Abstract
The androgen receptor (AR) is a ligand-regulated transcription factor that stimulates cell growth and differentiation in androgen-responsive tissues. The AR N terminus contains two activation functions (AF-1a and AF-1b) that are necessary for maximal transcriptional enhancement by the receptor; however, the mechanisms and components regulating AR transcriptional activation are not fully understood. We sought to identify novel factors that interact with the AR N terminus from an androgen-stimulated human prostate cancer cell library using a yeast two-hybrid approach designed to identify proteins that interact with transcriptional activation domains. A 157-amino acid protein termed ART-27 was cloned and shown to interact predominantly with the AR153–336, containing AF-1a and a part of AF-1b, localize to the nucleus and increase the transcriptional activity of AR when overexpressed in cultured mammalian cells. ART-27 also enhanced the transcriptional activation by AR153–336 fused to the LexA DNA-binding domain but not other AR N-terminal subdomains, suggesting that ART-27 exerts its effect via an interaction with a defined region of the AR N terminus. ART-27 interacts with AR in nuclear extracts from LNCaP cells in a ligand-independent manner. Interestingly, velocity gradient sedimentation of HeLa nuclear extracts suggests that native ART-27 is part of a multiprotein complex. ART-27 is expressed in a variety of human tissues, including sites of androgen action such as prostate and skeletal muscle, and is conserved throughout evolution. Thus, ART-27 is a novel cofactor that interacts with the AR N terminus and plays a role in facilitating receptor-induced transcriptional activation.
INTRODUCTION
The androgen receptor (AR) is a member of the steroid receptor (SR) family of transcriptional regulatory proteins that transduces the signaling information conveyed by androgens (Wilson et al., 1991; Chang et al., 1995). On androgen binding, the AR is released from the repressive effects of an Hsp90-based regulatory complex, allowing the receptor to either activate or inhibit transcription of target genes (Picard et al., 1990; Jenster et al., 1991, 1992; Duina et al., 1996; Fang et al., 1996, 1998; Segnitz and Gehring, 1997). In addition to its role in male sex determination, AR mediates normal prostate development as well as malignant growth by regulating genes and signaling pathways involved in cellular proliferation (Brinkmann et al., 1992; Hakimi et al., 1996; Trapman and Brinkmann, 1996; Dorkin and Neal, 1997; Dorkin et al., 1997; Jenster, 1999).
The mechanisms underlying the specificity of AR regula-tion of gene expression remain enigmatic. The DNA-binding domain of AR is highly conserved among SRs and recognizes the same hormone response element (HRE) as does the glucocorticoid receptor (GR). Although subtle preferences for particular HREs may contribute to the specificity of AR-mediated transcriptional response (Nelson et al., 1999; Schoenmakers et al., 2000), recent evidence suggests that the AR cell- and promoter-specific regulation is generated through interactions with regulatory proteins termed coactivators and corepressors (Cleutjens et al., 1997; Scheller et al., 1998). For instance, agonist binding to the AR C-terminal activation function-2 (AF-2) promotes a conformational change and the formation of a surface for protein-protein contacts between AF-2 and additional transcriptional regulatory factors, which, in turn, modulate the transcriptional activity of target genes (Onate et al., 1995; Smith et al., 1996; Voegel et al., 1996; Yeh and Chang, 1996; Chen et al., 1997; Hong et al., 1997; Li et al., 1997; Torchia et al., 1997; Kang et al., 1999). Because the growing number of SR coactivators and corepressors appear to function widely across the SR family with conserved AF-2 regions (Glass and Rosenfeld, 2000), it is unlikely that these factors alone determine specificity of receptor transcriptional regulation. In contrast, the N-terminal transcriptional regulatory regions, which are diverse throughout the SR family, may represent an important determinant of SR specificity, conceivably through the recruitment of distinct coregulators. Indeed, Hittelman et al. (1999) recently identified DRIP150 as a GR N-terminal coactivator that does not interact with the N termini of other SRs, including AR. However, the mechanisms of transcriptional activation by the AR N terminus are not well understood, and although the list of proteins proposed to bind to the AR N terminus is expanding (Gelman et al., 1999; Hsiao and Chang, 1999; Ma et al., 1999; Lee et al., 2000), it is likely that additional AR-binding partners remain to be identified.
Regions of the AR N terminus important for transcriptional activation have been identified by expressing and analyzing receptor deletion derivatives or fusion proteins in mammalian cells and in cell-free systems. At least two distinct activation domains within the AR N terminus have been identified, AF-1a (residues 154–167) and AF-1b (residues 295–459), both of which are required for full transcriptional activation mediated by the receptor (Chamberlain et al., 1996). The AR N-terminal residues 142–485 have also been shown to activate a minimal promoter construct in a cell-free transcription system and to selectively interact with the transcription factors TFIIF and the TATA-binding protein (TBP), suggesting a direct contact with the general transcription factors (McEwan and Gustafsson, 1997). Protein-protein interaction studies have recently suggested contacts between the AR N terminus and the TATA-element modulating factor, or ARA160, which increases AR transcriptional activity when overexpressed in certain cell types (Hsiao and Chang, 1999).
Interestingly, the growth of both normal and tumoregenic prostate cells is regulated by AR. A number of prostate cell lines display elevated AR-dependent transcriptional activation relative to nonprostatic cell lines; the AR N terminus appears responsible for this enhanced receptor activity (Gordon et al., 1995), suggesting the existence of cofactors that modulate transcriptional activation by the AR N-terminal activation domain in prostate epithelial cells. Together, the current data support the notion that the AR N terminus contains multiple surfaces capable of interaction with general transcription factors and possibly additional adapter proteins. Recently, a patient with androgen insensitivity syndrome was described whose cells lack AR transcriptional activity, probably through the loss of an as yet unidentified AR N-terminal cofactor (Adachi et al., 2000), underscoring the importance of the AR N terminus and associated factors in human disease.
In a yeast two-hybrid screen designed to identify factors that interact with transcriptional activation domains, we isolated from an androgen-stimulated prostate cancer cell library a novel factor that associates with the AR N terminus. We examined the specificity and molecular determinants of this interaction and characterized the effects on SR-dependent transcriptional activation in mammalian cells.
MATERIALS AND METHODS
Construction of Plasmids
Yeast expression vectors for the LexA-AR fusion protein, LexA-AR18–500, was created by digesting the rat AR N terminus with EcoRI-XhoI and subcloned into the pEG202 vector that was digested with EcoRI-XhoI. The subregions of the rat AR N terminus (LexA-AR18–156, LexA-AR153–336, and LexA-AR336–500) were generated as follows: for LexA-AR18–156, pEG202:AR18–500 was digested with EcoRI-PvuII and the insert was ligated into pEG202, which was digested with NotI, the 5′ overhang filled in with Klenow fragment, and EcoRI; for LexA-AR153–336, pEG202:AR18–500 was digested with BstYI-AflII, filled in with Klenow, and ligated into pEG202, which was digested with BamHI-XhoI with ends filled in; for LexA-AR336–500, pEG202:AR18–500 was digested with BstYI-XhoI and the insert was ligated into pEG202, which was digested with BamHI-XhoI. For mammalian expression, the LexA DNA-binding domain AR N-terminal fusions were excised with HindIII-XhoI, and the insert was ligated into pcDNA3. Yeast two-hybrid “bait” proteins, B42-AR18–156, B42-AR153–336, B42-AR336–500, and B42-AR18–500 were constructed by subcloning respective EcoRI-XhoI fragments from pEG202 into the corresponding sites in pJG4-5. The LexA-LNCaP cell cDNA library was purchased from Origene Technologies (Rockville, MD). The AR579–901 was PCR amplified using the following primers: (forward with a BglII site) 5′-AGATCTTAAGCAGAAATGATTGCACCATTG-3′ and (reverse with an XhoI site) 5′-GTAGATAAAGGTGTGTGTCACTGAGCTC-3′, ligated into pGEM:T-easy (Promega, Madison, WI), excised with BglII-XhoI, and the insert was ligated into pEG202, which was digested with BamHI-XhoI. pEG202:AR579–901 was then digested with EcoRI-XhoI and the insert was ligated into pJG4-5.
The LexA–ART-27 (androgen receptor trapped clone 27) C-terminal truncations 1–45, 1–67, and 1–127 were constructed by digesting pEG202:ART-27 with PvuII, BspMI, and StyI, respectively, filling in their 5′-overhangs, digesting with MluI, and ligating the inserts into pEG202, which was digested with NotI/Klenow-MluI. The LexA–ART-27 N-terminal truncations 46–157, 68–157, and 127–157 were constructed as follows: for LexA–ART-2746–157, pEG202:ART-27 was digested with PvuII-XhoI and ligated into pEG202, which was digested with BamHI/Klenow and XhoI; for LexA–ART-2768–157, pEG202:ART-27 was digested with BspMI/Klenow and XhoI, and the insert was ligated into pEG202, which was digested with BamHI/Klenow and XhoI; for LexA–ART-27127–157, pEG202:ART-27 was digested with StyI/Klenow and XbaI, and the insert was ligated into pEG202, which was digested with EcoRI/Klenow and XbaI. For LexA–ART-271–45/127–157, PCR primers were designed as follows: ART-271–45 (forward pEG202 primer) 5′-TTGGGGTTATTCGCAACGG-3′, (reverse with BamHI site) 5′-GAACTGGATCCCTGCTCATATACCTTGTCTCGATG-3′; ART-27127–157 (forward with BamHI site) 5′-GAACTGGATCCACCAAGGACTCCATG-3′, (reverse pEG202 primer) 5′-CGGAATTAGCTTGGCTGC-3′. The two separate fragments were PCR amplified, and the resulting products were digested as follows: ART-271–45 with EcoRI-BamHI, ART-27127–157 with BamHI-XhoI, and the two inserts were ligated together into pEG202, which was digested with EcoRI-XhoI. For mammalian expression, the pEG202:ART-27 was digested with EcoRI-XhoI and subcloned into a pcDNA3 vector that has an N-terminal hemagglutinin (HA) epitope (pcDNA3-HA).
pJG4–5:Sp1A83–262, pJG4–5:Sp1B263–542, pJG4–5:TAF130270–700, and pJG4–5:CREB3–296 were provided by N. Tanese (New York University School of Medicine, New York). pJG4–5:SRC-1374–800 was provided by H. Samuels (New York University School of Medicine). pJG4–5:GR107–237, pJG4–5:GR107–237 30IIB, and pJG4–5:VP16 were previously described (Hittelman et al., 1999). The pJK103 reporter plasmid, which contains a single LexA operator linked to β-galactosidase, was used in activity assays of the LexA fusion proteins and in the modified two-hybrid assay. The pΔ4X-LALO-luciferase reporter plasmid, which contains four LexA operators upstream of a minimal Drosophila alcohol dehydrogenase promoter linked to luciferase, was used in mammalian activity assays to monitor the intrinsic transcriptional activity of the LexA fusion proteins. The pcDNA3:hAR expression plasmid was used to produce full-length human AR, pMMTV:luciferase reporter was used to assay AR transcriptional activity, and pCMV:LacZ constitutively expressed β-galactosidase, a marker for transfection efficiency. Other receptor expression plasmids include pcDNA3-human estrogen receptor (ER)-α, pCMV5-human ERβ (Su et al., 2001), and pRep4-human thyroid hormone receptor β-1 (TRβ-1). The ER (XETL) and TR (pGL3-DR4) reporter plasmids have been previously described (Sharif and Privalsky, 1992; Su et al., 2001).
Modified Yeast Two-Hybrid System
To identify proteins that interact with AR N terminus, we used a modification of the yeast two-hybrid system that allows for the selection of proteins that interact with transcriptional activators (Hittelman et al., 1999). Linking amino acids 18–500 of the rat AR to the B42 activation domain created the AR “bait” (pJG4–5:AR18–500). The “prey” is created by fusing an androgen-stimulated LNCaP cell cDNA library to the LexA DNA-binding domain (pEG202:LNCaP cell cDNA library) rather than to an activation domain. The yeast strain EGY188 was transformed by the lithium acetate method with 1) pJG4–5:AR18–500, 2) pEG202:LNCaP cell cDNA library, and 3) pJK103, a β-galactosidase reporter gene with a single LexA operator. Potential interacting proteins were selected by plating the cDNA library expressing transformants onto leu−/X-gal+ galactose plates. Library proteins that interact with AR stimulate expression of Lex-responsive Leu2 and β-galactosidase reporter genes. To eliminate library plasmids that may possess intrinsic activation potential, Leu2+/LacZ+ colonies were replica plated onto glucose plates, conditions in which the AR protein is not expressed. Colonies that activated β-galactosidase expression on galactose plates, when AR is expressed, but not glucose plates, were deemed true interactors and further analyzed.
Quantitative Liquid β-Galactosidase Assay
Yeasts were grown in selective liquid media containing 2% glucose for ∼12 h, pelleted, washed once with sterile H2O, normalized to cell number, and resuspended to an optical density (OD600) of 0.15 in 2% galactose/1% raffinose. β-Galactosidase assays were performed 12 h later as described previously (Garabedian and Yamamoto, 1992).
Mammalian Cell Culture and Transient Transfection Assays
A human cervical carcinoma cell line (HeLa), a human prostate cancer cell line (PC-3), and SV40 T-antigen expressing monkey kidney cells (COS-1) were obtained from the American Type Culture Collection and maintained in DMEM (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT), 50 U/ml each of penicillin and streptomycin (Invitrogen), and 2 mM l-glutamine (Invitrogen). The androgen-dependent prostate cancer cell line (LNCaP) was maintained in RPMI-1640 (Invitrogen) supplemented with 10% FBS, 50 U/ml each penicillin and streptomycin, and 2 mM l-glutamine. For transfections, HeLa cells were seeded in 35-mm dishes at a density of 1.3 × 105, washed once with serum-free medium, and transfected with 0.2 μg of pcDNA3:human AR, 0.1 μg of pMMTV-Luc, 0.05 μg of pCMV-LacZ, and the indicated amounts of pcDNA3:HA–ART-27 using 5 μl of Lipofectamine reagent (Invitrogen) in a total volume of 1 ml of serum-free, phenol red-free DMEM per 35-mm dish according to the manufacturer's instructions. Approximately 4 h posttransfection, the transfection mixture was removed, and the cells were refed with 2 ml of DMEM-10% FBS, allowed to recover for 3–5 h, and fed again with fresh DMEM-10% FBS supplemented with 100 nM R1881, dexamethasone, 17β-estradiol, triac, or an identical volume of 100% ethanol and incubated for 12 h. Transfected cells were washed once in phosphate-buffered saline (PBS) and harvested in 1X reporter lysis buffer (Promega) according to the manufacturer's instructions. PC-3 cells were seeded in 35-mm dishes at a density of 1.1 × 105 and transfected as above. To assay the LexA-AR N terminus, 0.5 μg of pcDNA3-LexA:AR derivatives, 1.0 μg of pcDNA3-HA:ART-27 or empty vector, 1.0 μg of pΔ4X-LALO reporter, and 0.25 μg of pCMV-LacZ were transfected into HeLa cells that were transfected as above using 6 μl of Lipofectamine per 35-mm dish. Luciferase activity was quantitated in a reaction mixture containing 25 mM glycylglycine, pH 7.8, 15 mM MgSO4, 1 mM ATP, 0.1 mg/ml bovine serum albumin (BSA), 1 mM dithiothreitol using a Lumen LB 9507 luminometer (EG&G Wallac, Gaithersburg, MD), and 1 mM d-luciferin (PharMingen, San Diego, CA) as substrate.
Preparation of the ART-27 Antibody
A 16-amino acid peptide that corresponds to the ART-27 C-terminal amino acids 142–157 (R-E-L-Q-G-L-Q-N-P-G-K-P-H-H) with an additional cysteine residue at the N terminus was synthesized by Anaspec (San Jose, CA), coupled to KLH, and used to immunize rabbits by Covance Research Products (Denver, PA). The IgG fraction was purified from the serum by protein A chromatography, and the ART-27 antibody was obtained by affinity purification using the ART-27 peptide immunogen coupled to Affi-gel 15 (Bio-Rad Laboratories, Hercules, CA) resin.
Northern Blotting
Cells were cultured in 100-mm dishes for the indicated periods of time with appropriate treatments (see figure legends), the media were aspirated, and the cells were lysed by adding 3 ml/dish of RNA STAT-60 reagent (Tel-Test, Friendswood, TX). Total RNA was isolated from cell homogenates according to the manufacturer's instructions, denatured at 65°C for 15 min, chilled on ice, and separated on a 1.2% agarose/6% formaldehyde denaturing gel (10 μg RNA/lane). Equivalent loading was verified by ethidium bromide staining of rRNA. RNA was transferred to Duralon paper (Stratagene, San Diego, CA), UV cross-linked to the membrane, and hybridized to a cDNA probe using QuikHyb hybridization mixture (Stratagene) as described by the manufacturer. A 0.9-kb cDNA fragment encoding ART-27 was labeled with [α-32P]dCTP (NEN, Boston, MA) using RediPrime random priming labeling kit (Amersham Pharmacia Biotech, Piscataway, NJ) using the manufacturer's instructions. Blots were washed and exposed to BioMax film (Kodak, Rochester, NY) at −80°C for autoradiography. Hybridization of ART-27 to a multiple tissue Northern blot membrane (CLONTECH, Palo Alto, CA) was performed according to the manufacturer's instructions.
Coimmunoprecipitation
Full-length AR and HA–ART-27 were translated in vitro using TNT Quick Coupled Transcription/Translation System (Promega) in the presence of [35S]methionine. The radiolabeled proteins were incubated as indicated in binding buffer (20 mM Tris, pH 7.9, 170 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.05% Nonidet P-40, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 4 mg/ml BSA) for 1 h at 4°C. α-HA (12CA5, 1 μg) antibody (Boehringer Mannheim, Indianapolis, IN) was incubated with the radiolabeled proteins for 1 h at 4°C. Protein A Sepharose Fast Flow beads (30 μl, Amersham Pharmacia Biotech) were incubated with the respective reaction mixtures for 1 h at 4°C. The beads were washed three times in lysis buffer, resuspended in 2× SDS sample buffer, and boiled for 3 min; the associated proteins were resolved by SDS-PAGE and visualized by autoradiography.
LNCaP-stable cell lines were generated that express ART-27 with a C-terminal FLAG epitope in an inducible manner (LNCaP-Tet-on-ART-27-FLAG cells). This line was created in two steps. First, LNCaP cells were transfected with the pTet-On vector using DOTAP (Roche, Summerville, NJ), and resistant colonies were selected at 500 μg/ml Geneticin (Invitrogen). Clones were transferred to a 24-well dish coated with fibronectin (10 μg/ml; Invitrogen), expanded, and screened for Tet-dependent activation by measuring pRevTRE-luciferase reporter gene activity in the absence and presence of 1 μg/ml doxycyclin (Sigma). A LNCaP clone displaying tight Tet-dependent regulation was transfected with pRevTRE:ART-27 (C-FLAG) as above, and resistant colonies were selected at 150 μg/ml hygromycin B (Invitrogen). Multiple clones were screened for Tet-dependent activation by immunoblotting for the FLAG epitope resident on ART-27 in the absence and presence of 1 μg/ml doxycyclin. Small-scale nuclear extracts were prepared as described by Lee et al. (1988) from two confluent 100-mm dishes of LNCaP-Tet-on-ART-27-FLAG cells that had been induced overnight with 1 μg/ml doxycyclin and either 10 nM R1881 or an equivalent volume of ethanol vehicle. The total protein concentration in the nuclear extracts was equalized with lysis buffer, and 10 μl of either preimmune or immune ART-27 antisera were added. After incubation at 4°C for 1.5 h, 40 μl of Protein A Sepharose Fast Flow beads (Amersham Pharmacia Biotech) were added and incubated for 1 h at 4°C. The immune complexes were washed three times in wash buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM NaF, 10% glycerol, 1% Triton X-100), resuspended in 2× SDS sample buffer, and boiled for 3 min; the associated proteins were resolved by SDS-PAGE, transferred to Immobilon paper (Millipore, Bedford, MA), and probed with either an AR polyclonal antibody (sc-816; Santa Cruz Biochemicals, Santa Cruz, CA) or an affinity-purified ART-27 antibody.
Immunoblotting
Yeast protein extracts were prepared from 2-ml cultures and lysed using glass beads as previously described (Knoblauch and Garabedian, 1999). Lysates from mammalian cells were prepared as described by Hittelman et al. (1999). For immunoblotting, HeLa cell nuclear extracts, which were untreated, phorbol 12-myrisate 13 acetate (TPA; 50 ng/ml) for 2 h before harvesting, or serum stimulated (cells cultured for 24 h in medium containing 0.5% serum and serum stimulated [20%] for 2 h before harvesting), and PC-3 cell nuclear extracts were purchased from Geneka Biotechnology (Montreal, Quebec, Canada). Large-scale HeLa cell nuclear extracts for the velocity gradient sedimentation analysis were prepared from the nuclei of 5 liters of HeLa cells (Cellex Biosciences, Minneapolis, MN) by the method of Dignam and Roeder (Dignam et al., 1983). Protein concentration in extracts was normalized by the Bradford assay (Bio-Rad), separated on SDS/4–20% polyacrylamide gels (Novex, San Diego, CA), and transferred to Immobilon paper. Membranes were probed with a polyclonal antibody against LexA (a gift from E. Golemis), a monoclonal antibody to HA (12CA5; Boehringer Mannheim), a monoclonal antibody to TBP (a gift from N. Tanese) or affinity-purified rabbit anti–ART-27 antibody. The blots were developed using horseradish peroxidase-coupled donkey anti-rabbit or sheep anti-mouse antibodies and enhanced chemiluminescence (ECL; Amersham-Pharmacia). Quantitative analysis of immunoblots was performed using the NIH image software package (version 1.62; National Institutes of Health, Bethesda, MD).
Immunofluorescence
HeLa cells were seeded onto poly-d-lysine–coated coverslips, transfected with pcDNA3-FLAG–ART-27, and, 24 h later, washed five times with PBS and fixed in 4% paraformaldehyde in PBS for 20 min at room temperature (RT). Cells were then permeabilized with 0.2% Triton X-100 in PBS for 20 min and incubated in 100 μl of the FLAG monoclonal antibody (M2) diluted to a concentration of 2 μg/ml in blocking solution (5% BSA/Tris-buffered saline [TBS] for 2 h at RT. Cells were washed five times in 1 ml of 0.1% Triton X-100 in PBS, followed by incubation with goat anti-mouse fluorescein-conjugated secondary antibody (Vector Labs, Burlingame, CA) diluted in blocking solution, for 1 h at RT. Secondary antibody was removed by washing the cells five times in 0.1% Triton X-100 in PBS and three times in PBS. Nuclei were stained with 1 μg/ml Hoechst dye H334211 for 10 min, followed by one wash with PBS. Coverslips were mounted onto Citifluor (Ted Pella, Redding, CA), and the fluorescein and Hoechst signals were visualized and photographed using an Axioplan 2 microscope (Zeiss, Thornwood, NY).
Immunohistochemistry
An indirect immunoperoxidase method was used to identify ART-27 in LNCaP and PC-3 tumors grown in nude mice (xenografts). The LNCaP and PC-3 tumor xenografts were removed from the mice and fixed for 2 h in 4% paraformaldehyde in PBS (pH 7.4) at RT. The tissue was dehydrated through ethanol, cleared in chloroform, and embedded in paraffin. Tissue sections (5-μm) were serially cut on a microtome and mounted on slides. Sections were dewaxed in xylene, rehydrated, and washed in TBS, pH 7.4. For antigen retrieval, paraffin sections were heated in a microwave oven for 15 min (900 W, high power) in Target Retrieval Solution (Dako, Carpinteria, CA), cooled, and treated with 3% H2O2 for 15 min, rinsed with H2O, and blocked with 20% normal goat serum for 30 min. Sections were incubated with affinity-purified ART-27 antibody (1:100 dilution) in 10% normal goat serum and washed in TBS; a rabbit secondary biotinylated antibody was added and an avidin-biotin complex formed and developed using diaminobenzidine substrate. Slides were counterstained with hematoxylin.
Velocity Gradient Sedimentation
HeLa cell nuclear extracts (∼100 μl of 13 mg/ml) were loaded on top of a 5-ml linear 15–35% (vol/vol) glycerol gradient, with or without 2.4 M urea and centrifuged at 4°C in a SW50.1 rotor (Beckman Coulter, Fullerton, CA) for 12 h at 40,000 rpm (Tanese, 1997). Ten 0.5-ml fractions were collected from the top of the tube and analyzed by immunoblotting using rabbit anti–ART-27 or anti-TBP mouse monoclonal antibody.
RESULTS
Cloning and Characterization of ART-27
To identify proteins that interact with the AR N terminus, we elected to screen for interacting proteins using a yeast two-hybrid system. However, the AR N terminus shows strong transcriptional activity in yeast when fused to the LexA DNA-binding domain, making it unsuitable as a bait in a conventional yeast two-hybrid screen. Therefore, we modified the two-hybrid system to allow for the selection of proteins that interact with transcriptional activators. With this approach, the AR N-terminal bait is created by linking amino acids 18–500 to a heterologous activation domain. The prey is created by fusing a cDNA library to a DNA-binding domain rather than an activation domain as done in a conventional yeast two-hybrid system. An androgen-stimulated LNCaP prostate cancer cell cDNA library fused to the LexA DNA-binding domain was screened for proteins that interact with the AR N terminus, expressed as a galactose-inducible fusion protein linked to the B42 activation domain. Library proteins that interact with the AR N terminus will serve to reconstitute transcriptional activity, stimulating expression of Lex-responsive Leu2 and β-galactosidase reporter genes. However, because some library plasmids may posses intrinsic activation potential, rendering them transcriptionally active when bound to DNA, a second screen was performed to eliminate these self-activating false positives by replica plating the Leu2+/LacZ+ colonies onto glucose plates, conditions in which the AR protein is not expressed. Clones that activated the β-galactosidase reporter gene on both glucose and galactose plates were discarded as false positives. Colonies that activated β-galactosidase expression on galactose plates, when the AR N terminus is expressed, but not glucose plates, were deemed true interactors and were further analyzed.
Several positive clones were identified, one of which was termed ART-27 (androgen receptor trapped clone 27). The clone from the yeast two-hybrid assay corresponded to the full-length cDNA and contained an insert of ∼900 bp, in which the largest open reading frame (ORF) encoded a protein of 157 amino acids with an estimated size of 18 kDa. A BLAST search of the GenBank database revealed that ART-27 is located on the X chromosome (Xp11.23–11.22) and is identical to a recently identified ORF of unknown function, termed ubiquitously expressed transcript (UXT; accession no. AF092737), which is prevalent in tumor tissues (Schroer et al., 1999). The ART-27 protein contains potential phosphorylation sites for protein kinase A, protein kinase C and casein kinase II but no other obvious motifs as determined by PROSITE (Figure 1B; Hofmann et al., 1999). Secondary structure prediction algorithms suggest that ART-27 is composed of multiple, successive α helices (Chou and Fasman, 1978; Garnier et al., 1978). ART-27 also appears to be conserved throughout evolution, with model organisms Mus musculus (accession no. AF092738), Arabidopsis thaliana (accession no. AC006535), Drosophila melanogaster (accession no. AE003412), and Caenorhabditis elegans (accession no. U40934), displaying 79, 55, 49, and 26% identity, respectively, with human ART-27. Interestingly, ART-27 showed no significant homology to Saccharomyces cerevisiae proteins, suggesting that ART-27 first arose in metazoans, as did nuclear receptors (Amero et al., 1992; Owen and Zelent, 2000).
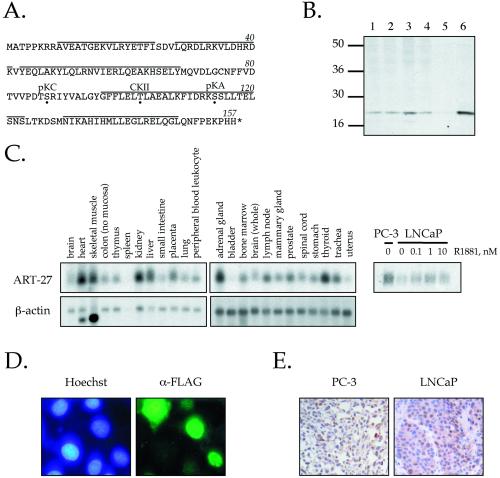
Figure 1.
Cloning and characterization of ART-27. (A) Amino acid sequence of ART-27. The amino acid sequence of ART-27 is shown with an asterisk, representing the stop codon. Potential phosphorylation sites for protein kinase C (pKC), casein kinase II (CKII), and protein kinase A (pKA) are marked by a dot below the target residue. Lines above the sequence represent predicted α-helical regions as determined by Chou-Fasman and GarnierOsguthorpe-Robson algorithms. (B) Expression of ART-27 protein. Equal amounts (50 μg) of nuclear extracts prepared from HeLa cells (lane 1), treated with TPA for 2 h (lane 2), serum-starved and stimulated with serum for 2 h (lane 3), PC-3 cells (lane 4), and whole cell extracts from COS1 cells transfected with pcDNA3 (lane 5) or pcDNA3:ART-27 (lanes 6) were analyzed by immunoblotting with affinity-purified anti–ART-27 antibody. No ART-27 immunoreactivity is observed with preimmune serum (Markus, Taneja, Logan, Li, Ha, Hittelman, Rogatsky, and Garabedian, unpublished results). (C) ART-27 mRNA expression in human tissues and prostate cancer cell lines. A human multiple tissue Northern blot (CLONTECH: human 12-lane MTN Blot I and II) containing 2 μg of poly(A+) mRNA from the indicated tissues was hybridized with 32P-labeled probes corresponding to ART-27 (top) and β-actin (bottom). Total RNA was extracted from PC-3 and LNCaP cells cultured in the absence or presence of the indicated doses of androgen (R1881) for 72 h. Equal amounts of RNA were separated on denaturing formaldehyde-agarose gels (see MATERIALS AND METHODS), transferred to a Duralon nylon membrane, and hybridized to 32P-labeled cDNA probes corresponding to ART-27 (right). Equal loading for each lane was determined by ethidium bromide staining of the 28S rRNA (Markus, Taneja, Logan, Li, Ha, Hittelman, Rogatsky, and Garabedian, unpublished results). (D) ART-27 subcellular localization. HeLa cells were transfected with an FLAG–ART-27 expression construct, fixed, permeabilized, and incubated with an anti-FLAG primary antibody and a corresponding fluorescein-conjugated secondary antibody, and the DNA in nucleus was stained with Hoechst dye H334211. Cells were visualized using a Zeiss Axioplan 2 fluorescence microscope. No signal above background was observed when the primary antibody was omitted (Markus, Taneja, Logan, Li, Ha, Hittelman, Rogatsky, and Garabedian, unpublished results) or in nontransfected cells. Note that the FLAG–ART-27 fluorescence is localized predominantly to the nucleus. (E) Expression pattern of endogenous ART-27 in human PC-3 and LNCaP prostate cancer cell xenografts. Immunohistochemical analysis of paraffin-embedded PC-3 (left) and LNCaP (right) human prostate cancer cell xenografts hybridized with affinity-purified ART-27 antibody is shown (200×). Immunoreactivity, seen as brown staining within the nuclei of these cells, is blocked by coincubation of the antibody with the ART-27 peptide immunogen (Markus, Taneja, Logan, Li, Ha, Hittelman, Rogatsky, and Garabedian, unpublished results).
A polyclonal antibody was made against the ART-27 C terminus. Immunoblot analysis of HeLa and PC-3 cell nuclear extracts show that the antibody recognizes a single endogenous protein with an estimated molecular mass of ∼18 kDa that migrates at the same apparent molecular weight as the cloned ART-27 expressed in COS-1 cells (Figure 1B, compare lanes 1, 4, and 6), confirming that the ORF predicted from sequence analysis was indeed ART-27. No difference in ART-27 expression was observed between untreated and TPA-treated HeLa nuclear extracts (Figure 1B, compare lanes 1 and 2), suggesting that activation of protein kinase C does not affect ART-27 expression or electrophoretic mobility indicative of phosphorylation. However, a slight increase in ART-27 expression was observed in serum-stimulated HeLa nuclear extracts as compared with untreated control cells (Figure 1B, compare lanes 1 and 3), suggesting that ART-27 expression may be regulated by extracellular signals or through the induction of cellular proliferation.
Northern hybridization analysis was carried out to determine the expression pattern of ART-27 mRNA in human tissues. A single transcript of ∼0.9 kb is present at variable levels in the human tissues examined, with the highest levels in the heart, skeletal muscle, kidney, liver, adrenal gland, lymph node, prostate, and thyroid and the lowest levels in bladder and uterus. We also performed Northern blot analysis on mRNA isolated from androgen-independent (PC-3) and androgen-dependent (LNCaP) prostate cancer cells, either untreated or treated for 72 h with the synthetic androgen R1881 at the indicated concentrations (Figure 1C). ART-27 steady-state mRNA expression is slightly higher in PC-3 relative to LNCaP cells and is weakly induced in LNCaP cells in response to androgen.
To investigate the subcellular localization of ART-27, we performed indirect immunofluorescence on HeLa cells transfected with FLAG-tagged ART-27. FLAG–ART-27 localized predominantly to the nucleus, although some diffuse staining was apparent in the cytoplasm of cells expressing high levels of the protein (Figure 1D). Immunohistochemical staining with affinity-purified ART-27 antibody performed on sections of human PC-3 and LNCaP cell xenograft tumors demonstrated strong nuclear staining for endogenous ART-27 (Figure 1E); staining was blocked by coincubation of the antibody with the ART-27 peptide immunogen (Markus, Taneja, Logan, Li, Ha, Hittelman, Rogatsky, and Garabedian, unpublished results), demonstrating the specificity of the antibody-antigen interaction. Interestingly, ART-27 appears to be expressed in a subset of cells from the xenografts. Although we did not investigate the reason for this heterogeneous staining of ART-27, it may reflect cell cycle regulation or other cellular parameters. Regardless, such predominant nuclear distribution of ART-27 is consistent with its role as a putative transcriptional regulatory protein.
ART-27 Interacts with AR In Vitro and in Cell Extracts
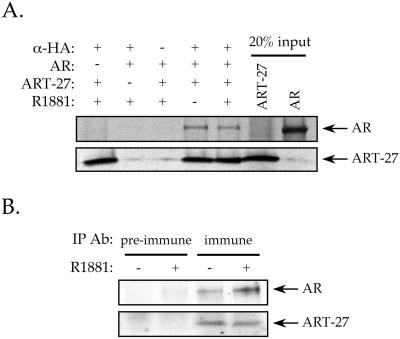
To confirm the results of the two-hybrid screen by an independent biochemical method, we examined whether AR and ART-27 proteins bind tightly enough to each other to be coimmunoprecipitated and whether the interaction was affected by androgen treatment. The ability of ART-27 and AR to interact was first tested in vitro. Full-length AR and HA–ART-27 were expressed in a coupled transcription/translation system and immunoprecipitated with an antibody against the HA epitope on ART-27. In vitro translated AR was precipitated with the HA antibody in the presence, but not in the absence, of HA–ART-27 in a hormone-independent manner (Figure 2A). We next tested the ability of ART-27 to interact with endogenous AR in LNCaP cells that express ART-27 in an inducible manner. Nuclear extracts from untreated or R1881-treated LNCaP cells were subjected to immunoprecipitation with anti–ART-27 antibody and analyzed by immunoblotting with antibodies to AR and ART-27. AR was coimmunoprecipitated by the ART-27 antibody but not by preimmune serum in both the absence and presence of hormone, with slightly more AR found in association with ART-27 upon hormone treatment (Figure 2B). As expected, ART-27 was also immunoprecipitated by the anti–ART-27 antibody in either the presence or absence of hormone (Figure 2B). These results demonstrate the presence of an AR–ART-27 complex in LNCaP cells and substantiate the interaction observed in the yeast two-hybrid system.
Figure 2.
ART-27 interacts with AR in vitro and in cell extracts. (A) Full-length human AR and HA–ART-27 were synthesized in vitro using a coupled transcription/translation system in the presence of [35S]methionine and in the absence or presence of 100 nM R1881, as indicated, and incubated with an antibody against HA. Bound proteins were collected on Protein A Sepharose beads, washed, eluted, resolved by SDS-PAGE, and visualized by autoradiography. Twenty percent of the input AR and ART-27 translation reaction are shown. (B) ART-27 interacts with endogenous AR in LNCaP cell extracts. Nuclear extracts from untreated and 100 nM R1881-treated LNCaP-Tet-on-ART-27-FLAG cells were prepared as described in MATERIALS AND METHODS and incubated with an antibody against either preimmune or ART-27 immune sera. Immune complexes were collected on protein A Sepharose beads, washed, eluted, resolved by SDS-PAGE, and transferred to a membrane. The filter was probed with antibody against AR and ART-27.
ART-27 Interaction Specificity
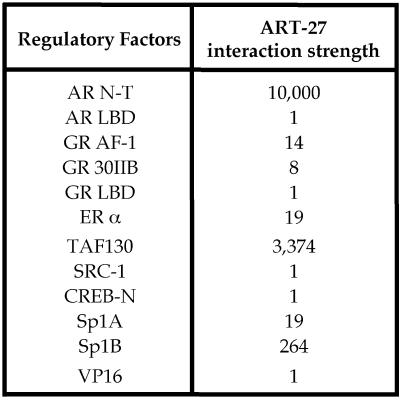
To analyze the specificity of AR:ART-27 interaction, we examined the capacity of ART-27 to associate with a panel of transcriptional regulatory proteins in the yeast two-hybrid system, including the AR C-terminal ligand-binding domain (LBD), the AF-1 region of the GR, ERα, the steroid receptor coactivator-1 (SRC-1), the TBP-associated factor 130 (TAFII130), the Sp1 (A and B domains), the cyclic AMP response element-binding protein (CREB), and VP16. As shown in Figure 3, ART-27 interacts with the AR N terminus and weakly with the GR N terminus, as well as with ERα, Sp1, and TAFII130 (rank order of interaction: AR N terminus18–500 > TAFII130 > Sp1B > ERα = Sp1A > GR N terminus) but not with SRC-1, CREB, VP16, or the AR or GR LBD in either the absence or presence of hormone. Proteins that did not interact with ART-27 have been shown to interact with other factors, suggesting that the lack of interaction with ART-27 is genuine. For example, SRC-1 has been shown to interact with ERα, whereas the AR and GR LBDs associate with GRIP-1, and CREB interacts with TAFII130 in the yeast two-hybrid system (Shibata et al., 1997; Saluja et al., 1998; Hong et al., 1999). Thus, our results indicate that ART-27 interacts not only with the AR N terminus but also with at least two other SRs and with certain other transcriptional regulators, including TAFII130.
Figure 3.
Specificity of AR:ART-27 interactions. Interaction of ART-27 with the AR N terminus (N-T; rat AR18–500), AR LBD (rat AR579–901), and other transcriptional regulatory factors was analyzed using the modified yeast two-hybrid assay. Regulatory factors examined include the GR AF-1 (rat GR107–237), GR 30IIB (rat GR107–237 E219K, F220L, W234R), SRC-1 (SRC-1374–800), GR LBD (rat GR525–795), TAFII130 (TAFII130270–700), Sp1 A (Sp183–262), Sp1 B (Sp1263–524), CREB-N (CREB3–296), ERα(ERα1–595), and VP16. The strength of interaction was determined by a quantitative liquid β-galactosidase assay after a 24-h incubation in galactose medium at 30°C and normalized to protein expression using the common HA epitope resident on each protein. The background from the vector-only control was subtracted from each sample. The interaction with ERα was observed in the presence, but not the absence (Markus, Taneja, Logan, Li, Ha, Hittelman, Rogatsky, and Garabedian, unpublished results), of 100 nM 17β-estradiol. No interaction was observed with AR LBD and the GR LBD in either the presence or absence of 100 nM R1881 and 10 μM deoxycorticosterone, the preferred GR ligand in yeast (Garabedian and Yamamoto, 1992).
ART-27 Interacts with AR153–336
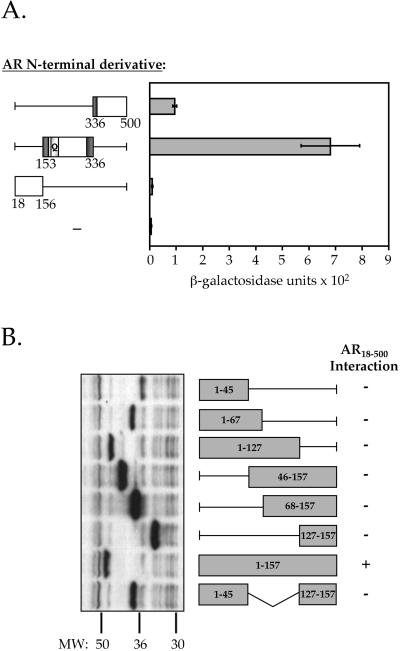
To characterize the interaction of AR with ART-27 in more detail, various AR N-terminal deletion constructs were produced and their ability to interact with ART-27 was tested in the yeast two-hybrid assay. As shown in Figure 4A, ART-27 interacts with two different regions of AR: the central region of the AR N terminus (AR153–336), encompassing all of AF-1a (residues 154–167), and a small part of AF-1b (residues 295–459), and it interacts strongly with ART-27; a weak interaction between ART-27 and the AR336–500 subdomain was also observed, whereas no interaction was detected between ART-27 and AR18–156. Immunoblot analysis of the AR18–156, AR153–336, and AR336–500 derivatives indicates that they are expressed at similar levels (Markus, Taneja, Logan, Li, Ha, Hittelman, Rogatsky, and Garabedian, unpublished results). These findings suggest that the AR153–336 region is the primary interaction site for ART-27.
Figure 4.
Domains of AR and ART-27 mediating interaction. (A) Quantitative analysis of ART-27 interaction with AR N-terminal derivatives. The relative affinity of ART-27 for the AR N-terminal subdomains 336–500, 153–336, and 18–156 was assessed using the yeast two-hybrid assay. The dark gray boxes show the location of the AR N-terminal activation functions AF-1a and AF-1b and the light gray box denotes the glutamine (Q) repeat region. Data represent the means of triplicate data points normalized to cell number. (B) Only full-length ART-27 interacts with the AR N terminus. ART-27 deletion derivatives fused to LexA were tested for their ability to interact with the AR N terminus (AR18–500) using a qualitative β-galactosidase plate assay after a 24-h incubation on galactose X-gal plates at 30°C. +, strong interactions (blue colonies); −, no interactions above background vector-only control (white colony). Left, an immunoblot of the ART-27 derivatives expressed in yeast and probed with an antibody against the LexA moiety common to all ART-27 truncations.
In an attempt to localize the region of ART-27 that interacts with the AR N terminus, we created a series of ART-27 deletion derivatives. ART-27 derivatives containing amino acids 1–45, 1–67, 1–127, 46–157, 68–157, 127–157, 1–157, and 1–45/127–157 were expressed as fusion proteins with LexA and analyzed for their ability to interact with AR18–500 in the modified yeast two-hybrid assay. Surprisingly, none of the deletion derivatives interacted with AR18–500 (Figure 4B), even though all of them were expressed (Figure 4B, left). This result suggests that either ART-27 requires multiple contacts for interaction with the AR N terminus or that the entire protein is involved in configuring a functional AR-interacting surface.
ART-27 Is Involved in Steroid and Thyroid Hormone Receptor Transcriptional Activation
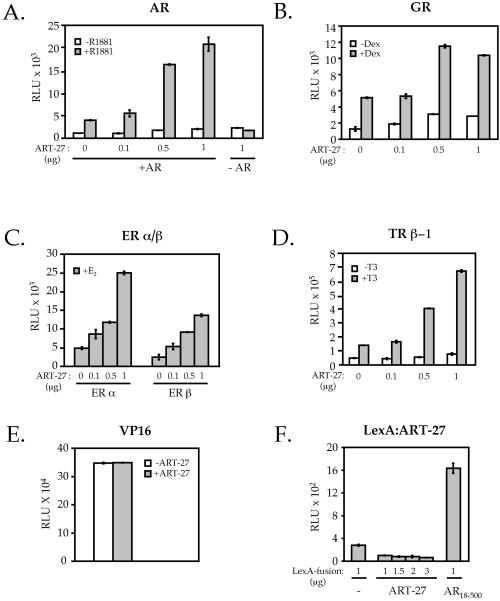
A transient transfection assay was used to examine the role of ART-27 in the regulation of AR transcription activation. AR-deficient HeLa cells were transfected with a constant amount of full-length AR and increasing concentrations of an expression vector encoding an HA–ART-27 along with an AR-responsive luciferase reporter gene. As shown in Figure 5A, the hormone-induced AR transcriptional activation was increased in a dose-dependent manner by overexpressed ART-27. This effect was AR-dependent, because in the absence of AR, ART-27 did not influence reporter gene activity (Figure 5A). This enhanced transcriptional activity did not result from increased AR protein production, because AR levels were not affected by ART-27 coexpression (Markus, Taneja, Logan, Li, Ha, Hittelman, Rogatsky, and Garabedian, unpublished results). The effect of ART-27 on AR was not restricted to a single cell type, because overexpression of ART-27 in PC-3 and COS-1 cells also increased AR transcriptional activity in a dose-dependent manner (Markus, Taneja, Logan, Li, Ha, Hittelman, Rogatsky, and Garabedian, unpublished results). Thus, ART-27 can act as a positive regulator of AR transcriptional activity in mammalian cells.
Figure 5.
ART-27 enhances steroid and thyroid receptor-dependent transcriptional activation. HeLa cells (1.2 × 105 cells/35-mm dish) were transiently transfected using Lipofectamine with paired receptor (0.2 μg/dish) and reporter constructs (0.1 μg/dish) for hAR + MMTV-Luc (A), GR + MMTV-Luc (B), ERα and ERβ + XETL (C), and human TRβ-1 + pGL3-DR4 (D) along with the indicated amount of ART-27 or empty expression vector to equalize the total amount of DNA per dish and 0.05 μg of pCMV-LacZ per dish as an internal control for transfection efficiency. Cells were treated with 100 nM R1881 (A), 100 nM dexamethasone (Dex; B), 10 nM 17β-estradiol (E2; C), 100 nM triac (T3, D), or the ethanol vehicle (white bars) for 12 h, and receptor transcriptional activation was assayed as described in MATERIALS AND METHODS, normalized to β-galactosidase activity, and expressed as relative luminescence units (RLU). The average of three independent experiments and SE is shown. (E) ART-27 does not affect transactivation by VP16. HeLa cells were transfected as above with an expression construct for GAL4-VP16 and a reporter plasmid containing five Gal4-binding sites upstream of the E1b promoter, in the absence (white bars) or presence (gray bars) of 1 μg of ART-27, and assayed for luciferase activity. (F) ART-27 failed to activate transcription when tethered to DNA. HeLa cells were transfected as above with expression constructs for LexA, LexA–ART-27, or LexA-AR18–500 and the pΔ4X-LALO reporter plasmid and assayed for luciferase activity.
The ability of ART-27 to affect transactivation by other members of the SR family, i.e., GR, ERα, ERβ, and the TRβ-1, was tested using transient transfection assays. Our results indicate that ART-27 increased the transcriptional activity of all four receptors in a dose-dependent manner (Figure 5, B–D). We next tested the effect of ART-27 on VP16-dependent transactivation. Recall that VP16 did not interact with ART-27 in the yeast two-hybrid assay. Consistent with this lack of interaction, ART-27 expression had no effect on GAL4-VP-16 activity from a reporter plasmid containing five Gal4-binding sites upstream of the E1b promoter (Figure 5E). Together, these results suggest that ART-27 increases transactivation by steroid and thyroid hormone receptors.
We also examined the ability of ART-27 to activate transcription when artificially recruited to promoters in mammalian cells. Our results indicate that recruitment of ART-27 to a promoter by fusing it to either the Gal4 (Markus, Taneja, Logan, Li, Ha, Hittelman, Rogatsky, and Garabedian, unpublished results) or LexA (Figure 5F) DNA-binding domain fails to activate transcription, even though the proteins were expressed (Markus, Taneja, Logan, Li, Ha, Hittelman, Rogatsky, and Garabedian, unpublished results). In fact, transcriptional activation of the LexA–ART-27 derivative was slightly reduced relative to LexA, suggesting that ART-27 may interact with and sequester factor(s) responsible for “basal” promoter activity. Thus, ART-27 lacks an intrinsic transactivation function. This may suggest that ART-27 is unable to overcome a rate-limiting step in transcription when artificially recruited to a promoter or, alternatively, that ART-27 is only one target of the AR and that the receptor requires multiple targets to recruit mammalian Pol II and initiate transcription.
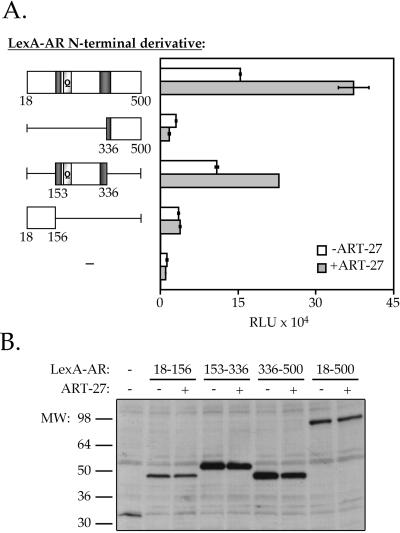
Enhanced AR-dependent Transcriptional Activation by ART-27 Is Mediated Through a Distinct Receptor N-Terminal Domain
Because ART-27 interacts strongest with the AR subdomain spanning amino acids 153–336 (Figure 4A), we expect that it would affect the transcriptional activation potential of this AR subdomain. To determine whether ART-27 could affect the function of the different AR subdomains, AR N-terminal derivatives containing amino acids 18–156, 153–336, 336–500, and 18–500 were expressed as fusion proteins with the LexA DNA-binding domain. HeLa cells were transfected with the LexA-AR N-terminal derivatives, along with a LexA-responsive luciferase reporter gene in the presence and absence of ART-27. In the absence of ART-27 coexpression, all four subdomains of the AR N terminus are capable of activating transcription of the LexA-luciferase reporter gene to varying degrees (Figure 6A). Importantly, overexpression of ART-27 enhances the transcriptional activity of two AR derivatives containing the ART-27 interaction region, LexA-AR153–336 and Lex-AR18–500, but not that of the derivatives lacking this region, LexA-AR18–156 and LexA-AR336–500. In fact, transcriptional activation of the LexA-AR336–500 derivative was slightly reduced by ART-27 overexpression, suggesting that ART-27 may interact with and sequester a factor responsible for AR transactivation via the 336–500 subdomain. Immunoblotting with an antibody against the LexA moiety common to all derivatives indicates that expression of these chimeras is unaffected by coexpression of ART-27 in HeLa cells (Figure 6B). These results suggest that the enhancement of AR transcriptional activation by ART-27 is mediated via the ART-27–interacting region.
Figure 6.
The effect of ART-27 on AR transcriptional activation is dependent on the AR-interacting region. (A) HeLa cells were transfected with 0.5 μg of the pcDNA3LexA:AR N-terminal derivatives indicated and either 1.0 μg of the empty expression vector (white bars) or 1.0 μg pcDNA3-HA–ART-27 (gray bars) along with 1.0 μg of the LexA responsive-luciferase reporter (pΔ4X-LALO) and 0.25 μg of pCMV-LacZ, and AR activity was determined as described in Figure 5. Data represent the means of duplicate data points normalized to β-galactosidase units. Error bars represent the range of the duplicate data points. (B) The expression of the LexA:AR derivatives was analyzed by immunoblotting from a parallel set of transfections using a polyclonal antibody to LexA.
ART-27 Is Part of a High Molecular Weight Complex
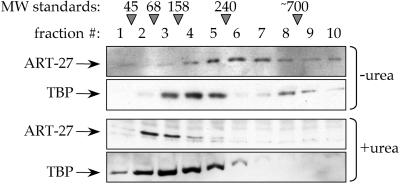
Several transcriptional regulatory cofactors have been identified as components of multiprotein complexes. We therefore examined whether ART-27 is part of a higher-order species using velocity gradient sedimentation. HeLa cell nuclear extracts were applied to 15–35% glycerol gradients, either containing or lacking 2.4 M urea. After centrifugation, fractions were collected and analyzed by immunoblotting with antibodies specific for ART-27 or TBP. Previous work has shown that TAFs and TBP exist in a complex, which can be dissociated with urea (Tanese et al., 1991). It has been shown that the larger, faster sedimenting species corresponds to TBP and TAFs binding together as TFIID, whereas the smaller, slower migrating peak represents additional non-TFIID TBP complexes (Tanese et al., 1991). As expected, under native conditions, TBP sedimented at the bottom, in the high-density region of the gradient in two discrete peaks. Under native conditions, a majority of ART-27 also migrated within the high-density region of the gradient (estimated range of molecular mass between 240 and 700 kDa; Figure 7). In the presence of 2.4 M urea, the sedimentation patterns of TBP and ART-27 were shifted to the top of the gradient (Figure 7). The change in mobility of TBP is in agreement with previous results demonstrating the sedimentation profile of dissociated TFIID (Tanese et al., 1991). These results are consistent with the idea that ART-27 is part of a high molecular weight complex, the components of which have yet to be identified.
Figure 7.
Native ART-27 is part of a high molecular weight complex. Nuclear extracts were prepared from HeLa cells and loaded onto linear 15–35%glycerol gradients with or without 2.4 M urea. Fractions were collected and analyzed by Western blot using affinity-purified ART-27 polyclonal or TBP monoclonal antibodies. Migration of molecular mass markers is indicated: 45- and 68-kDa albumin, 158-kDa aldolase, 240-kDa catalase. The molecular mass estimate of TFIID is 700 kDa (Tanese et al., 1991).
DISCUSSION
We have identified ART-27 as a protein that interacts with the AR N-terminal subdomain spanning amino acids 153–336, including AF-1a (154–167) and a part of AF-1b (295–459), and enhances AR transcriptional activation when overexpressed in mammalian cells. The ability of ART-27 to affect AR transcriptional activation is dependent on the ART-27:AR–interacting region, because only the AR N-terminal derivatives containing the interaction domain are enhanced by ART-27 coexpression. Thus, ART-27 represents a novel AR N terminus-associated coactivator.
ART-27 was originally identified as a novel transcript in a screen for genes that map to the human Xp11 locus, a region previously shown to contain an abundance of disease loci (Schroer et al., 1999). However, the designation of ART-27 as a ubiquitously expressed transcript is misleading because the level of ART-27 mRNA varies widely among tissues examined (Figure 2C). Interestingly, ART-27 and AR reside in an amplicon found in a subset of hormone-refractory prostate cancers, suggesting that ART-27 may play a role in AR-dependent prostate tumorigenesis (Visakorpi et al., 1995a,b).
Although we have localized the region of the AR N terminus (residues 153–336) necessary for interaction with ART-27, homology searches among the other receptors affected by ART-27 have not revealed a common ART-27 interaction motif. We are currently testing whether single amino acid substitutions in the AR N terminus that have been implicated in androgen insensitivity syndrome affect interaction with ART-27. The amino acid alternations in the AR N terminus identified in androgen insensitivity syndrome patients likely represent AR loss-of-function mutations, which have reduced AR activity possibly because of the loss of receptor-cofactor interactions. This approach might help pinpoint the residues of AR that interact with ART-27, which in turn will help define the corresponding sites in the other receptors.
Deletion analysis of ART-27 failed to define a distinct interaction surface within ART-27 that mediates association with the receptor. One plausible explanation is that ART-27 may associate with the AR N terminus through multiple low-affinity interactions, and removal of any one of these contacts renders ART-27 incapable of binding. Alternatively, the entire ART-27 may be involved in configuring a functional protein and its integrity may be compromised upon deletion of any region. Whether ART-27 contains independent interaction surfaces for AR or consists of multiple regions that function together to coordinate the tertiary structure of the protein will require a detailed structure-function analysis, which is currently underway.
The ART-27 primary amino acid sequence as well as the predicted secondary structure composed of tandem α-helices appears conserved from worms to humans. Although the function of the ART-27 orthologs has yet to be determined, our findings suggest a link to nuclear receptor function as a potential coactivator protein. Recently, the taiman gene of Drosophila, a protein involved in cell migration, was found to be homologous to the mammalian p160 coactivator AIB1 and to function as a coactivator for the ecdysone receptor (Bai et al., 2000). Thus, like taiman, ART-27 may represent an evolutionarily conserved transcriptional cofactor that regulates a wide array of responses controlled by nuclear receptors.
The mechanism by which ART-27 affects AR-mediated transcriptional activation remains to be defined. ART-27, a comparatively small protein with a predicted molecular mass of ∼18 kDa, has little transcriptional activation ability when tethered to DNA, suggesting that it does not harbor an intrinsic transactivation function. This may seem unusual for a transcriptional cofactor; however, recent studies by Dorris and Struhl (2000) demonstrate that several components of the RNA Pol II holoenzyme are not transcriptionally active when tethered to DNA. Because ART-27 migrates in velocity gradient sedimentation analysis as a large molecular weight species (Figure 7), we speculate that ART-27 may represent a subunit of a previously characterized (such as DRIP/TRAP/ARC; Fondell et al., 1996; Rachez et al., 1998, 1999; Naar et al., 1999) or novel multiprotein coactivator complex. Our preliminary findings suggest that the purified DRIP complex does not contain an ART-27 immunoreactive species by Western blot, suggesting that ART-27 is not part of the DRIP complex (S. M. Markus, L.P. Freedman, and M.J. Garabedian, unpublished observation). ART-27 also interacts with TAFII130 in the yeast two-hybrid assay, suggesting that ART-27 may communicate with at least one other transcriptional regulatory cofactor. Interestingly, TAFII130 itself appears to interact with and increase transcriptional activation of AR via a distinct N-terminal subreigon (Taneja and Garabedian, unpublished observation). Additional studies are needed to determine the components of this putative ART-27–containing complex as well as the mechanism of ART-27 function.
The AR N terminus appears to be a multifaceted platform capable of interacting with a variety of transcriptional regulatory proteins, including ART-27, which likely collaborate to regulate gene- and tissue-specific responses to AR. Consistent with this notion, the coactivators SRC-1, GRIP-1, and CBP have recently been shown to interact with the AR N terminus and modulate its activity (Ikonen et al., 1997; Alen et al., 1999; Bevan et al., 1999; Ma et al., 1999). Although ART-27 is expressed in a wide variety of tissues, distinct cofactor combinations in target tissues may result in cell-specific regulation of AR. Thus, based on its nuclear localization, its binding to the AR N terminus, and its ability to potentiate steroid and thyroid hormone receptor-dependent transcription in cultured cells, ART-27 represents a new class of coactivator proteins.
ACKNOWLEDGMENTS
We are grateful to Dr. Roger Miesfeld for the human and rat AR clones. We also thank Dr. Erica Golemis for the LexA antibody, Drs. Naoko Tanese and Herb Samuels for the activator constructs, Dr. Tanese for help with the glycerol gradient sedimentation analysis, and Dr. Tanese and Laura Su for critically reading the manuscript. We thank Dr. Serge Rome for immunohistochemical staining of prostate cancer xenografts and Dr. Herman Yee of the Kaplan Comprehensive Cancer Center Molecular Diagnostics Shared Resource for analysis (P30 CA-16087). This work was supported, in part, by National Institutes of Health Training grants 2T32 GM-07308 (to A.B.H.), 5T32 AI-07180 (to S.M.M. and I.R.), T32 DK-07775 (to W.L.), and K08 DK-02577 (to S.T.), by a grant from the Chemotherapy Foundation, and by a Merck/American Foundation for Urologic Disease Research Scholarship (to S.T.), as well as National Institutes of Health (R01 DK-58024) and Department of Defense Prostate Cancer Research Program (DAMD-17-00-1-0035) grants to M.J.G.
Footnotes
DOI:10.1091/mbc.01–10-0513.
REFERENCES
- Adachi M, Takayanagi R, Tomura A, Imasaki K, Kato S, Goto K, Yanase T, Ikuyama S, Nawata H. Androgen-insensitivity syndrome as a possible coactivator disease. N Engl J Med. 2000;343:856–862. doi: 10.1056/NEJM200009213431205. [DOI] [PubMed] [Google Scholar]
- Alen P, Claessens F, Verhoeven G, Rombauts W, Peeters B. The androgen receptor amino-terminal domain plays a key role in p160 coactivator-stimulated gene transcription. Mol Cell Biol. 1999;19:6085–6097. doi: 10.1128/mcb.19.9.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amero SA, Kretsinger RH, Moncrief ND, Yamamoto KR, Pearson WR. The origin of nuclear receptor proteins: a single precursor distinct from other transcription factors. Mol Endocrinol. 1992;6:3–7. doi: 10.1210/mend.6.1.1738368. [DOI] [PubMed] [Google Scholar]
- Bai J, Uehara Y, Montell DJ. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103:1047–1058. doi: 10.1016/s0092-8674(00)00208-7. [DOI] [PubMed] [Google Scholar]
- Bevan CL, Hoare S, Claessens F, Heery DM, Parker MG. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol. 1999;19:8383–8392. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann AO, Jenster G, Kuiper GG, Ris C, van Laar JH, van der Korput JA, Degenhart H J, Trifiro M A, Pinsky L, Romalo G, et al. The human androgen receptor: structure/function relationship in normal and pathological situations. J Steroid Biochem Mol Biol. 1992;41:361–368. doi: 10.1016/0960-0760(92)90362-m. [DOI] [PubMed] [Google Scholar]
- Chamberlain NL, Whitacre DC, Miesfeld RL. Delineation of two distinct type 1 activation functions in the androgen receptor amino-terminal domain. J Biol Chem. 1996;271:26772–26778. doi: 10.1074/jbc.271.43.26772. [DOI] [PubMed] [Google Scholar]
- Chang C, Saltzman A, Yeh S, Young W, Keller E, Lee HJ, Wang C, Mizokami A. Androgen receptor: an overview. Crit Rev Eukaryot Gene Expr. 1995;5:97–125. doi: 10.1615/critreveukargeneexpr.v5.i2.10. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Cleutjens CB, Steketee K, van Eekelen C C, van der Korput J A, Brinkmann A O, Trapman J. Both androgen receptor and glucocorticoid receptor are able to induce prostate-specific antigen expression, but differ in their growth-stimulating properties of LNCaP cells. Endocrinology. 1997;138:5293–5300. doi: 10.1210/endo.138.12.5564. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Dorkin TJ, Neal DE. Basic science aspects of prostate cancer. Semin Cancer Biol. 1997;8:21–27. doi: 10.1006/scbi.1997.0049. [DOI] [PubMed] [Google Scholar]
- Dorkin TJ, Robson CN, Neal DE. The molecular pathology of urological malignancies. J Pathol. 1997;183:380–387. doi: 10.1002/(SICI)1096-9896(199712)183:4<380::AID-PATH959>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Dorris DR, Struhl K. Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol Cell Biol. 2000;20:4350–4358. doi: 10.1128/mcb.20.12.4350-4358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina AA, Chang HC, Marsh JA, Lindquist S, Gaber RF. A cyclophilin function in Hsp90-dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- Fang Y, Fliss AE, Rao J, Caplan AJ. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Fliss AE, Robins DM, Caplan AJ. Hsp90 regulates androgen receptor hormone binding affinity in vivo. J Biol Chem. 1996;271:28697–28702. doi: 10.1074/jbc.271.45.28697. [DOI] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian MJ, Yamamoto KR. Genetic dissection of the signaling domain of a mammalian steroid receptor in yeast. Mol Biol Cell. 1992;3:1245–1257. doi: 10.1091/mbc.3.11.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J, Osguthorpe DJ, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gelman L, Zhou G, Fajas L, Raspe E, Fruchart JC, Auwerx J. p300 interacts with the N- and C-terminal part of PPARgamma2 in a ligand-independent and -dependent manner, respectively. J Biol Chem. 1999;274:7681–7688. doi: 10.1074/jbc.274.12.7681. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Gordon DA, Chamberlain NL, Flomerfelt FA, Miesfeld RL. A cell-specific and selective effect on transactivation by the androgen receptor. Exp Cell Res. 1995;217:368–377. doi: 10.1006/excr.1995.1099. [DOI] [PubMed] [Google Scholar]
- Hakimi JM, Rondinelli RH, Schoenberg MP, Barrack ER. Androgen-receptor gene structure and function in prostate cancer. World J Urol. 1996;14:329–337. doi: 10.1007/BF00184606. [DOI] [PubMed] [Google Scholar]
- Hittelman AB, Burakov D, Iniguez-Lluhi JA, Freedman LP, Garabedian MJ. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Darimont BD, Ma H, Yang L, Yamamoto KR, Stallcup MR. An additional region of coactivator GRIP1 required for interaction with the hormone-binding domains of a subset of nuclear receptors. J Biol Chem. 1999;274:3496–3502. doi: 10.1074/jbc.274.6.3496. [DOI] [PubMed] [Google Scholar]
- Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao PW, Chang C. Isolation and characterization of ARA160 as the first androgen receptor N-terminal-associated coactivator in human prostate cells. J Biol Chem. 1999;274:22373–22379. doi: 10.1074/jbc.274.32.22373. [DOI] [PubMed] [Google Scholar]
- Ikonen T, Palvimo JJ, Janne OA. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- Jenster G. The role of the androgen receptor in the development and progression of prostate cancer. Semin Oncol. 1999;26:407–421. [PubMed] [Google Scholar]
- Jenster G, van der Korput HA, van Vroonhoven C, van der Kwast TH, Trapman J, Brinkmann AO. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol. 1991;5:1396–404. doi: 10.1210/mend-5-10-1396. [DOI] [PubMed] [Google Scholar]
- Jenster G, van der Korput JA, Trapman J, Brinkmann AO. Functional domains of the human androgen receptor. J Steroid Biochem Mol Biol. 1992;41:671–675. doi: 10.1016/0960-0760(92)90402-5. [DOI] [PubMed] [Google Scholar]
- Kang HY, Yeh S, Fujimoto N, Chang C. Cloning and characterization of human prostate coactivator ARA54, a novel protein that associates with the androgen receptor. J Biol Chem. 1999;274:8570–8576. doi: 10.1074/jbc.274.13.8570. [DOI] [PubMed] [Google Scholar]
- Knoblauch R, Garabedian MJ. Role for Hsp90-associated cochaperone p23 in estrogen receptor signal transduction. Mol Cell Biol. 1999;19:3748–3759. doi: 10.1128/mcb.19.5.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Duan HO, Chang C. From androgen receptor to the general transcription factor TFIIH: identification of cdk activating kinase (CAK) as an androgen receptor NH(2)-terminal associated coactivator. J Biol Chem. 2000;275:9308–9313. doi: 10.1074/jbc.275.13.9308. [DOI] [PubMed] [Google Scholar]
- Lee KA, Bindereif A, Green MR. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- Li H, Gomes PJ, Chen JD. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Hong H, Huang SM, Irvine RA, Webb P, Kushner PJ, Coetzee GA, Stallcup MR. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol Cell Biol. 1999;19:6164–6173. doi: 10.1128/mcb.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan IJ, Gustafsson J. Interaction of the human androgen receptor transactivation function with the general transcription factor TFIIF. Proc Natl Acad Sci USA. 1997;94:8485–8490. doi: 10.1073/pnas.94.16.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- Nelson CC, Hendy SC, Shukin RJ, Cheng H, Bruchovsky N, Koop BF, Rennie PS. Determinants of DNA sequence specificity of the androgen, progesterone, and glucocorticoid receptors: evidence for differential steroid receptor response elements. Mol Endocrinol. 1999;13:2090–2107. doi: 10.1210/mend.13.12.0396. [DOI] [PubMed] [Google Scholar]
- Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Owen GI, Zelent A. Origins and evolutionary diversification of the nuclear receptor superfamily. Cell Mol Life Sci. 2000;57:809–827. doi: 10.1007/s000180050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Khursheed B, Garabedian MJ, Fortin MG, Lindquist S, Yamamoto KR. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- Rachez C, Suldan Z, Ward J, Chang CP, Burakov D, Erdjument-Bromage H, Tempst P, Freedman LP. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluja D, Vassallo MF, Tanese N. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/mcb.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller A, Hughes E, Golden KL, Robins DM. Multiple receptor domains interact to permit, or restrict, androgen- specific gene activation. J Biol Chem. 1998;273:24216–24222. doi: 10.1074/jbc.273.37.24216. [DOI] [PubMed] [Google Scholar]
- Schoenmakers E, Verrijdt G, Peeters B, Verhoeven G, Rombauts W, Claessens F. Differences in DNA binding characteristics of the androgen and glucocorticoid receptors can determine hormone-specific responses. J Biol Chem. 2000;275:12290–12297. doi: 10.1074/jbc.275.16.12290. [DOI] [PubMed] [Google Scholar]
- Schroer A, Schneider S, Ropers H, Nothwang H. Cloning and characterization of UXT, a novel gene in human Xp11, which is widely and abundantly expressed in tumor tissue. Genomics. 1999;56:340–343. doi: 10.1006/geno.1998.5712. [DOI] [PubMed] [Google Scholar]
- Segnitz B, Gehring U. The function of steroid hormone receptors is inhibited by the hsp90-specific compound geldanamycin. J Biol Chem. 1997;272:18694–18701. doi: 10.1074/jbc.272.30.18694. [DOI] [PubMed] [Google Scholar]
- Sharif M, Privalsky ML. V-erbA and c-erbA proteins enhance transcriptional activation by c-jun. Oncogene. 1992;7:953–960. [PubMed] [Google Scholar]
- Shibata H, Spencer TE, Onate SA, Jenster G, Tsai SY, Tsai MJ, O'Malley BW. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–164. [PubMed] [Google Scholar]
- Smith CL, Onate SA, Tsai MJ, O'Malley BW. CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LF, Knoblauch R, Garabedian MJ. Rho GTPases as modulators of the estrogen receptor transcriptional response. J Biol Chem. 2001;276:3231–3237. doi: 10.1074/jbc.M005547200. [DOI] [PubMed] [Google Scholar]
- Tanese N. Small-scale density gradient sedimentation to separate and analyze multiprotein complexes. Methods. 1997;12:224–234. doi: 10.1006/meth.1997.0475. [DOI] [PubMed] [Google Scholar]
- Tanese N, Pugh BF, Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Trapman J, Brinkmann AO. The androgen receptor in prostate cancer. Pathol Res Pract. 1996;192:752–760. doi: 10.1016/S0344-0338(96)80097-5. [DOI] [PubMed] [Google Scholar]
- Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi OP. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9:401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- Visakorpi T, Kallioniemi AH, Syvanen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi OP. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res. 1995;55:342–347. [PubMed] [Google Scholar]
- Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- Wilson EM, Simental JA, French FS, Sar M. Molecular analysis of the androgen receptor. Ann NY Acad Sci. 1991;637:56–63. doi: 10.1111/j.1749-6632.1991.tb27300.x. [DOI] [PubMed] [Google Scholar]
- Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]