Abstract
Vesicle transport is crucial for various cellular functions and development of multicellular organisms. ARF-GAP is one of the key regulators of vesicle transport and is diverse family of proteins. Arabidopsis has 15 ARF-GAP proteins and four members are classified as ACAP type ARF-GAP proteins. Our previous study identified that VASCULAR NETWORK DEFECTIVE3 (VAN3), an ACAP ARF-GAP, played crucial roles in leaf vascular formation. However, it remains question how other members of plant ACAP ARF-GAPs function in cellular and developmental processes. To characterize these, we analyzed spatial expression pattern and subcellular localization of VAN3 and three other ACAPs, so called VAN3-like proteins (VALs). Expression pattern analysis revealed that they were expressed in distinctive developmental processes. Subcellular localization analysis in protoplast cells indicated that in contrast to VAN3, which localizes on trans-Golgi networks/early endosomes (TGNs/EEs), VAL1 and VAL2 were localized on ARA6-labelled endosomes, and VAL3 resided mainly in the cytoplasm. These results indicated that VAN3 and VALs are differently expressed in a tissue level and function in different intracellular compartments, in spite of their significant sequence similarities. These findings suggested functional divergence among plant ACAPs. Cellular localizations of all members of animal ACAP proteins are identical. Therefore our findings also suggested that plant evolved ACAP proteins in plant specific manner.
Keywords: ACAP, ARF-GAP, VAN3, VAL
The trafficking of proteins by vesicle transport is essential for all eukaryotic cells. Membrane bound transport vesicles carry cargo proteins from one compartment to another, and discharge the cargos into a specific compartment by fusing with the target membrane. Formation of transport vesicles and selection of cargos from donor membrane are regulated by the adenosine diphosphate (ADP)-ribosylation factors (ARFs) (Donaldson and Klausner, 1994; Moss and Vaughan, 1995, 1998). ARF proteins function through a cycle of GTP-binding and GTP-hydrolysis, leading to the GTP-bound active form and GDP-bound inactive form of the proteins, respectively. Consistent with the cyclical nature of ARF action, ARF locked in either GTP- or GDP-bound forms blocks membrane traffic (Dascher and Balch 1994; Peters et al. 1995; Zhang et al. 1994). Because the GTPase cycle is critical for ARF function, a regulatory mechanism for ensuring the exchange between GTP-binding and GTP-hydrolysis of ARF protein appropriately is required for constitutive membrane traffic. These regulations are achieved through the ADP-ribosylation factor guanine nucleotide exchange factors (ARF-GEFs) which promote the formation of ARF-GTP and ADP-ribosylation factor GTPse activating proteins (ARF-GAPs) which recognize ARF-GTP and induce hydrolysis of GTP (Naramoto et al. 2010, 2014; Nie et al. 2003a).
The ARF-GAPs are a large family of proteins named for the ability to induce the hydrolysis of GTP bound to ARF to GDP. In mammals, sixteen ARF-GAPs have been identified and the ARF-GAP proteins have been categorized into three groups; ARF-GAP1 type, Git type, and AZAP type (Randazzo and Hirsch, 2004). The ARF-GAP1 type has the ARF-GAP domain at the immediate N terminus, while Git type, in addition to N-terminal GAP domain, has a unique C-terminal targeting domain that was composed of three ankyrin (ANK) repeats followed by the Spa-homology domain (SHD) and paxillin binding site (PBS). The third group, the AZAP type, contains Bin-Amphiphysin-Rvs (BAR) domains and pleckstrin homology (PH) domains at the immediate N-terminal of the ARF-GAP domain and ANK repeats at the immediate C-terminal. Each of these is further subdivided based on additional domains. Whole genome sequences revealed that Arabidopsis genome encodes fifteen ARF-GAPs, which can be classified into two groups; ARF-GAP1 type and AZAP type based on the similarity to mammalian ARF-GAP proteins. There are no Git type ARF-GAP proteins in Arabidopsis. From the literature of animals, AZAP type ARF-GAP proteins were divided into four subgroups, ACAPs, ASAPs, AGAPs, and ARAPs. The acronym refers to the domains within the proteins; AZAP for ARF GAP with ANK repeats and PH domains. The “Z” stands for domain that characterizes each subclass. ACAPs (coiled-coil domains) are localized on plasma membrane (PM)/endosome (Jackson et al. 2000), ASAPs (SH3 domain) also on PM/endosome (Randazzo et al. 2000), AGAPs (GLD domain) on lysosome/endosome (Nie et al. 2002, 2003b), and ARAPs (Rho GAP domain) on trans Golgi/TGN/PM (Miura et al. 2002) respectively, while all of the ARF-GAP1 type was localized are cis Golgi. Because single ARF protein functions at multiple organelles, intracellular site of ARF-GAP protein action plays a pivotal role in conferring the site specificity of ARF function. Based on these results, AZAP type ARF-GAP proteins were believed to be key players of regulating the membrane traffic of post Golgi transport pathway. Interestingly, however, there are no AZAP type ARF-GAP proteins other than ACAPs in plants. In Arabidopsis, there are four members of ACAPs, and one of them has been characterized as VAN3 that is involved in leaf vascular continuity (Aihara et al. 2014; Koizumi et al. 2005; Naramoto et al. 2009; Sieburth et al. 2006). VAN3 was localized on trans Golgi networks/early endosomes (TGNs/EEs), unknown organelles, and the plasma membrane (PM) in Arabidopsis cells, which presumably function together with ARF-GEF GNOM that is localized at Golgi apparatus and PMs (Koizumi et al. 2005; Naramoto et al. 2009, 2010, 2014). The van3 single mutant caused discontinuity of venation without affecting the continuity of primary veins, whereas quadruple mutation in VAN3 (At5g13300) and its homologus VAN3-like protein1 (VAL1) (At5g61980), VAL2 (At1g60860) and VAL3 (At1g10870) caused discontinuity of primary veins (Naramoto et al. 2010; Sieburth et al. 2006). These findings suggest the existence of functional redundancy in vascular formation among ACAPs in Arabidopsis. Although these previous works gave some conceptual framework of cellular and developmental function of plant ACAP proteins, subcellular localization of VAL proteins as well as the spatial expression patterns of VALs in planta are not reported yet.
To clarify these, we initially analyzed the spatial expression pattern of plant ACAPs. We studied the expression patterns of VAL genes in shoots and roots to further confirm the involvement of VALs in the vascular formation. As an index of root primodium development, we observed the developmental stage of lateral roots. For these purposes, we produced transgenic plants harboring GUS gene fused to a 2 kb upstream promoter region of each Arabidopsis ACAP type ARF-GAP gene. Ten independent transgenic lines were analyzed for each promoter::GUS fusion, and the expression patterns observed in the majority of the lines were shown. Detailed information of materials and methods in these experiments are shown in Supplemental data.
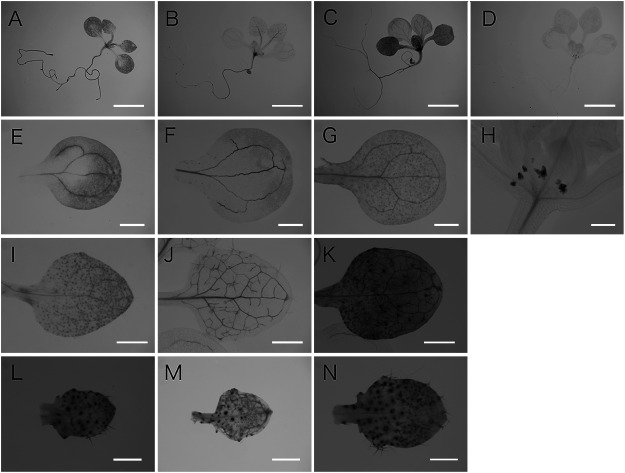
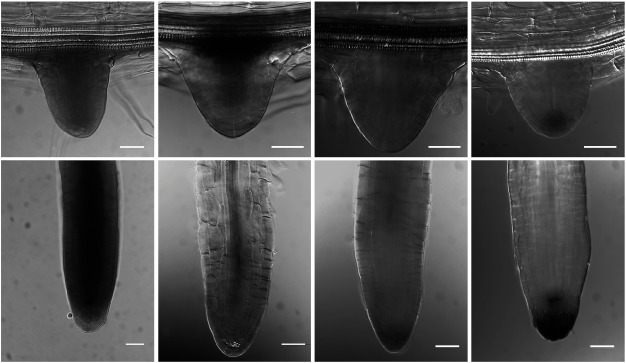
As we reported previously pVAN3::GUS expression was broadly detected throughout the plant body with stronger expression in vascular cells and its staining was gradually restricted to vascular cells in mature shoots and roots (Figure 1A, E, I, L, and Figure 2A, E) (Naramoto et al. 2009). In contrast to the ubiquitous expression of the VAN3, pVAL1::GUS expression was detected in the extremely restricted tissues of plants (Figure 1D). pVAL1::GUS expression was confined to stipules (Figure 1D, H) in aerial parts, and to QC cells and columella root cap cells in the underground (Figure 2H). The gradient in GUS activity with its maximum at the primordium root tip was gradually established during its development (Figure 2D, H). GUS staining patterns of transgenic Arabidopsis harboring the pVAL2::GUS construct showed preferential staining patterns in vascular cells throughout the plant body (Figure 1B). pVAL2::GUS was expressed ubiquitously in the leaf at the early stage of its development with a feature of strong GUS activity in the vasculature (Figure 1M). As the leaf developed, GUS activity was preferentially restricted to the vascular tissues (Figure 1B, F, J, M). GUS staining was also observed in developing trichomes and guard cells (Figure 1J, M). In the lateral root, throughout its development, strong GUS activity was observed in stele cells, whereas weak GUS activity was observed in the root cap (Figure 2B, F). Finally, we analyzed the expression pattern of pVAL3::GUS. pVAL3::GUS, similarly to pVAN3::GUS, was broadly expressed throughout the plant (Figure 1C). In leaves, pVAL3::GUS was ubiquitously expressed throughout its developmental process with a slight increase of GUS activity in trichomes and guard cells (Figure 1C, G, K, N). In the lateral root, GUS activity was detected from the earliest stages. Later, its activity was detected exclusively in primordium margins (Figure 2C). After the lateral root emerged, GUS activity was broadly detected in the regions other than meristematic cells (Figure 2G). These analyses identified that expression patterns of VAN3, VAL2, and VAL3 were overlapped in leaf vascular cells although their spatial expression patterns were in overall differently regulated in tissue specific manner.
Figure 1. Expression patterns of the VAN3 and the VAL genes in leaves and cotyledons of 11 day old Arabidopsis seedlings. Histochemical localization of pVAN3::GUS (A, E, I, L), pVAL2::GUS (B, F, J, M), pVAL3::GUS (C, G, K, N), and pVAL1::GUS (D, H) activities were examined in aerial part of 11-day-old Arabidopsis seedlings. Seedlings (A–D), the cotyledon (E–G), the first rosette leaf (I–K), and the third rosette leaf (L–N). Scale bars: 4 mm (A–D), 1 mm (E–G and I–K), 400 µm (H), and 250 µm (L–N).
Figure 2. Expression patterns of the VAN3 and the VAL genes during lateral root formation. Histochemical localization of pVAN3::GUS (A, E), pVAL2::GUS (B, F), pVAL3::GUS (C, G), and pVAL1::GUS (D, H) activities were examined in lateral roots of 11-day-old Arabidopsis seedlings. Lateral roots during emergence (A–D), and mature lateral roots (E–H). Scale bars: 20 µm.
Therefore, VAL2, VAL3, and VAN3 may redundantly regulate vascular formation. However, as it was reported previously, van3val2 and van3val3 double mutants as well as van3val2val3 triple mutants did not enhance the vascular discontinuity of van3 mutants. Strikingly, additional mutation in VAL1 to the van3val2val3 triple mutants enhanced the leaf vascular phenotypes (Naramoto et al. 2010; Sieburth et al. 2006), whereas VAL1 did not express in leaf vascular cells (Figure 1H). Notably, val1val2val3 triple mutants were reported to show weak but significant leaf vascular phenotypes (Sieburth et al. 2006). These findings indicated that relationship of VAN3 and VALs were complex and also imply that VALs did not necessarily function redundantly to VAN3 in the establishment of vascular continuity.
To examine whether plant ACAPs are functionally differentiated in plant cells or not, we next analyzed the subcellular localization of VAL proteins. For this purpose, we performed double labeling experiments of VALs and organelle markers. We transiently expressed a C-terminal sGFP fusion protein of VALs and a monomeric red fluorescent protein (mRFP) (Campbell et al. 2002) fusion protein of several organelle markers under the control of the CaMV 35S promoter in protoplasts prepared from Arabidopsis suspension cultured cells. Fluorescence of sGFP and mRFP-fusion proteins was examined with a confocal laser scanning microscopy. Detailed information of materials and methods in these experiments are shown in Supplemental data.
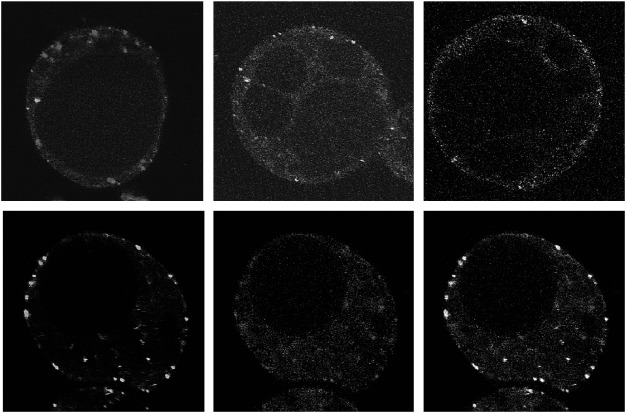
First, we examined the subcellular localization of VAL1. When sGFP tagged VAL1 was expressed in Arabidopsis suspension cells, green fluorescent dot like structures were observed. The localization of VAL1-sGFP did not overlap with the trans Golgi marker ST-mRFP or the TGN/EE marker mRFP-SYP41 (Bassham et al. 2000; Jin et al. 2001; Kim et al. 2001; Wee et al. 1998; Uemura et al. 2004) (Figure 3A, B) that are consistent with previous subcellular localization analysis in tobacco epidermal cells (Yoo et al. 2008). These results indicate that VAL1 is not located on the Golgi body or TGN/EE. Next, we examined whether VAL1 was localized on the multivesicular endosomes. As endosome markers, we used mRFP-ARA7 and ARA6-mRFP (Ueda et al. 2001, 2004). We did not observe clear colocalization between VAL1-sGFP and mRFP-ARA7 (Figure 3C), whereas some population of the green fluorescent signals of VAL1-sGFP clearly overlapped with the punctate red fluorescent signals of ARA6-mRFP (Figure 3D–F). Interestingly, however, other population of the VAL1-sGFP did not overlap the red fluorescent signals of ARA6 (Figure 3F arrows). These results show that VAL1 is localized on both ARA6-mRFP stained endosomes and uncharacterized organelles.
Figure 3. Subcellular localization of sGFP-tagged VAL1 (Green) and mRFP-tagged subcellular markers (Red) in Arabidopsis suspension cells. (A–C) Merged image of VAL1-sGFP and the Golgi body marker ST-mRFP (A), the TGN marker mRFP-SYP41 (B), and the endosome marker mRFP-ARA7 (C). (D–F) Localization of VAL1-sGFP (D), endosome marker mRFP-ARA6 (E), and a merged image of D and E (F). Arrows indicate the VAL1-sGFP stained organelles that were not marked by the endosome marker ARA6-sGFP. Scale bars: 5 µm.
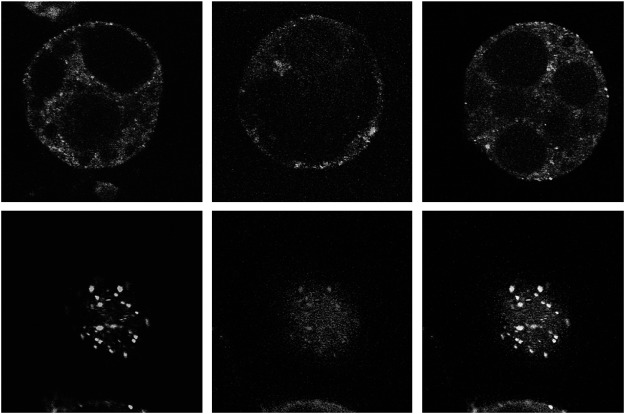
In addition, when sGFP tagged VAL2 is expressed in Arabidopsis suspension cells, it was localized on ARA6-positive endosomes (Figure 4D–F), whereas it did not colocalize to the trans Golgi marker ST-mRFP (Figure 4A), the TGN/EE marker mRFP-SYP41 (Figure 4B), or GFP-ARA7 positive endosomes (Figure 4C). However, all population of VAL2-sGFP did not overlap with ARA6-mRFP (Figure 4F arrows). These results indicate that VAL2 is, just like VAL1, localized on both ARA6-mRFP stained endosomes and uncharacterized organelles. As it was reported previously, VAN3 is not localized on ARA6 stained endosomes but on both SYP41 stained TGNs/EEs and unknown organelles (Koizumi et al. 2005). These findings implied that the intracellular localization of VAN3 is not identical to VAL1 and VAL2 in arabidopsis suspension protoplast cells, although there still remains the possibility that some population of VAN3 colocalizes with VAL1 and/or VAL2 in uncharacterized organelles.
Figure 4. Subcellular localization of sGFP-tagged VAL2 (Green) and mRFP-tagged subcellular markers (Red) in Arabidopsis suspension cells. (A–C) Merged image of VAL2-sGFP and the Golgi body marker ST-mRFP (A), the TGN marker mRFP-SYP41 (B), and the endosome marker mRFP-ARA7 (C). (D–F) Localization of VAL2-sGFP (D), endosome marker mRFP-ARA6 (E), and a merged image of D and E (F). Arrows indicate the VAL2-sGFP stained organelles that were not marked by the endosome marker ARA6-sGFP. Scale bars: 5 µm.
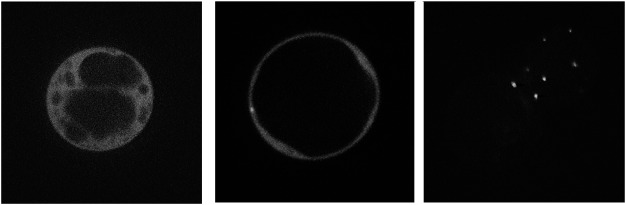
In contrast to the staining patterns of VAL1-sGFP and VAL2-sGFP, VAL3-sGFP showed the typical cytoplasmic distribution (Figure 5A), and sometimes dot like structures were observed (Figure 5B arrows). Because the VAL3 stained organelles are smaller than that of VAN3 (Figure 5C), VAL3 may be also localized on distinctive organelles from VAN3 stained organelles. In any case, VAL3 mainly localized at PMs, which differs from the subcellular localization of VAN3 proteins.
Figure 5. Subcellular localization of sGFP-tagged VAL3 and VAN3 in Arabidopsis suspension cells. (A, B) VAL3-sGFP was distributed in cytosol. An arrow indicates the punctate signal of VAL3-sGFP. (C) VAN3-VENUS showed punctate structures.
These results suggested that VAN3 and each of VALs regulate a distinctive intracellular membrane trafficking (Figures 3-5). Previous research on animal ACAPs indicated that both BAR domain and PH domain that recognize the membrane curvature and phospholipids, respectively can function cooperatively in its membrane targeting (Peter et al. 2004). In fact, we also showed that localization of VAN3 is regulated by BAR and PH domains (Naramoto et al. 2009). According to these reports, the difference of subcellular localization among plant ACAPs may be caused by the difference of the lipid binding specificity of PH domains or the membrane curvature sensing competence of BAR domains. The distinct subcellular localizations among plant ACAPs may contribute to divergent of developmental functions of them.
The divergent subcellular localization among plant ACAPs contrasts to the identical subcellular compartmentation of the animal ACAPs. In animals, each of the AZAP type ARF-GAP proteins, ACAPs, AGAPs, ARAPs, and ASAPs, is localized on distinct intracellular organelles and all together play indispensable roles in membrane trafficking in post Golgi transport pathway. In Arabidopsis genome, however, there are no AZAP type ARF-GAP proteins other than ACAPs. Therefore, plant divergent ACAPs may have evolved to compensate for the divergent functions of different AZAP type ARF-GAP subfamily members.
Now efforts are underway to further clarify the functional diversity of plant ACAPs. For this purpose, we are performing the promoter-swap experiments to VALs, in which the cDNAs of all plant ACAPs will be fused to the VAN3 cis-regulatory sequences and introduced into the van3 mutant, and then the complementation of the venation discontinuity will be examined. Simultaneous subcellular localization analysis of VAN3 and VAL proteins in Arabidopsis seedlings will be also indispensable because our transient expression system by using 35S promoter may cause their mislocalizations. Combinational analyses of these experiments will clearly indicate how molecular functions of plant ACAPs are divergent.
Acknowledgments
We gratefully acknowledge Tomohiro Uemura, Takashi Ueda and Akihiko Nakano for providing published materials and helpful discussions to pursue this work. This work was supported by Grant for Basic Science Research Projects from The Sumitomo Foundation to SN and the Japanese Society for Promotion of Science to SN (JSPS; 30612022).
Supplementary Data
References
- Aihara K, Naramoto S, Hara M, Mizoguchi T (2014) Increase in vascular pattern complexity caused by mutations in LHY and CCA1 in Arabidopsis thaliana under continuous light. Plant Biotechnol 31: 43–47 [Google Scholar]
- Bassham DC, Sanderfoot AA, Kovaleva V, Zheng H, Raikhel NV (2000) AtVPS45 complex formation at the trans-Golgi network. Mol Biol Cell 11: 2251–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher C, Balch WE (1994) Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J Biol Chem 269: 1437–1448 [PubMed] [Google Scholar]
- Donaldson JG, Klausner RD (1994) ARF: A key regulatory switch in membrane traffic and organelle structure. Curr Opin Cell Biol 6: 527–532 [DOI] [PubMed] [Google Scholar]
- Jackson TR, Brown FD, Nie Z, Miura K, Foroni L, Sun J, Hsu VW, Donaldson JG, Randazzo PA (2000) ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J Cell Biol 151: 627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I (2001) Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi K, Naramoto S, Sawa S, Yahara N, Ueda T, Nakano A, Sugiyama M, Fukuda H (2005) VAN3 ARF-GAP-mediated vesicle transport is involved in leaf vascular network formation. Development 132: 1699–1711 [DOI] [PubMed] [Google Scholar]
- Miura K, Jacques KM, Stauffer S, Kubosaki A, Zhu K, Hirsch DS, Resau J, Zheng Y, Randazzo PA (2002) ARAP1: A point of convergence for Arf and Rho signaling. Mol Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Moss J, Vaughan M (1995) Structure and function of ARF proteins: Activators of cholera toxin and critical components of intracellular vesicular transport processes. J Biol Chem 270: 12327–12330 [DOI] [PubMed] [Google Scholar]
- Moss J, Vaughan M (1998) Molecules in the ARF orbit. J Biol Chem 273: 21431–21434 [DOI] [PubMed] [Google Scholar]
- Naramoto S, Sawa S, Koizumi K, Uemura T, Ueda T, Friml J, Nakano A, Fukuda H (2009) Phosphoinositide-dependent regulation of VAN3 ARF-GAP localization and activity essential for vascular tissue continuity in plants. Development 136: 1529–1538 [DOI] [PubMed] [Google Scholar]
- Naramoto S, Kleine-Vehn J, Robert S, Fujimoto M, Dainobu T, Paciorek T, Ueda T, Nakano A, Van Montagu MC, Fukuda H, et al. (2010) ADP-ribosylation factor machinery mediates endocytosis in plant cells. Proc Natl Acad Sci USA 107: 21890–21895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S, Otegui MS, Kutsuna N, de Rycke R, Dainobu T, Karampelias M, Fujimoto M, Feraru E, Miki D, Fukuda H, et al. (2014) Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell 26: 3062–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hirsch DS, Randazzo PA (2003a) Arf and its many interactors. Curr Opin Cell Biol 15: 396–404 [DOI] [PubMed] [Google Scholar]
- Nie Z, Stanley KT, Stauffer S, Jacques KM, Hirsch DS, Takei J, Randazzo PA (2002) AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J Biol Chem 277: 48965–48975 [DOI] [PubMed] [Google Scholar]
- Nie Z, Boehm M, Boja ES, Vass WC, Bonifacino JS, Fales HM, Randazzo PA (2003b) Specific regulation of the adaptor protein complex AP-3 by the Arf GAP AGAP1. Dev Cell 5: 513–521 [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 303: 495–499 [DOI] [PubMed] [Google Scholar]
- Peters PJ, Hsu VW, Ooi CE, Finazzi D, Teal SB, Oorschot V, Donaldson JG, Klausner RD (1995) Overexpression of wild-type and mutant ARF1 and ARF6: Distinct perturbations of nonoverlapping membrane compartments. J Cell Biol 128: 1003–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo PA, Hirsch DS (2004) Arf GAPs: Multifunctional proteins that regulate membrane traffic and actin remodelling. Cell Signal 16: 401–413 [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Andrade J, Miura K, Brown MT, Long YQ, Stauffer S, Roller P, Cooper JA (2000) The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc Natl Acad Sci USA 97: 4011–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Muday GK, King EJ, Benton G, Kim S, Metcalf KE, Meyers L, Seamen E, Van Norman JM (2006) SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18: 1396–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A (2001) Ara6, a plant-unique novel type Rab GTPase, functions in endocytic pathway of Arabidopsis thaliana. EMBO Journal 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Uemura T, Sato MH, Nakano A (2004) Functional diversification of endosomes in Arabidopsis cells. The Plant Journal 40: 783–789 [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH (2004) Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct Funct 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Wee EG, Sherrier DJ, Prime TA, Dupree P (1998) Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell 10: 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CM, Wen J, Motes CM, Sparks JA, Blancaflor EB (2008) A class I ADP-ribosylation factor GTPase-activating protein is critical for maintaining directional root hair growth in Arabidopsis. Plant Physiol 147: 1659–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CJ, Rosenwald AG, Willingham MC, Skuntz S, Clark J, Kahn RA (1994) Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo. J Cell Biol 124: 289–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.