Abstract
Nitrogen limits crop yield, but application of nitrogen fertilizer can cause environmental problems and much fertilizer is lost without being absorbed by plants. Increasing nitrogen use efficiency in plants may help overcome these problems and is, therefore, an important and active subject of agricultural research. Here, we report that the expression of the chimeric repressor for the GATA4 transcription factor (35S:GATA4-SRDX) improved tolerance to nitrogen deficiency in Arabidopsis thaliana. 35S:GATA4-SRDX seedlings were significantly larger than wild type under nitrogen-sufficient and -deficient conditions (10 and 0.5 mM NH4NO3, respectively). 35S:GATA4-SRDX plants exhibited shorter primary roots, fewer lateral roots, and higher root hair density compared with wild type. The expression levels of NITRATE TRANSPORTER 2.1, ASPARAGINE SYNTHETASE and NITRATE REDUCTASE 1 were significantly higher in roots of 35S:GATA4-SRDX plants than in wild type under nitrogen-sufficient conditions. Under nitrogen-deficient conditions, the expression of genes for cytosolic glutamine synthetases was upregulated in shoots of 35S:GATA4-SRDX plants compared with wild type. This upregulation of nitrogen transporter and nitrogen assimilation-related genes might confer tolerance to nitrogen deficiency in 35S:GATA4-SRDX plants.
Keywords: Arabidopsis, chimeric repressor, nitrogen use efficiency, transcription factor, tolerance
Nitrogen is an essential macronutrient for plant growth and development, and a major factor limiting agricultural production. Plants absorb and utilize ammonium (NH4+) and nitrate (NO3−) from soil (Crawford and Forde 2002; Kronzucker et al. 1997; Marschner 1995). Nitrogen levels affect crop productivity by modulating the expression of genes that affect leaf development, root architecture, senescence, flowering, and metabolite biosynthesis (Diaz et al. 2008; Rubin et al. 2009; Stitt et al. 2002; Walch-Liu et al. 2000; Wang et al. 2004; Zhang and Forde 1998). Adding nitrogen fertilizer can boost crop yields, but such inputs are costly and plants fail to use 50–70% of nitrogen provided as fertilizer. Nitrogen loss can cause soil and water pollution, and may contribute to global warming. Therefore, increasing nitrogen use efficiency (NUE) remains a crucial issue for agriculture and plant nutrient research.
Ongoing work has identified factors that regulate nitrogen uptake, translocation, and assimilation (Masclaux-Daubresse et al. 2010), including enzymes such as NITRATE TRANSPORTERs (NRTs), AMMONIUM TRANSPORTERs (ATMs), GLUTAMINE SYNTHETASEs (GLNs) and ASPARAGINE SYNTHETASEs (ASNs) (Masclaux-Daubresse et al. 2010; Vidal and Gutiérrez 2008). In addition to these enzymes, the transcription factors that regulate the expression of nitrogen-responsive genes have been analyzed. NODULE INCEPTION (NIN) functions as a key regulator of the symbiotic nitrogen fixation pathway in legumes such as Lotus japonicus, and NIN-Like proteins were identified as transcription factors that interact with cis-elements conserved in promoters of nitrate-responsive genes in Arabidopsis thaliana (Konishi and Yanagisawa 2013).
Several studies have manipulated transcription factor expression in attempts to enhance NUE. For example, in Arabidopsis, ectopic expression of Dof1, which regulates the expression of genes related to organic acid metabolism, resulted in increased growth under low-nitrogen conditions through the accumulation of amino acids (Kurai et al. 2011; Yanagisawa et al. 2004). In this report, we attempted to identify novel transcription factors involved in tolerance to nitrogen deficiency by screening Arabidopsis lines that express chimeric repressors (CRES-T) for transcription factors. Many plant transcription factors are structurally and functionally redundant and a single-gene knock-out often fails to exhibit an informative phenotype. However, in the CRES-T gene silencing system, fusion to the SRDX repression domain (SUPERMAN Repression Domain X) converts a transcription factor to a strong repressor that dominantly represses the target genes, producing phenotypes similar to loss-of-function of its redundant transcription factor genes (Hiratsu et al. 2003).
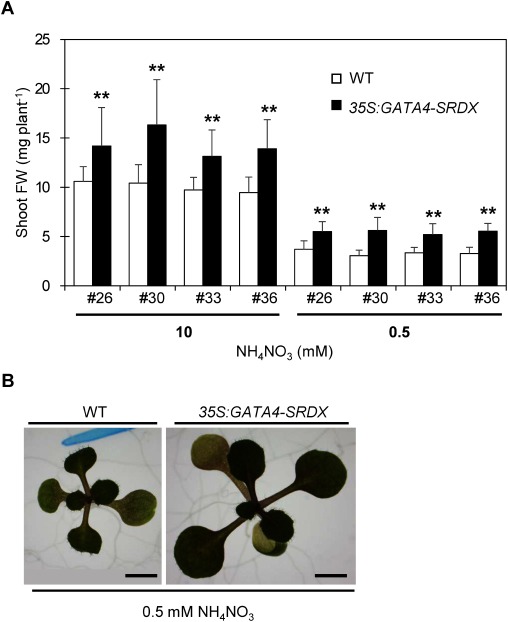
To screen our set of CRES-T lines for Arabidopsis transcription factors, we used Murashige-Skoog solid medium containing 0.5 mM NH4NO3 and 10 mM NH4NO3 as deficient and sufficient conditions, respectively. We screened for CRES-T lines that exhibit different sizes from wild type under nitrogen-deficient conditions. We identified a CRES-T line that produced bigger seedlings under nitrogen-deficient conditions. The fresh weight of 14-day-old seedlings of the CRES-T line was significantly higher than that of wild type under both nitrogen-sufficient and -deficient conditions (Figure 1). Genome PCR analysis revealed the transcription factor of the chimeric repressor to be GAT A4 (AT3G60530; 35S:GATA4-SRDX). Therefore, we considered that 35S:GATA4-SRDX plants are tolerant to nitrogen deficiency.
Figure 1. Phenotype at 14 days after sowing (DAS) of 35S:GATA4-SRDX and wild type seedlings. (A) Comparison of shoot fresh weight (FW) between wild type (WT) and 35S:GATA4-SRDX plants under nitrogen-sufficient (10 mM NH4NO3) and nitrogen-deficient (0.5 mM NH4NO3). Open and closed bars indicate wild type and 35S:GATA4-SRDX plants, respectively. An Arabic numeral with ‘#’ symbol under each closed bar represents an independent line of 35S:GATA4-SRDX plants. Values are means±SD (n=16–23). Double asterisks indicate significant differences at p<0.01 in t-test when compared to wild type grown in each nitrogen condition. (B) Photos of 14 DAS wild type and independent line #30 of 35S:GATA4-SRDX plants grown under nitrogen-deficient conditions. Scale bar; 2 mm.
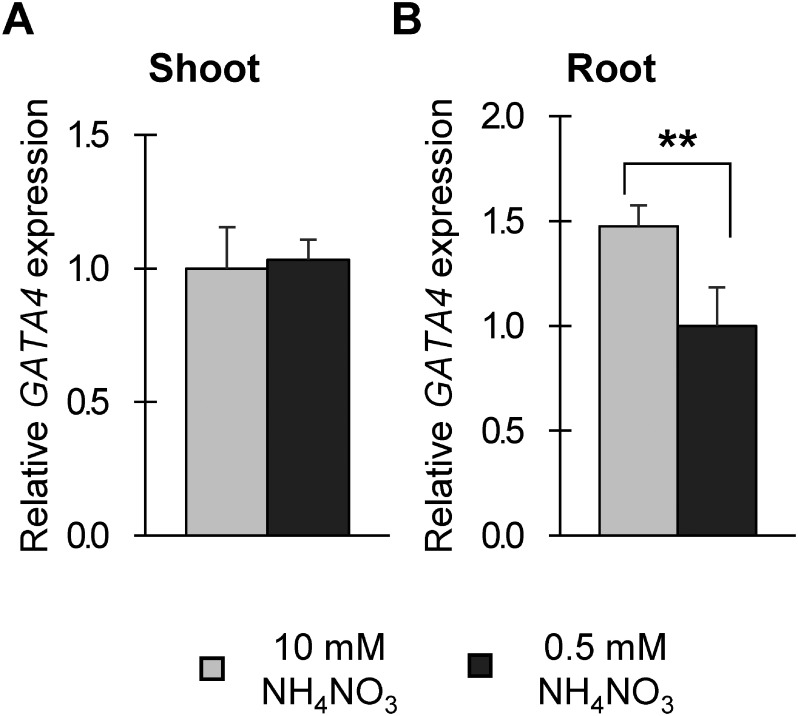
GAT A4 is a transcription factor that belongs to the GAT A family and possesses a C2C2 zinc finger DNA binding domain that binds to the (A/T)GAT A(A/G) motif (Orkin 1992). The Arabidopsis genome has 29 genes encoding GAT A transcription factors (Manfield et al. 2006; Riechmann et al. 2000). Arabidopsis GATA4 is expressed in all tissues including root, stem, leaf, flower, and silique at the reproductive stage and especially upregulated under darkness (Manfield et al. 2006). We analyzed whether nitrogen deficiency affects the expression level of GATA4 by qRT-PCR and found that the expression of GATA4 was moderately suppressed in roots under nitrogen-deficient conditions, but we observed no reduction in shoots (Figure 2).
Figure 2. GATA4 expression in response to different nitrogen conditions. Relative expression of GATA4 in shoots (A) and roots (B) of wild type at 13 DAS grown under different nitrogen conditions analyzed by qRT-PCR, and all expression levels were normalized to that of PP2AA3 (At1g3320), reference gene. Values are means±SD of four biological replicates. Double asterisks indicate significant difference at p< 0.01 in t-test when compared to plants grown in nitrogen-sufficient conditions (10 mM NH4NO3).
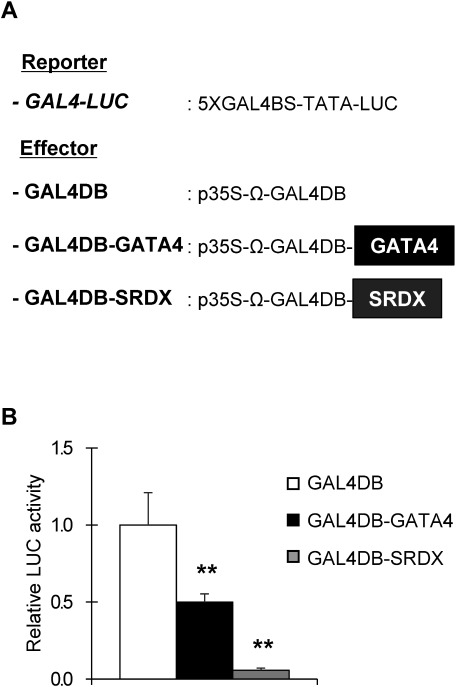
To analyze the molecular activity of GAT A4, we performed transient expression assays using a luciferase reporter gene (LUC) fused to a promoter containing the Gal4 DNA binding site (GAL4:LUC) and an effector in which the coding region of GATA4 was fused to the Gal4 DNA-binding domain (GAL4DB) driven by the CaMV 35S promoter (35S:GAL4BD-GATA4) in protoplasts isolated from leaf epidermal cells of Arabidopsis. The transient assay showed that the 35S:GAL4DB-GAT A4 effector significantly suppressed GAL4:LUC activity, compared with the control 35S:GAL4DB effector (Figure 3), suggesting that GAT A4 appears to function as a repressor.
Figure 3. Transient effector–reporter analysis of GATA4 transcriptional activity. (A) Schematic representation of the constructs used in transient assay in Arabidopsis leaf protoplasts. The reporter construct contains firefly luciferase (LUC) driven by the promoter containing 5×Gal4 DNA-binding sequence (GAL4BS) and a TATA sequence. Each effector construct contains a Gal4 DNA-binding domain (GALDB) and TMV Omega leader sequence (Ω) driven by the CaMV 35 S promoter (p35S). The effector construct with the SRDX-fused Gal4 DNA-binding domain (GALDB-SRDX) was used as a positive control for repression. (B) Relative LUC reporter activity when each effector was co-transfected into leaf protoplasts. Values are means±SD of six technical replicates. Double asterisks indicate significant difference at p< 0.001 in Dunnett’s test when compared with negative control (GAL4DB).
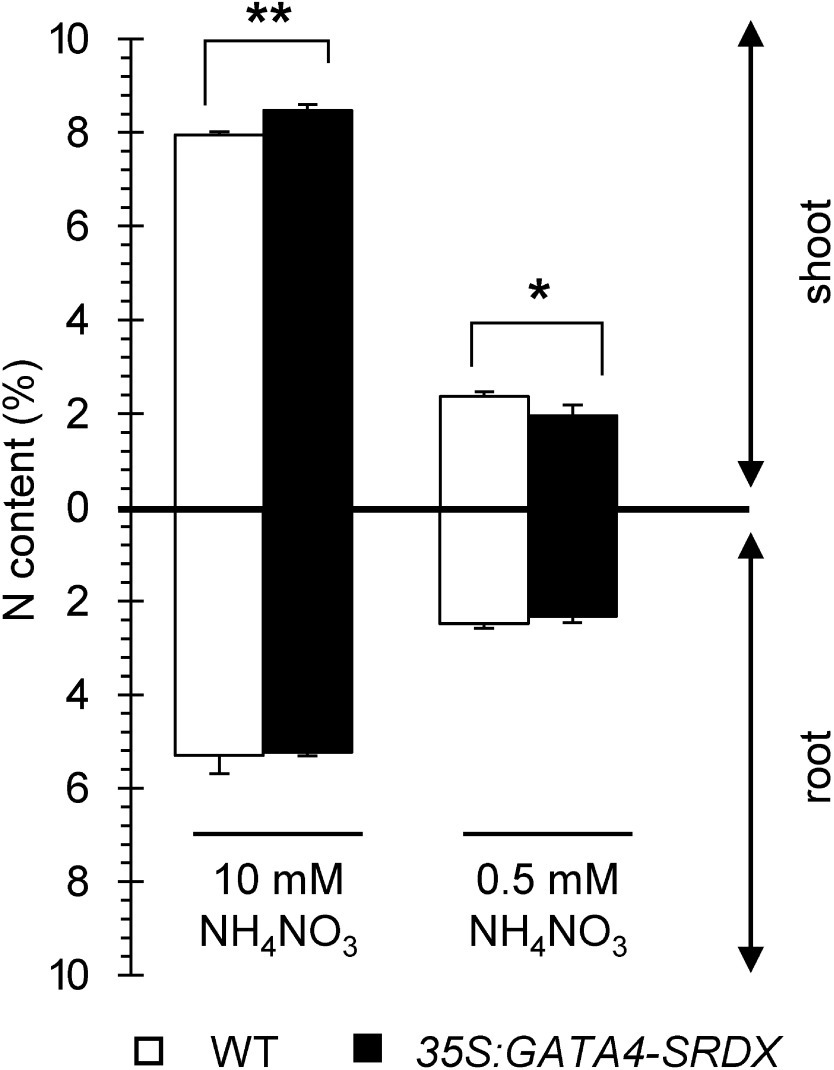
The nitrogen content of 35S:GATA4-SRDX shoots (about 8.5%) was significantly higher than that of wild type (about 7.9%) under nitrogen-sufficient conditions (Figure 4). By contrast, under nitrogen-deficient conditions, the nitrogen content of 35S:GATA4-SRDX shoots (about 2.0%) was lower than that of wild type (about 2.4%, Figure 4). These results suggest that NUE is enhanced in 35S:GATA4-SRDX plants to maintain larger shoot biomass, even under low nitrogen.
Figure 4. Nitrogen content in shoots and roots of wild type and 35S:GATA4-SRDX plants. Plants were grown under different nitrogen conditions for 15 DAS. Values are means±SD of four independent biological replicates. Single and double asterisks indicate significant difference at p<0.05 and 0.01, respectively, in t-test when compared to wild type grown in each nitrogen condition.
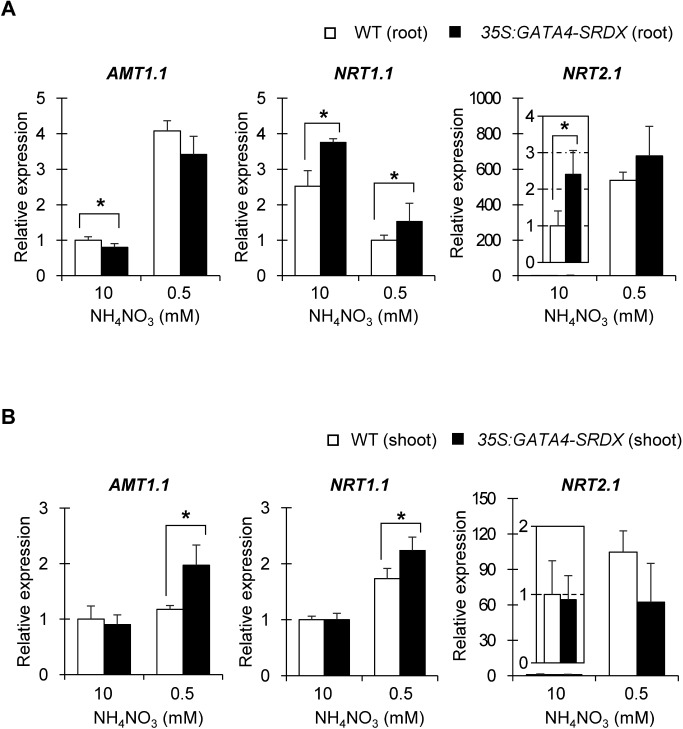
To examine the mechanisms by which 35S:GATA4-SRDX plants tolerate nitrogen deficiency, we analyzed the expression of genes related to nitrogen transport and assimilation. AMT1.1, which codes for a high-affinity ammonium transporter, is de-repressed in roots under nitrogen-deficient conditions, but upregulated in shoots under nitrogen-sufficient conditions (Engineer and Kranz 2007; Gazzarrini et al. 1999). Nitrate induces NRT1.1, which codes for a dual-affinity nitrate transporter, and NRT2.1, which codes for a high-affinity transporter, but they usually display opposite expression patterns under nitrogen-deficient conditions. In roots under nitrogen deficiency, NRT1.1 is down-regulated and NRT2.1 is dramatically induced (Forde 2000; Liu et al. 1999; Orsel et al. 2002; Wang et al. 1998). Our qRT-PCR analyses showed that NRT1.1 and NRT2.1 were highly expressed in roots of 35S:GATA4-SRDX plants compared to wild type under nitrogen-sufficient conditions (Figure 5A). NRT1.1 expression was higher in roots of 35S:GATA4-SRDX plants than in wild type, even though its expression was suppressed in wild-type and 35S:GATA4-SRDX roots by nitrogen deficiency (Figure 5A). NRT2.1 expression was highly increased by nitrogen deficiency in roots of wild-type and 35S:GATA4-SRDX plants, but did not show a significant difference between wild type and 35S:GTATA4-SRDX (Figure 5A). We also found that the expression of AMT1.1 was upregulated in shoots of 35S:GATA4-SRDX plants compared to wild type even under nitrogen-deficient conditions, and NRT1.1 was induced higher in shoots of 35S:GATA4-SRDX plants than in wild type (Figure 5B).
Figure 5. Expression of nitrogen transporter genes. Gene expression in roots (A) and shoots (B) of 13 DAS plants grown under different nitrogen conditions, measured by qRT-PCR, and all expression levels were normalized to that of PP2AA3 (At1g3320), reference gene. Values are means±SD of four independent biological replicates. Single and double asterisks indicate significant differences at p< 0.05 and 0.01, respectively, in t-test when compared to wild type grown in each nitrogen condition.
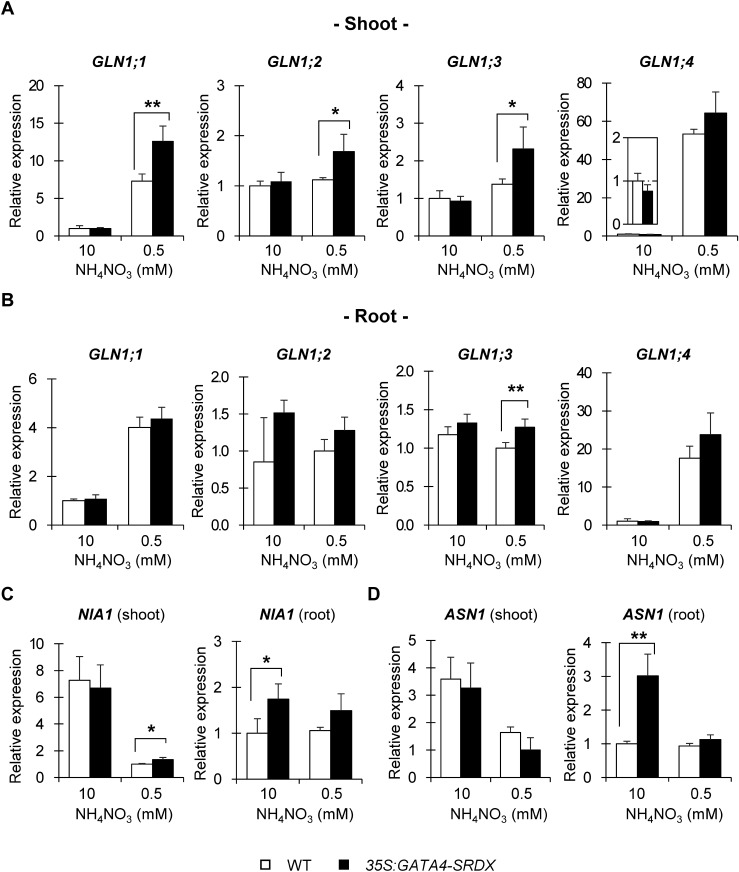
Glutamine synthetases assimilate ammonium to glutamine and re-assimilate ammonia released by photorespiration or protein modification during senescence (Fuentes et al. 2001; Krapp 2015; Xu et al. 2012). The expression of genes encoding the cytosolic glutamine synthetases GLN1;1, GLN1;2, GLN1;3, and GLN1;4, was induced to higher levels in shoots of 35S:GATA4-SRDX plants than in wild type in response to nitrogen deficiency (Figure 6A). In addition, the absorbed nitrate from soil to plant roots is reduced to nitrite by nitrate reductase, and NIA1 is one of isoforms of nitrate reductases in Arabidopsis (Wilkinson and Crawford 1993). The expression of NIA1 was higher in roots of 35S:GATA4-SRDX plants than in wild type when the plants were grown in nitrogen-sufficient conditions (Figure 6C). This expression pattern is similar to the upregulation of NRT1.1 and NRT2.1, which are induced by nitrate in roots of 35S:GATA4-SRDX plants grown in nitrogen-sufficient conditions (Figure 5B). ASN1, which encodes asparagine synthetase, plays an important role in nitrogen assimilation and translocation together with glutamine synthetase (Carvalho et al. 2003; Good et al. 2004; Miflin and Lea 1976), was highly upregulated in 35S:GATA4-SRDX roots, compared with wild type under nitrogen-sufficient conditions, but the two genotypes showed similar expression levels under nitrogen-deficient conditions (Figure 6D). These results indicate that the tolerance of 35S:GATA4-SRDX plants to nitrogen deficiency may be due to differential upregulation of genes related to nitrogen transport and nitrogen assimilation in response to nitrogen status.
Figure 6. Expression of nitrogen assimilation-related genes. Gene expression in shoots (A) and roots (B) of 13 DAS plants grown under different nitrogen conditions, measured by qRT-PCR, and all expression levels were normalized to that of PP2AA3 (At1g3320), reference gene. (A, B) Relative expression of GLN1;1, GLN1;2, GLN1;3, and GLN1;4. (C, D) Relative expression of NIA1 (C) and ASN1 (D) in shoots and roots. Values are means±SD of four independent biological replicates. Single and double asterisks indicate significant differences at p< 0.05 and 0.01, respectively, in t-test when compared to wild type grown in each nitrogen condition.
Our data base analyses showed that the AMT1.1, NRT1.1, GLN1;1-4, NIA and ASN1 genes have putative GAT A binding sites in their 5’ upstream regions. However, it is unlikely that those genes are the direct targets of GAT A4, because they were upregulated in 35S:GATA4-SRDX plants. GAT A4-SRDX might repress the expression of negative regulator(s) that suppress the expression of several nitrogen metabolism-related genes.
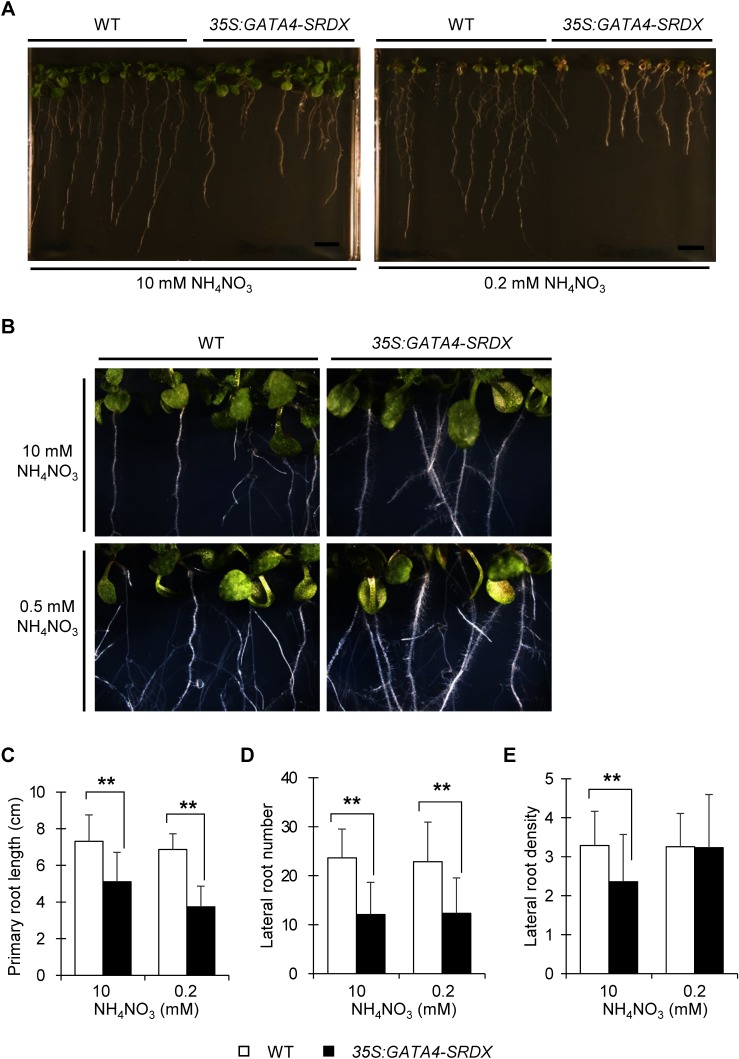
Root architecture can be modified based on nitrogen availability. Under mild nitrogen deficiency, the growth of lateral roots is promoted to expand the root surface area for nitrogen uptake, but under severe nitrogen-deficient conditions, primary and lateral root growth are suppressed (Giehl and von Wirén 2014; Gruber et al. 2013). 35S:GATA4-SRDX plants had shorter primary roots, fewer and shorter lateral roots, and more root hairs compared to wild type (Figure 7A, B). The growth of lateral and primary roots of 35S:GATA4-SRDX plants was suppressed compared with wild type in nitrogen-sufficient and deficient conditions (Figure 7A, C–E). In this experiment, we used more severe nitrogen deficient condition (0.2 mM instead of 0.5 mM), because such severe nitrogen deficiency clearly promoted the effect on root development and architecture of 35S:GATA4-SRDX plants.
Figure 7. Root architecture of wild type and 35S:GATA4-SRDX plants under different nitrogen conditions. (A) Photos of 15 DAS 35S:GATA4-SRDX plants and wild type grown vertically under nitrogen-sufficient (left; 10 mM NH4NO3) and nitrogen-deficient (right; 0.2 mM NH4NO3) conditions. Scale bar; 1 cm. (B) Root hair phenotype of 35S:GATA4-SRDX plants and wild type grown vertically under nitrogen-sufficient (top; 10 mM NH4NO3) and nitrogen-deficient (bottom; 0.5 mM NH4NO3) conditions, at 11 DAS. (C) Primary root length of 15 DAS seedlings of 35S:GATA4-SRDX plants and wild type. (D) Number of lateral root initiations. (E) Lateral root density (number of lateral roots divided by primary root length). Values are means±SD (n=35–50). Total replicates of 35S:GATA4-SRDX plants contain four independent lines. Double asterisks indicate significant difference at p< 0.01 in t-test when compared to wild type grown in each nitrogen condition.
These results suggest that the GAT A4 transcription factor is involved in the regulation of root development and GAT A4-SRDX altered root architectures that affect nitrogen availability for plants. Increased root hair density may extend root surface area, thus increasing absorption of nutrients and water from soil compared to lateral roots (Gilroy and Jones 2000; Marschner 1995). As we demonstrated above, the expression of ASN1 was highly increased in roots of 35S:GATA4-SRDX plants (Figure 6B). Considering that ASN1 is expressed in root hairs and in the elongation and maturation zones of the root (Brady et al. 2007; Schultz et al. 2017), the upregulation of ASN1 in the roots of 35S:GATA4-SRDX plants might be related to the root phenotype. Further experiments will be required to elucidate the molecular mechanisms responsible for relationship between nitrogen assimilation and root development including the roles of GAT A4 in root development and in nitrogen metabolism regardless of the alteration of root architecture.
Acknowledgments
We thank Ms. Yoshimi Sugimoto and Ms. Miyoko Yamada for their technical assistance.
References
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Denneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Carvalho HG, Lopes-Cardoso IA, Lima LG, Melo PM, Cullimore JV (2003) Nodule-specific modulation of glutamine synthetase in transgenic Medicago truncatula leads to inverse alterations in asparagine synthetase expression. Plant Physiol 133: 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Forde BG (2002) Molecular and developmental biology in inorganic nitrogen nutrition. Arabidopsis Book 1: e0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Lemaître T, Christ C, Azzopardi M, Kato Y, Sato F, Morot-Gaudry JF, Dily FL, Masclaux-Daubresse C (2008) Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol 147: 1437–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engineer CB, Kranz RG (2007) Reciprocal leaf and root expression of AtAmt1.1 and root architectural changes in response to nitrogen starvation. Plant Physiol 143: 236–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG (2000) Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta 1465: 219–235 [DOI] [PubMed] [Google Scholar]
- Fuentes S, Allen D, Ortiz-Lopez A, Hernández G (2001) Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J Exp Bot 52: 1071–1081 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wirén N (1999) Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11: 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl RFH, von Wirén N (2014) Root nutrient foraging. Plant Physiol 166: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Jones DL (2000) Through form to function: Root hair development and nutrient uptake. Trends Plant Sci 5: 56–60 [DOI] [PubMed] [Google Scholar]
- Good AG, Shrawat AK, Muench DG (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9: 597–605 [DOI] [PubMed] [Google Scholar]
- Gruber BD, Giehl RFH, Friedel S, von Wirén N (2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S (2013) Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun 4: 1617. [DOI] [PubMed] [Google Scholar]
- Krapp A (2015) Plant nitrogen assimilation and its regulation: A complex puzzle with missing pieces. Curr Opin Plant Biol 25: 115–122 [DOI] [PubMed] [Google Scholar]
- Kronzucker HJ, Siddiqi MY, Glass ADM (1997) Conifer root discrimination against soil nitrate and the ecology of forest succession. Nature 385: 59–61 [Google Scholar]
- Kurai T, Wakayama M, Abiko T, Yanagisawa S, Aoiki N, Ohsugi R (2011) Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biol J 9: 826–837 [DOI] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF (1999) CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11: 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield IW, Devlin PF, Jen CH, Westhead DR, Gilmartin PM (2006) Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol 143: 941–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants, 2nd ed. Academic Press, London
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010) Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann Bot (Lond) 105: 1141–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miflin RD, Lea PJ (1976) The pathway of nitrogen assimilation in plants. Phytochemistry 15: 873–885 [Google Scholar]
- Orkin SH (1992) GATA-binding transcription factors in hematopoietic cells. Blood 80: 575–581 [PubMed] [Google Scholar]
- Orsel M, Krapp A, Daniel-Vedele F (2002) Analysis of the NRT2 nitrate transporter family in Arabidopsis: Structure and gene expression. Plant Physiol 129: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz ER, Zupanska AK, Sng SJ, Paul AL, Rerl RJ (2017) Skewing in Arabidopsis roots involves disparate environmental signaling pathways. BMC Plant Biol 17: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Műller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible WR, Krapp A (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53: 959–970 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Gutiérrez RA (2008) A system view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol 11: 521–529 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Neumann G, Bangerth F, Engels C (2000) Rapid effects of nitrogen form on leaf morphogenesis in tobacco. J Exp Bot 51: 227–237 [DOI] [PubMed] [Google Scholar]
- Wang R, Liu D, Crawford N (1998) The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc Natl Acad Sci USA 95: 15134–15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Crawford NM (1993) Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol Gen Genet 239: 289–297 [DOI] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63: 153–182 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T (2004) Metabolic engineering with Dof transcription factor in plants: Improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA 101: 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]