Abstract
Patients with triple negative breast cancer (TNBC) have a poor survival rate following chemotherapy due to drug resistance. Notably, the molecular mechanism of drug resistance remains elusive. Between December 2011 and December 2014, 36 TNBC samples were obtained from Liaocheng People's Hospital. Three gemcitabine-resistant MDA-MB-231 cell lines (MDA-MB-231rGEM1, MDA-MB-231rGEM2 and MDA-MB-231rGEM3) were obtained by exposure of MDA-MB-231 cells to increasing concentrations of gemcitabine for >12 months. Reverse transcription-quantitative polymerase chain reaction was performed to detect the expression levels of specific genes, including microRNA (miR)-620, ATP-binding cassette sub-family B member 1 (ABCB1), ABCC10, cytidine monophosphate kinase, deoxycytidine monophosphate deaminase (DCTD), nucleoside diphosphate kinase 1 (NME1), ribonucleoside-diphosphate reductase large subunit (RRM1) and RRMB2. Western blot analysis was performed to assess the protein expression levels of DCTD. Furthermore, cell proliferation was assessed using a Cell Counting Kit-8 assay and cell apoptosis was detected using an Annexin V/Dead Cell Apoptosis kit. Interactions between miR-620 and DCTD were predicted using TargetScan and detected with the dual luciferase reporter assay. Elevation of miR-620 expression levels were detected in two of the assessed gemcitabine-resistant MDA-MB-231 cell lines compared with MDA-MB-231 cells. Gemcitabine induced significant elevation of miR-620 in MDA-MB-231 cells. An increase of DCTD at mRNA and protein expression levels in MDA-MB-231rGEM1 cells was observed compared with those in MDA-MB-231 cells. Results suggested that DCTD was directly regulated by miR-620. Inhibition of miR-620 and overexpression of DCTD reversed gemcitabine resistance in MDA-MB-231rGEM1 cells via inducing cell apoptosis and cell growth arrest. A negative correlation was identified between miR-620 and DCTD mRNA expression levels in patients with TNBC. The present results demonstrated that overexpression of miR-620 could contribute to the development of gemcitabine resistance in patients with TNBC via the direct downregulation of DCTD.
Keywords: microRNA-620, triple negative breast cancer, gemcitabine, deoxycytidine monophosphate deaminase
Introduction
Breast cancer remains a leading cause of fatality in women worldwide (1). According to the expression of estrogen receptor α (ERα), progesterone receptor (PR) and human epidermal growth factor receptor-2 (Her2), breast cancer can be classified into three different types, including ERα+, Her2+ and triple negative breast cancer (TNBC) (2). Due to the lack of ERα, PR and Her2 expression in patients with TNBC, gemcitabine-based chemotherapy is one of the major procedures for the treatment of TNBC (3). However, the survival of patients with TNBC receiving chemotherapy is compromised due to intrinsic or acquired resistance to gemcitabine (4). The molecular mechanism for gemcitabine resistance is common among various cancer types, including breast cancer, which is complicated and still poorly understood (5).
MicroRNAs (miRs) are small non-coding RNAs that were identified decades ago (6). Deregulation of miRs has been reported to contribute to various diseases, including cancer (7,8). During cancer progression, miRs regulate their target genes to promote or inhibit cancer cell proliferation, metastasis, apoptosis and drug resistance (9–11). In breast cancer, dysregulation of miR patterns is also associated with chemoresistance (12). Several miRs, including miR-34a, miR-21 and miR-489, have been identified to promote chemoresistance via targeting their specific target genes (13–15).
In breast cancer, a set of genes have been proved to contribute to gemcitabine resistance via deoxycytidine monophosphate deaminase (DCTD) (16). DCTD catalyzes the deamination of dCMP to dUMP, and its underexpression can induce dNTP pool imbalance, which affects DNA amplification (17). Thus, decreased DCTD would reduce gemcitabine self-potentiation so as to cause drug resistance (18). However, to the best of our knowledge the regulatory mechanism of DCTD in gemcitabine-resistant breast cancer has not been studied yet.
In the current study, miR-620 levels were assessed in gemcitabine-resistant TNBC cells. Luciferase assays, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis were performed to assess if DCTD, a gene associated with gemcitabine resistance, was directly regulated by miR-620 in TNBC cells. Furthermore, TNBC cells were assessed following the inhibition of miR-620 or silencing of DCTD in TNBC cells. The present study further extended current understandings on chemoresistance of TNBC and implied miR-620 as a promising prognostic and therapeutic target for patients with gemcitabine-resistant TNBC.
Materials and methods
Patients
Patients who received radiotherapy, chemotherapy or surgery were excluded from the current study. Between December 2011 and December 2014, 36 TNBC samples were obtained from patients with TNBC in Liaocheng People's Hospital (Liaocheng, China). The adjacent normal tissues were extracted ≥5 cm away from the tumor tissues. Patients (20 males and 16 females) were aged from 43 to 65 years old. A total of 21 gemcitabine-sensitive patients with TNBC [who had reached pCR (pathologic complete response) following gemcitabine-based neoadjuvant chemotherapy] and 15 patients with gemcitabine-resistant TNBC (tumor size was the same or larger at the time of surgical removal) were enrolled in the research. The detailed clinicopathologic features are indicated in Table I. All patients provided written consent prior to surgery, and the present study was approved by the Ethics Committee of Liaocheng People's Hospital. Tissues were immediately frozen at liquid nitrogen with the temperature of −196°C for RNA extraction.
Table I.
The clinicopathologic features of patients with triple negative breast cancer.
| Clinicopathologic features | Case (n) | Percent (%) |
|---|---|---|
| Age (years) | ||
| ≥50 | 25 | 69.44 |
| <50 | 11 | 30.56 |
| Response to emcitabine | ||
| Sensitive | 21 | 58.33 |
| Resistant | 15 | 41.67 |
| Tumor size (3 cm) | ||
| ≥3 | 12 | 33.33 |
| <3 | 24 | 66.67 |
| TNM stage | ||
| I–II | 26 | 72.22 |
| III–IV | 10 | 27.78 |
| Menopause state | ||
| Yes | 13 | 36.11 |
| No | 23 | 63.89 |
Cell culture
Human TNBC MDA-MB-231 and BT549 cell lines were obtained from American Type Culture Collection (Manassas, VA, USA). Three gemcitabine-resistant MDA-MB-231 cell lines (MDA-MB-231rGEM1, MDA-MB-231rGEM2 and MDA-MB-231rGEM3) were generated by continuous exposure of MDA-MB-231 cells to increasing concentrations of gemcitabine for >12 months as described previously study (19). All cell lines were cultured in Dulbecco's Minimum Essential Medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (Hyclone, Logan, UT, USA) in an incubator containing 5% CO2. For analysis of the gemcitabine response, increased concentrations of gemcitabine were added into the culture medium of MDA-MB-231 cells and gemcitabine-resistant MDA-MB-231 cells for 0, 24, 48 and 72 h. Subsequently, cells were subjected to further experiments.
Inhibition and overexpression of miR-620
miR-NC inhibitor, miR-620 inhibitor, miR-NC mimic and miR-620 mimic were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences were as follows: miR-NC inhibitor, 5′-UUCUCCGAACGUGUCACGUUU-3′; miR-620 inhibitor, 5′-AUUUCUAUAUCUCCAUUU-3′; miR-NC mimic, 5′-UCGCUUGGUGCAGGUCGGG-3′; miR-620 mimic, 5′-AUGGAGAUAGAUAUAGAAAUUU-3′. The control group was treated with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 30 nM miR-NC inhibitor, miR-620 inhibitor, miR-NC mimic and miR-620 mimic were transfected with Lipofectamine 2000 into cells at 37°C 24 h prior to subsequent experiments.
RNA extraction and RT-qPCR
Total RNA of tissues and cells was isolated using the miRNeasy mini kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's instructions. cDNA was synthesized using the M-MLV kit (Thermo Fisher Scientific, Inc.). Following this, qPCR was performed to detect the expression levels of specific genes using SYBR Premix Ex Taq (Takara Bio, Inc., Otsu, Japan). GAPDH and U6 served as internal controls for mRNA and miR, respectively. The primer sequences were as follows: Stem loop primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATTTCTA-3′; miR-620-forward (F): 5′-GCCGAGATGGAGATAGATAT-3′; miR-620-reverse (R), 5′-CTCAACTGGTGTCGTGGA-3′; DCTD-F, 5′-TGCAAGAAACGGGACGACTAT-3′; DCTD-R, 5′-ATCACTGCACCCATTTGGCAT-3′; U6-F, 5′-CTCGCTTCGGCAGCACA-3′, and U6-R, 5′-AACGCTTCACGAATTTGCGT-3′; GAPDH-F, 5′-GAAATCCCATCACCATCTTCCAGG-3′ and GAPDH-R, 5′-GAGCCCCAGCCTTCTCCATG-3′. The PCR conditions were as follows: 95°C for 5 min followed by 40 cycles of amplification at 95°C for 30 sec, 57°C for 30 sec and 72°C for 30 sec. The relative expression levels of indicated genes were calculated using the 2−ΔΔCq method (20).
Western blot analysis
GAPDH antibody was purchased from Zhejiang Kangchen Biotech Co., Ltd. (Wuhan, China). DCTD antibody was bought from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Secondary antibodies against rabbit and mouse were bought from Abcam (Cambridge, MA, USA). Cell lysates were prepared using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) with protease inhibitor (Sigma Aldrich; Merck KGaA, Darmstadt, Germany). Protein concentration was determined using the BCA method. Proteins (15 µg/lane) were separated by SDS-PAGE using an 8% gel and transferred to a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). Following blocking in 5% non-fat milk at room temperature for 2 h, the membranes were washed with Tris-buffered saline with Tween-20 and incubated with the primary antibodies against DCTD (cat. no. ab183607) and GAPDH (cat. no. ab181602; both 1:1,000; Abcam) overnight at 4°C. On the following day, the membranes were incubated in horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (cat. no. ab6721; 1:2,000; Abcam) for 1 h at room temperature. The bands were developed using an enhanced chemiluminescence detection agent (Thermo Fisher Scientific, Inc.) and the images were obtained with a densitometer (GS-700; Bio-Rad Laboratories, Inc., Hercules, CA, USA). GAPDH was used as an internal control. Densitometry was achieved by Image J version 1.8.0 (National Institutes of Health, Bethesda, MD, USA).
Cell proliferation assay and calculation of IC50
The cell proliferation assay was performed with Cell Counting Kit-8 (CCK8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan). MDA-MB-231, MDA-MB-231rGEM1 and MDA-MB-231rGEM2 cells (1×103/well) were seeded into 96-well plates with DMEM with 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) containing vehicle (0.9% NaCl in water) or various concentrations of gemcitabine (0, 0.1, 1, 10, 100 and 100 nM; Selleck Chemicals, Houston, TX, USA) and sustained at 37°C for 72 h. Following this, 10 µl CCK8 solution was added into each well and incubated at 37°C for 1 h. For each well, the absorbance at 450 nm was measured using a microplate reader (Bio-Rad Laboratories, Inc.). The IC50 (the cell proliferation that was inhibited by 50% compared with cells in control group) was calculated using CompuSyn software (version 1.0; ComboSyn, Inc., Paramus, NJ, USA).
Cell apoptosis assay
Cell apoptosis was detected using the Annexin V/Dead Cell Apoptosis Kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells were trypsinized and suspended in annexin binding buffer. Following this, propidium iodide (PI) and annexin v-fluorescein isothiocyanate (FITC) were added into the cell suspension and incubated for 15 min. The stained cells were analyzed on a BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). Cells that were positive for annexin V-FITC staining and negative staining for PI were considered as early apoptotic cells, whereas cells that were positive for Annexin V-FITC staining and PI staining were considered as late apoptotic cells. The results were analyzed using FlowJo software (version 10.3; FlowJo LLC, Ashland, OR, USA).
Construction and transfection of plasmid
The full length of DCTD was amplified from the cDNA of 293 cells and ligated into pcDNA3 (YouBio, Changsha, China). For the overexpression of DCTD, 2 µg pcDNA3-DCTD was mixed with Lipofectamine 2000 in Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.) for 15 min and then added into the culture medium in each well of the 6-well plates. The control group was transfected with 2 µg pcDNA3 plasmid with the same method. Following 24 h of incubation, the cells were subjected to further experiments.
Luciferase reporter assay
The binding site between DCTD and miR-620 was first predicted by TargetScan (http://www.targetscan.org/vert_71/). The 3′-untranslated region (UTR) of DCTD was amplified from cDNA of 293 and inserted into pGL-3 (Promega Corporation, Madison, WI, USA). The mutated DCTD 3′-UTR was generated using site-directed mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA) according to the manufacturer's protocol. 293 cells were cotransfected with pGL3-DCTD 3′-UTR wild-type (WT) or pGL3-DCTD 3′-UTR mutant (Mut), miR-620 mimics or miR-NC mimics and internal control Renilla plasmid using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Following a total of 24 h, the activity of luciferase and Renilla activity was detected using a Dual Luciferase Reporter Assay Kit (Promega Corporation) according to the manufacturer's protocol.
Statistical analysis
All data were analyzed using GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA), and results were presented as the mean ± standard deviation. Differences between two groups were compared using a Student's t-test. Differences among three or more groups were analyzed with one-way analysis of variance followed by Newman-Keuls analysis. The correlation between miR-620 and DCTD mRNA expression was analyzed by a Spearman's correlation coefficient test. P<0.05 was considered to indicate a statistically significant difference.
Results
Generation of gemcitabine-resistant MDA-MB-231 cell lines
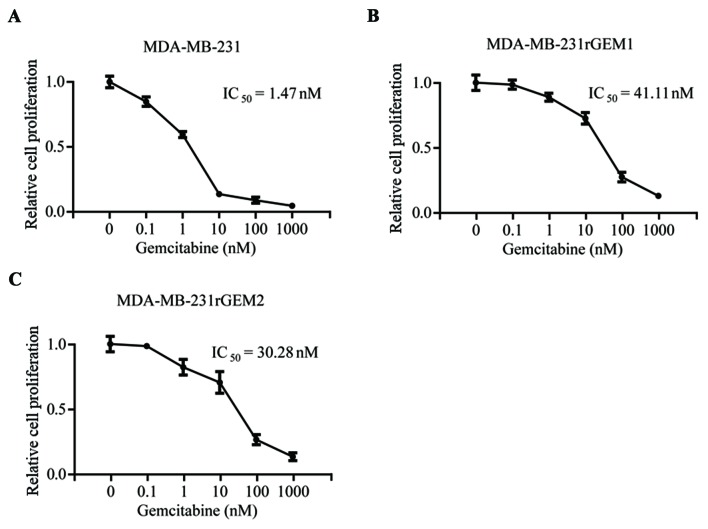
To explore the molecular mechanism of gemcitabine resistance in TNBC, gemcitabine-resistant MDA-MB-231 cell lines were developed by continuous exposure of MDA-MB-231 cells to gemcitabine. As indicated in Fig. 1, MDA-MB-231 cells were sensitive towards gemcitabine (IC50=1.47 nM), gemcitabine-resistant MDA-MB-231 cells (MDA-MB-231rGEM1 and MDA-MB-231rGEM2) were relatively insensitive towards gemcitabine (IC50=41.11 nM and IC50=30.28 nM, respectively). These results indicated that MDA-MB-231rGEM1 and MDA-MB-231rGEM2 were useful gemcitabine-resistant TNBC models.
Figure 1.
Establishment of gemcitabine-resistant MDA-MB-231 cell line models. MDA-MB-231 cells were sensitive towards gemcitabine treatment. (A) IC50 was calculated via measuring the cell proliferation of MDA-MB-231 cells exposed to increased concentrations of gemcitabine. (B) MDA-MB-231rGEM1 (gemcitabine resistant MDA-MB-231 cell line 1) cells were insensitive towards gemcitabine as a higher IC50 (41.11 nM) was revealed compared with MDA-MB-231 cells (1.47 nM). (C) MDA-MB-231rGEM2 (gemcitabine resistant MDA-MB-231 cell line 2) cells were insensitive towards gemcitabine as a higher IC50 (30.28 nM) was indicated compared with MDA-MB-231 cells (1.47 nM).
DCTD is upregulated in gemcitabine-resistant MDA-MB-231 cells
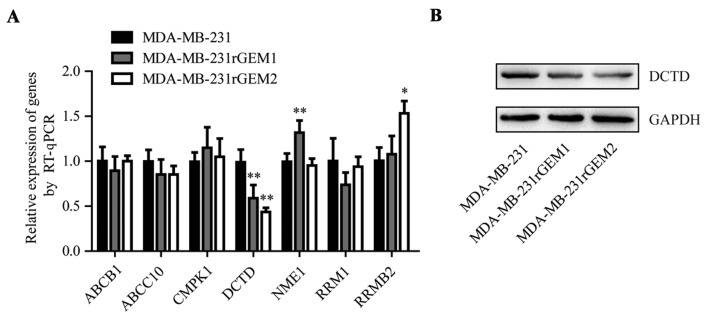
Previous research identified that deregulation of 7 genes (ABCB1, ABCC10, CMPK1, DCTD, NME1, RRM1 and RRMB1) was involved in the development of gemcitabine resistance (16). Consequently, the mRNA levels of these 7 genes were detected in MDA-MB-231, MDA-MB-231rGEM1 and MDA-MB-231rGEM2 cells. RT-qPCR data revealed that DCTD mRNA expression levels were significantly decreased in MDA-MB-231rGEM1 and MDA-MB-231rGEM2 cells compared with that in MDA-MB-231 cells (Fig. 2A). In addition, DCTD protein expression levels were also downregulated in MDA-MB-231rGEM1 and MDA-MB-231rGEM2 cells when compared with MDA-MB-231 cells (Fig. 2B).
Figure 2.
DCTD is decreased in MDA-MB-231rGEM1 and MDA-MB-231rGEM2 cells compared with parental MDA-MB-231 cells. mRNA expression levels of ABCB1, ABCC10, CMPK1, DCTD, NME1, RRM1 and RRMB1 were detected in MDA-MB-231, MDA-MB-231rGEM1 and MDA-MB-231rGEM2 cells. (A) Compared with the DCTD mRNA expression levels of MDA-MB-231 cells, DCTD was decreased in MDA-MB-231rGEM1 and MDA-MB-231rGEM2 cells. (B) Western blot analysis indicated that DCTD protein expression levels were reduced in MDA-MB-231rGEM1 and MDA-MB-231rGEM2 cells compared with MDA-MB-231 cells. *P<0.05 and **P<0.01 vs. MDA-MB-231 cells. RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Upregulation of miR-620 in gemcitabine-resistant MDA-MB-231 cells negatively regulates DCTD
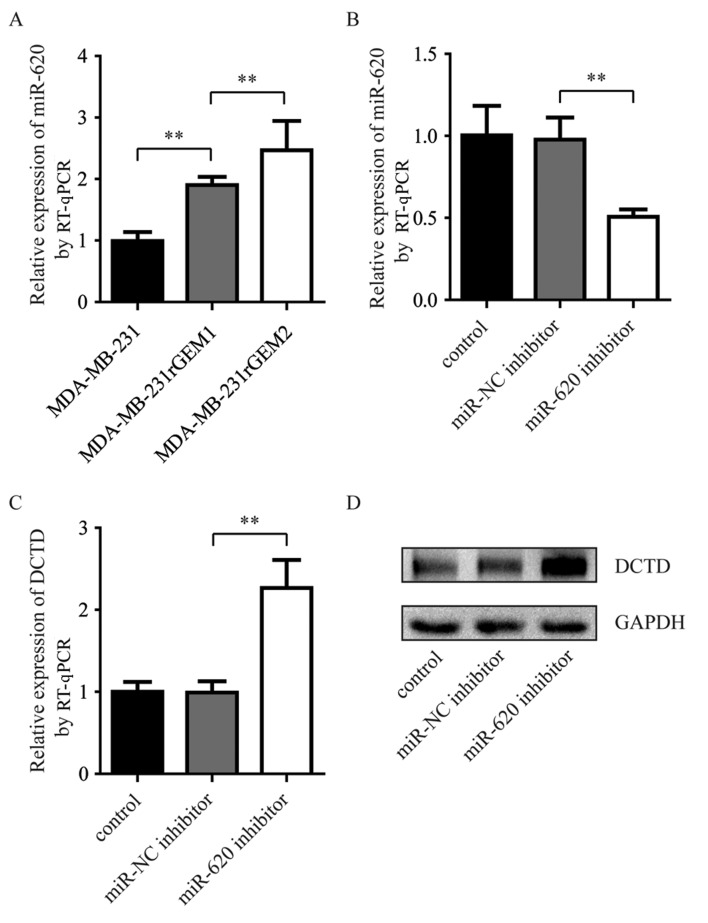
miR-620 expression levels were measured in MDA-MB-231rGEM1, MDA-MB-231rGEM2 cells and their parental MDA-MB-231 cells. miR-620 expression levels were significantly upregulated in MDA-MB-231rGEM1 compared with parental MDA-MB-231 cells. In addition, miR-620 expression levels were significantly upregulated in MDA-MB-231rGEM2 cells compared with MDA-MB-231rGEM1 cells (Fig. 3A). To investigate whether elevation of miR-620 contributed to the deregulation of DCTD in gemcitabine-resistant MDA-MB-231 cells, an miR-620 inhibitor was used to explore the association between miR-620 and DCTD. Compared with MDA-MB-231rGEM1 cells transfected with miR-NC inhibitor, transfection with miR-620 inhibitor significantly decreased miR-620 expression levels and increased DCTD mRNA expression levels (Fig. 3B and C). Additionally, inhibition of miR-620 upregulated DCTD protein expression levels in MDA-MB-231rGEM1 cells. These data suggested that miR-620 may contribute to gemcitabine resistance via regulation of DCTD.
Figure 3.
miR-620 negatively regulates DCTD in gemcitabine-resistant MDA-MB-231 cells. (A) Compared with the miR-620 expression levels of MDA-MB-231 cells, miR-620 was increased in MDA-MB-231rGEM1 and MDA-MB-231rGEM2 cells. (B) miR-620 inhibitor significantly decreased miR-620 expression levels compared with miR-NC inhibitor in MDA-MB-231rGEM1 cells. (C) DCTD mRNA expression levels were significantly elevated in MDA-MB-231rGEM1 cells transfected with miR-620 inhibitor compared with cells transfected with miR-NC inhibitor. (D) DCTD protein expression levels were also elevated in MDA-MB-231rGEM1 cells transfected with miR-620 inhibitor compared with cells transfected with miR-NC inhibitor. **P<0.01 as indicated. miR, microRNA; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
DCTD is directly regulated by miR-620
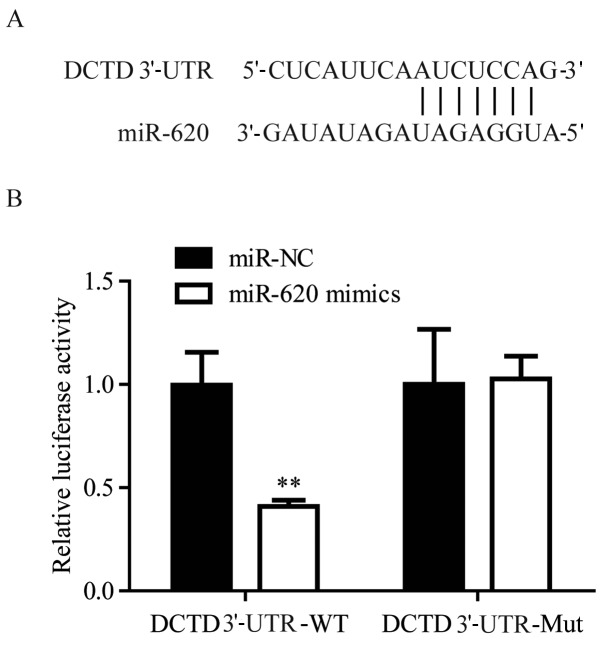
Using TargetScan, it was predicted DCTD was a target gene of miR-620 (Fig. 4A). To validate the regulatory association between miR-620 and DCTD, dual luciferase activity was performed. In 293, miR-620 mimics significantly suppressed the luciferase activity of cells transfected with DCTD 3′-UTR-WT, but not DCTD 3′-UTR-Mut, when compared with miR-NC mimics (Fig. 4B). These results confirmed that miR-620 directly regulated DCTD mRNA expression levels via binding to its 3′-UTR.
Figure 4.
miR-620 directly regulates DCTD expression levels by binding to its 3′-UTR. (A) Sequence alignment of the human DCTD mRNA 3′-UTR and complementary miR-620 sequence. 293 cells were cotransfected with DCTD 3′-UTR-WT or DCTD 3′-UTR-Mut and miR-620 mimics or miR-NC mimics. (B) The luciferase activity of cells transfected with DCTD 3′-UTR-WT and miR-620 mimics was significantly reduced compared with cells transfected with DCTD 3′-UTR-WT and miR-NC mimics. **P<0.01 vs. the DCTD 3′-UTR-WT miR-NC group. miR, microRNA; UTR, untranslated region; WT, wild-type; Mut, mutant; NC, negative control.
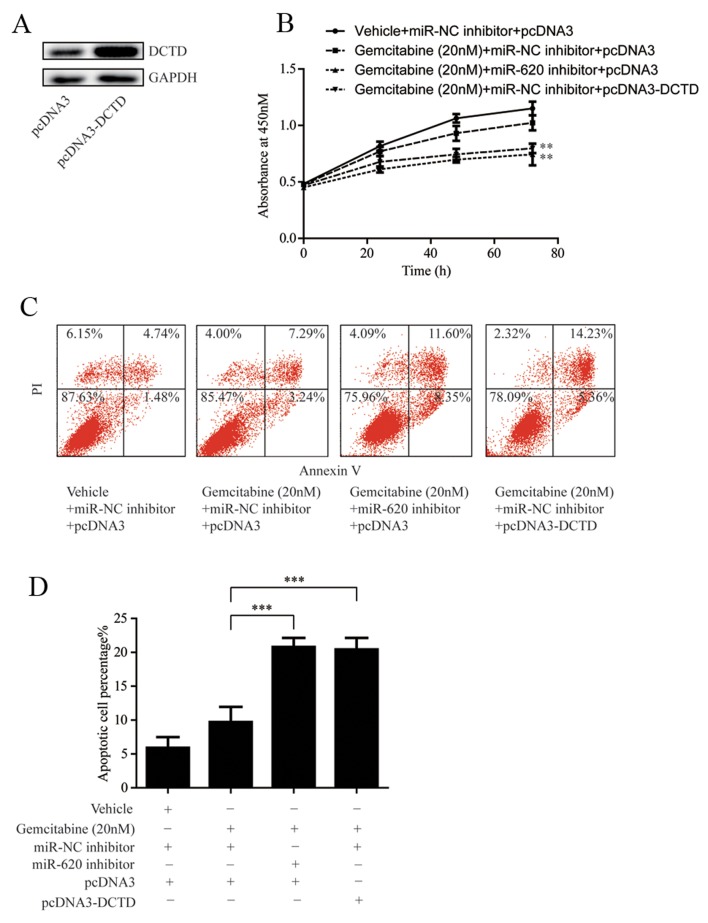
miR-620 contributes to gemcitabine resistance through DCTD
To verify whether miR-620 and DCTD were involved in the development of gemcitabine resistance, DCTD was overexpressed in MDA-MB-231rGEM1 cells via transfection of pcDNA3-DCTD (Fig. 5A). Subsequently, cell proliferation of MDA-MB-231rGEM1 cells was determined following 20 nM gemcitabine treatment with or without transfection of miR-620 inhibitor or pcDNA3-DCTD. As indicated in Fig. 5B, miR-620 inhibition or overexpression of DCTD significantly enhanced the sensitivity of MDA-MB-231rGEM1 cells to gemcitabine (Fig. 5B). Consistently, transfection with miR-620 inhibitor or pcDNA3-DCTD significantly induced cell apoptosis in MDA-MB-231rGEM1 cells in response to 20 nM gemcitabine treatment (Fig. 5C and D). These results confirmed the hypothesis that upregulation of miR-620 conferred gemcitabine resistance in TNBC cells via regulation of DCTD.
Figure 5.
miR-620 inhibition or overexpression of DCTD reverses gemcitabine resistance of MDA-MB-231rGEM1 cells. (A) Compared with cells transfected with pcDNA3, transfection of pcDNA3-DCTD markedly increased DCTD protein expression levels in MDA-MB-231rGEM1 cells. (B) Transfection with miR-620 inhibitor or pcDNA3-DCTD sensitized MDA-MB-231rGEM1 towards gemcitabine treatment by inhibiting cell proliferation. **P<0.01 vs. gemcitabine + miR-NC inhibitor + pcDNA3. (C) Transfection of miR-620 inhibitor or pcDNA3-DCTD sensitized MDA-MB-231rGEM1 towards gemcitabine treatment by inducing cell apoptosis. (D) Quantitative analysis of cell apoptosis. ***P<0.001 as indicated. miR, microRNA; NC, negative control; PI, propidium iodide.
Altered expression of miR-620 and DCTD contributes to gemcitabine resistance in patients with TNBC
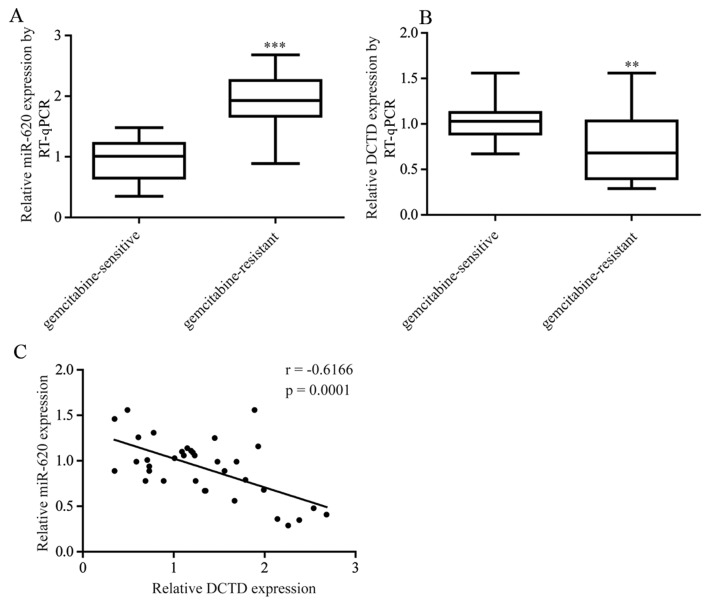
The expression of miR-620 and DCTD was assessed in tumor tissues obtained from 21 patients with gemcitabine-sensitive TNBC and pCR (pathologic complete response) following gemcitabine-based neoadjuvant chemotherapy and 15 patients with gemcitabine-resistant TNBC (the tumor size was the same or larger at the time of surgical removal). As expected, miR-620 expression levels were significantly increased in tumor tissues from patients with gemcitabine-resistant TNBC whereas DCTD mRNA expression levels were significantly decreased (Fig. 6A and B). Furthermore, a negative correlation between miR-620 and DCTD mRNA expression was observed (Fig. 6C).
Figure 6.
Altered expression of miR-620 and DCTD contributes to gemcitabine resistance in patients with TNBC. (A) Compared with tumor tissues from patients with gemcitabine-sensitive TNBC, miR-620 expression levels were elevated in tumor tissues from patients with gemcitabine-resistant TNBC. (B) DCTD expression levels were increased in tumor tissues from patients with gemcitabine-resistant TNBC patients. (C) A negative correlation between miR-620 and DCTD mRNA expression levels in tumor tissues from patients with TNBC. **P<0.01 and ***P<0.001 vs. gemcitabine-sensitive. TNBC, triple negative breast cancer; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
As a first-line drug for chemotherapy in patients with recurrent or metastatic cancer, gemcitabine can improve prognosis of patients with TNBC with a response rate as high as 78.6% (21). However, TNBC cells gradually bypass the desired cell apoptosis and develop chemoresistance, which typically leads to patient fatality (22). A previous study indicated that the metabolic pathway of gemcitabine is involved in gemcitabine resistance (23). In the present study, it was revealed that miR-620 could negatively regulate DCTD, which is a key gene in gemcitabine metabolism, to contribute to gemcitabine resistance in TNBC.
Various miRs, such as miR-608 and miR-145, have been indicated to promote gemcitabine resistance (24,25). In breast cancer, prostate cancer and pancreatic cancer cells, miR-620 upregulation could promote cell proliferation and decrease the number of cells in the G2/M phase so as to contribute to radiation resistance (26). In the present work, an elevation of miR-620 in gemcitabine-resistant MDA-MB-231 cells was indicated. Additionally, inhibition of miR-620 evoked cell apoptosis and cell growth arrest in MDA-MB-231rGEM1 cells treated with gemcitabine, which indicated that downregulation of miR-620 could reverse gemcitabine resistance in MDA-MB-231rGEM1 cells. The data uncovered a critical role for miR-620 in driving gemcitabine resistance in TNBC.
DCTD promotes catalysis of deamination and converts dCMP to dUMP (17). In the gemcitabine metabolic pathway, DCTD transfers gemcitabine monophosphate to difluorodeoxyuridine monophosphate to inhibit thymidylate synthetase (27). Decreased DCTD expression leads to the reduction of gemcitabine self-potential by interference of dNTP pool, and multi-factorial, principal component analysis suggested that the low expression of DCTD was associated with gemcitabine resistance in breast cancer (16). High expression of DCTD has also been linked to shortened overall survival in patients with gliomas (28). In the present study, a decrease of DCTD in gemcitabine-resistant MDA-MB-231 cells was indicated. Further study revealed that DCTD was directly regulated by miR-620 in MDA-MB-231rGEM1 cells. In addition, DCTD overexpression could reverse gemcitabine resistance in MDA-MB-231rGEM1 cells, inducing cell growth arrest and cell apoptosis. Furthermore, there was a negative correlation between miR-620 and DCTD in patients with TNBC. The present data extended current knowledge on the regulation of DCTD, and further implied a DCTD-miR-620 interplay during the development of gemcitabine resistance.
In conclusion, the present findings suggested miR-620 as a novel factor in promoting gemcitabine resistance in TNBC. Furthermore, miR-620 may contribute to gemcitabine resistance via directly targeting DCTD. Thus, the present work indicated that miR-620 could be a predictor for gemcitabine sensitivity of patients with TNBC and a therapeutic target for patients with gemcitabine-resistant TNBC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contribution
AZ and TT collected the clinical samples. CW, TT and YW acquired and interpreted the data. CW and ZS wrote the manuscript. ZS designed and supervised the study.
Ethics approval and consent to participate
The current study was supervised by the Ethics Committee of Liaocheng People's Hospital (Liaocheng, China). All patients provided written consent prior to surgery.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 3.Wu ZH, Lin C, Liu MM, Zhang J, Tao ZH, Hu XC. Src inhibition can synergize with gemcitabine and reverse resistance in triple negative breast cancer cells via the AKT/c-Jun pathway. PLoS One. 2016;11:e0169230. doi: 10.1371/journal.pone.0169230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen M, He M, Song Y, Chen L, Xiao P, Wan X, Dai F, Shen P. The cytoprotective role of gemcitabine-induced autophagy associated with apoptosis inhibition in triple-negative MDA-MB-231 breast cancer cells. Int J Mol Med. 2014;34:276–282. doi: 10.3892/ijmm.2014.1772. [DOI] [PubMed] [Google Scholar]
- 5.de Sousa Cavalcante L, Monteiro G. Gemcitabine: Metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur J Pharmacol. 2014;741:8–16. doi: 10.1016/j.ejphar.2014.07.041. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hesse M, Arenz C. MicroRNA maturation and human disease. Methods Mol Biol. 2014;1095:11–25. doi: 10.1007/978-1-62703-703-7_2. [DOI] [PubMed] [Google Scholar]
- 8.Gommans WM, Berezikov E. Controlling miRNA regulation in disease. Methods Mol Biol. 2012;822:1–18. doi: 10.1007/978-1-61779-427-8_1. [DOI] [PubMed] [Google Scholar]
- 9.Selcuklu SD, Donoghue MT, Rehmet K, de Souza Gomes M, Fort A, Kovvuru P, Muniyappa MK, Kerin MJ, Enright AJ, Spillane C. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem. 2012;287:29516–29528. doi: 10.1074/jbc.M111.335943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu M, Xu Y, Ge M, Gui Z, Yan F. Regulation of UHRF1 by microRNA-9 modulates colorectal cancer cell proliferation and apoptosis. Cancer Sci. 2015;106:833–839. doi: 10.1111/cas.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pouliot LM, Chen YC, Bai J, Guha R, Martin SE, Gottesman MM, Hall MD. Cisplatin sensitivity mediated by WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer Res. 2012;72:5945–5955. doi: 10.1158/0008-5472.CAN-12-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv J, Xia K, Xu P, Sun E, Ma J, Gao S, Zhou Q, Zhang M, Wang F, Chen F, et al. miRNA expression patterns in chemoresistant breast cancer tissues. Biomed Pharmacother. 2014;68:935–942. doi: 10.1016/j.biopha.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Park EY, Chang E, Lee EJ, Lee HW, Kang HG, Chun KH, Woo YM, Kong HK, Ko JY, Suzuki H, et al. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014;74:7573–7582. doi: 10.1158/0008-5472.CAN-14-1140. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Bourguignon LY. Hyaluronan-CD44 interaction promotes c-Jun signaling and miRNA21 expression leading to Bcl-2 expression and chemoresistance in breast cancer cells. Mol Cancer. 2014;13:52. doi: 10.1186/1476-4598-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Wang YW, Xing AY, Xiang S, Shi DB, Liu L, Li YX, Gao P. Suppression of SPIN1-mediated PI3K-Akt pathway by miR-489 increases chemosensitivity in breast cancer. J Pathol. 2016;239:459–472. doi: 10.1002/path.4743. [DOI] [PubMed] [Google Scholar]
- 16.Dorman SN, Baranova K, Knoll JH, Urquhart BL, Mariani G, Carcangiu ML, Rogan PK. Genomic signatures for paclitaxel and gemcitabine resistance in breast cancer derived by machine learning. Mol Oncol. 2016;10:85–100. doi: 10.1016/j.molonc.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eriksson S, Skog S, Tribukait B, Jäderberg K. Deoxyribonucleoside triphosphate metabolism and the mammalian cell cycle. Effects of thymidine on wild-type and dCMP deaminase-deficient mouse S49 T-lymphoma cells. Exp Cell Res. 1984;155:129–140. doi: 10.1016/0014-4827(84)90774-2. [DOI] [PubMed] [Google Scholar]
- 18.Xu YZ, Plunkett W. Modulation of deoxycytidylate deaminase in intact human leukemia cells. Action of 2′,2′-difluorodeoxycytidine. Biochem Pharmacol. 1992;44:1819–1827. doi: 10.1016/0006-2952(92)90077-V. [DOI] [PubMed] [Google Scholar]
- 19.Tsaur I, Makarević J, Juengel E, Gasser M, Waaga-Gasser AM, Kurosch M, Reiter M, Wedel S, Bartsch G, Haferkamp A, et al. Resistance to the mTOR-inhibitor RAD001 elevates integrin α2- and β1-triggered motility, migration and invasion of prostate cancer cells. Br J Cancer. 2012;107:847–855. doi: 10.1038/bjc.2012.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Lee SY, Im SA, Park YH, Woo SY, Kim S, Choi MK, Chang W, Ahn JS, Im YH. Genetic polymorphisms of SLC28A3, SLC29A1 and RRM1 predict clinical outcome in patients with metastatic breast cancer receiving gemcitabine plus paclitaxel chemotherapy. Eur J Cancer. 2014;50:698–705. doi: 10.1016/j.ejca.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 22.O'Reilly EA, Gubbins L, Sharma S, Tully R, Guang MH, Weiner-Gorzel K, McCaffrey J, Harrison M, Furlong F, Kell M, McCann A. The fate of chemoresistance in triple negative breast cancer (TNBC) BBA Clin. 2015;3:257–275. doi: 10.1016/j.bbacli.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosell R, Cobo M, Isla D, Camps C, Massuti B. Pharmacogenomics and gemcitabine. Ann Oncol. 2006;17(Suppl 5):v13–v16. doi: 10.1093/annonc/mdj942. [DOI] [PubMed] [Google Scholar]
- 24.Rajabpour A, Afgar A, Mahmoodzadeh H, Radfar JE, Rajaei F, Teimoori-Toolabi L. MiR-608 regulating the expression of ribonucleotide reductase M1 and cytidine deaminase is repressed through induced gemcitabine chemoresistance in pancreatic cancer cells. Cancer Chemother Pharmacol. 2017;80:765–775. doi: 10.1007/s00280-017-3418-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhuang J, Shen L, Yang L, Huang X, Lu Q, Cui Y, Zheng X, Zhao X, Zhang D, Huang R, et al. TGFβ1 promotes gemcitabine resistance through regulating the LncRNA-LET/NF90/miR-145 signaling axis in bladder cancer. Theranostics. 2017;7:3053–3067. doi: 10.7150/thno.19542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Taeb S, Jahangiri S, Korpela E, Cadonic I, Yu N, Krylov SN, Fokas E, Boutros PC, Liu SK. miR-620 promotes tumor radioresistance by targeting 15-hydroxyprostaglandin dehydrogenase (HPGD) Oncotarget. 2015;6:22439–22451. doi: 10.18632/oncotarget.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueno H, Kiyosawa K, Kaniwa N. Pharmacogenomics of gemcitabine: Can genetic studies lead to tailor-made therapy? Br J Cancer. 2007;97:145–151. doi: 10.1038/sj.bjc.6603860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu H, Wang Z, Li M, Zeng F, Wang K, Huang R, Wang H, Yang F, Liang T, Huang H, Jiang T. Gene expression and methylation analyses suggest DCTD as a prognostic factor in malignant glioma. Sci Rep. 2017;7:11568. doi: 10.1038/s41598-017-11962-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.