Summary
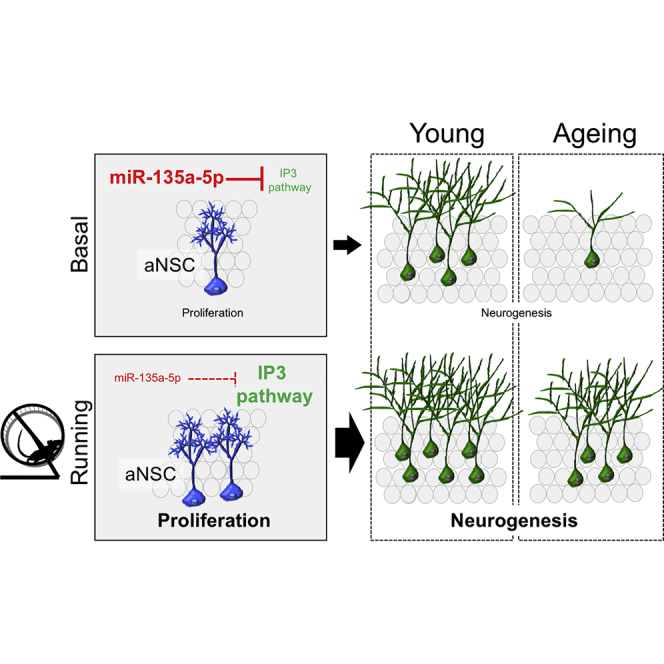
Physical exercise stimulates adult hippocampal neurogenesis and is considered a relevant strategy for preventing age-related cognitive decline in humans. The underlying mechanisms remains controversial. Here, we show that exercise increases proliferation of neural precursor cells (NPCs) of the mouse dentate gyrus (DG) via downregulation of microRNA 135a-5p (miR-135a). MiR-135a inhibition stimulates NPC proliferation leading to increased neurogenesis, but not astrogliogenesis, in DG of resting mice, and intriguingly it re-activates NPC proliferation in aged mice. We identify 17 proteins (11 putative targets) modulated by miR-135 in NPCs. Of note, inositol 1,4,5-trisphosphate (IP3) receptor 1 and inositol polyphosphate-4-phosphatase type I are among the modulated proteins, suggesting that IP3 signaling may act downstream miR-135. miR-135 is the first noncoding RNA essential modulator of the brain's response to physical exercise. Prospectively, the miR-135-IP3 axis might represent a novel target of therapeutic intervention to prevent pathological brain aging.
Keywords: adult neurogenesis; running; aging; miR-135a; inositol 1,4,5-trisphosphate (IP3) pathway; ITPR1; INPP4A
Graphical Abstract
Highlights
-
•
miR-135a-5p mediates running-induced neurogenesis in adult mouse hippocampus
-
•
Downregulation of miR-135a-5p stimulates NPC proliferation in aged mice
-
•
miR-135 modulates phosphatidylinositol signaling
Pons-Espinal, Gasperini, and colleagues report that running induces NPC proliferation and neurogenesis via downregulation of miR-135a-5p in the mouse hippocampus. Remarkably, downregulation of miR-135a stimulates proliferation in the hippocampus of aged mice. ITPR1 and INPP4A, involved in IP3 signaling, are modulated by miR-135 in NPCs. Prospectively, therapeutic exploitation of the miR-135-IP3 axis might represent a novel intervention strategy for successful aging.
Introduction
In most mammalian species, the postnatal subgranular zone (SGZ) of the hippocampal dentate gyrus (DG) maintains a population of neural precursor cells (NPCs) retaining the lifelong capability to generate new neurons and astrocytes. However, this process inexorably declines with age (Eriksson et al., 1998, Spalding et al., 2013, Spalding et al., 2005 and; Knoth et al., 2010, Dennis et al., 2016, Mathews et al., 2017, Kempermann et al., 2018), and this decline has been correlated with the loss of cognitive abilities and the occurrence of several brain pathologies (Bond et al., 2015, Urban and Guillemot, 2014). Currently, many translational concepts for preserving cognitive abilities in the aging brain thus aim at sustaining, or even increasing, the potential for cognitive plasticity and flexibility that is contributed by the adult-generated neurons.
Environmental enrichment and physical activity (e.g., voluntary running in a wheel) potentiate adult neurogenesis in rodents (Farioli-Vecchioli et al., 2014, Fischer et al., 2014, Kronenberg et al., 2006, Kronenberg et al., 2003, Lugert et al., 2010, Overall et al., 2013, van Praag et al., 1999). The positive response of adult neurogenesis to these stimuli is maintained into old age and counteracts the age-associated cognitive decline in rodents and likely in humans (Kempermann et al., 2002, Kempermann et al., 1998, Kempermann et al., 2018, Kronenberg et al., 2006, van Praag, 2005). However, the cellular and molecular mechanisms underlying homeostasis of adult neurogenesis and its response to environmental stimuli remain elusive (Encinas and Fitzsimons, 2017, Overall et al., 2016). We hypothesize that exploiting these mechanisms is relevant for preventing age-related cognitive decline in humans and that our animal models can contribute to providing evidence-based recommendations for an active lifestyle for successful aging.
The molecular control of adult neurogenesis is highly polygenic (Kempermann, 2011) and very likely regulated at multiple levels, including epigenetic, post-transcriptional as well as post-translational (Encinas and Fitzsimons, 2017, Stricker and Götz, 2018). Single-cell RNA sequencing efforts start to reveal molecular cascades underlying adult neurogenesis (Shin et al., 2015). However, it remains unclear how the subtle changes in transcript abundance can be translated to biologically relevant protein levels. This enormous complexity thus hinders the identification of the proteins and pathways that are at the top of the molecular control of adult neurogenesis and its response to the environment.
MicroRNAs (miRNAs) are small noncoding RNAs which, by post-transcriptional repression of hundreds of target messenger RNAs (mRNAs) in parallel, tune the entire cell proteome (Selbach et al., 2008). The functional synergism of few miRNAs achieves gene regulation essential for proliferation, cell fate determination, and survival in embryonic (Barca-Mayo and De Pietri Tonelli, 2014) and adult NPCs (Encinas and Fitzsimons, 2017, Pons-Espinal et al., 2017, Stappert et al., 2018). Interestingly, running stimulates hippocampal NPC proliferation (Overall et al., 2016) and alters miRNA expression in rodents (Bao et al., 2014, Cosín-Tomás et al., 2014, Hu et al., 2015, Pan-Vazquez et al., 2015). Hence, we hypothesize that investigating miRNAs involved in running-induced neurogenesis would allow the identification of the most prominent pathways that constrain NPC proliferative potential in the adult mouse hippocampus. With this approach, we aim to uncover the proteins and pathways acting within this circuit-level context, hence providing a system-level biological understanding of scientific and therapeutic value.
Results
Running-Induced Proliferation Downregulates miRNA Expression in Nestin+ Adult Hippocampal NPCs In Vivo
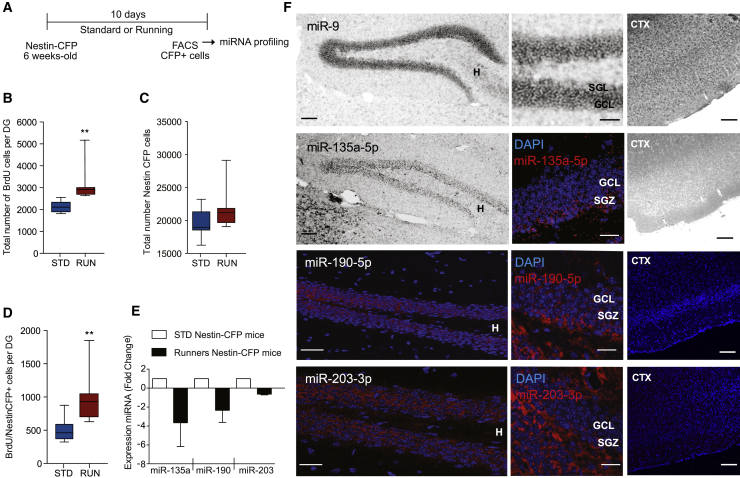
To investigate whether running alters miRNAs expression in adult hippocampal NPCs, 6-week-old mice, expressing the fluorescent protein CFPnuc under control of the Nestin promoter (Nestin-CFPnuc), were housed under standard conditions or equipped with a running wheel for 10 days (Figure 1A). As expected, in the hippocampal SGZ of running mice, we found a statistically significant increase in the number of bromodeoxyuridine (BrdU)-positive cells (Figure 1B; p = 0.005) and BrdU/Nestin-CFPnuc double-positive NPCs (Figure 1D; p = 0.007), and a slight increase (not statistically significant) in the total number of Nestin-CFPnuc-positive NPCs (Figure 1C; p = 0.136), suggesting an expansion of the proliferative NPC pool.
Figure 1.
Running-Induced Proliferation Downregulates miRNA Expression in Nestin-Positive Adult Hippocampal NPCs In Vivo
(A) Schematic representation of the experiment.
(B–D) (B) Total number of BrdU and (C) Nestin-CFPnuc positive adult neural progenitor cells (NPCs), or (D) proportion of BrdU/Nestin-CFPnuc double-positive cells counted from the hippocampal SGZ of Nestin-CFPnuc mice under standard (STD) or running (RUN) conditions for 10 days; n = 8 mice per group.
(E) Quantification of relative expression levels of miR-135-5p, miR-190-5p, and miR-203-3p by TaqMan low density array (TLDA) in sorted Nestin-CFPnuc NPCs from the hippocampus of adult mice in STD or RUN conditions.
(F) Representative micrographs showing expression of miR-9-5p (positive control), miR-135-5p, miR-190-5p, and miR-203-3p by in situ hybridization in the DG and cortex (CTX) of 6-week-old C57Bl6J mice. H, hilus, GCL, granular cell layer; SGZ, subgranular zone.
Data are expressed as means ± SEM, n = 3 independent experiments. One-way ANOVA Bonferroni as post hoc: ∗∗p < 0.01. Scale bar, 100 μm (large panel), 50 μm (small panel).
We sorted Nestin-CFPnuc+ NPCs from the DG of resting and running mice and found eight miRNAs that were reproducibly downregulated in runners compared with resting mice (Figure S1, TaqMan low density array [TLDA], n = 3 independent biological replicates each containing a pool of Nestin-CFPnuc+ cells isolated from eight mice per condition). Of relevance, none of the miRNAs in the TLDA were reproducibly induced in Nestin-CFPnuc+ NPC upon running (Table S1). The three most downregulated miRNAs in Nestin-CFPnuc+ NPCs from the DG of running mice were mmu-miR-135a-5p (miR-135a), mmu-miR-190-5p (miR-190), and mmu-miR-203-3p (miR-203) (Figure 1E). By in situ hybridization, we found that expression of these miRNAs was enriched in the DG of adult resting mice (Figure 1F) and that only miR-135a and miR-190 were preferentially enriched in the hippocampal SGZ, where NPCs are located in vivo (Figure 1F). These results indicate that running decreases miRNA expression in hippocampal NPCs in vivo, opening the possibility that some of these miRNAs might be involved in the mechanism underlying running-induced proliferation of adult NPCs.
miR-135a Inhibits Cell-Cycle Progression of Cultured Adult NPCs and Mediates Running-Induced Proliferation in the Hippocampal SGZ In Vivo
To investigate this possibility, we compared expression of the three miRNAs in cultures of primary hippocampal NPCs (Babu et al., 2011) in quiescence and proliferative conditions. Quiescence is operationally defined here as “non-proliferative” and induced in vitro by the addition of bone morphogenetic protein 4 (BMP4, Martynoga et al., 2013) to the culture medium containing fibroblast growth factor 2 (FGF2/bFGF). Proliferation medium was supplemented with both FGF2 and epidermal growth factor (EGF) (Pons-Espinal et al., 2017, Figures 2A–2C). As expected, the proportion of BrdU-positive NPCs in proliferative medium was higher than in quiescence medium (Figures 2A and 2B; p < 0.001) and, consistent with miRNA profiling of running mice (Figure 1), proliferating NPCs had significantly lower levels of miR-135a, miR-190, and miR-203 compared with cells in quiescence (Figure 2C; p < 0.001).
Figure 2.
miR-135a Levels in Adult NPCs Are Cell-Cycle Dependent and Its Modulation Affects Cell Proliferation In Vitro
(A and B) Representative micrographs showing (A) BrdU-positive and (B) quantification in primary hippocampal adult NPCs cultured in proliferative medium (EGF + bFGF), or quiescence medium (BMP4 + bFGF).
(C) Relative miRNA fold change expression.
(D and E) Representative micrographs showing (D) Ki67-positive NPCs and (E) quantification cultured in proliferative medium (EGF + bFGF) upon transfection with 50 nM scrambled control, miR-203-3p, miR-190-5p, miR-135a-5p mimics, or upon transduction with lentivirus transcribing a sponge for miR-135a (sponge miR-135a, i.e., loss of function) or virus expressing control (scrambled) RNAs.
Data are expressed as means ± SEM, n = 3 independent experiments containing three replicates. One-way ANOVA Bonferroni as post hoc. ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 50 μm.
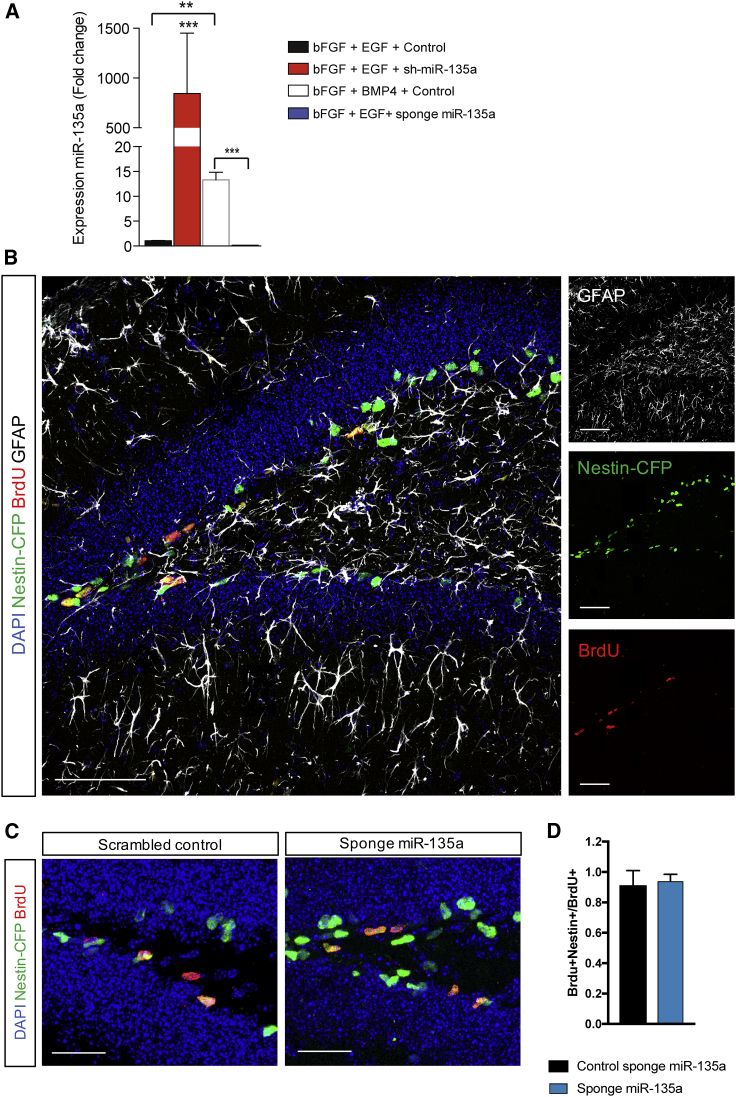
To ascertain whether miR-135a, miR-190, or miR-203 affects adult NPC proliferation, we transfected synthetic miRNA mimics or scrambled control into NPCs in vitro and quantified the proliferative marker Ki67 (Bruno and Darzynkiewicz, 1992). Overexpression of miR-135a, but not miR-203 or miR-190, was sufficient to reduce the proliferation of NPCs (Figures 2D and 2E; p < 0.001). Conversely, inhibition of miR-135a, upon transduction with a virus expressing a sponge (loss of function, see also Figure 3A), led to a significant increase in NPC proliferation (Figures 2D and 2E). These results indicate an anti-proliferative function of miR-135a in adult hippocampal NPCs in vitro.
Figure 3.
Validation of Lentiviruses to Overexpress/Downregulate miR-135a in NPCs In Vitro and In Vivo
(A) Relative expression levels of mature miR-135a in primary NPCs transduced in vitro with lentivirus transcribing the immature short-hairpin precursor of miR-135a (sh-miR-135a, i.e., gain of function), or a sponge for miR-135a (sponge miR-135a, i.e., loss of function), or control viruses expressing scrambled RNAs.
(B) Representative micrographs showing immunostaining for Nestin-CFPnuc (green), BrdU-positive cells (red), and GFAP-positive cells (white), and nuclear DNA with DAPI (blue) in the hippocampal SGZ of 6- to 8-week-old Nestin-CFPnuc mice, injected with lentiviruses (same used in A), kept 10 days under standard conditions and subjected to three injections of BrdU 24 h before sacrifice.
(C) Representative micrographs showing immunostaining for Nestin-CFPnuc (green), BrdU-positive cells (red) and nuclear DNA with DAPI (blue) in the hippocampal SGZ of 6- to 8-week-old Nestin-CFPnuc mice, injected with miR-135a sponge or scrambled-sponge lentivirus, kept 10 days under standard conditions, and subjected to three injections of BrdU 24 h before sacrifice.
(D) Percentage of BrdU and Nestin-CFPnuc double-positive cells over total BrdU+ cells in the SGZ of mice injected with lentiviruses.
Data are expressed as means ± SEM, n = 7 mice per group. One-way ANOVA Bonferroni as post hoc. ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 50 μm (B) and 25 μm (C).
To confirm this result in vivo, we transduced lentiviruses expressing either a short-hairpin precursor of miR-135a (sh-miR-135, gain of function), or a sponge for miR-135a, or scrambled control RNA sequences in NPC cultures (Figure 3A). Expression of miR-135a in NPCs was higher upon transduction with sh-miR-135a and significantly reduced upon transduction with sponge for miR-135a compared with controls (Figure 3A). We injected the miR-135a sponge or a scrambled control lentivirus in the DG of 6- to 8-week-old Nestin-CFPnuc mice housed under standard (resting) conditions. Ten days after injection, we found a higher percentage of Nestin-CFPnuc+ NPCs upon miR-135 inhibition compared with mice injected with the scrambled control using flow cytometry (sponge 1.6%, Figure S2B; control 1.2%, Figure S2A). In another set of experiments (Figures 3, 4, and 5), we administered BrdU (three injections every 2 h) 10 days after virus injection and killed the mice 24 h after the first BrdU administration. We found in both control and miR-135a sponge-injected mice that >90% of the BrdU-positive cells also expressed Nestin-CFPnuc (Figures 3B–3D) and glial fibrillary acidic protein (GFAP) (Figure 3B), indicating that the majority of BrdU-positive cells in the SGZ of these mice were bona fide NPCs.
Figure 4.
miR-135a Mediates Running-Induced Proliferation in the Hippocampal SGZ In Vivo
(A) Schematic representation of the experiment.
(B) Representative micrographs showing BrdU (green), Ki67 (red), or double-positive cells (yellow arrowheads) in the hippocampal SGZ of 6- to 8-week-old C57BL/6 mice, injected with scrambled, sponge miR-135a, or sh-miR-135a virus under standard or running conditions for 10 days and subjected to three injections of BrdU 24 h before sacrifice. White arrowheads, BrdU+ Ki67− cells; yellow arrowheads, BrdU+ Ki67+ cells.
(C) Number of BrdU-positive cells per DG volume (μm3).
(D) Percentage of BrdU+Ki67− over total BrdU+ cells as a measure of cell-cycle exit.
Data are expressed as means ± SEM, n = 6 mice per group. One-way ANOVA Bonferroni as post hoc. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 50 μm.
Figure 5.
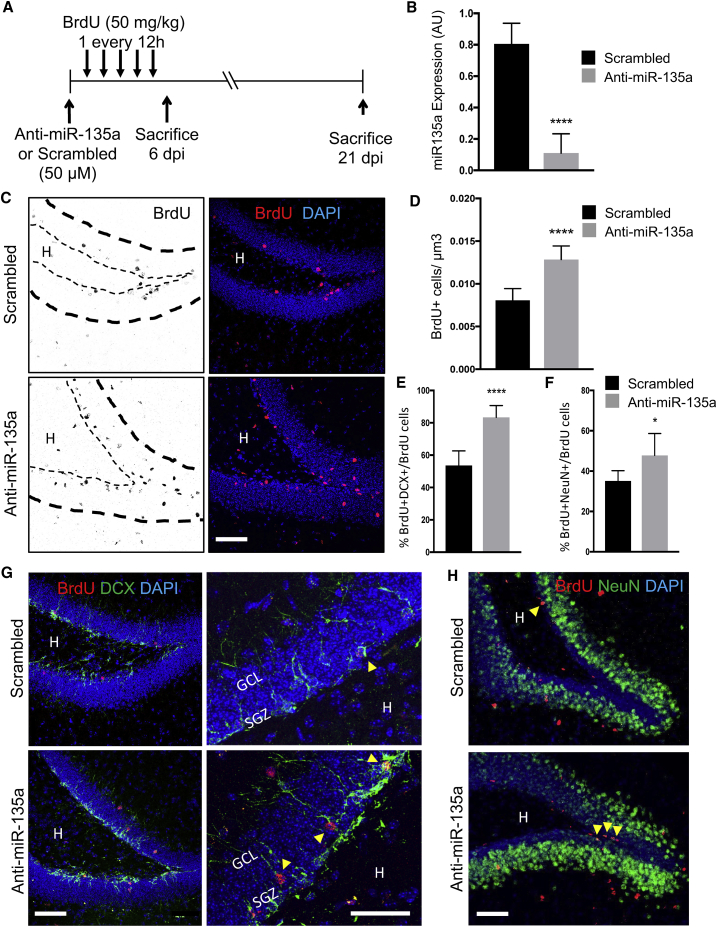
Transient miR-135a Inhibition Stimulates Hippocampal Neurogenesis In Vivo
(A) Schematic representation of the experiment.
(B) Relative expression levels of mature miR-135a in hippocampal DG of mice injected with control scrambled or anti-miR-135a 48 h after the injection.
(C and D) (C) Representative micrographs showing BrdU (black or red) cells 6 days after injection of control (scrambled) or anti-miR-135a and (D) the number of BrdU+ NPCs per volume (μm3) in the hippocampal SGZ of 6-week-old C57BL/6 mice.
(E and F) Percentage of BrdU+DCX+ (E), BrdU+NeuN+ (F) over total BrdU+ cells in the hippocampal SGZ of 6-week-old C57BL/6 mice 3 weeks after the injection with scrambled or anti-miR-135a antagomiRs.
(G and H) Representative micrographs showing staining for BrdU (red), DCX (G, green) or NeuN (H, green); and nuclear DNA with DAPI (blue); arrowheads indicate double-positive cells. H, hilus, GCL, granular cell layer; SGZ, subgranular zone.
Data are expressed as means ± SEM, n = 7 mice per group. One-way ANOVA Bonferroni as post hoc. ∗p < 0.05, ∗∗∗∗p < 0.0001. Scale bars, 50 μm and 25 μm (G, high magnification).
Next, we assessed the effect of miR-135a manipulation on NPC proliferation in vivo. We injected viruses expressing sh-miR-135, sponge, or scrambled controls into the DG of 8-week-old C57BL/6 mice and placed them in standard cages or in cages equipped with a running wheel for 10 days, followed by BrdU administration (Figure 4A). In resting mice, miR-135a inhibition led to a significant increase in the number of BrdU-positive cells in the SGZ compared with controls (Figures 4B and 4C; p < 0.01). In contrast, no significant differences in the number of BrdU-positive cells upon overexpression of miR-135a were observed (Figures 4B and 4C). The latter result could be explained by saturation of the system due to the high expression levels of the endogenous miR-135a in NPCs (Figure 1). Importantly, we found that overexpression of miR-135a prevented running-induced NPC proliferation in the SGZ (Figures 4B and 4C; p < 0.05). To corroborate these results, we analyzed the proportion of cells exiting the cell cycle upon miR-135a manipulation in resting and running mice by quantifying BrdU-positive cells that were negative for Ki67 in the SGZ (Figures 4B–4D). Consistent with the antiproliferative function of miR-135a, we found a significant decrease in cell-cycle exit upon injection with the miR-135a sponge in the DG of mice housed under standard conditions (Figure 4D; p < 0.05). In contrast, overexpression of miR-135a significantly increased cell-cycle exit in running mice (Figure 4D; p < 0.01). These results indicate an antiproliferative function of miR-135a in NPCs and that its downregulation is necessary for the running-induced proliferation in the SGZ of adult mice.
Transient miR-135a Inhibition Stimulates Hippocampal Neurogenesis, but Not Astrogliogenesis In Vivo
Next, we asked whether an increased proportion of proliferating NPCs, upon miR-135a inhibition, would also increase neurogenesis (Figure 5). Since constitutive inhibition of miR-135a prevents neuronal differentiation of NPCs (Pons-Espinal et al., 2017), we used synthetic “antagomiRs” to transiently inhibit miR-135a (anti-miR-135a) or scrambled control inhibitors in DG of resting mice and followed the fate of NPCs with BrdU (Figure 5A). As expected, injection of anti-miR-135a dramatically reduced endogenous miR-135a levels compared with control mice (Figure 5B; p < 0.0001) and increased NPC proliferation, as indicated by a higher number of BrdU+ cells in SGZ of mice (Figures 5C and 5D; p < 0.0001). Next, to evaluate the fate of NPCs, we quantified the proportion of cells co-expressing BrdU and the immature neuronal marker doublecortin (DCX) or the postmitotic neuronal marker NeuN in the DG 3 weeks after antagomiRs injection. Interestingly, we found that inhibition of miR-135a increased the proportions of BrdU+DCX+ (Figures 5E and 5G; p < 0.0001) and BrdU+NeuN+ neurons compared with control mice (Figures 5F and 5H; p < 0.05). This result indicated that increased NPC proliferation, upon transient inhibition of miR-135a, leads to enhanced neurogenesis, thus phenocopying running (van Praag et al., 1999). In contrast, miR-135a inhibition did not alter the proportion of BrdU+ cells expressing astrocyte markers such as GFAP (Figures S3A and S3B), glutamate transporter GLT-1 (i.e., solute carrier family 1 member 2 [SLC1A2]) (Figures S3C and S3D), or glutamate aspartate transporter (GLAST) (i.e., solute carrier family 1 glial high-affinity glutamate transporter member 3 [SLC1A3]) (Figures S3E and S3F). The latter result is consistent with our previous finding that adult hippocampal NPCs can undergo astrogliogenesis in absence of miRNAs (Pons-Espinal et al., 2017).
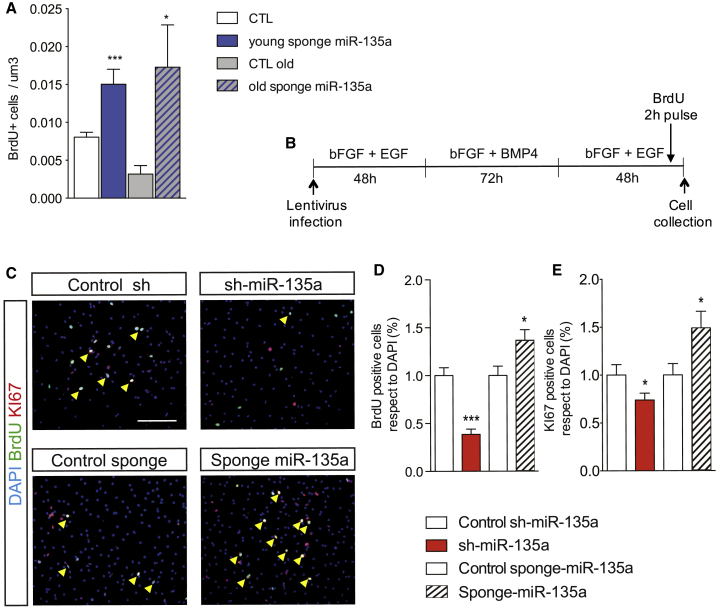
Inhibition of miR-135a Re-activates Proliferation in the Hippocampal SGZ of Aged Mice and Stimulates Cell-Cycle Re-entry of Quiescent NPCs
The age-associated reduction in adult neurogenesis has been attributed to exhaustion of the NPC pools and/or to increased NPC quiescence (Encinas et al., 2011, Jaskelioff et al., 2011, Lugert et al., 2010, Seib et al., 2013). Environmental stimuli have been shown to counteract the age-associated loss of adult neurogenesis in rodents, suggesting that cell-cycle arrest of aged NPCs is reversible, at least to some extent. To verify whether miR-135a inhibition could restore NPC proliferation in the SGZ of aged mice, we injected a sponge for miR-135a in the DG of 8-week-old (young) or 90-week-old (aged) C57BL/6 mice housed under standard conditions, followed by BrdU administration (as in Figures 3 and 4). Remarkably, inhibition of miR-135a significantly increased the number of BrdU-positive cells in the SGZ of both aged and young mice compared with control mice (Figure 6A). This result indicates that inhibition of miR-135a is sufficient to restore NPC proliferation in the hippocampal SGZ of aged mice, opening the possibility that it might occur through cell-cycle re-entry of quiescent NPC pools. To ascertain this possibility, we overexpressed or inhibited miR-135a in cultured NPCs (Figures 6B–6E). To simulate “aged” adult neurogenesis, we first cultured NPCs in quiescence medium for 72 h, followed by 48 h in fresh proliferative medium (Figure 6B) and measured their capacity to re-enter into proliferative state (Figures 6C–6E) by quantifying BrdU (2 h pulse, Figures 6C and 6D) or Ki67 (Figures 6C–6E). Overexpression of miR-135a impaired NPC proliferation re-entry, as shown by the lower percentage of BrdU and Ki67 positive NPCs (Figures 6C–6E) compared with control conditions (Figures 6C–6E; p < 0.001, normalized to the short-hairpin control). Interestingly, inhibition of miR-135a stimulated re-entry of NPCs into a proliferative state, as shown by a higher percentage of BrdU and Ki67 positive NPCs compared with the scrambled control (Figures 6C–6E; p < 0.05, normalized to the scrambled sponge). Cell-cycle analysis using propidium iodide and fluorescence-activated cell sorting (FACS) revealed that inhibition of miR-135a significantly increased the proportion of cells in the S phase (10.24% versus 20.63%, p < 0.05) and G2/M phase (9.63% versus 12.61%; Figures S4A and S4B) at the expense of the G1/G0 phase (80.12% versus 66.55%; Figures S4A and S4B; p < 0.01) compared with scrambled control NPCs. Moreover, inhibition of miR-135a in NPCs led to increased mRNA expression of the proliferative markers Ki67, Mcm2, and cyclins A and cyclin E (Figure S4C). Together, these results indicate that inhibition of miR-135a is sufficient to reactivate NPC proliferation in the SGZ of aged mice, suggesting this might occur by stimulating quiescent NPC re-entry into the proliferative state.
Figure 6.
Inhibition of miR-135a Re-activates Proliferation in the Hippocampal SGZ of Aged Mice and Stimulates Cell-Cycle Re-entry of Quiescent NPCs
(A) Number of BrdU+ NPCs per DG volume (μm3) in the hippocampal SGZ of 8-week-old (young) and 90-week-old (aged) C57BL/6 mice, injected with lentiviral sponge for miR-135a (loss of function), housed for 10 days under standard conditions, and subjected to BrdU administration (three injections every 2 h) 24 h before sacrifice.
(B) Schematic representation of the in vitro experiment. Primary hippocampal NPCs were allowed to re-enter the cell cycle after 72 h in quiescence medium and fixed 2 h after BrdU administration.
(C) Representative micrographs showing BrdU (green) and Ki67 (red) double-positive cells (yellow, arrowheads) of hippocampal NPCs infected with control sh-scrambled RNA, sh-miR-135a (gain of function), control sponge, or sponge miR-135a lentiviruses.
(D) Percentage of BrdU+ cells relative to total cells (DAPI) normalized to controls.
(E) Percentage of Ki67-positive cells relative to total cells (DAPI) normalized to controls.
Data are expressed as means ± SEM, n = 3 independent experiments containing three replicates. One-way ANOVA Bonferroni as post hoc. ∗p < 0.05, ∗∗∗p < 0.001. Scale bars, 50 μm.
Phosphatidylinositol Signaling Proteins Are Modulated by miR-135a in NPCs
To dissect the underlying mechanism of miR-135a in adult hippocampal NPCs, we manipulated its levels in vitro and performed shotgun label-free proteomic analysis. Overexpression of miR-135a upregulated 431 proteins (threshold >1.5-fold; Table S2) and downregulated 101 proteins (threshold <0.5-fold; Figure 7A and Table S2); while miR-135a inhibition upregulated 326 proteins (>1.5-fold; Figure 7A and Table S3) and downregulated 109 (<0.5-fold; Table S3). Gene Ontology (GO) analysis with DAVID software (Huang et al., 2009) indicated that the biological functions affected by miR-135a in NPCs were protein transport (GO: 001503; p value sh-miR-135a = 5.59 × 10−6; p value miR-135 sponge = 3.29 × 10−4); vesicle-mediated transport (GO: 0016192; p value sh-miR-135 = 5.34 × 10−5; p value miR-135 sponge = 2.10 × 10−3); transport (GO: 0006810; p value sh-miR-135 = 9.65 × 10−3; p value miR-135 sponge = 5.10 × 10−3), and nervous system development (GO: 0007399; p value sh-miR-135 = 9.79 × 10−3; p value miR-135 sponge = 2.81 × 10−2) (Tables S2 and S3).
Figure 7.
Phosphatidylinositol Signaling Proteins Are Modulated by miR-135a in NPCs
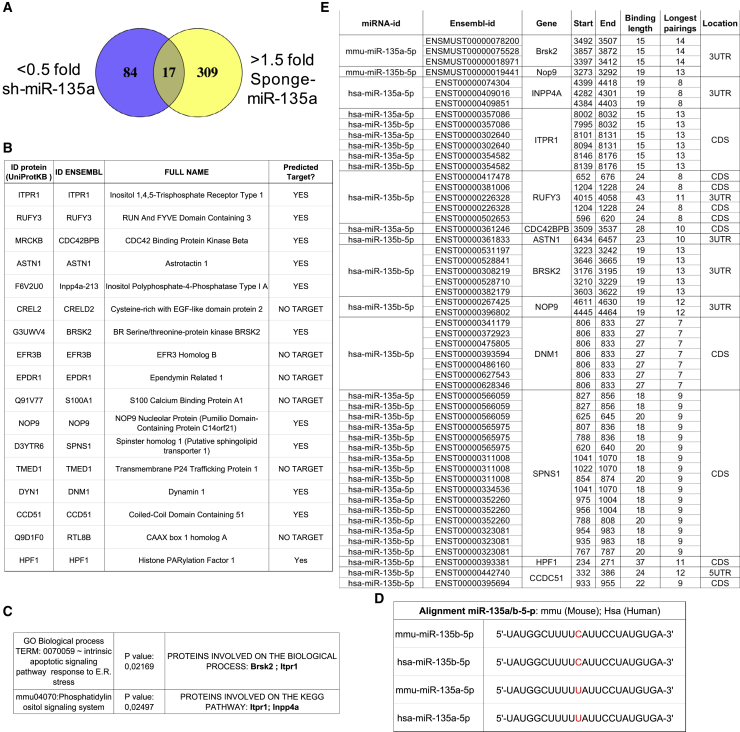
(A and B) Venn diagram indicating the number of downregulated proteins (purple, <0.5-fold) upon overexpression of miR-135a (sh-miR-135a), or upregulated proteins (yellow, >1.5-fold) upon inhibition of miR-135a (Sponge-miR-135a), and the 17 differently expressed proteins (table in B) found in both datasets, in cultured primary hippocampal NPCs.
(C) In silico GO analysis and KEGG pathway analysis.
(D) Alignment of mouse and human miR-135a-5p and miR-135b-5p.
(E) Predicted targets of miR135 according to MiRWalk. Position and length of predicted target sites of miR-135a-5p and miR-135b-5p are shown for each transcript (Ensembl ID).
miRNAs are mostly post-transcriptional repressors. Hence, to identify potential miR-135a targets, we focused on the downregulated proteins upon miR-135a overexpression and compared them with those upregulated upon miR-135a inhibition in NPCs. Seventeen proteins were consistently modulated by miR-135a levels in both datasets (Figures 7A and 7B). These proteins are involved in an intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress (GO: 0070059; p = 0.0021, Figure 7C) and a phosphatidylinositol signaling system (mmu04070, p value 0.0249; Figure 7C). The proteins predicted to be involved in these processes were inositol 1,4,5-trisphosphate (IP3) receptor 1 (ITPR1) and BR serine threonine kinase (BRSK2), ITPR1 and inositol polyphosphate-4-phosphatase, type I (INPP4A), respectively (Figure 7C). Interestingly, the human INPP4A transcript is a validated target of miR-135a-5p (DIANA-Tarbase v8.0; Karagkouni et al., 2018) in the brain cortex (Boudreau et al., 2014) and encodes for an enzyme involved in phosphatidylinositol metabolism. Remarkably, 11 of the 17 hits (64%) are predicted targets of miR-135 (miR-Walk 3.0; Dweep and Gretz, 2015; both miR-135a-5p and miR-135b-5p, collectively defined as miR-135, were considered in the analysis as they share identical seed regions that are conserved in mouse and human) (Figures 7D and 7E), suggesting these proteins could be directly modulated by miR-135. As a control, we repeated the same analysis for the proteins upregulated (>1.5-fold; Table S2) upon miR-135a overexpression and downregulated upon miR-135a inhibition (<0.5-fold; Table S3). However, only 4 (17%) of the 23 proteins consistently modulated by miR-135a in both datasets were predicted targets of either human or mouse miR-135 (not shown), suggesting these proteins are likely indirectly modulated by miR-135.
Discussion
In this study, we identify miR-135a as the first noncoding RNA essential modulator of the brain response to physical exercise. We report that overexpression of miR-135a in the DG prevents running-induced NPC proliferation. On the other hand, miR-135a inhibition stimulates NPC proliferation leading to increased neurogenesis, but not astrogliogenesis, in DG of resting mice. Remarkably, miR-135a inhibition re-activates NPC proliferation in DG of aged mice, likely by stimulating quiescent NPC pools to re-enter the cell cycle.
Several studies reported altered hippocampal miRNA expression in response to physical exercise (Bao et al., 2014, Cosín-Tomás et al., 2014, Hu et al., 2015, Pan-Vazquez et al., 2015) or pathological conditions (Encinas and Fitzsimons, 2017). However, to our knowledge, this is the first study reporting a functional role of an miRNA underlying the exercise-mediated increase in adult neurogenesis. Functions of the two members of the miR-135 family are poorly described in the mammalian CNS. In postmitotic neurons, miR-135 regulates axon growth/regeneration and mediates long-term depression (Hu et al., 2014, van Battum et al., 2017). miR-135a expression is high in the amygdala of stressed mice (Mannironi et al., 2013), and in the mouse raphe nucleus (functionally connected to the hippocampus) it is a key regulator of serotoninergic networks and antidepressants action (Issler et al., 2014). However, since miR-135 association with depression- and anxiety-related phenotypes in patients is very variable (Zurawek et al., 2017), its role in the pathological mechanism of these diseases remains unclear. In the adult mouse hippocampus, miR-135 is rapidly upregulated after prolonged kainic acid-induced seizures (Schouten et al., 2015). Moreover, we recently found that miR-135a is one of the 11 miRNAs required and sufficient to sustain the neurogenic lineage fate of NPCs (Pons-Espinal et al., 2017). Hence, miR-135 is required to regulate multiple aspects of adult hippocampal neurogenesis, suggesting that further studies on miR-135 in the brain response to physiological and pathological conditions are warranted.
Physical exercise is a potent trigger of adult hippocampal neurogenesis in both young and aged mice, but cellular and molecular mechanisms underlying this phenomenon remain controversial. Cellular mechanisms include recruitment of quiescent neural stem cells, acceleration of the cell cycle of NPCs, increased number of cell divisions, and reduction of cell death (Overall et al., 2016). At the molecular level, physical exercise has been shown to increase levels of growth factors BDNF, IGF, FGF-2, and VEGF, leading to the activation of MAPK/ERK and PI3K-Akt signaling pathways (Cotman at al., 2007). We report that INPP4A, a key enzyme for phosphatidylinositol metabolism and known target of miR-135 in the cortex (Boudreau et al., 2014), is one of the top proteins modulated by miR-135 in NPCs. ITPR1, another differentially expressed protein identified in our analysis, is also a key player in IP3 signaling. Hence, phosphatidylinositol signaling could represent a prominent constraint to NPC proliferative potential. This hypothesis is consistent with previous studies indicating that the PI3K-Akt signaling pathway is activated by exercise in rodents (Bruel-Jungerman et al., 2009, Chen and Russo-Neustadt, 2005). However, while these studies concluded that the PI3K-Akt pathway primarily mediates the effect of exercise on the survival of newly generated DG neurons and the associated increase in synaptic plasticity, our results suggest that miR-135/phosphatidylinositol signaling could mediate exercise-induced proliferation of NPCs. Together, this evidence opens the possibility that the miR-135-IP3-axis might represent a novel target of therapeutic intervention to stimulate adult neurogenesis.
One unanswered question arising from our study is how running decreases miR-135 levels in adult NPCs. miR-135 is a tumor suppressor (Cheng et al., 2017, Xu et al., 2016), which is downregulated in several cancers (Guo et al., 2018, Mao et al., 2015, Pei et al., 2015, Wu et al., 2012, Zubieta et al., 2017). A Wnt/miR-135a auto-regulatory loop has been identified in brain development, which could modulate differentiation of forebrain (Caronia-Brown et al., 2016) and dopaminergic (Anderegg et al., 2013, De Gregorio et al., 2018) neurons. Moreover, Wnt/TGFβ/BMP pathways are known to influence long-term maintenance of NPC pools in the adult hippocampus, age-associated cognitive decline, and brain dysfunctions (Inestrosa and Arenas, 2010, Urban and Guillemot, 2014). This evidence provides possible mechanisms to explain the rescue of proliferation in the hippocampal stem cell niche of aged mice upon miR-135 downregulation.
In summary, we propose that the therapeutic exploitation of miR-135 might offer intriguing perspectives to delay or prevent pathological brain aging.
Experimental Procedures
Animal Model
Mice (C57BL/6J, Jackson Lab no. 000664; Td-Tomatoflox/wt knockin reporter mice, Jackson Lab no. 007908; Madisen et al., 2010; Nestin-CFPnuc, Encinas et al., 2006) were housed under standard laboratory conditions at Istituto Italiano di Tecnologia (IIT), the Center for Regenerative Therapies Dresden (CRTD), or the Medizinische-Theoretisches Zentrum (MTZ) Dresden. Experiments were approved by the Italian and German authorities (permit nos. 056/2013, 214/2015-PR, and 24-9168.11-1/2013-15) and conducted in accordance with the Guide for the Care and Use of Laboratory Animals of the European Community Directives. Mice were maintained under a 12 h light/dark cycle with food and water ad libitum.
NPC Culture
Hippocampal NPCs were prepared as previously published (Babu et al., 2011) from ten 6-week-old C57BL/6J mice and cultured as previously described (Pons-Espinal et al., 2017). For quiescence, 1.2 × 104 cells/cm2 were plated into normal proliferation medium; after 16 h, quiescence medium (50 ng/mL BMP4 [R&D Systems] and 20 ng/mL bFGF; Martynoga et al., 2013) was added.
Running
Nine-week-old Nestin-CFPnuc mice or WT C57BL/6J were double housed under standard conditions or in cages equipped with a running wheel (TSE System, animal facility of CRTD; or ENV-044 [Med Associates], Animal facility of IIT) for 10 days before sacrifice. Twenty-four hours before sacrifice, mice received one administration of 50 mg/kg BrdU (B9285, Sigma).
miRNA Manipulation
Mimics and antagomiRs used are listed in Supplemental Experimental Procedures. Scrambled miRNA mimics (50 nM; QIAGEN) or miRNA antagomiRs (150 nM; QIAGEN) were nucleofected (Amaxa) in proliferating or quiescent NPC cultures. Then, 48 h after nucleofection, 10 μM BrdU was added to the medium for 2 h, followed by fixation (4% PFA) or RNA/protein extraction. Virus and synthetic oligos were stereotaxically injected in the hippocampus (coordinates: −2.0 A/P, ±1.6 M/L, and −1.9 to −2.1 D/V relative to bregma [mm]) as previously described (Pons-Espinal et al., 2017). Codes for viruses and oligos are listed in Supplemental Experimental Procedures. After virus injection, mice received three BrdU intraperitoneal injections per day (100 mg/kg, every 2 h) and sacrificed 24 h later. After injection of oligos, mice received two BrdU intraperitoneal injections per day (50 mg/kg) for 5 days and sacrificed 2 h (6 days post injection of oligos) or 2 weeks after the last BrdU injection (21 days post injection of oligos).
Immunofluorescence and Cell Quantification
Immunofluorescence staining on brain slices (40 μm) was performed in one of every six sections of the hippocampus. A list of primary antibodies and the detailed protocol is provided in the Supplemental Experimental Procedures. Confocal stack images of brain slices were obtained with the Confocal A1 Nikon Inverted SFC with 40× objective and the Zeiss Spinning Disc with a 20× objective. Cell quantification and analysis were performed using NIS-Elements software (Nikon) and ZenBlue (Zeiss). Immunofluorescence on cell cultures was performed as previously described (Pons-Espinal et al., 2017). Images were obtained using a Nikon Eclipse microscope at 20× or 40× magnification, and quantification was performed using a cell-counter plugin in Fiji (Fiji is just ImageJ; Schindelin et al., 2012).
FACS and RNA Analysis
For RNA extraction and cDNA preparation, 6–10 Nestin-CFPnuc mice per condition were euthanized at the indicated times. Cells were dissociated with the Neural Tissue Dissociation Kit P (Miltenyi), FACS sorted, and immediately processed for RNA extraction (Walker et al., 2016). Total RNA was extracted with QIAzol (QIAGEN), RNA was purified with an RNeasy Kit, or miRNeasy Kit (QIAGEN) following the manufacturer's instructions. Quantification of RNA was performed as in Pons-Espinal et al. (2017).
In Situ Hybridization
In situ hybridization was performed as previously published (De Pietri Tonelli et al., 2014) with minor modifications. Brain slices (18–20 μm) were permeabilized and post-fixed in 4% PFA. Slides were blocked with 0.25% acetic anhydride (Sigma), and a pre-hybridization solution was added, followed by incubation with a hybridization solution containing 160 nM (miR-135, -190, -203) or 100 nM (miR-9) of the DIG-labeled LNA probe (Exiqon) overnight. The anti-DIG antibody (Roche; 1:2000) was incubated overnight at 4°C. For development of the color reactions, two different alkaline phosphatase substrates were used: NBT/BCIP (Roche) or Fast Red TR/Naphthol AS-MX solution (Sigma), following the manufacturers' instructions. Sections were mounted and imaged using conventional bright-field microscopy or a confocal microscope with the Cy-3 filter.
Proteomics
NPCs (three independent experiments) were lysed with RIPA buffer, and 50 μg of proteins was collected from all the samples and processed as previously described (Braccia et al., 2018). Protein pools were processed for liquid chromatography-tandem mass spectrometry analysis.
Statistical Analysis
Data are presented as means ± SEM and were analyzed using Prism 6 (GraphPad). Statistical details of the experiments can be found in the Results and figure legends. Statistical significance was assessed with a two-tailed unpaired t test for two experimental groups. For experiments with three or more groups, one-way ANOVA with Bonferroni's multiple comparison test post hoc was used. Mean differences were considered to be statistically significant when p < 0.05.
Author Contributions
M.P.E., C.G., and D.D.P.T. co-designed the experiments. M.P.E. and C.G. performed most of the experiments, analyzed the data, prepared the figures, and drafted the manuscript. T.L.W. and A.P. carried out experiments and analyzed the data of Figures 1A–1D and S2 under the supervision of K.F. and G.K. M.J.M. performed quantification and analysis of miRNA expression (Figures 1E and S1 and Table S1) under the supervision of F.N. C.B. prepared the SWATH library and performed mass spectrometry under the supervision of A.A. D.D.P.T. conceived and coordinated the project and wrote the manuscript. All authors edited and approved the final version of the manuscript.
Acknowledgments
We are grateful to Dr. G. Enikolopov (Stony Brook University, New York) for kindly providing the Nestin-CFPnuc mouse line. We thank R. Pelizzoli for generation of the viruses and help with NPC cultures and western blot, the technical staff of IIT (M. Pesce, imaging; E. Albanesi, FACS facility) and of DZNE (N Rund, FACS; and P. Oloth, histology of Figures 1B and 1C) for excellent assistance. Images for Figure 1D were acquired and processed using equipment of the DZNE-Imaging facility. We thank the staff of IIT Animal Facility and M. Morini for excellent assistance with animal experiments. This study was supported by intramural funds of Fondazione Istituto Italiano di Tecnologia (IIT) and partly by Fondazione Cariplo grant no. 2015-0590 to D.D.P.T. and F.N. and AIRC IG 2017 ID 20106 to D.D.P.T.
Published: May 23, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.04.020.
Accession Numbers
All the RAW data files used for protein quantification acquired for the present work are freely available through the ProteomeExchange database (Vizcaíno et al., 2014, Deutsch et al., 2017) with identifier ProteomeXchange: PXD009845.
Supplemental Information
References
- Anderegg A., Lin H.P., Chen J.A., Caronia-Brown G., Cherepanova N., Yun B., Joksimovic M., Rock J., Harfe B.D., Johnson R. An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool. PLoS Genet. 2013;9:e1003973. doi: 10.1371/journal.pgen.1003973. [DOI] [PMC free article] [PubMed] [Google Scholar]; Anderegg, A., Lin, H.P., Chen, J.A., Caronia-Brown, G., Cherepanova, N., Yun, B., Joksimovic, M., Rock, J., Harfe, B.D., Johnson, R., et al. (2013). An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool. PLoS Genet. 9, e1003973. [DOI] [PMC free article] [PubMed]
- Babu H., Claasen J.H., Kannan S., Rünker A.E., Palmer T., Kempermann G. A protocol for isolation and enriched monolayer cultivation of neural precursor cells from mouse dentate gyrus. Front. Neurosci. 2011;5:89. doi: 10.3389/fnins.2011.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]; Babu, H., Claasen, J.H., Kannan, S., Runker, A.E., Palmer, T., and Kempermann, G. (2011). A protocol for isolation and enriched monolayer cultivation of neural precursor cells from mouse dentate gyrus. Front. Neurosci. 5, 89. [DOI] [PMC free article] [PubMed]
- Bao T.H., Miao W., Han J.H., Yin M., Yan Y., Wang W.W., Zhu Y.H. Spontaneous running wheel improves cognitive functions of mouse associated with miRNA expressional alteration in hippocampus following traumatic brain injury. J. Mol. Neurosci. 2014;54:622–629. doi: 10.1007/s12031-014-0344-1. [DOI] [PubMed] [Google Scholar]; Bao TH, Miao W, Han JH, Yin M, Yan Y, Wang WW, Zhu YH(2014). Spontaneous running wheel improves cognitive functions of mouse associated with mirna expressional alteration in hippocampus following traumatic brain injury. J. Mol. Neurosci. 54, 622-629. [DOI] [PubMed]
- Barca-Mayo O., De Pietri Tonelli D. Convergent microRNA actions coordinate neocortical development. Cell. Mol. Life Sci. 2014;71:2975–2995. doi: 10.1007/s00018-014-1576-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Barca-Mayo, O., and De Pietri Tonelli, D. (2014). Convergent microRNA actions coordinate neocortical development. Cell. Mol. Life Sci. 71, 2975-2995. [DOI] [PMC free article] [PubMed]
- Bond A.M., Ming G.L., Song H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell. 2015;17:385–395. doi: 10.1016/j.stem.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bond, A.M., Ming, G.L., and Song, H. (2015). Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell 17, 385-395. [DOI] [PMC free article] [PubMed]
- Boudreau R.L., Jiang P., Gilmore B.L., Spengler R.M., Tirabassi R., Nelson J.A., Ross C.A., Xing Y., Davidson B.L. Transcriptome-wide discovery of microRNA binding sites in Human Brain. Neuron. 2014;81:294–305. doi: 10.1016/j.neuron.2013.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]; Boudreau, R.L., Jiang, P., Gilmore, B.L., Spengler, R.M., Tirabassi, R., Nelson, J.A., Ross, C.A., Xing, Y., and Davidson, B.L. (2014). Transcriptome-wide discovery of microRNA binding sites in Human Brain. Neuron 81, 294-305. [DOI] [PMC free article] [PubMed]
- Braccia C., Espinal M.P., Pini M., De Pietri Tonelli D., Armirotti A. A new SWATH ion library for mouse adult hippocampal neural stem cells. Data Brief. 2018;18:1–8. doi: 10.1016/j.dib.2018.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]; Braccia, C., Espinal, M.P., Pini, M., De Pietri Tonelli, D., and Armirotti, A. (2018). A new SWATH ion library for mouse adult hippocampal neural stem cells. Data Brief 18, 1-8. [DOI] [PMC free article] [PubMed]
- Bruel-Jungerman E., Veyrac A., Dufour F., Horwood J., Laroche S., Davis S. Inhibition of PI3K-Akt signaling blocks exercise-mediated enhancement of adult neurogenesis and synaptic plasticity in the dentate gyrus. PLoS One. 2009;4:e7901. doi: 10.1371/journal.pone.0007901. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bruel-Jungerman, E., Veyrac, A., Dufour, F., Horwood, J., Laroche, S., and Davis, S. (2009). Inhibition of PI3K-Akt signaling blocks exercise-mediated enhancement of adult neurogenesis and synaptic plasticity in the dentate gyrus. PLoS One 4, e7901. [DOI] [PMC free article] [PubMed]
- Bruno S., Darzynkiewicz Z. Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Prolif. 1992;25:31–40. doi: 10.1111/j.1365-2184.1992.tb01435.x. [DOI] [PubMed] [Google Scholar]; Bruno, S., and Darzynkiewicz, Z. (1992). Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Prolif.. 25, 31-40. [DOI] [PubMed]
- Caronia-Brown G., Anderegg A., Awatramani R. Expression and functional analysis of the Wnt/beta-catenin induced mir-135a-2 locus in embryonic forebrain development. Neural Dev. 2016;11:9. doi: 10.1186/s13064-016-0065-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Caronia-Brown, G., Anderegg, A., and Awatramani, R. (2016). Expression and functional analysis of the Wnt/beta-catenin induced mir-135a-2 locus in embryonic forebrain development. Neural Dev. 11, 9. [DOI] [PMC free article] [PubMed]
- Chen M.J., Russo-Neustadt A.A. Exercise activates the phosphatidylinositol 3-kinase pathway. Mol. Brain Res. 2005;135:181–193. doi: 10.1016/j.molbrainres.2004.12.001. [DOI] [PubMed] [Google Scholar]; Chen, M.J., and Russo-Neustadt, A.A. (2005). Exercise activates the phosphatidylinositol 3-kinase pathway. Mol. Brain Res. 135, 181-193. [DOI] [PubMed]
- Cheng Z., Liu F., Zhang H., Li X., Li Y., Li J., Liu F., Cao Y., Cao L., Li F. miR-135a inhibits tumor metastasis and angiogenesis by targeting FAK pathway. Oncotarget. 2017;8:31153–31168. doi: 10.18632/oncotarget.16098. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cheng, Z., Liu, F., Zhang, H., Li, X., Li, Y., Li, J., Liu, F., Cao, Y., Cao, L., and Li, F. (2017). miR-135a inhibits tumor metastasis and angiogenesis by targeting FAK pathway. Oncotarget 8, 31153-31168. [DOI] [PMC free article] [PubMed]
- Cosín-Tomás M., Alvarez-López M.J., Sanchez-Roige S., Lalanza J.F., Bayod S., Sanfeliu C., Pallàs M., Escorihuela R.M., Kaliman P. Epigenetic alterations in hippocampus of SAMP8 senescent mice and modulation by voluntary physical exercise. Front. Aging Neurosci. 2014;6:51. doi: 10.3389/fnagi.2014.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cosin-Tomas, M., Alvarez-Lopez, M.J., Sanchez-Roige, S., Lalanza, J.F., Bayod, S., Sanfeliu, C., Pallas, M., Escorihuela, R.M., and Kaliman, P. (2014). Epigenetic alterations in hippocampus of SAMP8 senescent mice and modulation by voluntary physical exercise. Front. Aging Neurosci. 6, 51. [DOI] [PMC free article] [PubMed]
- Cotman C.W., Berchtold N.C., Christie L.A. Exercise builds brain health: key roles of growth factor cascades and inflammation. Trends Neurosci. 2007;30:464–472. doi: 10.1016/j.tins.2007.06.011. [DOI] [PubMed] [Google Scholar]; Cotman, C.W., Berchtold, N.C., and Christie, L.A. (2007). Exercise builds brain health: key roles of growth factor cascades and inflammation. Trends Neurosci. 30, 464-472. [DOI] [PubMed]
- De Gregorio R., Pulcrano S., De Sanctis C., Volpicelli F., Guatteo E., von Oerthel L., Latagliata E.C., Esposito R., Piscitelli R.M., Perrone-Capano C. miR-34b/c regulates Wnt1 and enhances mesencephalic dopaminergic neuron differentiation. Stem Cell Reports. 2018;10:1237–1250. doi: 10.1016/j.stemcr.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; De Gregorio, R., Pulcrano, S., De Sanctis, C., Volpicelli, F., Guatteo, E., von Oerthel, L., Latagliata, E.C., Esposito, R., Piscitelli, R.M., Perrone-Capano, C., et al. (2018). miR-34b/c regulates Wnt1 and enhances mesencephalic dopaminergic neuron differentiation. Stem Cell Reports 10, 1237-1250. [DOI] [PMC free article] [PubMed]
- Dennis C.V., Suh L.S., Rodriguez M.L., Kril J.J., Sutherland G.T. Human adult neurogenesis across the ages: an immunohistochemical study. Neuropathol. Appl. Neurobiol. 2016;42:621–638. doi: 10.1111/nan.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dennis, C. V., Suh, L.S., Rodriguez, M.L., Kril, J.J., and Sutherland, G.T. (2016). Human adult neurogenesis across the ages: An immunohistochemical study. Neuropathol. Appl. Neurobiol. 42, 621-638. [DOI] [PMC free article] [PubMed]
- De Pietri Tonelli D., Clovis Y.M., Huttner W.B. Detection and monitoring of microRNA expression in developing mouse brain and fixed brain cryosections. Methods Mol. Biol. 2014;1092:31–42. doi: 10.1007/978-1-60327-292-6_3. [DOI] [PubMed] [Google Scholar]; De Pietri Tonelli, D., Clovis, Y.M., Huttner, W.B. (2014). Detection and monitoring of microRNA expression in developing mouse brain and fixed brain cryosections. Methods Mol. Biol. 1092, 31-42. [DOI] [PubMed]
- Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017;45:D1100–D1106. doi: 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]; Deutsch, E.W., Csordas, A., Sun, Z., Jarnuczak, A., Perez-Riverol, Y., Ternent, T., Campbell, D.S., Bernal-Llinares, M., Okuda, S., Kawano, S., et al. (2017). The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45, D1100-D1106. [DOI] [PMC free article] [PubMed]
- Dweep H., Gretz N. MiRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]; Dweep, H., and Gretz, N. (2015). MiRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 12, 697. [DOI] [PubMed]
- Encinas J.M., Vaahtokari A., Enikolopov G. Fluoxetine targets early progenitor cells in the adult brain. Proc. Natl. Acad. Sci. U S A. 2006;103:8233–8238. doi: 10.1073/pnas.0601992103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Encinas, J.M., Vaahtokari, A., and Enikolopov, G. (2006). Fluoxetine targets early progenitor cells in the adult brain. Proc. Natl. Acad. Sci. U S A 103, 8233-8238. [DOI] [PMC free article] [PubMed]
- Encinas J.M., Michurina T.V., Peunova N., Park J.H., Tordo J., Peterson D.A., Fishell G., Koulakov A., Enikolopov G. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8:566–579. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Encinas, J.M., Michurina, T. V., Peunova, N., Park, J.H., Tordo, J., Peterson, D.A., Fishell, G., Koulakov, A., and Enikolopov, G. (2011). Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell 8, 566-579. [DOI] [PMC free article] [PubMed]
- Encinas J.M., Fitzsimons C.P. Gene regulation in adult neural stem cells. Current challenges and possible applications. Adv. Drug Deliv. Rev. 2017;120:118–132. doi: 10.1016/j.addr.2017.07.016. [DOI] [PubMed] [Google Scholar]; Encinas, J.M., and Fitzsimons, C.P. (2017). Gene regulation in adult neural stem cells. Current challenges and possible applications. Adv. Drug Deliv. Rev. 120, 118-132. [DOI] [PubMed]
- Eriksson P.S., Perfilieva E., Björk-Eriksson T., Alborn A.-M., Nordborg C., Peterson D.A., Gage F.H. Neurogenesis in the adult human hippocampus. Nat. Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]; Eriksson, P.S., Perfilieva, E., Bjork-Eriksson, T., Alborn, A.-M., Nordborg, C., Peterson, D.A., and Gage, F.H. (1998). Neurogenesis in the adult human hippocampus. Nat. Med. 4, 1313-1317. [DOI] [PubMed]
- Farioli-Vecchioli S., Mattera A., Micheli L., Ceccarelli M., Leonardi L., Saraulli D., Costanzi M., Cestari V., Rouault J.P., Tirone F. Running rescues defective adult neurogenesis by shortening the length of the cell cycle of neural stem and progenitor cells. Stem Cells. 2014;32:1968–1982. doi: 10.1002/stem.1679. [DOI] [PubMed] [Google Scholar]; Farioli-Vecchioli, S., Mattera, A., Micheli, L., Ceccarelli, M., Leonardi, L., Saraulli, D., Costanzi, M., Cestari, V., Rouault, J.P., and Tirone, F. (2014). Running rescues defective adult neurogenesis by shortening the length of the cell cycle of neural stem and progenitor cells. Stem Cells 32, 1968-1982. [DOI] [PubMed]
- Fischer T.J., Walker T.L., Overall R.W., Brandt M.D., Kempermann G. Acute effects of wheel running on adult hippocampal precursor cells in mice are not caused by changes in cell cycle length or S phase length. Front. Neurosci. 2014;8:314. doi: 10.3389/fnins.2014.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fischer, T.J., Walker, T.L., Overall, R.W., Brandt, M.D., and Kempermann, G. (2014). Acute effects of wheel running on adult hippocampal precursor cells in mice are not caused by changes in cell cycle length or S phase length. Front. Neurosci. 8, 314. [DOI] [PMC free article] [PubMed]
- Guo L., Ding G., Xu W., Ge H., Jiang Y., Chen X., Lu Y. MiR-135a-5p represses proliferation of HNSCC by targeting HOXA10. Cancer Biol. Ther. 2018;19:1–28. doi: 10.1080/15384047.2018.1450112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Guo, L., Ding, G., Xu, W., Ge, H., Jiang, Y., Chen, X., and Lu, Y. (2018). MiR-135a-5p represses proliferation of HNSCC by targeting HOXA10. Cancer Biol. Ther. 1-28. [DOI] [PMC free article] [PubMed]
- Hu T., Zhou F.J., Chang Y.F., Li Y.S., Liu G.C., Hong Y., Chen H.L., Xiyang Y.B., Bao T.H. miR21 is associated with the cognitive improvement following voluntary running wheel exercise in TBI mice. J. Mol. Neurosci. 2015;57:114–122. doi: 10.1007/s12031-015-0584-8. [DOI] [PubMed] [Google Scholar]; Hu, T., Zhou, F.J., Chang, Y.F., Li, Y.S., Liu, G.C., Hong, Y., Chen, H.L., Xiyang, Y. Bin, and Bao, T.H. (2015). miR21 is associated with the cognitive improvement following voluntary running wheel exercise in TBI mice. J. Mol. Neurosci. 57, 114-122. [DOI] [PubMed]
- Hu Z., Yu D., Gu Q.H., Yang Y., Tu K., Zhu J., Li Z. MiR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat. Commun. 2014;5:3263. doi: 10.1038/ncomms4263. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hu, Z., Yu, D., Gu, Q.H., Yang, Y., Tu, K., Zhu, J., and Li, Z. (2014). MiR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat. Commun. 5, 3263. [DOI] [PMC free article] [PubMed]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]; Huang, D.W., Sherman, B.T., and Lempicki, R.A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57. [DOI] [PubMed]
- Inestrosa N.C., Arenas E. Emerging roles of Wnts in the adult nervous system. Nat. Rev. Neurosci. 2010;11:77–86. doi: 10.1038/nrn2755. [DOI] [PubMed] [Google Scholar]; Inestrosa, N.C., and Arenas, E. (2010). Emerging roles of Wnts in the adult nervous system. Nat. Rev. Neurosci. 11, 77-86. [DOI] [PubMed]
- Issler O., Haramati S., Paul E.D., Maeno H., Navon I., Zwang R., Gil S., Mayberg H.S., Dunlop B.W., Menke A. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron. 2014;83:344–360. doi: 10.1016/j.neuron.2014.05.042. [DOI] [PubMed] [Google Scholar]; Issler, O., Haramati, S., Paul, E.D., Maeno, H., Navon, I., Zwang, R., Gil, S., Mayberg, H.S., Dunlop, B.W., Menke, A., et al. (2014). MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 83, 344-360. [DOI] [PubMed]
- Jaskelioff M., Muller F.L., Paik J.H., Thomas E., Jiang S., Adams A.C., Sahin E., Kost-Alimova M., Protopopov A., Cadiñanos J. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature. 2011;469:102–107. doi: 10.1038/nature09603. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jaskelioff, M., Muller, F.L., Paik, J.H., Thomas, E., Jiang, S., Adams, A.C., Sahin, E., Kost-Alimova, M., Protopopov, A., Cadiñanos, J., et al. (2011). Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 469, 102-107. [DOI] [PMC free article] [PubMed]
- Karagkouni D., Paraskevopoulou M.D., Chatzopoulos S., Vlachos I.S., Tastsoglou S., Kanellos I., Papadimitriou D., Kavakiotis I., Maniou S., Skoufos G. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46:D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]; Karagkouni, D., Paraskevopoulou, M.D., Chatzopoulos, S., Vlachos, I.S., Tastsoglou, S., Kanellos, I., Papadimitriou, D., Kavakiotis, I., Maniou, S., Skoufos, G., et al. (2018). DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 46, D239-D245. [DOI] [PMC free article] [PubMed]
- Kempermann G., Kuhn H.G., Gage F.H. Experience-induced neurogenesis in the senescent dentate gyrus. J. Neurosci. 1998;18:3206–3212. doi: 10.1523/JNEUROSCI.18-09-03206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kempermann, G., Kuhn, H.G., and Gage, F.H. (1998). Experience-induced neurogenesis in the senescent dentate gyrus. J. Neurosci. 18, 3206-3212. [DOI] [PMC free article] [PubMed]
- Kempermann G. Seven principles in the regulation of adult neurogenesis. Eur. J. Neurosci. 2011;33:1018–1024. doi: 10.1111/j.1460-9568.2011.07599.x. [DOI] [PubMed] [Google Scholar]; Kempermann, G. (2011). Seven principles in the regulation of adult neurogenesis. Eur. J. Neurosci. 33, 1018-1024. [DOI] [PubMed]
- Kempermann G., Gage F.H., Aigner L., Song H., Curtis M.A., Thuret S., Kuhn H.G., Jessberger S., Frankland P.W., Cameron H.A. Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell. 2018;23:25–30. doi: 10.1016/j.stem.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kempermann, G., Gage, F.H., Aigner, L., Song, H., Curtis, M.A., Thuret, S., Kuhn, H.G., Jessberger, S., Frankland, P.W., Cameron, H.A., et al. (2018). Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell 23, 25-30. [DOI] [PMC free article] [PubMed]
- Kempermann G., Gast D., Gage F.H. Neuroplasticity in old age: sustained fivefold induction of hippocampal neurogenesis by long-term environmental enrichment. Ann. Neurol. 2002;52:135–143. doi: 10.1002/ana.10262. [DOI] [PubMed] [Google Scholar]; Kempermann, G., Gast, D., and Gage, F.H. (2002). Neuroplasticity in old age: Sustained fivefold induction of hippocampal neurogenesis by long-term environmental enrichment. Ann. Neurol. 52, 135-143. [DOI] [PubMed]
- Knoth R., Singec I., Ditter M., Pantazis G., Capetian P., Meyer R.P., Horvat V., Volk B., Kempermann G. Murine features of neurogenesis in the human hippocampus across the lifespan from 0 to 100 years. PLoS One. 2010;5:e8809. doi: 10.1371/journal.pone.0008809. [DOI] [PMC free article] [PubMed] [Google Scholar]; Knoth, R., Singec, I., Ditter, M., Pantazis, G., Capetian, P., Meyer, R.P., Horvat, V., Volk, B., and Kempermann, G. (2010). Murine features of neurogenesis in the human hippocampus across the lifespan from 0 to 100 years. PLoS One 5, e8809. [DOI] [PMC free article] [PubMed]
- Kronenberg G., Bick-Sander A., Bunk E., Wolf C., Ehninger D., Kempermann G. Physical exercise prevents age-related decline in precursor cell activity in the mouse dentate gyrus. Neurobiol. Aging. 2006;27:1505–1513. doi: 10.1016/j.neurobiolaging.2005.09.016. [DOI] [PubMed] [Google Scholar]; Kronenberg, G., Bick-Sander, A., Bunk, E., Wolf, C., Ehninger, D., and Kempermann, G. (2006). Physical exercise prevents age-related decline in precursor cell activity in the mouse dentate gyrus. Neurobiol. Aging 27, 1505-1513. [DOI] [PubMed]
- Kronenberg G., Reuter K., Steiner B., Brandt M.D., Jessberger S., Yamaguchi M., Kempermann G. Subpopulations of proliferating cells of the adult hippocampus respond differently to physiologic neurogenic stimuli. J. Comp. Neurol. 2003;467:455–463. doi: 10.1002/cne.10945. [DOI] [PubMed] [Google Scholar]; Kronenberg, G., Reuter, K., Steiner, B., Brandt, M.D., Jessberger, S., Yamaguchi, M., and Kempermann, G. (2003). Subpopulations of proliferating cells of the adult hippocampus respond differently to physiologic neurogenic stimuli. J. Comp. Neurol. 467, 455-463. [DOI] [PubMed]
- Lugert S., Basak O., Knuckles P., Haussler U., Fabel K., Götz M., Haas C.A., Kempermann G., Taylor V., Giachino C. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]; Lugert, S., Basak, O., Knuckles, P., Haussler, U., Fabel, K., Gotz, M., Haas, C.A., Kempermann, G., Taylor, V., and Giachino, C. (2010). Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell 6, 445-456. [DOI] [PubMed]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]; Madisen, L., Zwingman, T.A., Sunkin, S.M., Oh, S.W., Zariwala, H.A., Gu, H., Ng, L.L., Palmiter, R.D., Hawrylycz, M.J., Jones, A.R., et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133-140. [DOI] [PMC free article] [PubMed]
- Mannironi C., Camon J., De Vito F., Biundo A., De Stefano M.E., Persiconi I., Bozzoni I., Fragapane P., Mele A., Presutti C. Acute stress alters amygdala microRNA miR-135a and miR-124 expression: inferences for corticosteroid dependent stress response. PLoS One. 2013;8:e73385. doi: 10.1371/journal.pone.0073385. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mannironi, C., Camon, J., De Vito, F., Biundo, A., De Stefano, M.E., Persiconi, I., Bozzoni, I., Fragapane, P., Mele, A., and Presutti, C. (2013). Acute stress alters amygdala microRNA miR-135a and miR-124 expression: inferences for corticosteroid dependent stress response. PLoS One 8, e73385. [DOI] [PMC free article] [PubMed]
- Mao X.P., Zhang L.S., Huang B., Zhou S.Y., Liao J., Chen L.W., Qiu S.P., Chen J.X. Mir-135a enhances cellular proliferation through post-transcriptionally regulating PHLPP2 and FOXO1 in human bladder cancer. J. Transl. Med. 2015;13:86. doi: 10.1186/s12967-015-0438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mao, X.P., Zhang, L.S., Huang, B., Zhou, S.Y., Liao, J., Chen, L.W., Qiu, S.P., and Chen, J.X. (2015). Mir-135a enhances cellular proliferation through post-transcriptionally regulating PHLPP2 and FOXO1 in human bladder cancer. J. Transl. Med. 13, 86. [DOI] [PMC free article] [PubMed]
- Martynoga B., Mateo J.L., Zhou B., Andersen J., Achimastou A., Urbán N., van den Berg D., Georgopoulou D., Hadjur S., Wittbrodt J. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 2013;27:1769–1786. doi: 10.1101/gad.216804.113. [DOI] [PMC free article] [PubMed] [Google Scholar]; Martynoga, B., Mateo, J.L., Zhou, B., Andersen, J., Achimastou, A., Urban, N., van den Berg, D., Georgopoulou, D., Hadjur, S., Wittbrodt, J., et al. (2013). Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 27, 1769-1786. [DOI] [PMC free article] [PubMed]
- Mathews K.J., Allen K.M., Boerrigter D., Ball H., Shannon Weickert C., Double K.L. Evidence for reduced neurogenesis in the aging human hippocampus despite stable stem cell markers. Aging Cell. 2017;16:1195–1199. doi: 10.1111/acel.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mathews, K.J., Allen, K.M., Boerrigter, D., Ball, H., Shannon Weickert, C., and Double, K.L. (2017). Evidence for reduced neurogenesis in the aging human hippocampus despite stable stem cell markers. Aging Cell 16, 1195-1199. [DOI] [PMC free article] [PubMed]
- Overall R.W., Walker T.L., Fischer T.J., Brandt M.D., Kempermann G. Different mechanisms must be considered to explain the increase in hippocampal neural precursor cell proliferation by physical activity. Front. Neurosci. 2016;10:362. doi: 10.3389/fnins.2016.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]; Overall, R.W., Walker, T.L., Fischer, T.J., Brandt, M.D., and Kempermann, G. (2016). Different mechanisms must be considered to explain the increase in hippocampal neural precursor cell proliferation by physical activity. Front. Neurosci. 10, 362. [DOI] [PMC free article] [PubMed]
- Overall R.W., Walker T.L., Leiter O., Lenke S., Ruhwald S., Kempermann G. Delayed and transient increase of adult hippocampal neurogenesis by physical exercise in DBA/2 mice. PLoS One. 2013;8:e83797. doi: 10.1371/journal.pone.0083797. [DOI] [PMC free article] [PubMed] [Google Scholar]; Overall, R.W., Walker, T.L., Leiter, O., Lenke, S., Ruhwald, S., and Kempermann, G. (2013). Delayed and transient increase of adult hippocampal neurogenesis by physical exercise in DBA/2 mice. PLoS One 8, e83797. [DOI] [PMC free article] [PubMed]
- Pan-Vazquez A., Rye N., Ameri M., McSparron B., Smallwood G., Bickerdyke J., Rathbone A., Dajas-Bailador F., Toledo-Rodriguez M. Impact of voluntary exercise and housing conditions on hippocampal glucocorticoid receptor, miR-124 and anxiety. Mol. Brain. 2015;8:40. doi: 10.1186/s13041-015-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pan-Vazquez, A., Rye, N., Ameri, M., McSparron, B., Smallwood, G., Bickerdyke, J., Rathbone, A., Dajas-Bailador, F., and Toledo-Rodriguez, M. (2015). Impact of voluntary exercise and housing conditions on hippocampal glucocorticoid receptor, miR-124 and anxiety. Mol. Brain 8, 40. [DOI] [PMC free article] [PubMed]
- Pei H., Jin Z., Chen S., Sun X., Yu J., Guo W. MiR-135b promotes proliferation and invasion of osteosarcoma cells via targeting FOXO1. Mol. Cell. Biochem. 2015;400:245–252. doi: 10.1007/s11010-014-2281-2. [DOI] [PubMed] [Google Scholar]; Pei, H., Jin, Z., Chen, S., Sun, X., Yu, J., and Guo, W. (2015). MiR-135b promotes proliferation and invasion of osteosarcoma cells via targeting FOXO1. Mol. Cell. Biochem. 400, 245-252. [DOI] [PubMed]
- Pons-Espinal M., de Luca E., Marzi M.J., Beckervordersandforth R., Armirotti A., Nicassio F., Fabel K., Kempermann G., De Pietri Tonelli D. Synergic functions of miRNAs determine neuronal fate of adult neural stem cells. Stem Cell Reports. 2017;8:1046–1061. doi: 10.1016/j.stemcr.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pons-Espinal, M., de Luca, E., Marzi, M.J., Beckervordersandforth, R., Armirotti, A., Nicassio, F., Fabel, K., Kempermann, G., and De Pietri Tonelli, D. (2017). Synergic functions of miRNAs determine neuronal fate of adult neural stem cells. Stem Cell Reports 8, 1046-1061. [DOI] [PMC free article] [PubMed]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., et al. (2012). Fiji: an open-source platform for biological-image analysis, Nat. methods 9, 676-682. [DOI] [PMC free article] [PubMed]
- Schouten M., Fratantoni S.A., Hubens C.J., Piersma S.R., Pham T.V., Bielefeld P., Voskuyl R.A., Lucassen P.J., Jimenez C.R., Fitzsimons C.P. MicroRNA-124 and -137 cooperativity controls caspase-3 activity through BCL2L13 in hippocampal neural stem cells. Sci. Rep. 2015;5:12448. doi: 10.1038/srep12448. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schouten, M., Fratantoni, S.A., Hubens, C.J., Piersma, S.R., Pham, T. V., Bielefeld, P., Voskuyl, R.A., Lucassen, P.J., Jimenez, C.R., and Fitzsimons, C.P. (2015). MicroRNA-124 and -137 cooperativity controls caspase-3 activity through BCL2L13 in hippocampal neural stem cells. Sci. Rep. 5, 12448. [DOI] [PMC free article] [PubMed]
- Seib D.R.M., Corsini N.S., Ellwanger K., Plaas C., Mateos A., Pitzer C., Niehrs C., Celikel T., Martin-Villalba A. Loss of dickkopf-1 restores neurogenesis in old age and counteracts cognitive decline. Cell Stem Cell. 2013;12:204–214. doi: 10.1016/j.stem.2012.11.010. [DOI] [PubMed] [Google Scholar]; Seib, D.R.M., Corsini, N.S., Ellwanger, K., Plaas, C., Mateos, A., Pitzer, C., Niehrs, C., Celikel, T., and Martin-Villalba, A. (2013). Loss of dickkopf-1 restores neurogenesis in old age and counteracts cognitive decline. Cell Stem Cell 12, 204-214. [DOI] [PubMed]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]; Selbach, M., Schwanhausser, B., Thierfelder, N., Fang, Z., Khanin, R., and Rajewsky, N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58-63. [DOI] [PubMed]
- Shin J., Berg D.A., Zhu Y., Shin J.Y., Song J., Bonaguidi M.A., Enikolopov G., Nauen D.W., Christian K.M., Ming G.L. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shin, J., Berg, D.A., Zhu, Y., Shin, J.Y., Song, J., Bonaguidi, M.A., Enikolopov, G., Nauen, D.W., Christian, K.M., Ming, G.L., et al. (2015). Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17, 360-372. [DOI] [PMC free article] [PubMed]
- Spalding K.L., Bergmann O., Alkass K., Bernard S., Salehpour M., Huttner H.B., Boström E., Westerlund I., Vial C., Buchholz B.A. XDynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Spalding, K.L., Bergmann, O., Alkass, K., Bernard, S., Salehpour, M., Huttner, H.B., Bostrom, E., Westerlund, I., Vial, C., Buchholz, B.A., et al. (2013). XDynamics of hippocampal neurogenesis in adult humans. Cell 153, 1219-1227. [DOI] [PMC free article] [PubMed]
- Spalding K.L., Bhardwaj R.D., Buchholz B.A., Druid H., Frisén J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]; Spalding, K.L., Bhardwaj, R.D., Buchholz, B.A., Druid, H., and Frisen, J. (2005). Retrospective birth dating of cells in humans. Cell 122, 133-143. [DOI] [PubMed]
- Stappert L., Klaus F., Brüstle O. MicroRNAs engage in complex circuits regulating adult neurogenesis. Front. Neurosci. 2018;12:707. doi: 10.3389/fnins.2018.00707. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stappert, L., Klaus, F., and Brustle, O. (2018) MicroRNAs engage in complex circuits regulating adult neurogenesis. Front. Neurosci. 12:707. [DOI] [PMC free article] [PubMed]
- Stricker S.H., Götz M. DNA-methylation: master or slave of neural fate decisions? Front. Neurosci. 2018;12:5. doi: 10.3389/fnins.2018.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stricker, S.H., and Gotz, M. (2018). DNA-methylation: Master or slave of neural fate decisions? Front. Neurosci. 12, 5. [DOI] [PMC free article] [PubMed]
- Urban N., Guillemot F. Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front. Cell. Neurosci. 2014;8:396. doi: 10.3389/fncel.2014.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]; Urban, N., and Guillemot, F. (2014). Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front. Cell. Neurosci. 8, 396. [DOI] [PMC free article] [PubMed]
- van Battum E.Y., Verhagen M.G., Vangoor V.R., Fujita Y., Derijck A.A.H.A., O’Duibhir E., Giuliani G., de Gunst T., Adolfs Y., Lelieveld D. An Image-Based miRNA screen identifies miRNA-135s as regulators of CNS axon growth and regeneration by targeting Krüppel-like factor 4. J. Neurosci. 2017;38:0662–717. doi: 10.1523/JNEUROSCI.0662-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]; van Battum, E.Y., Verhagen, M.G., Vangoor, V.R., Fujita, Y., Derijck, A.A.H.A., O’Duibhir, E., Giuliani, G., de Gunst, T., Adolfs, Y., Lelieveld, D., et al. (2017). An Image-Based miRNA screen identifies miRNA-135s as regulators of CNS axon growth and regeneration by targeting Kruppel-like factor 4. J. Neurosci. 38, 0662-17. [DOI] [PMC free article] [PubMed]
- van Praag H. Exercise enhances learning and hippocampal neurogenesis in aged mice. J. Neurosci. 2005;25:8680–8685. doi: 10.1523/JNEUROSCI.1731-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; van Praag, H. (2005). Exercise enhances learning and hippocampal neurogenesis in aged mice. J. Neurosci. 25, 8680-8685. [DOI] [PMC free article] [PubMed]
- van Praag H., Kempermann G., Gage F.H. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 1999;2:266–270. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]; van Praag, H., Kempermann, G., and Gage, F.H. (1999). Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 2, 266-270. [DOI] [PubMed]
- Vizcaíno J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J.A., Sun Z., Farrah T., Bandeira N. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vizcaino, J.A., Deutsch, E.W., Wang, R., Csordas, A., Reisinger, F., Rios, D., Dianes, J.A., Sun, Z., Farrah, T., Bandeira, N., et al. (2014). ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223-226. [DOI] [PMC free article] [PubMed]
- Walker T.L., Overall R.W., Vogler S., Sykes A.M., Ruhwald S., Lasse D., Ichwan M., Fabel K., Kempermann G. Lysophosphatidic acid receptor is a functional marker of adult hippocampal precursor cells. Stem Cell Reports. 2016;6:552–565. doi: 10.1016/j.stemcr.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Walker, T.L., Overall, R.W., Vogler, S., Sykes, A.M., Ruhwald, S., Lasse, D., Ichwan, M., Fabel, K., and Kempermann, G. (2016). Lysophosphatidic acid receptor is a functional marker of adult hippocampal precursor cells. Stem Cell Reports 6, 552-565. [DOI] [PMC free article] [PubMed]
- Wu H., Huang M., Cao P., Wang T., Shu Y., Liu P. MiR-135a targets JAK2 and inhibits gastric cancer cell proliferation. Cancer Biol. Ther. 2012;13:281–288. doi: 10.4161/cbt.18943. [DOI] [PubMed] [Google Scholar]; Wu, H., Huang, M., Cao, P., Wang, T., Shu, Y., and Liu, P. (2012). MiR-135a targets JAK2 and inhibits gastric cancer cell proliferation. Cancer Biol. Ther. 13, 281-288. [DOI] [PubMed]
- Xu B., Tao T., Wang Y., Fang F., Huang Y., Chen S., Zhu W., Chen M. hsa-miR-135a-1 inhibits prostate cancer cell growth and migration by targeting EGFR. Tumor Biol. 2016;37:14141–14151. doi: 10.1007/s13277-016-5196-6. [DOI] [PubMed] [Google Scholar]; Xu, B., Tao, T., Wang, Y., Fang, F., Huang, Y., Chen, S., Zhu, W., and Chen, M. (2016). hsa-miR-135a-1 inhibits prostate cancer cell growth and migration by targeting EGFR. Tumor Biol. 37, 14141-14151. [DOI] [PubMed]
- Zubieta D.M.G., Hamood M.A., Beydoun R., Pall A.E., Kondapalli K.C. MicroRNA-135a regulates NHE9 to inhibit proliferation and migration of glioblastoma cells. Cell Commun. Signal. 2017;15:1–12. doi: 10.1186/s12964-017-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zubieta, D.M.G., Hamood, M.A., Beydoun, R., Pall, A.E., and Kondapalli, K.C. (2017). MicroRNA-135a regulates NHE9 to inhibit proliferation and migration of glioblastoma cells. Cell Commun. Signal. 15, 1-12. [DOI] [PMC free article] [PubMed]
- Zurawek D., Kusmider M., Faron-Gorecka A., Gruca P., Pabian P., Solich J., Kolasa M., Papp M., Dziedzicka-Wasylewska M. Reciprocal MicroRNA expression in mesocortical circuit and its interplay with serotonin transporter define resilient rats in the chronic mild stress. Mol. Neurobiol. 2017;54:5741–5751. doi: 10.1007/s12035-016-0107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zurawek, D., Kusmider, M., Faron-Gorecka, A., Gruca, P., Pabian, P., Solich, J., Kolasa, M., Papp, M., and Dziedzicka-Wasylewska, M. (2017). Reciprocal MicroRNA expression in mesocortical circuit and its interplay with serotonin transporter define resilient rats in the chronic mild stress. Mol. Neurobiol. 54, 5741-5751. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.