs against FHF2 were first tested in HEK293 cells using qPCR. FHF2A expression was suppressed by miRNA-FHF2A and miRNA-FHF2core to high levels, while the scrambled sequence, miRNA-Scr, was relatively low (75.6% ± 4.9%, 80.7% ± 4.6% and 7.2% ± 2.1% respectively) (Fig. 6D).
Likewise, FHF2B was suppressed well by miRNA-FHF2core (76.4% ± 0.6%) and poorly by miRNA-FHF2A and miRNA-Scr (6.2% and 9.5% ± 2.9% respectively) (Fig. 6E).
As a control, since FHF1 is the other FHF expressed in DRG cells, and FHF1A is the longest isoform of that protein, we tested for cross-reactivity of the miRNAs with FHF1A. Their knockdown efficiency was low, and similar to that of the negative control, miRNA-Scr (11.2% ± 1.9%, and 11.9% ± 1.1% for miRNA-FHF2A and miRNA-FHF2core respectively) (Fig. 6F).
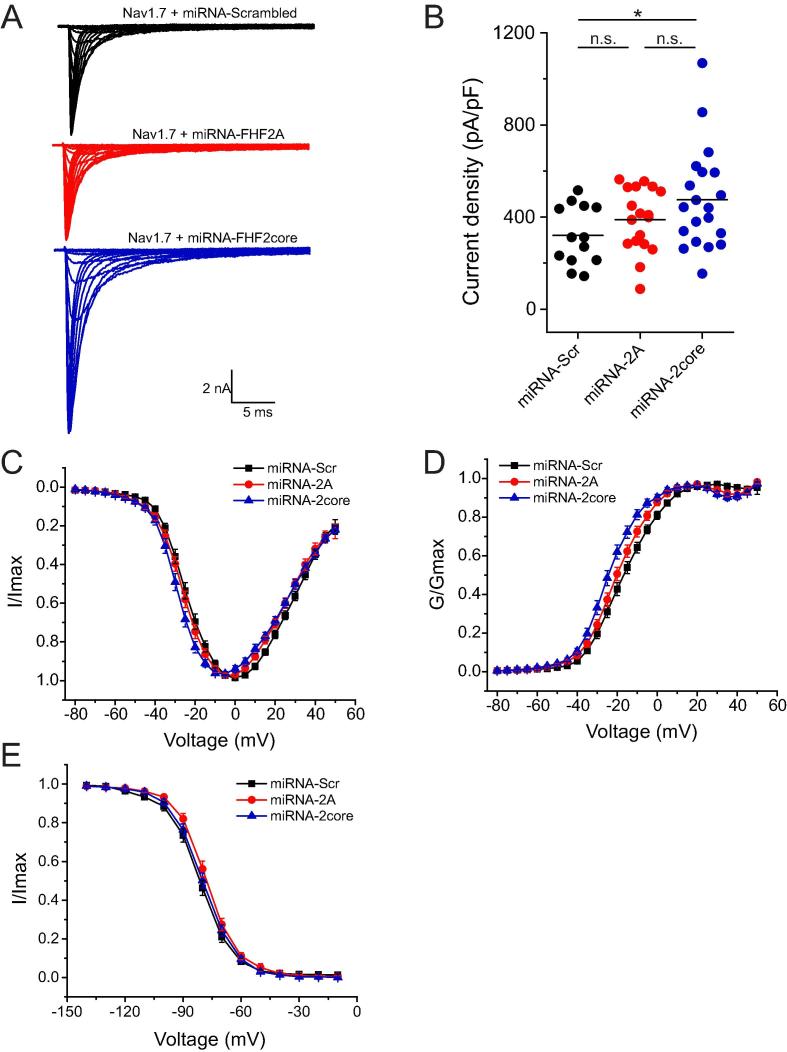
3.10. Knockdown of FHF2 in DRGs modulates voltage-dependent activation, current density and enhances repriming but does not affect fast-inactivation
Endogenous Nav1.7 currents were elicited by voltage steps from −80 mV to +45 mV in 5 mV increments for 100 ms from a holding potential of −100 mV in small DRG neurons lacking endogenous Nav1.6 and Nav1.8 channels. Representative Nav1.7 current traces recorded from cells transfected with miRNA-scrambled, miRNA-FHF2A and miRNA-FHF2core are shown in Fig. 7A. Knock-down of FHF2A did not affect current density but knock-down of FHF2A and FHF2B combined by miRNA-2core resulted in an increased current density of endogenous Nav1.7 channels in small DRG neurons (miRNA-scrambled: −320 ± 36 pA/pF; miRNA-FHF2A: −389 ± 33 pA/pF, p = 0.525; miRNA-FHF2core: −475 ± 49 pA/pF, p = 0.0396) (Table1, Fig. 7B). Voltage-dependence of activation of Nav1.7 channels was significantly hyperpolarized by 7.4 mV after knockdown of both FHF2A and FHF2B using miRNA-2core; however, knocking down FHF2A alone hyperpolarized the midpoint of activation by 3.4 mV, which did not reach statistical significance (miRNA-scrambled: −16.9 ± 1.3 mV; miRNA-FHF2A: −20.3 ± 1.4 mV, p = 0.246; miRNA-FHF2core: −24.3 ± 1.3 mV, p = 0.00182) (Table 1, Fig. 7C and D). Knocking down both FHF2A and FHF2B also steepened the activation curve, as indicated by a significant reduction of the slope factor (miRNA-scrambled: 9.19 ± 0.36 mV; miRNA-FHF2A: 8.10 ± 0.36 mV, p = 0.151; miRNA-FHF2core: 7.16 ± 0.41 mV, p = 0.00219) (Table 1, Fig. 7D). Steady-state fast-inactivation of Nav1.7 currents was measured by a 40 ms test pulse at −10 mV following a series of 500 ms prepulses ranging from −140 mV to −10 mV in 10 mV increments. Voltage-dependence of steady-state fast-inactivation was unchanged by knocking down either FHF2A alone or FHF2A together with FHF2B (miRNA-scrambled: −81.4 ± 1.4 mV; miRNA-FHF2A: −77.7 ± 1.3 mV, p = 0.209; miRNA-FHF2core: −79.7 ± 1.4 mV, p = 0.672) (Table 1, Fig. 7E). Recovery of Nav1.7 channels from fast inactivation was examined using a two-pulse protocol with interpulse intervals varying from 1 to 1025 ms. Recovery rates were measured by normalizing peak current elicited by the test pulse (10 ms depolarization to −10 mV) to that of the prepulse (20 ms at −10 mV) at conditioning voltages of −100 mV, −90 mV, −80 mV, −70 mV and −60 mV, respectively, followed by single-exponential fits of the normalized currents against recovering intervals.
Fig. 7.
Knockdown of both FHF2A and 2B together increases current density and hyperpolarizes voltage-dependent activation of Nav1.7 channels in small DRG neurons.
Table 1.
Biophysical properties of Nav1.7 channels in the presence of miRNA-FHF2A, miRNA-FHF2core and scrambled miRNA.
| Nav1.7 | Current Density (pA/pF) |
Reversal Potential (mV) |
Activation (mV) |
Steady-state fast inactivation (mV) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | n | V1/2,act | k | n | V1/2,fast | k | n | |||
| miRNA-Scrambled | −320 ± 36 | 13 | 57.2 ± 1.4 | 13 | −16.9 ± 1.3 | 9.19 ± 0.36 | 13 | −81.4 ± 1.4 | 7.90 ± 0.42 | 13 |
| miRNA-FHF2A | −389 ± 33 | 18 | 58.0 ± 2.0 | 18 | −20.3 ± 1.4 | 8.10 ± 0.36 | 18 | −77.7 ± 1.3 | 7.65 ± 0.26 | 13 |
| miRNA-FHF2core | −475 ± 49* | 20 | 60.6 ± 1.4 | 20 | −24.3 ± 1.3** | 7.16 ± 0.41** | 20 | −79.7 ± 1.4 | 7.80 ± 0.28 | 19 |
*p < 0.05, **p < 0.01 versus miRNA-Scrambled.
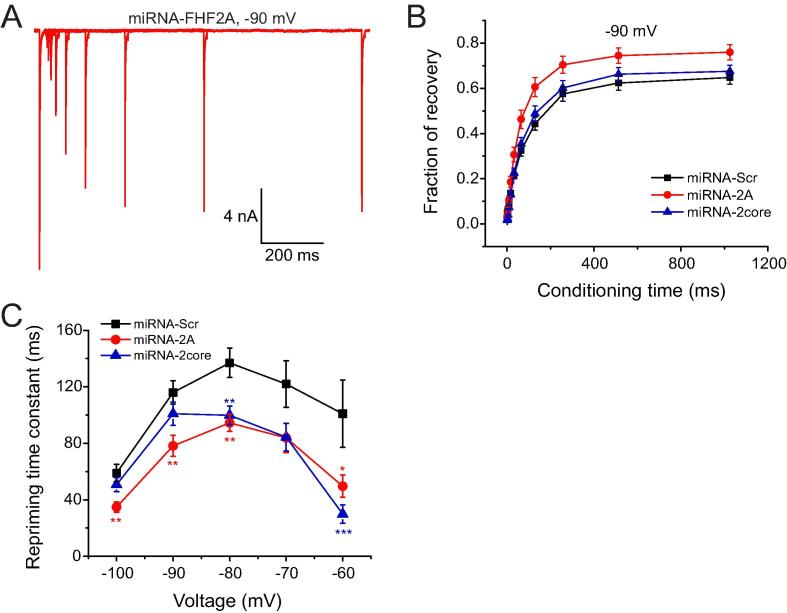
Fig. 8A shows representative traces of repriming of endogenous Nav1.7 currents at a conditioning voltage of −90 mV from a holding potential of −100 mV in a small DRG neuron transfected with miRNA against FHF2A. Fraction of recovery from fast-inactivation at steady state was enhanced by knock-down of FHF2A at −90 mV (Fig. 8B). In addition, both miRNA-FHF2A and miRNA-FHF2core accelerated repriming of Nav1.7 channels significantly (Fig. 8C).
Fig. 8.
Knocking down FHF2 accelerates repriming of Nav1.7 currents in small DRG neurons.
4. Discussion
We show here that Nav1.7, a threshold channel in small DRG neurons which regulates their excitability, interacts with the two splice variants of the intracellular members of the FGF family, FGF13A and B (FHF2A and B), yielding isoform- and cell-type-dependent effects on channel properties. Our immunohistochemical data show colocalization of FHF2A with Nav1.7 in small diameter DRG neurons, most of which are nociceptors, and immunoprecipiation experiments revealed an interaction between FHF2 and Nav1.7 in HEK293 cells, supporting the conclusion that the two proteins may interact in vivo. We assessed the effects of FHF2A and B on Nav1.7 currents both in a heterologous expression system (HEK293 cells) and in native DRG neurons where Nav1.7 is preferentially expressed. Our data shows that some effects are conserved in the two cell backgrounds, while others are not, highlighting the need to study Nav1.7/FHF interaction in a native cell environment.
Using the whole-cell voltage-clamp method, we studied the effects of FHF2A and FHF2B on Nav1.7 in isolation in the heterologous expression system HEK293 cells (overexpression) and in native small DRG neurons. Since Nav1.6 and Nav1.7 produce roughly 30% and 60–70% percent of the TTX-S current in small DRGs, respectively (Shields et al., 2012, Vasylyev et al., 2014, Chen et al., 2018), we studied modulation of endogenous TTX-S currents by FHF2 proteins in DRG neurons from Nav1.6 and 1.8 double knockout animals, which ensured that the TTX-S current measured was primarily that of Nav1.7. The absence of Nav1.8 currents makes the analysis of the residual currents easier due to the feasibility of separating Nav1.9 currents from TTX-S currents because of their distinct gating properties (Cummins et al., 1999, Cummins et al., 2009). This allowed us to address the possibility that FHF might be affected by the cell background in which the sodium current recordings are conducted, and to avoid the confound of supra-physiological levels of targets in a heterologous overexpression system.
The differences in the data we obtained after channel expression in HEK293 cells, as compared to DRG neurons, probably reflect differences in cell background (including interaction with partner molecules and modulatory factors) and underscore the importance of assessing channel properties and functions within native neuronal cell backgrounds when possible (Cummins et al., 2001). Although the reduction of FHF2A levels using an FHF2A-specific miRNA had no effect on Nav1.7 current density, reduction of all FHF2 levels via miRNA-FHF2core led to a statistically significant increase in current density, which suggests that FHF2B might be responsible for suppressing levels of Nav1.7 at the plasma membrane in native neurons, an antiexcitatory effect. This result contrasts with that of overexpressing FHF2 and Nav1.7 in HEK293 cells where no effect was observed. The depletion of FHF2A levels in DRG neurons did not alter the voltage-dependence of activation of Nav1.7, which is in agreement with the observations from the studies in HEK293 cells. When miRNA-FHF2core was used to knockdown all the FHF2 isoforms in DRG neurons we observed a ∼7.4 mV hyperpolarizing shift in the voltage-dependence of activation, which is a proexcitatory effect. This is in agreement with our previous studies of Nav1.7 genetic variants which display similarly hyperpolarizing shifts in activation and increase DRG neuronal excitability (Estacion and Waxman, 2017). Given that the midpoint of activation is depolarized when FHF2B is overexpressed in HEK293 cells, the observed hyperpolarization when FHF2B is knocked down is congruent, and strongly suggests that FHF2B might be a key factor in modulating the activation of Nav1.7 in DRG neurons as well as in HEK293 cells. Neither knockdown of FHF2A alone nor knockdown of all isoforms of FHF2 changed the voltage-dependence of fast-inactivation of Nav1.7, which is in stark disagreement with the effects in HEK293 cells where FHF2A and FHF2B depolarized voltage-dependence of fast-inactivation.
Knockdown of FHF2A yielded a statistically significant increase in the rate of repriming (recovery from fast inactivation) at almost every voltage tested. When all isoforms of FHF2 were knocked down, no further enhancement of repriming was observed. Together, these data confirm that FHF2A is important in controlling the repriming rate and use-dependence inhibition of Nav1.7 in an antiexcitatory manner and suggest that FHF2A might be a dominant factor in this process, with similar effect in DRG neurons and in HEK293 cells. This effect on repriming suggests that the FHF2A-mediated enhanced use-dependent inhibition that we observed in HEK293 cells is probably conserved in DRG neurons.
The differences in Nav1.7 behavior in HEK293 and DRG neurons could be due to factors that modulate the effects of FHF2 on the channel or because of supra-physiological levels of both Nav1.7 and FHF2 proteins in the overexpression system in HEK293 cells. It is possible that in the relatively non-neuronal HEK293 cells the presence of FHF2A and B are sufficient to modulate fast inactivation, whereas in the more complex DRG neuronal environment, lowering the concentration of FHF2 was insufficient to effect a change in fast-inactivation because of the presence of cellular proteins that might compete with FHF2 proteins for binding to Nav1.7 and regulating Nav1.7 fast-inactivation. According to this schema, the observed changes in fast inactivation in HEK293 cells are a consequence of a binding equilibrium favoring FHF2A/B because of the supra-physiological levels generated in overexpression systems, and perhaps the absence of other competing proteins. However, as noted above, the effect of overexpression of FHF2 isoforms on Nav1.7 repriming in HEK293 cells was in agreement with the effect of FHF2 knockdown in DRG neurons, suggesting that FHF2 might be the dominant cellular factor controlling Nav1.7 repriming.
TTX-S current in axotomized DRG neurons reprimes faster than in naïve neurons, but neither voltage-dependence nor current density are affected (Cummins and Waxman, 1997). TTX-S current in adult DRG sensory neurons can be comprised of combinations of Nav1.1, Nav1.6 or Nav1.7, although the contribution of Nav1.1 to the TTX-S current in small DRG neurons is debatable (Black et al., 1996, Vasylyev et al., 2014, Usoskin et al., 2015); all of these channels have been shown to interact with members of the FHF family (Wittmack et al., 2004, Goetz et al., 2009, Wang et al., 2011, Yang et al., 2017). Nav1.3 channels which are characterized by fast repriming (Cummins et al., 2001), re-emerge in DRG neurons after axotomy (Waxman et al., 1994, Dib-Hajj et al., 1996) and could contribute to the faster repriming of the TTX-S currents in these neurons. However, after axotomy it was shown that FHF2 expression levels are down-regulated in DRG neurons (Li et al., 2002). The reduction of FHF2 levels after axotomy did not cause a hyperpolarizing shift in channel activation or an increase in current density as would be predicted from the knockdown experiments, but the reasons are not known. Knockdown of FHF2 in DRG neurons, however, enhanced repriming of Nav1.7 (this study) and Nav1.6 channels (Rush et al., 2006) which suggest that these two channels could also contribute to the faster repriming of the TTX-S current in injured neurons. Thus the down-regulation of FHF2 proteins in axotomized DRG neurons could contribute to the increase in excitability and repetitive firing, as was reported for DRG neurons after peripheral nerve injury (Cummins and Waxman, 1997, Chen et al., 2018).
A recent study from Yang et al utilized a mouse with FHF2 knocked-out in DRG neurons to assess the role of FHF2 in thermally induced pain (Yang et al., 2017). Their conclusion was that FHF2B enables thermal nociception in mice by stabilizing Nav1.7 at the cell membrane. In in vitro experiments in HEK293 cells, they observed an increase in the current density of Nav1.7 in the presence of FHF2B. A similar observation was made for current density of total TTX-sensitive current in DRG neurons taken from their knock-out mouse. This study did not report any other changes in the gating properties of Nav1.7 in HEK293 cells or in the TTX-sensitive current of the DRG neurons. In behavior testing of the knock-out mice, a decrease in the sensitivity to noxious thermal stimuli was observed relative to wildtype. The study did not address the effects of FHF2A. These observations are incongruent with our data. While our study did not investigate the behavioral effects of FHF2 knockdown, the in vitro data can be compared. In our study, irrespective of whether FHF2A was knocked down alone or with FHF2B, gain-of-function changes in Nav1.7 activity were observed. Given this, we would predict heightened sensitivity in vivo to noxious stimuli rather than reduced sensitivity as observed by Yang et al. It is possible that because a knockout mouse was used, compensatory mechanisms to the absence of FHF2 could have been established during the animal’s development. In addition, for the in vitro experiments of DRG neurons in the Yang study, the total TTX-sensitive current was studied, whereas in our study we were able to investigate the Nav1.7 current in relative isolation.
Another difference between our study and that of Yang et al is the type of small diameter DRG neurons that were analyzed. The majority of neurons studied here were 20–30 µm in diameter, whereas in Yang et al, 10–20 µm neurons were used. In both mouse and rat, DRG neurons in the 10–20 µm range are predominantly IB4 positive, while neurons in the size range used in the present study are predominantly IB4 negative (Fang et al., 2006, Stucky and Lewin, 1999). While IB4 staining was not used in either this study or in Yang et al, and IB4 immunostatus might not explain the difference in electrophysiological results observed, it does provide evidence that it is possible that the two studies looked at different subpopulations of small DRG neurons. Interestingly, since IB4 positive neurons are known to be involved in mechanical pain, and IB4 negative in thermal pain, the thermal pain regulation observed in Yang et al is even more surprising.
In summary, this study has shown that the two main variants of FHF2 not only associate with Nav1.7, but also modulate its current properties in a cell environment-dependent manner. Our results demonstrate that knockdown of FHF2A and FHF2B in DRG neurons both lead to gain-of-function changes in Nav1.7 activity, albeit by different mechanisms. These results, combined with other published data, suggest that FHF2 might be involved in a mechanism linking axotomy to neuronal hyperexcitability.
Acknowledgments
Acknowledgements
The authors thank Lynda Tyrrell and Bart Toftness for excellent technical assistance.
Grants
This work was supported in part by Grants from the Rehabilitation Research Service and Medical Research Service, Department of Veterans Affairs, the National Multiple Sclerosis Society and the Erythromelalgia Association. P. E. was supported by T32 training grant no. 5T32GM086287. A.L. was supported by the Robert Bosch Foundation (Fast Track). The Center for Neuroscience and Regeneration Research is a Collaboration of the Paralyzed Veterans of America and the United Spinal Association with Yale University.
Conflict of interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynpai.2019.100029.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Barbosa C., Xiao Y., Johnson A.J., Xie W., Strong J.A., Zhang J.M., Cummins T.R. FHF2 isoforms differentially regulate Nav1.6-mediated resurgent sodium currents in dorsal root ganglion neurons. Pflugers Arch. 2017;469:195–212. doi: 10.1007/s00424-016-1911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J.A., Dib-Hajj S., McNabola K., Jeste S., Rizzo M.A., Kocsis J.D., Waxman S.G. Spinal sensory neurons express multiple sodium channel alpha-subunit mRNAs. Brain Res. Mol. Brain Res. 1996;43:117–131. doi: 10.1016/s0169-328x(96)00163-5. [DOI] [PubMed] [Google Scholar]
- Boudreau R.L., Davidson B.L. Generation of hairpin-based RNAi vectors for biological and therapeutic application. Methods Enzymol. 2012;507:275–296. doi: 10.1016/B978-0-12-386509-0.00014-4. [DOI] [PubMed] [Google Scholar]
- Chen L., Huang J., Zhao P., Persson A.K., Dib-Hajj F.B., Cheng X., Tan A., Waxman S.G., Dib-Hajj S.D. Conditional knockout of NaV1.6 in adult mice ameliorates neuropathic pain. Sci. Rep. 2018;8:3845. doi: 10.1038/s41598-018-22216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins T.R., Waxman S.G. Downregulation of tetrodotoxin-resistant sodium currents and upregulation of a rapidly repriming tetrodotoxin-sensitive sodium current in small spinal sensory neurons after nerve injury. J. Neurosci. 1997;17:3503–3514. doi: 10.1523/JNEUROSCI.17-10-03503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins T.R., Rush A.M., Estacion M., Dib-Hajj S.D., Waxman S.G. Voltage-clamp and current-clamp recordings from mammalian DRG neurons. Nat. Protoc. 2009;4:1103–1112. doi: 10.1038/nprot.2009.91. [DOI] [PubMed] [Google Scholar]
- Cummins T.R., Dib-Hajj S.D., Black J.A., Akopian A.N., Wood J.N., Waxman S.G. A novel persistent tetrodotoxin-resistant sodium current in SNS-null and wild-type small primary sensory neurons. J. Neurosci. 1999;19:RC43. doi: 10.1523/JNEUROSCI.19-24-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins T.R., Aglieco F., Renganathan M., Herzog R.I., Dib-Hajj S.D., Waxman S.G. Nav1.3 sodium channels: rapid repriming and slow closed-state inactivation display quantitative differences after expression in a mammalian cell line and in spinal sensory neurons. J. Neurosci. 2001;21:5952–5961. doi: 10.1523/JNEUROSCI.21-16-05952.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib-Hajj S.D., Waxman S.G. Isoform-specific and pan-channel partners regulate trafficking and plasma membrane stability; and alter sodium channel gating properties. Neurosci. Lett. 2010;486:84–91. doi: 10.1016/j.neulet.2010.08.077. [DOI] [PubMed] [Google Scholar]
- Dib-Hajj S.D., Geha P., Waxman S.G. Sodium channels in pain disorders: pathophysiology and prospects for treatment. Pain. 2017;158(Suppl. 1):S97–S107. doi: 10.1097/j.pain.0000000000000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib-Hajj S.D., Hinson A.W., Black J.A., Waxman S.G. Sodium channel mRNA in the B104 neuroblastoma cell line. FEBS Lett. 1996;384:78–82. doi: 10.1016/0014-5793(96)00273-6. [DOI] [PubMed] [Google Scholar]
- Dib-Hajj S.D., Yang Y., Black J.A., Waxman S.G. The Na(V)1.7 sodium channel: from molecule to man. Nat. Rev. Neurosci. 2013;14:49–62. doi: 10.1038/nrn3404. [DOI] [PubMed] [Google Scholar]
- Dib-Hajj S.D., Choi J.S., Macala L.J., Tyrrell L., Black J.A., Cummins T.R., Waxman S.G. Transfection of rat or mouse neurons by biolistics or electroporation. Nat. Protoc. 2009;4:1118–1126. doi: 10.1038/nprot.2009.90. [DOI] [PubMed] [Google Scholar]
- Estacion M., Waxman S.G. Nonlinear effects of hyperpolarizing shifts in activation of mutant Nav1.7 channels on resting membrane potential. J. Neurophysiol. 2017;117:1702–1712. doi: 10.1152/jn.00898.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Djouhri L., McMullan S., Berry C., Waxman S.G., Okuse K., Lawson S.N. Intense isolectin-B4 binding in rat dorsal root ganglion neurons distinguishes C-fiber nociceptors with broad action potentials and high Nav1.9 expression. J. Neurosci. 2006;26:7281–7292. doi: 10.1523/JNEUROSCI.1072-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz R., Dover K., Laezza F., Shtraizent N., Huang X., Tchetchik D., Eliseenkova A.V., Xu C.F., Neubert T.A., Ornitz D.M., Goldfarb M., Mohammadi M. Crystal structure of a fibroblast growth factor homologous factor (FHF) defines a conserved surface on FHFs for binding and modulation of voltage-gated sodium channels. J. Biol. Chem. 2009;284:17883–17896. doi: 10.1074/jbc.M109.001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog R.I., Cummins T.R., Ghassemi F., Dib-Hajj S.D., Waxman S.G. Distinct repriming and closed-state inactivation kinetics of Nav1.6 and Nav1.7 sodium channels in mouse spinal sensory neurons. J. Physiol. 2003;551:741–750. doi: 10.1113/jphysiol.2003.047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klugbauer N., Lacinova L., Flockerzi V., Hofmann F. Structure and functional expression of a new member of the tetrodotoxin-sensitive voltage-activated sodium channel family from human neuroendocrine cells. EMBO J. 1995;14:1084–1090. doi: 10.1002/j.1460-2075.1995.tb07091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert A., Dib-Hajj S.D., Tyrrell L., Waxman S.G. Size matters: Erythromelalgia mutation S241T in Nav1.7 alters channel gating. J. Biol. Chem. 2006;281:36029–36035. doi: 10.1074/jbc.M607637200. [DOI] [PubMed] [Google Scholar]
- Lampert A., O'Reilly A.O., Dib-Hajj S.D., Tyrrell L., Wallace B.A., Waxman S.G. A pore-blocking hydrophobic motif at the cytoplasmic aperture of the closed-state Nav1.7 channel is disrupted by the erythromelalgia-associated F1449V mutation. J. Biol. Chem. 2008;283:24118–24127. doi: 10.1074/jbc.M802900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.D., Wo Y., Zhong M.F., Zhang F.X., Bao L., Lu Y.J., Huang Y.D., Xiao H.S., Zhang X. Expression of fibroblast growth factors in rat dorsal root ganglion neurons and regulation after peripheral nerve injury. Neuroreport. 2002;13:1903–1907. doi: 10.1097/00001756-200210280-00014. [DOI] [PubMed] [Google Scholar]
- Lou J.Y., Laezza F., Gerber B.R., Xiao M., Yamada K.A., Hartmann H., Craig A.M., Nerbonne J.M., Ornitz D.M. Fibroblast growth factor 14 is an intracellular modulator of voltage-gated sodium channels. J. Physiol. 2005;569:179–193. doi: 10.1113/jphysiol.2005.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Sanjuan I., Smallwood P.M., Nathans J. Isoform diversity among fibroblast growth factor homologous factors is generated by alternative promoter usage and differential splicing. J. Biol. Chem. 2000;275:2589–2597. doi: 10.1074/jbc.275.4.2589. [DOI] [PubMed] [Google Scholar]
- Olsen S.K., Garbi M., Zampieri N., Eliseenkova A.V., Ornitz D.M., Goldfarb M., Mohammadi M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J. Biol. Chem. 2003;278:34226–34236. doi: 10.1074/jbc.M303183200. [DOI] [PubMed] [Google Scholar]
- Pitt G.S., Lee S.Y. Current view on regulation of voltage-gated sodium channels by calcium and auxiliary proteins. Protein Sci. 2016;25:1573–1584. doi: 10.1002/pro.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush A.M., Wittmack E.K., Tyrrell L., Black J.A., Dib-Hajj S.D., Waxman S.G. Differential modulation of sodium channel Na(v)1.6 by two members of the fibroblast growth factor homologous factor 2 subfamily. Eur. J. Neurosci. 2006;23:2551–2562. doi: 10.1111/j.1460-9568.2006.04789.x. [DOI] [PubMed] [Google Scholar]
- Schoorlemmer J., Goldfarb M. Fibroblast growth factor homologous factors and the islet brain-2 scaffold protein regulate activation of a stress-activated protein kinase. J. Biol. Chem. 2002;277:49111–49119. doi: 10.1074/jbc.M205520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields S.D., Cheng X., Uceyler N., Sommer C., Dib-Hajj S.D., Waxman S.G. Sodium channel Na(v)1.7 is essential for lowering heat pain threshold after burn injury. J. Neurosci. 2012;32:10819–10832. doi: 10.1523/JNEUROSCI.0304-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucky C.L., Lewin G.R. Isolectin B(4)-positive and -negative nociceptors are functionally distinct. J. Neurosci. 1999;19:6497–6505. doi: 10.1523/JNEUROSCI.19-15-06497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D., Furlan A., Islam S., Abdo H., Lonnerberg P., Lou D., Hjerling-Leffler J., Haeggstrom J., Kharchenko O., Kharchenko P.V., Linnarsson S., Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- Vasylyev D.V., Han C., Zhao P., Dib-Hajj S., Waxman S.G. Dynamic-clamp analysis of wild-type human Nav1.7 and erythromelalgia mutant channel L858H. J. Neurophysiol. 2014;111:1429–1443. doi: 10.1152/jn.00763.2013. [DOI] [PubMed] [Google Scholar]
- Wang C., Wang C., Hoch E.G., Pitt G.S. Identification of novel interaction sites that determine specificity between fibroblast growth factor homologous factors and voltage-gated sodium channels. J. Biol. Chem. 2011;286:24253–24263. doi: 10.1074/jbc.M111.245803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman S.G., Kocsis J.D., Black J.A. Type III sodium channel mRNA is expressed in embryonic but not adult spinal sensory neurons, and is reexpressed following axotomy. J. Neurophysiol. 1994;72:466–470. doi: 10.1152/jn.1994.72.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmack E.K., Rush A.M., Craner M.J., Goldfarb M., Waxman S.G., Dib-Hajj S.D. Fibroblast growth factor homologous factor 2B: association with Nav1.6 and selective colocalization at nodes of Ranvier of dorsal root axons. J. Neurosci. 2004;24:6765–6775. doi: 10.1523/JNEUROSCI.1628-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Dong F., Yang Q., Yang P.F., Wu R., Wu Q.F., Wu D., Li C.L., Zhong Y.Q., Lu Y.J., Cheng X., Xu F.Q., Chen L., Bao L., Zhang X. FGF13 Selectively Regulates Heat Nociception by Interacting with Nav1.7. Neuron. 2017;93:806–821. doi: 10.1016/j.neuron.2017.01.009. e809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.