Abstract
The unfolded protein response (UPR) mitigates stress caused by accumulation of unfolded proteins in the endoplasmic reticulum (ER). Inositol-requiring enzyme 1 (IRE1) is the most conserved sensor of the UPR with ribonuclease activity that mediates cytoplasmic splicing and decay of mRNA encoding secretory and membrane proteins. In the present study, we demonstrate that the Arabidopsis mutant defective in two IRE1 genes exhibit retarded growth of primary roots under moderate salt stress, although such grow retardation is not observed in wild type plants. Microscopic observation showed decrease in the number of meristematic cells in the mutant under salt stress. This finding suggests that IRE1 plays a role in the maintenance of root meristems under salt stress. Possible connections between the function of IRE1 and the salt sensitivity are discussed.
Keywords: Arabidopsis, endoplasmic reticulum, salt stress, unfolded protein response
Endoplasmic reticulum (ER) is a cellular factory producing one third of proteins transported to their final destination via vesicle transport. Before leaving from the ER, proteins need to be correctly folded. The system monitoring protein folding and correcting misfolded proteins is known as ER quality control (ERQC) (Kleizen and Braakman 2004). If ERQC is disturbed, the unfolded protein response (UPR) is activated to restore ERQC by inducing a set of genes encoding ER chaperones and folding enzymes. The most conserved sensor molecule of the UPR is inositol-requiring enzyme 1 (IRE1) consisting of three domains; the sensor domain in the ER, and the kinase and RNase domains in the cytoplasm. IRE1 catalyzes unconventional cytoplasmic splicing and activates bZIP transcription factors (Walter and Ron 2011). A resulting bZIP transcription factor induces expression of genes for ERQC. In addition to cytoplasmic splicing of bZIP transcription factors, IRE1 mediates degradation of mRNA encoding secretory and membrane proteins, which are translated in ribosomes on the ER membrane. This degradation of mRNA is known as regulated IRE1-dependent decay (RIDD) and considered to alleviate the overloading of proteins into the stressed ER (Hollien et al. 2009).
Arabidopsis has two IRE1 homologs, IRE1A and IRE1B (Koizumi et al. 2001), which target bZIP60 mRNA for cytoplasmic splicing to generate the active transcription factor bZIP60 (Deng et al. 2011; Nagashima et al. 2011). In addition, IRE1A and IRE1B function in RIDD (Mishiba et al. 2013) as in other organisms. A broad range of mRNAs encoding secretory and membrane proteins are destabilized through RIDD in Arabidopsis. Importantly, the ire1a ire1b double mutant, which is defective in both RIDD and cytoplasmic splicing, which exhibits higher sensitivity to ER stress-inducing drugs than does the bzip60 mutant, suggesting the importance of RIDD to alleviate ER stress.
Since the identification of signaling molecules of the plant UPR, its physiological importance has been reported (Chen and Brandizzi 2013; Howell 2013; Iwata and Koizumi 2012). Some external stimuli such as heat stress, salicylic acid treatment, and viral infection have been reported to activate bZIP60 and bZIP28 (Deng et al. 2011; Gao et al. 2008; Moreno et al. 2012; Nagashima et al. 2014; Ye et al. 2011; Zhang et al. 2015). Furthermore, it was reported that an Arabidopsis mutant defective in both IRE1A and IRE1B genes grown under high temperature contains less viable pollen grains, indicating the importance of the UPR in pollen development under high temperature (Deng et al. 2016). However, whether the UPR plays important roles in salt stress tolerance has not been investigated, although there has been some reports showing the connection between salt stress tolerance and ER-associated degradation, which is part of ERQC machineries induced during the UPR (Cui et al. 2012; Liu et al. 2011).
Arabidopsis thaliana Col-0 and T-DNA insertion mutants in Col-0 backgrounds were used in this study. bzip60-1 (SALK_050203), ire1a-2 (SALK_018112), ire1b-1 (GABI_638B07), and the double mutant ire1a-2 ire1b-1 were previously described (Iwata et al. 2008; Nagashima et al. 2011). Sterilized Arabidopsis seeds were sown on half strength MS plates with 1% sucrose and 0.8% agar containing the indicated concentration of NaCl, LiCl, or mannitol. After two days at 4°C they were incubated vertically at 23°C under 16-h light/8-h dark for the indicated period.
Plants were photographed and length of primary roots was measured using ImageJ (https://imagej.nih.gov/ij/). Root tips were stained with 50 µg ml−1 FM4-64 and observed under the confocal laser microscope LSM 700 (Zeiss). Cells showing no signs of rapid elongation were considered as root meristematic cells.
For complementation experiments, genomic fragments of IRE1A and IRE1B were amplified with primers (5′-CAC CCG TCT AGG ACG CCT AGG CAC-3′ and 5′-CAG AAA CGA TGG ATG TTT TCC CG-3′ for IRE1A and 5′-CAC CGT TGA TAC TCA CGG AAG TCG G-3′ and 5′-GGG TAC GGG TCT TTC AGA TTG-3′ for IRE1B) using wild-type genomic DNA as template and cloned into pENTR/D-TOPO vector (Invitrogen). They were then transferred to pSMAB binary vector using Gateway LR Clonase II Enzyme Mix (Invitrogen) and used for transformation of the ire1a-2 ire1b-1 mutants by the floral dip method (Zhang et al. 2006).
We investigated the effect of salt stress on root growth and morphology in ire1a ire1b. When wild-type and bzip60-1, ire1a-2, ire1b-1, and ire1a-2 ire1b-1 mutant plants were grown on medium containing 50 mM NaCl, growth of primary roots of ire1a-2 ire1b-1 were severely inhibited (Figure 1A). However, such growth inhibition was not observed in wild-type, bzip60, ire1a-2 and ire1b-1. Length of primary roots of ire1a-2 ire1b-1 mutants was shorter than the wild-type plants (72%) even without NaCl in our growth condition, consistent with the previous observation that the ire1a ire1b double mutants exhibit shorter primary roots compared to the wild-type plants (Chen and Brandizzi 2012). With 50 mM NaCl, length of primary root of ire1a-2 ire1b-1 mutants was 22% of that of wild-type plants whereas other mutants did not show significant difference with wild-type plants (Figure 1B). Since only ire1a-2 ire1b-1 mutants showed distinct phenotype, we used wild-type and ire1a-2 ire1b-1 mutant plants for later experiments. In addition to inhibition of elongation of primary roots, the ire1a-2 ire1b-1 mutant exhibited bushy roots with 50 mM NaCl (Figure 1C).
Figure 1. Inhibition of elongation of primary roots by NaCl. (A) Seeds of wild-type (WT), bzip60-1 (bzip60), ire1a-2 (ire1a), ire1b-1 (ire1b), and ire1a-2 ire1b-1 (ire1ab) plants were sown on medium containing 0 mM and 50 mM NaCl, grown for 10 days at 22°C, and photographed. (B) Quantification of primary root length. The growth condition was same as in (A). Values are means with standard errors from three independent experiments. Asterisks indicate significant differences between the wild-type and the indicated mutant plants (Student’s t-test, p<0.01, n=6). (C) Enlarged picture of 10-day-old ire1a-2 ire1b-1 seedlings grown on medium containing 0 mM and 50 mM NaCl.
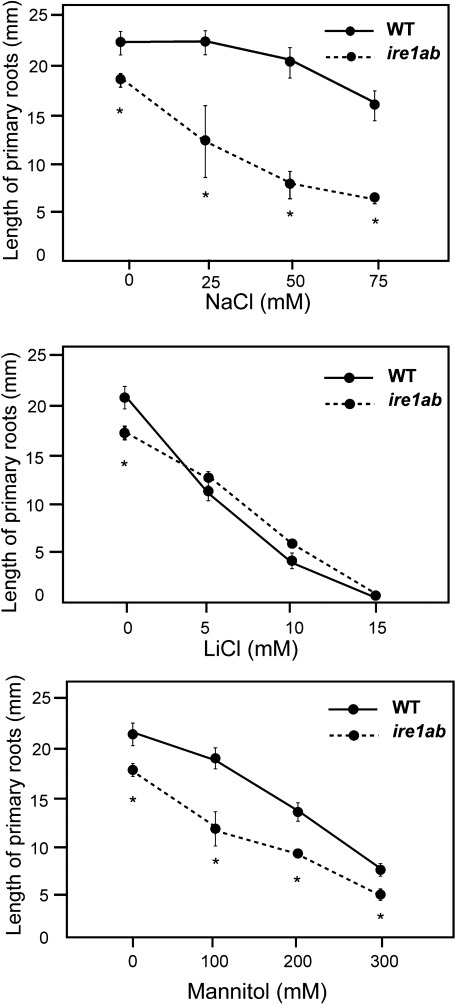
Growth of plants on NaCl-containing medium is affected by both ionic and osmotic stresses. Therefore, we examined the effects of LiCl and mannitol as ionic and osmotic stress, respectively, on growth of primary roots. As shown in Figure 2, mannitol inhibited elongation of primary roots of ire1a-2 ire1b-1 mutant seedlings more severely than that of wild-type seedlings as did NaCl, although the effect of mannitol is weaker than that of NaCl. In contrast, growth of ire1a-2 ire1b-1 mutant roots in LiCl was similarly inhibited as that of wild-type roots. It indicates that osmotic stress is the cause of the growth retardation of ire1a-2 ire1b-1 roots.
Figure 2. Effects of NaCl, LiCl and mannitol on elongation of primary roots. The length of primary roots of wild-type (WT) and ire1a-2 ire1b-1 (ire1ab) on medium containing indicated concentrations of NaCl, LiCl, or mannitol were measured 7 days after germination. Values are means with standard errors from three independent experiments. Asterisks indicate significant differences between the mutant and the wild-type (Student’s t-test, p<0.01, n=6).
Since elongation of roots was inhibited by moderate NaCl stress, root tips were observed using a differential interference contrast microscope. As shown in Figure 3A, the cell number of the meristematic zone in ire1a-2 ire1b-1 mutant roots grown on 50 mM NaCl was significantly less than that of wild-type roots. Furthermore, the length of the meristematic zone in ire1a-2 ire1b-1 mutant roots grown on 50 mM NaCl was 73% that of wild-type roots (Figure 3B, C). We conclude that it is the cell number rather than the cell size that decreased in the meristematic zone of salt-stressed ire1a-2 ire1b-1 roots, although the cell size in the elongation zone might also be affected in the mutant, which could also account for the short-root phenotype.
Figure 3. Observation of root tip. (A) Representative pictures of root tip of wild-type (WT) and ire1a-2 ire1b-1 (ire1ab) on medium containing 0 mM and 50 mM NaCl. Cells marked by blue and red were considered as meristematic and elongated cells, respectively. The photo was taken 7 days after germination under a differential interference contrast microscope. (B) Representative pictures of root tip as in (A) using a confocal laser microscope. The photo was taken 7 days after germination. White arrowheads indicate the end of the meristem. (C) Quantification of length of meristematic regions in (B). Values are means with standard errors from three independent experiments. Asterisks indicate significant differences between the two adjacent groups (Student’s t-test, p<0.01, n=5).
To verify NaCl sensitive phenotype is due to loss of both IRE1 genes, a genomic fragment of IRE1A or IRE1B was introduced into ire1a-2 ire1b-1 mutants. As shown in Figure 4, the salt sensitive phenotype observed in ire1a-2 ire1b-1 mutants was recovered in these complementation lines, showing that NaCl sensitivity was caused by loss of both IRE1 genes.
Figure 4. Complementation of ire1a-2 ire1b-1 mutant phenotypes with IRE1A and IRE1B. (A) Seedlings of wild-type (WT), ire1a-2 ire1b-1 (ire1ab), and its complementation lines on 50 mM NaCl plates. Seeds of indicated genotypes were sown on medium containing 50 mM NaCl, grown at 22°C for 10 days, and photographed. (B) Quantification of length of primary roots. The growth condition was same as in (A). Values are means with standard errors from three independent experiments. Asterisks indicate significant differences between the wild-type and the mutant or complementation plants (Student’s t-test, p<0.01, n=6).
In the present study, we investigated whether mutants deficient in IRE1 genes show enhanced salt stress sensitivity since the ERQC has been implicated in salt stress responses in plant (Cui et al. 2012; Liu et al. 2011). In our growth condition, ire1a-2 ire1b-1 mutant seedlings showed shorter roots, consistent with the previous report (Chen and Brandizzi 2012). We observed significant inhibition of primary root elongation in ire1a-2 ire1b-1 seedlings grown on medium containing 50 mM NaCl. Furthermore, the ire1a-2 ire1b-1 seedlings exhibited bushy root architecture on NaCl-containing medium. In contrast to the root phenotype, no obvious growth retardation was observed in shoots. These characteristics are distinct from those observed in well-characterized salt overly sensitive mutants, sos1 (Shi et al. 2000), sos2 (Guo et al. 2001) and sos3 (Ishitani et al. 2000), which exhibit growth arrest of shoots under salt stress but do not exhibit bushy root architecture.
It has been reported that salt stress can inhibit root elongation under moderate salt conditions (Wang et al. 2009). It is well known that auxin plays a critical role in orchestrating root development. A recent study reported that salt stress affects Arabidopsis root meristem maintenance, in part, through changes in redox status and auxin transport (Jiang et al. 2016). Therefore, our observation that the ire1a-2 ire1b-1 mutant exhibits retarded primary root growth and bushy root architecture could be attributed to altered auxin distribution in roots. Indeed, connection of IRE1 and auxin transport was reported previously in Arabidopsis roots (Chen et al. 2014). Other plant hormones might also be involved, because abscisic acid and gibberellic acid have been reported to play roles in root growth inhibition under salt stress (Duan et al. 2013).
In Arabidopsis, IRE1 mediates cytoplasmic splicing of bZIP60 and RIDD. Since changes in root architecture under moderate salt stress were not observable in bzip60-1, it is likely that a defect in RIDD accounts for the observed phenotypes. Nevertheless, we do not exclude a possibility that salt-sensitive phenotype of ire1a-2 ire1b-1 is due to undiscovered functions of IRE1 such as phosphorylation of the JNK protein reported in mammalian cells (Urano et al. 2000).
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 17K07450 to N.K.
References
- Chen Y, Aung K, Rolčík JC, Walicki K, Friml J, Brandizzi F (2014) Inter-regulation of the unfolded protein response and auxin signaling. Plant J 77: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Brandizzi F (2013) IRE1: ER stress sensor and cell fate executor. Trends Cell Biol 23: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Brandizzi F (2012) AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J 69: 266–277 [DOI] [PubMed] [Google Scholar]
- Cui F, Liu L, Zhao Q, Zhang Z, Li Q, Lin B, Wu Y, Tang S, Xie Q (2012) Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24: 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Humbert S, Liu J-X, Srivastava R, Rothstein SJ, Howell SH (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA 108: 7247–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Quilichini TD, Dong H, Bao Y, Horner HT, Howell SH (2016) IRE1, a component of the unfolded protein response signaling pathway, protects pollen development in Arabidopsis from heat stress. Plant J 88: 193–204 [DOI] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR (2013) Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Brandizzi F, Benning C, Larkin RM (2008) A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 16398–16403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu JK (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13: 1383–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS (2009) Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186: 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SH (2013) Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol 64: 477–499 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12: 1667–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N (2008) Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N (2012) Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci 17: 720–727 [DOI] [PubMed] [Google Scholar]
- Jiang K, Moe-Lange J, Hennet L, Feldman LJ (2016) Salt stress affects the redox status of Arabidopsis root meristems. Front Plant Sci 7: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleizen B, Braakman I (2004) Protein folding and quality control in the endoplasmic reticulum. Curr Opin Cell Biol 16: 343–349 [DOI] [PubMed] [Google Scholar]
- Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ (2001) Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases 1. Plant Physiol 127: 949–962 [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cui F, Li Q, Yin B, Zhang H, Lin B, Wu Y, Xia R, Tang S, Xie Q (2011) The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res 21: 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiba K, Nagashima Y, Suzuki E, Hayashi N, Ogata Y, Shimada Y, Koizumi N (2013) Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc Natl Acad Sci USA 110: 5713–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, Chen Y, Brandizzi F, Dong X, Orellana A, et al. (2012) IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One 7: e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y, Iwata Y, Ashida M, Mishiba K, Koizumi N (2014) Exogenous salicylic acid activates two signaling arms of the unfolded protein response in Arabidopsis. Plant Cell Physiol 55: 1772–1778 [DOI] [PubMed] [Google Scholar]
- Nagashima Y, Mishiba K-I, Suzuki E, Shimada Y, Iwata Y, Koizumi N (2011) Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287: 664–666 [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D (2011) The unfolded protein response: From stress pathway to homeostatic regulation. Science 334: 1081–1086 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li K, Li X (2009) Auxin redistribution modulates plastic development of root system architecture under salt stress in Arabidopsis thaliana. J Plant Physiol 166: 1637–1645 [DOI] [PubMed] [Google Scholar]
- Ye C, Dickman MB, Whitham SA, Payton M, Verchot J (2011) The unfolded protein response is triggered by a plant viral movement protein 1. Plant Physiol 156: 741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chen H, Brandizzi F, Verchot J, Wang A (2015) The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet 11: e1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin S-S, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]