Abstract
Stathmin is a ubiquitous regulatory phosphoprotein, the generic element of a family of neural phosphoproteins in vertebrates that possess the capacity to bind tubulin and interfere with microtubule dynamics. Although stathmin and the other proteins of the family have been associated with numerous cell regulations, their biological roles remain elusive, as in particular inactivation of the stathmin gene in the mouse resulted in no clear deleterious phenotype. We identified stathmin phosphoproteins in Drosophila, encoded by a unique gene sharing the intron/exon structure of the vertebrate stathmin and stathmin family genes. They interfere with microtubule assembly in vitro, and in vivo when expressed in HeLa cells. Drosophila stathmin expression is regulated during embryogenesis: it is high in the migrating germ cells and in the central and peripheral nervous systems, a pattern resembling that of mammalian stathmin. Furthermore, RNA interference inactivation of Drosophila stathmin expression resulted in germ cell migration arrest at stage 14. It also induced important anomalies in nervous system development, such as loss of commissures and longitudinal connectives in the ventral cord, or abnormal chordotonal neuron organization. In conclusion, a single Drosophila gene encodes phosphoproteins homologous to the entire vertebrate stathmin family. We demonstrate for the first time their direct involvement in major biological processes such as development of the reproductive and nervous systems.
INTRODUCTION
During development and in the adult, cell proliferation, differentiation, and activities involve numerous intracellular events among which cytoskeleton dynamics under the control of integrated cell signaling plays major roles. In neurons, microtubules play specific roles in process outgrowth, pathfinding, and synapse formation, as well as in axonal and dendritic transport.
Stathmin (Sobel, 1991), also named Op18 (Hailat et al., 1990), is a ubiquitous cytosolic phosphoprotein interfering with microtubule assembly and highly expressed in the early embryo, gonads, and the nervous system. Stathmin has been originally characterized as an intracellular relay integrating diverse signaling pathways through combinatorial phosphorylation on four sites within its N-terminal regulatory region (reviewed by Sobel, 1991; Lawler, 1998). It also possesses a C-terminal interaction domain made of two partially repeated stretches predicted as α-helical coiled-coil–forming structures (Maucuer et al., 1990). In agreement with this structural model, several potential stathmin-interacting proteins have been identified (Maucuer et al., 1995; Manceau et al., 1999), among which tubulin appeared recently as a major stathmin target (Belmont and Mitchison, 1996). Stathmin overexpression inhibits the microtubule network in vivo, this effect being impaired by phosphorylation (Marklund et al., 1996; Horwitz et al., 1997; Gavet et al., 1998). Although it was originally proposed that microtubule destabilization by stathmin results from direct catastrophe promotion (Belmont and Mitchison, 1996), it is actually due at least in part to sequestration of free tubulin, because stathmin interacts with tubulin in a phosphorylation-dependent manner, to form a T2S complex of one stathmin (S) and two tubulin (T) α/β heterodimers (Curmi et al., 1997; Jourdain et al., 1997; Gigant et al., 2000).
Stathmin is the generic element of a phosphoprotein family, including neural proteins SCG10, SCLIP, and RB3/RB3′/RB3", conserved in vertebrates and highly expressed in the nervous system (Maucuer et al., 1993; Ozon et al., 1997, 1998). They share a stathmin-like domain that interacts with tubulin (Charbaut et al., 2001), and are able to impair microtubule assembly in vitro and in vivo (Antonsson et al., 1998; Gavet et al., 1998). Each stathmin-related protein further contains a variable N-terminal extension able to target it to vesicular, Golgi-like membranes (Stein et al., 1988; Di Paolo et al., 1997; Gavet et al., 1998). The various members of the stathmin family most likely play specific, possibly complementary roles in the development, maturation, and functional regulation of the nervous system (Curmi et al., 1999), because they display specific expression and regulatory patterns (Himi et al., 1994; Beilharz et al., 1998; Ozon et al., 1998, 1999). Although it is clear that the actions of stathmin family proteins are at least in part mediated by their interaction with tubulin, their biological roles are not fully characterized in vertebrates. In particular, stathmin gene inactivation in the mouse did not induce any major phenotype (Schubart et al., 1996), probably because of redundancies and compensation phenomena.
To further investigate the function of stathmin family proteins in relation with their role in the control of microtubule dynamics, we identified and characterized the single stathmin-related gene in Drosophila, that we designate the D-stathmin gene. D-stathmin proteins display characteristic molecular and biochemical properties of vertebrate stathmin, they are phosphorylated in vivo when expressed in HeLa cells, and interfere with microtubule assembly in vitro and in vivo. During Drosophila embryogenesis, expression of D-stathmin is mainly restricted to two cell types: migrating germ cells and neurons of the central and peripheral nervous system. Furthermore, impairment of D-stathmin expression by RNA interference allows to demonstrate for the first time a direct and essential role of stathmin in germ cell migration and in the formation of the nervous system, possibly through its microtubule-destabilizing activity.
MATERIALS AND METHODS
cDNA Library Screening
A 900-base pair 32P random multiprime-labeled cDNA probe, obtained by EcoRI/XhoI digestion from Expressed Sequence Tag (EST) clone GM04023 (GenBank accession number AA802209) from the Berkeley Drosophila Genome Project, was used to screen 5 × 105 phages of a λgt11 0-16H Drosophila embryo cDNA library (Hovemann et al., 1991) according to standard protocols (Sambrook et al., 1989), with a final wash at 68°C in 0.1× SSC, 0.1% SDS for 30 min. Phage DNAs were extracted and the Drosophila cDNA inserts were subcloned in the Bluescript plasmid. The clones were analyzed by restriction and sequenced in both directions.
RNA Preparation and Northern Blot
Adult Drosophila flies were homogenized and total RNA was prepared as described (Chomczynski and Sacchi, 1987). Total RNA (20 μg) was electrophoresed on a denaturing formaldehyde agarose gel, transferred to a Hybond N+ membrane (Amersham plc, Little Chalfont, Buckinghamshire, UK) in 20× SSC and stained with methylene blue. Prehybridization was performed in hybridization buffer [0.25 mg/ml salmon DNA, 0.01 M piperazine-N,N′-bis(2-ethanesulfonic acid) pH 6.4, 0.01 M EDTA, 5× SSC, 5× Denhardt's solution, 1% SDS, 50% formamide] at 45°C for 2 h. Multiprime-labeled fragments of the EST clone AA802209 were added at 106 cpm/ml in the hybridization buffer and hybridization was allowed to proceed overnight. The final wash was performed at 68°C in 0.1× SSC, 0.1% SDS for 30 min.
Eukaryotic and Prokaryotic Expression
For eukaryotic expression, D-stathmin cDNAs were amplified by polymerase chain reaction with 5′ and 3′ primers containing KpnI and BamHI restriction sites, respectively, to allow their insertion into the pcDNA3-myc vector, in phase with the myc epitope (Lawler et al., 1998). For bacterial expression, D-stathmin cDNAs were amplified by polymerase chain reaction with 5′ and 3′ primers containing NcoI and BamHI restriction sites, respectively, to allow the insertion of the coding sequence into the pET-8c vector (Novagen, Madison, WI). The resulting cDNA clones were verified by sequencing and used to produce the corresponding proteins in the BL-21(DE3) Escherichia coli strain. After induction, cells were collected by centrifugation and sonicated in 10 mM Tris-HCl, pH 7.4, with an antiprotease cocktail (Complete; Roche Molecular Biochemicals, Mannheim, Germany). Differential centrifugations were performed as described (Ozon et al., 1997). Briefly, the S1/C1, S2/C2 and S3/C3 are, respectively, supernatant and pellet of a 1000 × g centrifugation for 10 min of the initial homogenate, of a 100,000-rpm centrifugation in a Beckman TL-100 for 6 min of S1, and of S2 heated at 100°C for 5 min.
In Vitro Protein Expression
Bluescript plasmid clone 14 (1 μg) was used for in vitro transcription and translation with the TNT Coupled Reticulocyte Lysate System (Promega, Madison, WI) and [35S]methionine as described by the manufacturer. Five microliters of 25 μl of total transcription/translation mix was analyzed by gel electrophoresis.
Antiserum Production
Rabbits were immunized with 1 mg of synthetic KLH-coupled peptide (Neosystem, Strasbourg, France) corresponding to the C-terminal sequence of D-stathmin-A (GQQSAIASSG), or with 0.1 mg of partially purified recombinant D-stathmin-ΔC in complete Freund's adjuvant. They were boosted every 3 wk with half the initial amount of peptide or protein in incomplete Freund's adjuvant.
PAGE (SDS-PAGE) and Western Blotting
One-dimensional gel electrophoresis was performed on 13% polyacrylamide gels (Laemmli, 1970). Two-dimensional gels were performed as described (Ozon et al., 1997). The isoelectric focusing gel contained 2% ampholines pH 3.5–10.5, and the second dimension was run on 13% polyacrylamide gels. The gels were transferred to nitrocellulose in a semidry electroblotting apparatus and probed with diluted antiserum [antipeptide COOH-terminal antiserum (1:1000), anti-D-stathmin-ΔC antiserum (1:10,000), anti-myc monoclonal antibody (mAb) (1:2000) (DAKO, Glostrup, Denmark)]. Bound antibodies were detected with a goat antirabbit or antimouse antiserum coupled to peroxidase (1:10,000) (DAKO) and the chemiluminescent ECL kit (Amersham plc).
Cell Culture and DNA Transfection
Human HeLa cells were grown as monolayers in DMEM containing 10% (vol/vol) fetal calf serum (Invitrogen Corporation, Carlsbad, CA) at 37°C in 5% CO2. Transfections were performed using LipofectAMINE (Invitrogen) according to the manufacturer's instructions. To prepare interphasic cell extracts (see below), transfected cells were collected 24 h posttransfection (Gavet et al., 1998). For mitotic cell extracts, cells were collected 48 h posttransfection after an incubation period of 16 h with 1 μM taxol (Paclitaxel; Aventis, Strasbourg, France).
Cell and Embryo Extracts
Embryos and cells for fractionation were homogenized in a Dounce homogenizer in 10 volumes of 10 mM Tris-HCl pH 7.5, 1 mM EDTA, 25 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin and fractionated by differential centrifugation as described above. For phosphorylation studies, transfected cells were extracted with homogenization buffer [80 mM piperazine-N,N′-bis(2-ethanesulfonic acid) KOH pH 6.8, 5 mM MgCl2, 1 mM EGTA, 150 mM NaCl, 0.2% NP-40, Complete protease inhibitor cocktail] for 10 min at 4°C and centrifuged at 10,000 × g for 10 min. For dephosphorylation reactions, 100 μg of proteins was incubated with 1600 U of Lambda phosphatase (New England Biolabs, Beverly, CA) in the corresponding buffer for 1 h at 30°C. Efficiency of dephosphorylation reactions was checked on endogenous human stathmin. Tranfected myc-tagged proteins were analyzed by Western blot with 1:1000 monoclonal anti-myc antibody 9E10 (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunofluorescence Analysis
HeLa cells were fixed with phosphate-buffered saline (PBS) + 2% paraformaldehyde and 30 mM saccharose for 10 min at 23°C. They were successively treated with PBS + 0.1% Triton X-100 for 10 min, PBS + 50 mM NH4Cl for 10 min, and PBS + 3% bovine serum albumin for 15 min before incubation for 1 h with primary antibodies (1:300 monoclonal anti-α tubulin N356; Amersham plc; 1:100 polyclonal anti-myc sc-789; Tebu). The primary antibodies were revealed with appropriate rhodamine or fluorescein-conjugated anti-rabbit (1:300) and anti-mouse (1:300) secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA). The cells were mounted with AF1 antifade mountant solution (Citifluor, Leicester, UK) and examined with a Provis Olympus fluorescence photomicroscope equipped with a Princeton Instruments camera.
In Vitro Microtubule Polymerization
Tubulin was purified from calf brain by two cycles of polymerization/depolymerization followed by chromatography on phosphocellulose and an additional cycle of polymerization/depolymerization as described (Curmi et al., 1997). Tubulin polymerization was monitored turbidimetrically at 350 nm in an Ultrospec 3000 spectrophotometer (Amersham plc) thermostated at 37°C. Experiments were carried out as described (Jourdain et al., 1997; Charbaut et al., 2001) in buffer M [50 mM 2-(N-morpholino)ethanesulfonic acid-KOH pH 6.8, 30% glycerol, 0.5 mM EGTA, 6 mM MgCl2, and 0.5 mM GTP]. Increasing amounts of D-stathmin-A or -ΔC S3 extracts were added to the polymerization buffer M. The negative control is an S3 from BL21 bacteria transformed with an empty pET vector. The concentrations of D-stathmins were estimated by Coomassie blue staining comparison with an amino acid analysis-assayed RB3SLD protein (Charbaut et al., 2001).
Embryo and Egg Chamber In Situ Hybridization and Immunohistochemical Staining
RNA in situ hybridization was performed as described (Tautz and Pfeifle, 1989). Briefly, Dig-U–labeled RNA was synthesized from a linearized plasmid template containing cDNA insert of EST GM04023. Fixed embryos were hybridized with Dig-U–labeled RNA overnight at 55°C and then incubated with alkaline phosphatase-conjugated anti-digoxigenin antibodies. The signal was developed using the alkaline phosphatase reaction. For examination, egg chambers and embryos were mounted in Aqua-Polymount (Polysciences, Warrington, PA). For sectioning, embryos were mounted in araldite according to Roth et al. (1989).
Immunostaining was performed according to Roth et al. (1989) with 1:10 22C10 mAb (Fujita et al., 1982), or 1:2000 D-stathmin polyclonal antibody for which we checked that the immunoreactive signal was inhibited by an excess of the antigen. After RNA interference, embryos were briefly washed in n-heptane and fixed in 8% paraformaldehyde saturated with heptane in PBS. Vitelline membranes were removed by hand and embryos were washed with PBS 0.1% Triton X-100 and 100% methanol.
RNA Interference
EST GM04023 has been used as DNA template for transcription of sense and antisense mRNAs with the T3 and T7 mMessage mMachine kit (Ambion, Austin, TX). For RNA interference, D-stathmin double-stranded RNA (dsRNA) was produced and injected in cleavage stage embryos as described (Kennerdell and Carthew, 1998), on the dorsal side to minimize the risk of mechanical interference with the development of the nervous system and the gonads. The same volume of buffer was injected in 570 control embryos: among the 270 recovered after fixation, 130 were examined for pole cells and 150 for central nervous system (CNS). Approximately 650 embryos were injected with D-stathmin dsRNA. Among them, 150 were left to develop, 200 were processed to follow the fate of germ cells, and 220 that of the nervous system, of which, respectively, 55 and 90 were actually examined after discarding embryos that were severely damaged either during the injection or sample preparation. The dsRNA-injected embryos considered to follow the development of the nervous system were first immunolabeled with anti-stathmin and then with the anti-Futsch 22C10 mAb to reveal the nervous system.
RESULTS
We identified Drosophila cDNA clones and the corresponding gene displaying sequence identities with vertebrate stathmin. Their sequence analysis, together with the subsequent identification of the corresponding proteins and the characterization of their biochemical and functional properties clearly identified these proteins as stathmin-like proteins. We therefore refer below to the proteins as “D-stathmins,” and to the gene as the “D-stathmin gene” (“stai”).
Identification and Sequence Analysis of Stathmin cDNAs and Gene in Drosophila
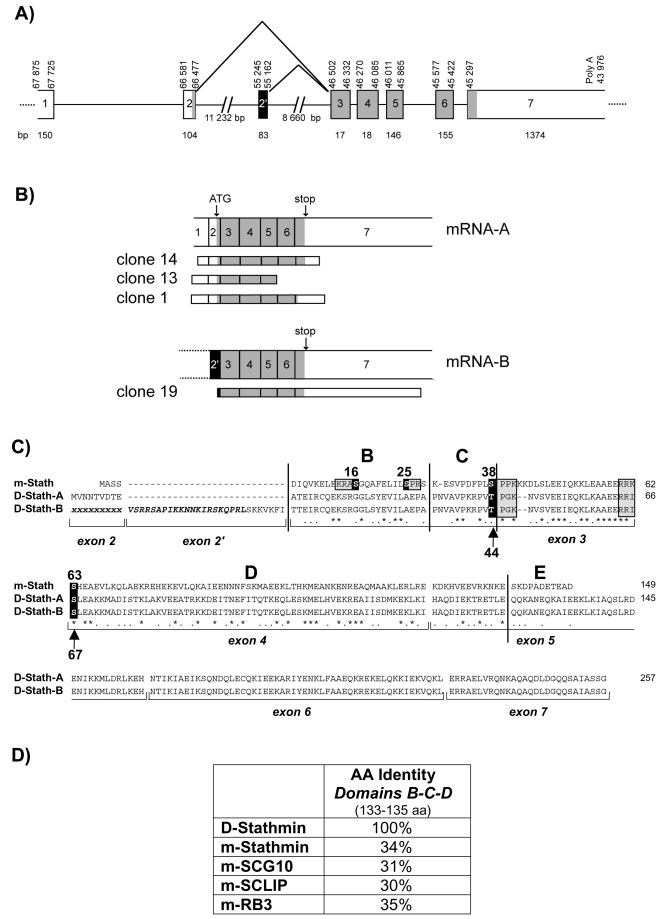
We isolated four stathmin-related cDNA clones from a 0–16-h Drosophila embryo cDNA library (Hovemann et al., 1991), corresponding to the same genomic stathmin clone (GenBank accession number AC004639) (Figure 1, A and B). Two of the clones (13 and 14) correspond to the transcription of all or part of predicted exons 1–7, containing an open reading frame coding for a protein, D-stathmin-A, of 257 amino acids (29,582 Da). Remarkably, clone 1 contains toward the end of exon 6 a stretch of eight instead of nine adenosines, which may have resulted from a transcription mistake inducing a reading frame shift and the coding of a protein of 234 amino acids (27,048 Da) not further characterized in this study.
Figure 1.
D-stathmin gene, cDNA clones, and protein sequence analyses. (A) Schematic representation of the D-stathmin gene showing the noncoding (white) and coding (gray) exons, as well as the alternatively spliced exon 2′ (black), as identified by comparison with the cDNA clones and with the acceptor/donor splice motifs. Numbers delimiting the exons correspond to nucleotide numeration of clone AAC004639. (B) mRNA-A and -B transcribed from the D-stathmin gene as predicted from both the gene and cDNAs are shown with the structures of the corresponding cDNA clones. (C) Amino acid sequence alignment of D-stathmin-A and -B with mouse (m-) stathmin [identities (∗); homologs (•)]. Exon limits and the previously described domains B, C, D, and E (Maucuer et al., 1990) are indicated. Consensus phosphorylation sites known in m-stathmin are boxed and the corresponding conserved sites in D-stathmins are indicated by arrows. (D) Amino acid identities between D-stathmin and mouse stathmin-related proteins in their aligned B-C-D domains.
Clone 19, although shorter in its 5′ end, codes for seven additional amino acids upstream from exon 3. The corresponding sequence is part of the region 55,245–55,162 of the gene, predicted with a high probability to code for an exon, numbered 2′, and coding for a stretch of 28 inserted amino acids. The corresponding protein, D-stathmin-B, is thus likely derived from the same gene by alternative splicing.
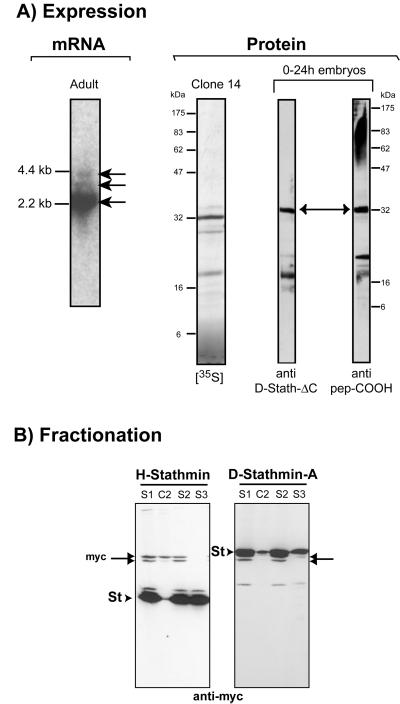
The D-stathmin gene (Figure 1A), located in the region 26B9–26C1 of chromosome 2L, contains at least eight identified exon sequences and at least seven possible polyadenylation consensus sites. Accordingly, we detected several stathmin-positive bands on adult fly RNA Northern blots, at 2.2, 3, and 4 kb (Figure 3A), indicating the likely use of various polyA sites as in mammals (Maucuer et al., 1990). Finally, like in mammals (Luo et al., 1991), we found no TATA box in the D-stathmin gene.
Figure 3.
D-stathmin expression and subcellular localization. (A) mRNA: northern blot analysis showing a major D-stathmin transcript of ∼2.2 kb and two minor ones of ∼3 and 4 kb in total adult Drosophila RNA. Protein: a major 32-kDa protein is the product of the in vitro transcription/translation of clone 14 ([35S], left) and corresponds to a major 32-kDa protein detected in 0–24-h embryo homogenates by Western blot analysis with both anti-D-stathmin antisera (right). (B) HeLa cell homogenates overexpressing human (H-) and Drosophila (D-) stathmin-myc fusion proteins were fractionated by differential centrifugation (see MATERIALS AND METHODS): low-speed supernatant (S1); 100,000 × g supernatant (S2) and pellet (C2); 100°C supernatant (S3). Equivalent amounts of each fraction were analyzed by Western blotting with anti-myc antiserum. H- and D-stathmins (arrowheads) and endogenous c-myc (double-headed arrows) are indicated.
Interestingly, the stathmin genes of Drosophila and mammals share the same intron/exon structure organization (Melhem et al., 1991; Okazaki et al., 1993): a noncoding first exon, and the same intron/exon junctions for exons 2, 3, 4, and 5 (Figure 1), also conserved for the vertebrate stathmin-related genes SCG10, SCLIP, and RB3 (Okazaki et al., 1993; Beilharz et al., 1998; Bai et al., 2000).
Drosophila Stathmin Proteins
Sequence Analysis.
The predicted amino acid sequences of D-stathmin-A and -B contain a stathmin-like domain corresponding to subdomains B-C-D of vertebrate stathmin (Maucuer et al., 1993), an additional COOH-terminal domain and, in the case of D-stathmin-B, a very basic 28 amino acid insert encoded by exon 2′ (Figure 1). Interestingly, D-stathmins are not significantly closer to any given member of the vertebrate stathmin family (Ozon et al., 1998) because they share 31–35% amino acid identity with their “stathmin-like” B-C-D domains (Figure 1D).
The regulatory and interaction domains of vertebrate stathmin can also be identified in D-stathmins.
The regulatory domain is characterized by the presence of several consensus phosphorylation sites, threonine 44 and serine 67 corresponding, respectively, to serines 38 and 63 of mammalian stathmin. They are similarly located within phosphorylation consensus sites for proline-directed kinases and protein kinase A, respectively.
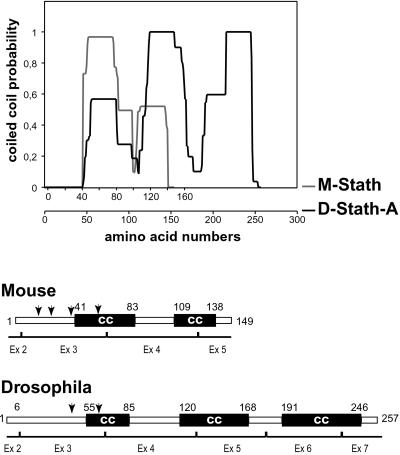
The vertebrate interaction domain is characterized by two predicted α-helical coiled-coil–forming regions involved in tubulin binding (Redeker et al., 2000). In Drosophila, two similar adjacent coiled-coil subdomains are also predicted within the overlapping region encoded by exons 2–5 (Figure 2). The second subdomain is longer on its C-terminal side and is followed by a third predicted coiled-coil subdomain, encoded by exons 6 and 7, which may reflect specific interaction properties of D-stathmins.
Figure 2.
Structural predictions for D- and m-stathmin. Coiled coil (CC) formation probability of mouse (M-) and Drosophila (D-) stathmins determined by the “picoil” software, and the deduced schematic representation of vertebrate and Drosophila stathmins.
Biochemical Properties.
To identify D-stathmins in vivo, we produced two rabbit polyclonal antisera directed, respectively, against a peptide of the C-terminal domain and against recombinant D-stathmin-ΔC (see below). In Drosophila embryos (0–24 h), a major 32-kDa protein is recognized specifically by both antisera on Western blots (Figure 3A). This protein is probably D-stathmin-A because it has the same apparent molecular mass as the major in vitro-translated product from clone 14 (Figure 3A) or bacterially expressed recombinant D-stathmin-A (our unpublished data).
We expressed in HeLa cells C-terminal myc-tagged D-stathmin-A, -ΔC (amino acids 1–142 sharing similarities with vertebrate stathmin), -Ex2–5 (amino acids 1–175 encoded by exons 2–5, conserved between Drosophila and mammals), and -ΔN (amino acids 143–257) (Figure 5). As previously described for vertebrate stathmin, all proteins were present in the cytosolic fractions after differential centrifugation (Figure 3B; our unpublished data). Furthermore, all these proteins remained soluble after boiling in the presence of 100 mM NaCl (Figure 3B), showing that they are, like vertebrate stathmin, cytosolic heat-stable proteins.
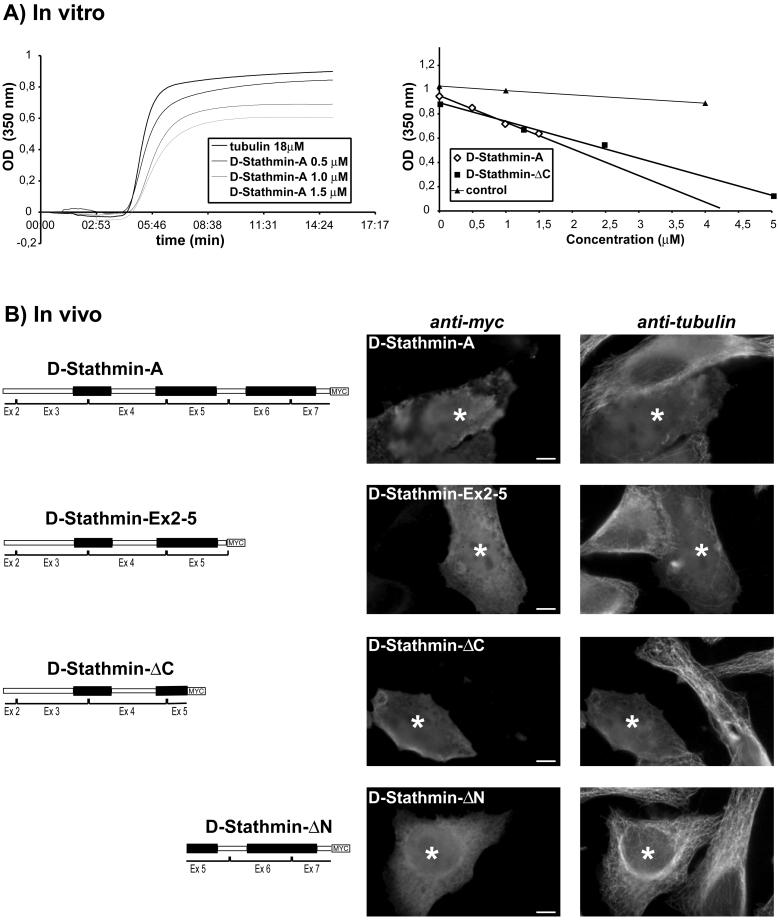
Figure 5.
Microtubule destabilization by D-stathmins in vitro and in vivo. (A) Left, increasing amounts of D-stathmin-A were added to 18 μM tubulin, inhibiting the amount of microtubule polymerized at equilibrium as detected by turbidimetry at 350 nm. Right, linear dose-dependent decrease of the amount of microtubules polymerized at equilibrium in the presence of increasing amounts of D-stathmin-A, or -ΔC. (B) Immunocytochemical detection of overexpressed D-stathmin-A, -Ex2–5, -ΔC, and -ΔN myc-tagged proteins in HeLa cells with an anti-myc antiserum. Double labeling with anti-α-tubulin showed a destabilization of the microtubule network in the presence of all proteins but D-stathmin-ΔN. For each D-stathmin form, a field containing one nontransfected cell and one significant D-stathmin overexpressing cell (asterisk) is shown. A high magnification of the field is presented in order to visualize the microtubule network.
In Vivo Phosphorylation of D-stathmins Expressed in HeLa Cells.
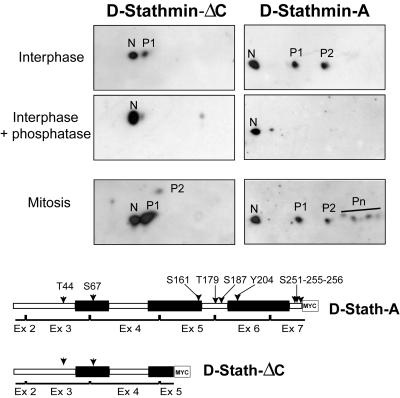
We examined whether D-stathmins-A and -ΔC were phosphorylated in vivo, when expressed in HeLa cells. In interphasic cell extracts, both D-stathmin-A and -ΔC migrate on two-dimensional gels as several spots of decreasing pI (Figure 4). After phosphatase treatment, only the most basic spot (N) was detected, indicating that D-stathmin-A and -ΔC were effectively phosphorylated and that spots N and P1/P2 are, respectively, the unphosphorylated and phosphorylated forms of the proteins. Furthermore, both proteins were more phosphorylated in mitosis, as previously shown for vertebrate stathmin. The truncated form D-stathmin-ΔC migrated as two spots N/P1 in interphase and as three spots N/P1/P2 in mitosis. Because the truncated D-stathmin-ΔC form contains the two residues, Thr 44 and Ser 67, present in consensus phosphorylation sites conserved with mammalian stathmin, it suggests that these two residues were effectively phosphorylated. Full-length D-stathmin-A displayed two phosphoforms (P1/P2) in interphase and at least four (P1/P2/Pn) in mitosis. Thus, it probably contains additional phosphorylated residues in the region 143–257.
Figure 4.
Phosphorylation of D-stathmin-A and -ΔC during the cell cycle. D-stathmin-ΔC-myc and D-stathmin-A-myc overexpressing interphase or mitotic HeLa cell homogenates were analyzed by two-dimensional PAGE-Western blotting with an anti-myc antiserum. Interphase HeLa cell proteins were also treated with phosphatase to identify their phosphorylated and nonphosphorylated forms. The nonphosphorylated (N) or phosphorylated forms of each protein on one, two, or more sites (P1, P2, Pn) are labeled. The likely (arrowheads) and other possible (arrows) phosphorylated residues are indicated on the schematic representation of the D-stathmin constructs.
D-stathmins Inhibit Microtubule Assembly In Vitro and In Vivo.
The addition of bacterially produced, partially purified recombinant D-stathmin-A or -ΔC to purified bovine brain tubulin resulted in a decrease of microtubule polymerization, in a linear manner with increasing concentrations of d-stathmins (Figure 5A), similarly to vertebrate stathmin family proteins (Jourdain et al., 1997; Charbaut et al., 2001). Furthermore, overexpression of D-stathmin-A, -ΔC, and -Ex2–5 in HeLa cells resulted in the effective disruption of the interphasic microtubule network in vivo (Figure 5B). The overexpression of different forms of D-stathmin containing the region conserved with mammalian stathmin is thus sufficient to induce microtubule depolymerization in vivo. Finally, no effect was observed with the -ΔN form.
Expression, Regulation, and Functions of D-stathmin during Drosophila Embryogenesis
The molecular characterization of D-stathmins indicates that they are homologs of vertebrate stathmin and stathmin-related proteins. To understand their physiological roles and action in Drosophila, we examined the dynamic expression of their mRNAs and of the proteins during embryogenesis, and looked for defects induced by their impaired expression.
D-stathmin Expression Is Mostly Restricted to the Nervous System and to Pole Cells.
During embryogenesis (Figure 6) maternal D-stathmin mRNA (Figure 6, A–I) is very abundant at egg deposition (our unpublished data) and then decreases but remains evenly distributed in the embryo to stage 4 (Figure 6A). During gastrulation (Figure 6B), D-stathmin mRNA has disappeared from the embryo except for the pole cells, where it is restricted from stage 6–11 and remains expressed until the end of embryogenesis. Because it was shown that gene expression in pole cells is not initiated before stage 10 (Williamson and Lehmann, 1996), D-stathmin mRNA in pole cells must be of maternal origin, at least up to stage 10. Then, D-stathmin mRNA begins to be weakly expressed in neuroblasts at stage 12 (Figure 6D) and more strongly in the developing CNS around stage 13 (Figure 6, E and F). At stage 14 (Figure 6, G–I), it is strongly present in the forming brain and in the ventral nerve cord, and starts also to be expressed in the sensory neurons. In the CNS, D-stathmin becomes expressed toward the end of germ band retraction, when the differentiation of neurons begins, whereas in the peripheral nervous system, it is only expressed once the sensory organs have started to differentiate.
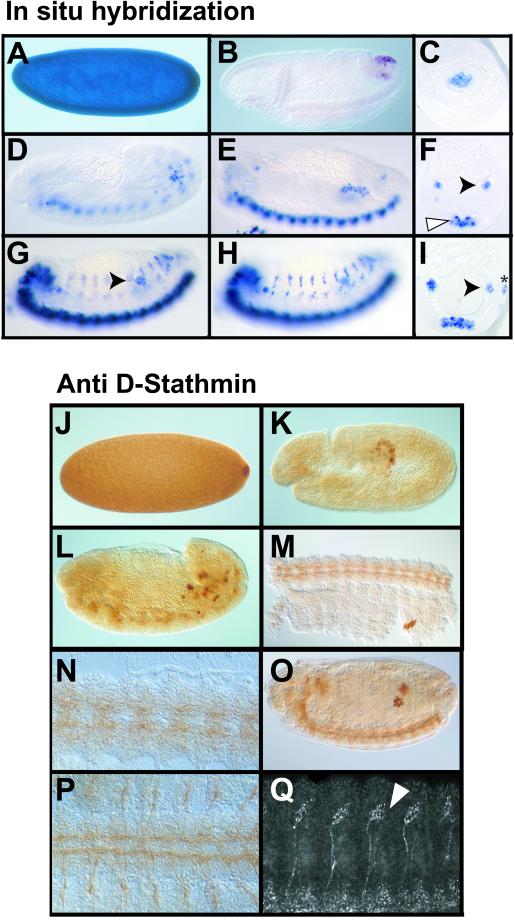
Figure 6.
Expression of D-stathmin during embryonic development. The orientation is anterior to the left and dorsal to the top. (A–I) Sagittal (A and B, D and E, and G and H) and cross-section (C, F, and I) in situ hybridization views of D-stathmin mRNA distribution at successive stages of Drosophila embryonic development. Strong expression of D-stathmin mRNA in a stage 4 embryo (A). In the gastrulating embryo (B) D-stathmin expression is restricted to the migrating pole cells and at stage 9 (C) to pole cells in the posterior midgut. At stages 12 (D) and 13 (E and F, same embryo) D-stathmin starts to be expressed in the developing CNS (white arrowhead) and is still expressed in the migrating pole cells (black arrowhead). At stage 14 (G and H, two optical sagittal sections; and I, cross section of the same embryo) D-stathmin is expressed in the pole cells within the already formed gonads (black arrowhead) as well as in the CNS, and starts to be expressed in the developing peripheral nervous system (asterisk). (J–Q) Immunohistochemical distribution of D-stathmin protein at successive stages of embryonic development. At stage 4 (J) D-stathmin is evenly distributed throughout the embryo, but starts to accumulate in the forming pole cells where it becomes restricted at stage 10 (K). At stage 12 (L–P) it is present in migrating pole cells and begins to be expressed in the developing CNS, as best visualized on the ventral view of a dissected embryo (M, and at higher magnification N). At stage 14 (O–Q) expression in the CNS and in the pole cells is illustrated in a sagittal optical section (O) and a ventral view of a dissected embryo (P). Expression in the peripheral nervous system becomes also clearly visible, as shown in a lateral confocal section (Q) in the chordotonal organs (white arrowhead).
The D-stathmin protein was revealed by immunohistochemistry: because both anti-D-stathmin antisera gave similar results, those with the anti-pep-COOH are shown in Figure 6, J–Q. Like its mRNA, the D-stathmin protein is evenly distributed during the cleavage stages of the embryo (Figure 6J). Interestingly, the protein accumulates at the posterior pole of the embryo when pole cells start to bud. During gastrulation, the protein level decreases except in the pole cells (Figure 6K). However, the persistence of likely maternal D-stathmin protein is slightly longer than that of its mRNA. Until stage 11, D-stathmin protein expression is restricted to the migrating germ cells. Toward stage 12–14 (Figure 6, L–Q), it starts to be expressed, like its mRNA, within neuronal cells in the central and later in the peripheral nervous system. Its expression pattern actually overlaps that of Futsch (Figure 8A), a “microtubule-associated protein” present in axons and dendrites of all differentiated neuronal cells (Fujita et al., 1982; Hummel et al., 2000).
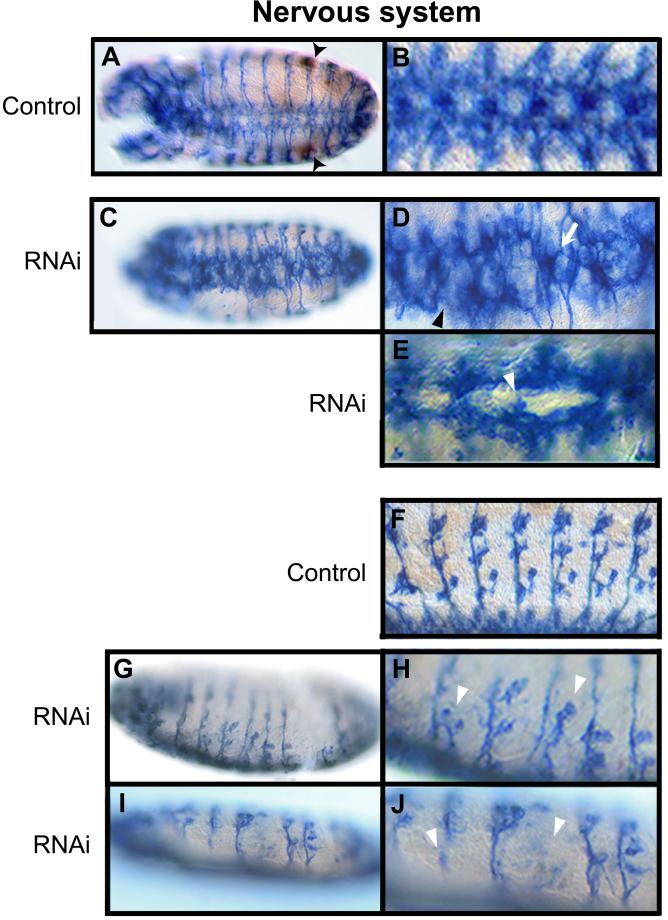
Figure 8.
RNA interference with D-stathmin dsRNA affects nervous system development. (A–J) Whole mount antibody staining of Drosophila stage 14 embryos, in brown for D-stathmin, and in blue for Futsch with the 22C10 antibody revealing the entire nervous system. (A and C) Ventral views of stage 14 control (A) and dsRNA-injected (C) embryos. The pole cells at the gonads appear clearly labeled for stathmin in the control embryo (A, arrowheads). (B, D, and E) Magnified views of the CNS in control (B) and dsRNA-injected embryos (D and E): fuzzy commissures, defects in the separation of the commissures (arrow), and disruptions of the longitudinal connective (arrowhead) are visible in (D), and failure in the establishment of a commissure (arrowhead) can be seen in E. (G and I) Sagittal views of dsRNA-injected embryos show defects in the peripheral nervous system, visualized with more detail on the magnified views (H and J) showing defects in sensory neuron migration and/or differentiation (arrowheads) compared with the normal embryo (F).
D-stathmin Is Required for Normal Germ Cell Migration and Nervous System Development.
The D-stathmin gene is located on the second chromosome at position 26B9. Because no Drosophila mutation is available for D-stathmin, we inhibited D-stathmin expression by RNA interference (RNAi), injecting D-stathmin dsRNA (Kennerdell and Carthew, 1998). All dsRNA-injected embryos examined until the end of embryogenesis failed to hatch and died, 70% of them without any cuticular defect.
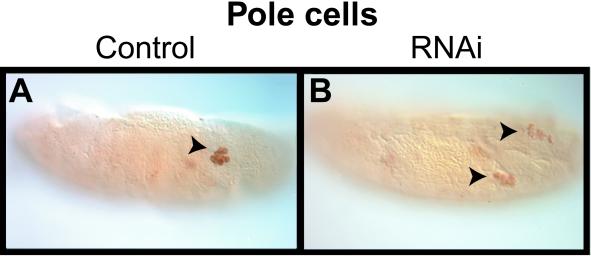
The level of D-stathmin in germ cells of the dsRNA-injected embryos examined was reduced to various extents, but it remained detectable and was therefore used to follow pole cells during development of the injected embryos (Figure 7, A and B). The process of pole cell formation did not appear to be affected, either because D-stathmin is not involved or because the maternal protein already present at the time of injection is sufficient for this process. However, at later stages, migration of pole cells was affected in 29 (53%) of the 55 dsRNA-injected embryos with reduced D-stathmin expression examined. Germ cells arrested their migration around stage 14, lined up along parasegments 12–10 as small groups that failed to coalesce into the embryonic gonads.
Figure 7.
RNA interference with D-stathmin dsRNA affects pole cell migration. (A and B) Sagittal views of whole mount D-stathmin antibody staining of stage 15 embryos: a control embryo injected with buffer, with the labeled pole cells at the forming gonad (A, arrowhead); and dsRNA-injected embryo with strongly reduced D-stathmin in the pole cells, which failed to coalesce to form the gonad (B, arrowheads).
To follow the development of the nervous system in D-stathmin dsRNA-injected embryos, they were stained with the anti-Futsch 22C10 antibody, which labels all differentiated neuronal cells (Hummel et al., 2000). Before anti-Futsch staining, the embryos were immunolabeled for D-stathmin, which revealed an extensive decrease of the expression of the protein within the developing nervous system of D-stathmin dsRNA-injected embryos, and was used as an internal control for the efficiency of RNA interference (our unpublished data). In control embryos (Figure 8, A, B, and F), the development of the nervous system was normal. In the ventral nerve cord of embryos at stage 14, the majority of the axons are organized around the midline into a metameric pattern of commissures, each being subdivided into an anterior and a posterior part and spaced by a longitudinal connective (Figure 8, A and B). In 54 (64%) of the 90 dsRNA-injected embryos examined, the organization of the axons in the neuromeres of the CNS was disturbed, with the appearance of various defects (Figure 8, C–E): commissures along the ventral cord appeared fuzzy with separation defects, and longitudinal connectives were disrupted. Furthermore, the formation of the peripheral nervous system also was perturbed in dsRNA-injected embryos (Figure 8, G–J), because the chordotonal sensory neurons appeared missing or defective. The various degree of penetrance of the phenotype in dsRNA-injected embryos is probably due to variable residual amounts of D-stathmin protein. Together, it appears that D-stathmin activity is required for the correct assembly of the central and peripheral nervous systems of Drosophila.
DISCUSSION
Microtubule dynamics is an essential process involved in numerous functional activities of cells as well as at various stages of their proliferation and differentiation. Stathmin and its protein family in vertebrates interact with tubulin and hence interfere with microtubule dynamics, which most likely underlies their involvement in diverse intracellular regulations, in particular in the nervous system. In the present study, we identified a stathmin gene in Drosophila, and characterized D-stathmin proteins at the molecular level and demonstrated their functional activity toward microtubules. We also demonstrate for the first time the physiological importance of stathmin family proteins in essential biological processes such as germ cell migration and formation of the nervous system in the embryo.
Stathmin Family Proteins in Invertebrates
In addition to their significant sequence identities, the D-stathmin protein sequences display characteristic features of vertebrate stathmin family proteins, such as several consensus phosphorylation sites and similar secondary structure prediction of α-helical, potentially coiled-coil–forming interaction domains. Furthermore, biochemical, functional, and physiological characteristics, such as cytosolic solubility, inhibition of microtubule polymerization, and high expression in the oocyte and then in the gonads and the nervous system, also advocate for the belonging of D-stathmins to the genuine stathmin family.
As opposed to the stathmin family in vertebrates (Ozon et al., 1997, 1998), a single stathmin-related gene is found in the Drosophila genome. The phylogenetic relationship between Drosophila and vertebrate stathmin family proteins is further ascertained by the conservation of the intron/exon junctions of the respective genes within their common regions, indicating that they have a common ancestor. Furthermore, like the RB3 gene in vertebrates, the Drosophila stathmin gene encodes for several splice variants, including a domain coded by exon 2′ that is highly basic like the A" domain of the neural members of the vertebrate stathmin family. The functional role of this alternative splicing has to be further evaluated as well as the possible existence of other as yet unidentified exons and splice variants. Taking into account the functional and physiological properties of the corresponding proteins as well as the similar level of sequence identity with all members of the vertebrate stathmin family, the D-stathmin gene could be actually considered as an invertebrate homolog to the whole stathmin family, which arose with evolution probably only in vertebrates.
In a search for stathmin homologs in invertebrates we identified EST clones homologous to D-stathmins-A and -B in another insect, Bombyx mori (our unpublished data), but not in nematodes or yeast, nor in prokaryotes, whose complete sequences are available. Together, it appears that stathmin as such may not exist in the lower evolutionary species examined, which does not exclude, however, the existence of as yet unidentified functional homologs.
Functional Properties and Regulation of D-stathmins
The best characterized functional property of vertebrate stathmin and stathmin family proteins is their capacity to interfere with microtubule assembly as a result of tubulin sequestration and possibly also through direct catastrophe promotion. D-stathmin also interferes with microtubule assembly both in vitro and in vivo. Its inhibition of tubulin polymerization in vitro is compatible with a sequestration mechanism, because the level of microtubules formed decreases proportionally to the concentration of D-stathmin. Stathmin-like domains of the vertebrate stathmin family form a complex with two tubulin molecules (Charbaut et al., 2001) in which each tubulin interacts with an α-helical region, including one of the two predicted coiled-coil–forming domains of the stathmin-like domain (Gigant et al., 2000). A particular feature of D-stathmins is the presence of a third predicted coiled-coil–forming domain on the C-terminal side of the molecule coded by exons 6 and 7. Interestingly, the two first coiled-coil–forming domains of D-stathmin suffice to confer microtubule-interfering properties both in vitro and in vivo, whereas the third domain of D-stathmin does not interfere with microtubules in vivo when expressed alone. It remains however to determine whether this third coiled-coil–forming domain within D-stathmin can bind additional tubulin(s), because our present assays do not allow to distinguish between a 2:1 or higher tubulin/D-stathmin stoichiometry. Alternatively, the additional domain might contribute to the stability and/or regulation of the stathmin–tubulin complex.
In vertebrates, stathmin is regulated by combinatorial phosphorylation on several sites, in response to extracellular regulators as well as during the cell cycle. Phosphorylation of stathmin regulates its tubulin-interacting properties, resulting in particular in its conversion to a mitosis permissive form (Marklund et al., 1996; Gavet et al., 1998). The fact that D-stathmin-A becomes phosphorylated on several, most likely homologous sites in mitosis when overexpressed in HeLa cells argues for a similar regulation of stathmin in Drosophila.
Expression of Stathmin in Drosophila
D-stathmin is involved in the control of microtubule dynamics, an essential process for a variety of cellular functions. Its high expression in the early Drosophila embryo is similar to that of stathmin in the mouse or Xenopus (Doye et al., 1992; Maucuer et al., 1993; Koppel et al., 1999). D-stathmin is thus highly and uniformly expressed at the very beginning of embryogenesis, before cellularization, when mitosis is very active and microtubules are highly dynamic. This can be related to the likely checkpoint function of stathmin (Gavet et al., 1998; Lawler et al., 1998), because only fully phosphorylated stathmin is permissive for mitosis in vertebrates (Marklund et al., 1996; Lawler, 1998). During embryogenesis, after the end of the blastoderm stage, D-stathmin is only expressed in germ cells and cells of the central and peripheral nervous systems, where stathmin is also most highly expressed in vertebrates, the other members of the stathmin family being restricted to the nervous system. It is interesting to note that D-stathmin is then expressed in essentially postmitotic cells that will either migrate or undergo morphological changes, indicating that stathmin is also related to differentiation and mature cell activities beside its suggested implication in the control of cell proliferation. Together, the expression patterns of D-stathmin and vertebrate stathmin family proteins are thus comparable, which further underlines their likely functional homology.
D-stathmin Fulfills Essential Roles during Embryogenesis
Stathmin gene invalidation has been achieved in the mouse but resulted in no significant phenotype (Schubart et al., 1996), which could suggest either that stathmin function is not essential or that it can be compensated. In Drosophila, the easy access to the embryo and the existence of a single D-stathmin gene allowed to reveal striking germ cell and neural phenotypes as a result of inhibition of D-stathmin expression by RNAi.
The formation and initial migration of pole cells was not affected by D-stathmin dsRNA injection in the embryo, possibly because of the high levels of maternal D-stathmin mRNA and protein sustained until stage 10. However, after that stage, when expression of zygotic genes begins in pole cells and RNAi is potentially effective, the germ cells in dsRNA-treated embryos failed to coalesce and to form the gonads (Moore et al., 1998). This result might be due to a defect in germ cell migration and/or differentiation.
The most striking defects induced by RNAi concern the differentiation of the nervous system. In the normal embryo, most of the axons of the ventral nerve cord are organized in a repetitive pattern of commissures. The development of commissures is established in a stepwise manner, where both attractive and repulsive cues orient the growth cones of the elongating axons. In dsRNA-injected embryos, the formation of the commissures failed to occur, whereas in other cases some commissures appeared fuzzy with defects in their separation, and defects also in the longitudinal connectives. The proper formation of the peripheral nervous system was also partially impaired. The phenotypes observed suggest that D-stathmin is important at various stages of differentiation and maturation of the nervous system, mostly in a way related to neurite elongation and/or guidance.
Together, our results demonstrate for the first time the essential roles of stathmin family proteins in the control of the formation and maturation of the nervous system, most likely as a result of their action on the regulation of microtubule dynamics.
ACKNOWLEDGMENTS
We thank F. Schweisguth for valuable help and discussions at the initiation of this work; B. Hovemann for the gift of the Drosophila cDNA library; E. Charbaut for help with tubulin experiments; P. Curmi and S. Lachkar for purified tubulin; and E. Charbaut, P. Curmi, A. Maucuer, F. Schweisguth, and M. Vigny for critical reading of the manuscript. This work was supported by funds from Institut National de la Santé et de la Recherche Médicale, Association pour la Recherche sur le Cancer, Association Française contre le Myopathies, and European Molecular Biology Organization.
Abbreviations used:
- CNS
central nervous system
- dsRNA
double-stranded RNA
- RNAi
RNA interference
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–07-0362. Article and publication date are at www.molbiolcell.org/cgi/10.1091/mbc.01–07-0362.
REFERENCES
- Antonsson B, Kassel D, Di Paolo G, Lutjens R, Riederer BM, Grenningloh G. Identification of in vitro phosphorylation sites in the growth cone protein SCG10. Effect of phosphorylation site mutants on microtubule-destabilizing activity. J Biol Chem. 1998;273:8439–8446. doi: 10.1074/jbc.273.14.8439. [DOI] [PubMed] [Google Scholar]
- Bai C, Connolly B, Metzker ML, Liu X, Sandig V, Soderman A, Galloway SM, Liu Q, Austin CP, Caskey CT. Overexpression of M68/DcR3 in human gastrointestinal tract tumors independent of gene amplification and its location in a four-gene cluster. Proc Natl Acad Sci USA. 2000;97:1230–1235. doi: 10.1073/pnas.97.3.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz EJ, Zhukovsky E, Lanahan AA, Worley PF, Nikolich K, Goodman LJ. Neuronal activity induction of the stathmin-like gene RB3 in the rat hippocampus: possible role in neuronal plasticity. J Neurosci. 1998;18:9780–9789. doi: 10.1523/JNEUROSCI.18-23-09780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Charbaut E, Curmi PA, Ozon S, Lachkar S, Redeker V, Sobel A. Stathmin family proteins display specific molecular and tubulin binding properties. J Biol Chem. 2001;276:16146–16154. doi: 10.1074/jbc.M010637200. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Curmi PA, Andersen SSL, Lachkar S, Gavet O, Karsenti E, Knossow M, Sobel A. The stathmin/tubulin interaction in vitro. J Biol Chem. 1997;272:25029–25036. doi: 10.1074/jbc.272.40.25029. [DOI] [PubMed] [Google Scholar]
- Curmi P, Gavet O, Charbaut E, Ozon S, Lachkar-Colmerauer S, Manceau V, Siavoshian S, Maucuer A, Sobel A. stathmin and its phosphoprotein family: general properties, biochemical and functional interaction with tubulin. Cell Struct Funct. 1999;24:345–357. doi: 10.1247/csf.24.345. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, Lutjens R, Pellier V, Stimpson SA, Beuchat MA, Catsicas M, Grenningloh G. Targeting of SCG10 to the area of the Golgi complex is mediated by its NH2-terminal region. J Biol Chem. 1997;272:5175–5182. doi: 10.1074/jbc.272.8.5175. [DOI] [PubMed] [Google Scholar]
- Doye V, Kellermann O, Buc-Caron MH, Sobel A. High expression of stathmin in multipotential teratocarcinoma and normal embryonic cells versus their early differentiated derivatives. Differentiation. 1992;50:89–96. doi: 10.1111/j.1432-0436.1992.tb00489.x. [DOI] [PubMed] [Google Scholar]
- Fujita SC, Zipursky SL, Benzer S, Ferrus A, Shotwell SL. Monoclonal antibodies against the Drosophila nervous system. Proc Natl Acad Sci USA. 1982;79:7929–7933. doi: 10.1073/pnas.79.24.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Ozon S, Manceau V, Lawler S, Curmi P, Sobel A. The stathmin phosphoprotein family. Intracellular localization and effects on the microtubule network. J Cell Sci. 1998;111:3333–3346. doi: 10.1242/jcs.111.22.3333. [DOI] [PubMed] [Google Scholar]
- Gigant B, Curmi PA, Martin-Barbey C, Charbaut E, Lachkar S, Lebeau L, Siavoshian S, Sobel A, Knossow M. The 4 A X-ray structure of a tubulin:stathmin-like domain complex. Cell. 2000;102:809–816. doi: 10.1016/s0092-8674(00)00069-6. [DOI] [PubMed] [Google Scholar]
- Hailat N, Strahler JR, Melhem RF, Zhu XX, Brodeur G, Seeger RC, Reynolds CP, Hanash SM. N-myc gene amplification in neuroblastoma is associated with altered phosphorylation of a proliferation related polypeptide (Op 18) Oncogene. 1990;5:1615–1618. [PubMed] [Google Scholar]
- Himi T, Okazaki T, Wang H, McNeill TH, Mori N. Differential localization of SCG10 and p19/stathmin messenger RNAs in adult rat brain indicates distinct roles for these growth-associated proteins. Neuroscience. 1994;60:907–926. doi: 10.1016/0306-4522(94)90271-2. [DOI] [PubMed] [Google Scholar]
- Horwitz SB, Shen H-J, He L, Dittmar P, Neef R, Chen J, Schubart UK. The microtubule-destabilizing activity of metablastin (p19) is controlled by phosphorylation. J Biol Chem. 1997;272:8129–8132. doi: 10.1074/jbc.272.13.8129. [DOI] [PubMed] [Google Scholar]
- Hovemann BT, Dessen E, Mechler H, Mack E. Drosophila snRNP associated protein P11 which specifically binds to heat shock puff 93D reveals strong homology with hnRNP core protein A1. Nucleic Acids Res. 1991;19:4909–4914. doi: 10.1093/nar/19.18.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Krukkert K, Roos J, Davis G, Klämbt C. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron. 2000;26:357–370. doi: 10.1016/s0896-6273(00)81169-1. [DOI] [PubMed] [Google Scholar]
- Jourdain L, Curmi P, Sobel A, Pantaloni D, Carlier MF. Stathmin: a tubulin-sequestering protein which forms a ternary T2S complex with two tubulin molecules. Biochemistry. 1997;36:10817–10821. doi: 10.1021/bi971491b. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Koppel J, Rehák P, Baran V, Veselá J, Hlinka D, Manceau V, Sobel A. Cellular and subcellular localization of stathmin during oocyte and preimplantation embryo development. Mol Reprod Dev. 1999;53:306–317. doi: 10.1002/(SICI)1098-2795(199907)53:3<306::AID-MRD6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawler S. Microtubule dynamics: if you need a shrink try stathmin/Op18. Curr Biol. 1998;8:R212–R214. doi: 10.1016/s0960-9822(98)70128-9. [DOI] [PubMed] [Google Scholar]
- Lawler S, Gavet O, Rich T, Sobel A. Stathmin overexpression in 293 cells affects signal transduction and cell growth. FEBS Lett. 1998;421:55–60. doi: 10.1016/s0014-5793(97)01519-6. [DOI] [PubMed] [Google Scholar]
- Luo X-N, Arcasoy MO, Brickner HE, Schechter AD, Atweh GF. Regulated expression of p18, a major phosphoprotein of leukemic cells. J Biol Chem. 1991;31:21004–21010. [PubMed] [Google Scholar]
- Manceau V, Gavet O, Curmi P, Sobel A. Stathmin interaction with HSC70 family proteins. Electrophoresis. 1999;20:409–417. doi: 10.1002/(SICI)1522-2683(19990201)20:2<409::AID-ELPS409>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Marklund U, Larsson N, Melander Gradin H, Brattsand G, Gullberg M. Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics. EMBO J. 1996;15:5290–5298. [PMC free article] [PubMed] [Google Scholar]
- Maucuer A, Camonis JH, Sobel A. Stathmin interaction with a novel putative kinase and coiled-coil forming protein domains. Proc Natl Acad Sci USA. 1995;92:3100–3104. doi: 10.1073/pnas.92.8.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maucuer A, Doye V, Sobel A. A single amino acid difference distinguishes the human and the rat sequences of stathmin, a ubiquitous intracellular phosphoprotein associated with cell regulations. FEBS Lett. 1990;264:275–278. doi: 10.1016/0014-5793(90)80266-l. [DOI] [PubMed] [Google Scholar]
- Maucuer A, Moreau J, Mechali M, Sobel A. The stathmin gene family: phylogenetic conservation and developmental regulation in Xenopus. J Biol Chem. 1993;268:16420–16429. [PubMed] [Google Scholar]
- Melhem RF, Zhu XX, Hailat N, Strahler JR, Hanash SM. Characterization of the gene for a proliferation-related phosphoprotein (oncoprotein 18) expressed in high amounts in acute leukemia. J Biol Chem. 1991;266:17747–17753. [PubMed] [Google Scholar]
- Moore LA, Broihier HT, Van Doren M, Lehmann R. Gonadal mesoderm and fat body initially follow a common developmental path in Drosophila. Development. 1998;125:837–844. doi: 10.1242/dev.125.5.837. [DOI] [PubMed] [Google Scholar]
- Okazaki T, Yoshida BN, Avraham KB, Wang H, Wuenschell CW, Jenkis NA, Copeland NG, Anderson DJ, Mori N. Molecular diversity of the SCG10/stathmin gene family in the mouse. Genomics. 1993;18:360–373. doi: 10.1006/geno.1993.1477. [DOI] [PubMed] [Google Scholar]
- Ozon S, Byk T, Sobel A. SCLIP: a novel SCG10-like protein of the stathmin family expressed in the nervous system. J Neurochem. 1998;70:2386–2396. doi: 10.1046/j.1471-4159.1998.70062386.x. [DOI] [PubMed] [Google Scholar]
- Ozon S, El Mestikawy S, Sobel A. Differential, regional and cellular expression of the stathmin family transcripts in the adult rat brain. J Neurosci Res. 1999;56:553–564. doi: 10.1002/(SICI)1097-4547(19990601)56:5<553::AID-JNR11>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ozon S, Maucuer A, Sobel A. The stathmin family: molecular and biological characterization of novel mammalian proteins expressed in the nervous system. Eur J Biochem. 1997;248:794–806. doi: 10.1111/j.1432-1033.1997.t01-2-00794.x. [DOI] [PubMed] [Google Scholar]
- Redeker V, Lachkar S, Siavoshian S, Charbaut E, Rossier J, Sobel A, Curmi P. Probing the native structure of stathmin and its interaction domains with tubulin. J Biol Chem. 2000;275:6841–6849. doi: 10.1074/jbc.275.10.6841. [DOI] [PubMed] [Google Scholar]
- Roth S, Stein D, Nusslein-Volhard C. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell. 1989;59:1189–1202. doi: 10.1016/0092-8674(89)90774-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schubart UK, Yu JH, Amat JA, Wang ZQ, Hoffmann MK, Edelmann W. Normal development of mice lacking metablastin (P19), a phosphoprotein implicated in cell cycle regulation. J Biol Chem. 1996;271:14062–14066. doi: 10.1074/jbc.271.24.14062. [DOI] [PubMed] [Google Scholar]
- Sobel A. Stathmin: a relay phosphoprotein for multiple signal transduction? Trends Biochem Sci. 1991;16:301–305. doi: 10.1016/0968-0004(91)90123-d. [DOI] [PubMed] [Google Scholar]
- Stein R, Mori N, Matthews K, Lo LC, Anderson DJ. The NGF-inducible SCG10 mRNA encodes a novel membrane-bound protein present in growth cones and abundant in developing neurons. Neuron. 1988;1:463–476. doi: 10.1016/0896-6273(88)90177-8. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Williamson A, Lehmann R. Germ cell development in Drosophila. Annu Rev Cell Dev Biol. 1996;12:365–391. doi: 10.1146/annurev.cellbio.12.1.365. [DOI] [PubMed] [Google Scholar]