Summary
Mitochondria not only supply the energy for cell function, but also take part in cell signaling. This review describes the dysfunctions of mitochondria in aging and neurodegenerative diseases, and the signaling pathways leading to mitochondrial biogenesis (including PGC‐1 family proteins, SIRT1, AMPK) and mitophagy (parkin‐Pink1 pathway). Understanding the regulation of these mitochondrial pathways may be beneficial in finding pharmacological approaches or lifestyle changes (caloric restrict or exercise) to modulate mitochondrial biogenesis and/or to activate mitophagy for the removal of damaged mitochondria, thus reducing the onset and/or severity of neurodegenerative diseases.
Keywords: aging, mitochondria, mitochondrial biogenesis, mitophagy, neurodegenerative diseases
1. MITOCHONDRIA IN THE NORMAL CONDITION
The mitochondrion is a double‐membrane‐bound organelle in most eukaryotic cell. Generally, mitochondria are between 0.75 and 3 μm in diameter but differ in lengths and structure. Hundreds of mitochondria form cable‐like structures and supply the energy demand of cells through the oxidative phosphorylation (OXPHOS) process.
Mitochondria constantly divide, fuse, and alter their size and shape, forming a dynamic network to maintain their integrity and quantity. Normal mammalian cells maintain a balance between fusion and fission. Mitofusin 1 and 2 (Mfn1/2) proteins are involved in fusion of the outer mitochondrial membrane, and optic atrophy 1 (OPA1) protein mediates fusion of the inner mitochondrial membranes. Mitochondrial fission 1 (Fis1) and dynamin‐related protein 1 (Drp1) are associated with mitochondrial fission. Fis1 localizes primarily on the outer mitochondrial membrane. Drp1is a cytoplasmic protein that translocates to mitochondria and interacts with Fis1 to enhance fission.1 Figure 1 illustrates these general features of mitochondrial fission and fusion.
Figure 1.
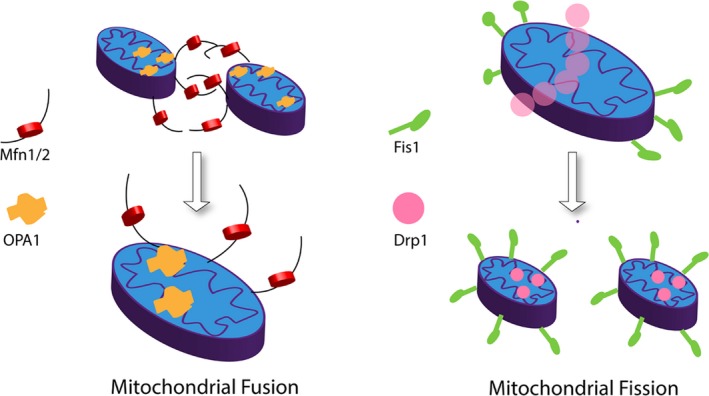
General features of mitochondria fusion and fission. Left: Outer mitochondrial membrane proteins Mfn1 and Mfn2 mediate fusion of the outer mitochondrial membrane while the inner membrane protein OPA1 regulates the fusion of the inner mitochondrial membranes. Right: Fis1 and Drp1 contribute to mitochondrial fission process
A positive mitochondrial membrane potential (Δψm) of 120‐200 mV is fundamental for the normal performance and survival of cells, especially those that have a high‐energy requirement. Thus, loss of Δψm is an indicator of reduced cell health. The collapse of Δψm due to the opening of a high‐conductance pore in the inner mitochondrial membrane is part of the molecular mechanism in apoptosis. The mitochondrial uncoupler carbonyl cyanide‐m‐chlorophenylhydrazone (CCCP) depolarizes the inner mitochondrial membrane, reducing Δψm and ATP production, thus increasing the level of AMP and the phosphorylated (active) AMP‐activated protein kinase (pAMPK). This sequence of events elevates the level of reactive oxygen species (ROS), leading to oxidative damage.
While mitochondria are critical in regulating cellular energy balancem, they also have well‐described roles in the maintenance of essential cellular functions. These include processes such as cellular differentiation, regulation of the cellular growth cycle, and cell death.2
2. MITOCHONDRIAL BIOGENESIS AND ITS REGULATORS
Mitochondrial biogenesis is regulated to adapt the mitochondrial population to a cell's energy demands in response to the conditions of growth, cell division, and changes in oxidative stimuli and hormones. Mitochondrial biogenesis depends on the coordinated expression of nuclear and mitochondrial DNA. Mitochondrial transcription factor A (TFAM) and nuclear respiratory factors 1 and 2 (NRF1 and NRF2) regulate the biogenesis of mitochondria.3, 4 The PPARγ coactivator‐1 family of transcription coactivators, sirtuins and AMPK, also are involved in regulating gene expression during mitochondria biogenesis (Figure 2).
Figure 2.
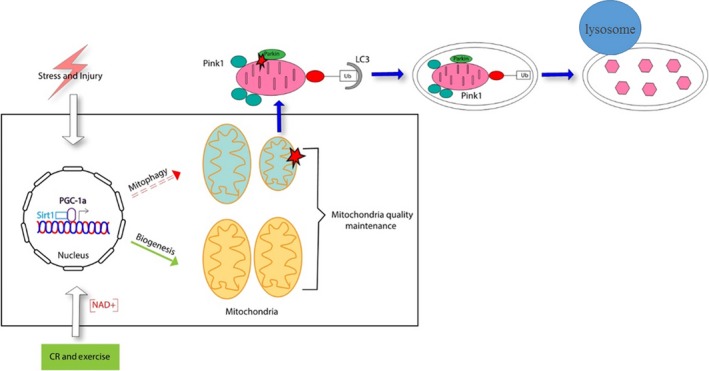
The proteins involved in the process of mitochondrial biogenesis and mitophagy that help to maintain the mitochondria quality. Caloric restriction (CR), physical exercise, and energetic status elevate SIRT1 or NAD+ levels that promote the nuclear translocation of PGC‐1α, which would enhance the transcription of genes associated with the mitochondrial function and biogenesis. Supplementing with NAD+ also enhance SIRT1 activity and trigger mitophagy
2.1. PGC‐1 families
There are three members in the PPARγ coactivator‐1 (PGC‐1) family: PGC‐1α, PGC‐1β, and PGC‐1‐related coactivator (PRC). The PGC‐1 coactivators respond to different stimuli to promote mitochondrial gene expression.5, 6 PGC‐1α, the master regulator of mitochondrial biogenesis, directly interacts with NRF1 to increase the transcription of the genes regulated by NRF1. Overexpression of PGC‐1α in cerebellar neurons increases mitochondrial density by 30% and protects the neurons against mutant synuclein A53T‐α or mutant Huntingtin gene (Htt)‐induced degeneration.7 The brains of mice lacking PGC‐1α presents microvacuolation and neuronal loss, which highlights the important role of PGC‐1α in the nervous system.8, 9 In fact, the common character of several neurodegenerative diseases, including Alzheimer's disease, Parkinson's disease, and Huntington's disease, is the impaired function of PGC‐1α.
PGC‐1β plays a role in the maintenance of basal mitochondrial functions.7 PRC, which is expressed ubiquitously, also enhances the gene transcription of NRF1. PRC mainly regulates gene expression in proliferating cells and in growth‐regulated mitochondrial biogenesis.10 PRC depletion results in the aggregation of atypical mitochondria and severe respiratory chain dysfunction.11
2.2. Sirtuins
Sirtuins are class III protein deacetylases that consume one molecule of NAD+ during each deacetylation cycle. Evidence form the past research indicates that sirtuins regulate the aging process and extend life span.12, 13 Sirtuin levels are influenced by diet, exercise, and environmental stress. There are seven mammalian sirtuins, SIRT1‐7. In addition to deacetylating histones or other specific transcription factors to regulate gene expression, the mammalian sirtuins also regulate the activities of metabolic enzymes to response the calorie restriction or other stresses.
SIRT1 is the most studied sirtuin. Research has demonstrated that overexpression of SIRT1 can alleviate diseases, including neurodegenerative diseases, diabetes, and liver steatosis.14, 15 SIRT1 deacetylates various transcription factors and coactivators, including PGC‐1α, the tumor suppressor p53, and FOXO to enhance the transcription of genes regulated by these factors.16 In the energy‐deficient condition caused by disease or injury, SIRT1 activation has neuroprotective effects by promoting mitochondrial biogenesis and triggering the turnover of damaged mitochondria.17
Unlike SIRT1, which mostly localizes to the nucleus, SIRT3 localizes to mitochondria and is essential to mediate the response to oxidative stress by activating superoxide dismutase‐2 (SOD2).In response to oxidative stress, SIRT3 deacetylates SOD2 to enhance its ability to scavenge ROS.18 Normally, the endogenous SOD2 is inactivated by acetylation and it is important for antioxidant enzymes to counteract cellular ROS. SIRT3 deficiency dramatically exacerbates the degeneration of nigro‐striatal dopaminergic neurons in 1‐methyl‐4‐phenyl‐1,2,3,6‐tetrahydropyridine (MPTP)‐induced Parkinson Disease mice.19 Overexpression of SIRT3 decreases the excitotoxicity of N‐methyl‐d‐aspartic acid (NMDA) to the cultured mouse cortical neurons.20
2.3. AMP‐activated protein kinase
AMP‐activated protein kinase (AMPK) is an important regulator of cellular metabolism in eukaryotes. AMPK is activated by the increased AMP/ATP ratio which leads to enhanced glucose transport, fatty acid oxidation, and so on.21 AMPK also regulates mitochondria function. On one hand, AMPK can stimulate mitochondria biogenesis through increasing the gene transcription regulated by PGC‐1α; on the other hand, AMPK can acutely trigger the destruction of existing defective mitochondria through Unc‐51‐like autophagy activating kinase (ULK1)‐dependent mitophagy. The dual processes controlled by AMPK have the net effect of replacing existing defective mitochondria with new functional mitochondria. AICAR (5‐aminoimidazole‐4‐carboxamide ribonucleoside), the agonist of AMPK, has neuroprotective effects by reducing β‐amyloid peptide (Aβ) production in neuronal cell culture in the AD cellular model.22
3. MITOCHONDRIAL ANOMALIES WITH AGING
Aging induces many potentially interconnected defects and is a common risk factor for adult human diseases. During the aging process, multiple mitochondrial anomalies may occur, including bioenergetic deficiency, the increased oxidative stress from respiratory chain, and the accumulation of the dysfunctional mitochondria.23, 24 Damaged mitochondrial DNA (mtDNA) and the accumulation of injured mitochondria are considered major contributors to aging.
Mutation in mtDNA reduces the lifespan of mice and humans.27 According to the mathematical models, there is only a limited clonal expansion of somatic mtDNA mutations that can occur in short‐lived organisms like fruit flies. The experimentally obtained very high mtDNA mutation levels, which are unlikely to be found in nature, also reduce the lifespan of fruit flies. Additionally, adulthood is less sensitive to mtDNA mutations than is embryonic development.28
3.1. Mitochondrial damage and mutation caused by ROS and replication errors
Impairment of mitochondrial dynamics can result in reduced oxidative phosphorylation and cell death. Mitochondria produce ATP for cellular energy requirements, and ROS are toxic by‐products generated during OXPHOS in mitochondria. Mitochondria generate about 90% of the ROS present in cells. Excess ROS may cause oxidative damage to both nuclear DNA (nDNA) and mtDNA, which may contribute to many age‐related disease states.29 Compared to nDNA, mtDNA is prone to be damaged by oxidative stress because it is not protected by associated histones and other chromatin proteins and is near the ROS‐generating respiratory chain.
Besides ROS, replication errors and failure of the repair mechanisms may be the more important reason for accumulation of mtDNA mutations.30 mtDNA replication is independent of cellular division, so the replication rate is higher than that of nDNA.31 In the meanwhile, there is a decrease in the DNA repair enzymes such as DNA polymerase 1 and endonuclease in aged tissue.26 Ultra‐deep sequencing was used to study genome‐wide mtDNA mutation load; the sequence analysis showed that most somatic mtDNA mutations occur as replication errors during development and do not result from damage accumulation in adult life.32
3.2. Accumulation of dysfunctional mitochondria
Fusion and fission proteins control mitochondrial dynamics. Dysfunctional mitochondria are marked and selectively removed by the specific autophagic process called mitophagy. Through the process of fission, the dysfunctional mitochondria are selected for mitochondrial fragmentation, then turnovered by mitophagy and degraded in lysosomes. Therefore, fission is important for keeping mitochondrial quality and integrity. The level of both autophagy and mitophagy decline with aging,33, 34 which results in an accumulation of dysfunctional mitochondria, advanced oxidative stress, and increased cell apoptosis. Dysfunctional mitochondrial accumulation occurs in all tissues during aging, including skeletal muscle, liver, and brain.
4. MITOCHONDRIAL ABNORMALITIES LEAD TO NEURODEGENERATIVE DISEASES
Mitochondrial dysfunction occurs in most neurodegenerative diseases, including Parkinson's diseases (PD), Alzheimer's (AD), Huntington's disease (HD), Friedreich's ataxia (FRDA), and amyotrophic lateral sclerosis (ALS).
4.1. Parkinson's disease
Parkinson's disease is a widespread neurodegenerative disease. The primary hallmark of PD is the loss of dopaminergic neurons of the substantia nigra (SN). Several important genes including PARK7 (encoding DJ‐1), α‐synuclein, parkin, PINK1, or LRRK2 have pathogenic mutations in PD which cause defects in mitochondrial dynamics and function.36 Specifically, mutation of α‐synuclein leads to its aggregation and these α‐synuclein aggregates delay fusion of phagosomes with lysosomes during the mitophagic process. Meanwhile, PINK1 deletion results in increased oxidative stress within mitochondria.37 In addition, environmental toxins cause mitochondrial dysfunction and are regarded as risk factors for PD as implicated in PD animal or cell models. For example, the complex I inhibitors MPTP, rotenone, pyridaben, and fenpyroximate can mimic the pathological features of PD at low doses and lead to neurodegeneration in flies, rodents, and mammalian cell culture models. Recently, the MitoPark mouse, a model that specifically lacks the gene for TFAM, has become the new genetic model for PD. The mitochondria dysfunction in dopaminergic neurons in MitoPark mice mimics many distinct characteristics of PD, including progressive and selective loss of dopamine neurons, motor deficits, and accumulation of inclusion bodies.38
4.2. Alzheimer's disease
Alzheimer's disease is a common form of dementia that is associated with aging. The disease‐defining appearance of h Aβ aggregates and Tau pathologies correlate with mitochondrial dysfunctions in neurons. Aβ is generated through the cleavage of Aβ precursor protein39 by α‐, β‐, and γ‐secretases. The increased oxidative stress resulting from mitochondrial dysfunction generates the lipid peroxidation product 4‐hydroxynonenal, which covalently modifies the γ‐secretase complex and leads to amplified secretase activity. Increased γ‐secretase activity results in accelerated Aβ accumulation.40, 41 Hyperphosphorylation of Tau protein (pTau) is another hallmark of AD along with Aβ.42 Elevated Ca2+ and ROS levels during mitochondrial dysfunction both contribute to the accumulation of pTau aggregates.43, 44 Melov et al45 showed that mitochondrial SOD2 deficiency can result in pTau aggregates in mice, a symptom that is reversible by the administration of antioxidants. Most recently, Sorrentino et al46 also demonstrated that amyloid aggregation in cells can be reduced by pharmacologically and genetically targeting the mitophagy process.
4.3. Huntington's disease
Huntington's disease is a neurodegenerative disease caused by CAG repeat expansion in the mutant HTT (or IT15) gene, which increases the size of the polyglutamine (polyQ) tract in the N‐terminal of the Huntington (Htt) protein. The mutant Htt protein with expanded polyQ forms aggregates. This aggregation recruits other proteins and mitochondria, adversely affecting the mitochondrial fission‐fusion process and disrupting the mitochondrial transportation along axons and dendrites.47, 48 Mutant Htt may interact directly with the outer mitochondrial membrane and destabilize the membrane while increasing the sensitivity to Ca2+ or other apoptotic inducers.50 Meanwhile, ROS production also increases in HD patients and the mouse models, causing mitochondrial impairment.51 In HD patients and mouse models, mitochondrial fragmentation increases while the motility and respiration decrease. 3‐Nitropropionic acid (3‐NP), an inhibitor of the mitochondrial citric acid cycle, produces the selective striatal degeneration and mimics the progressive locomotor deterioration of HD.52, 53 Glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) is a key molecule in the glycolytic pathway. In normal cells, oxidized inactive GAPDH (iGAPDH) helps initiate the engulfment of the damaged mitochondrion into the lysosome for degradation. However, Hwang et al55 have shown that expanded polyglutamine repeats in HD cell models abnormally interacted with GAPDH, which stalled the GAPDH‐mediated mitophagy.
4.4. Friedreich's ataxia
Similar to HD, Friedreich's ataxia (FRDA) is an inherited neurodegenerative disorder caused by homozygous DNA repeat expansion mutation. FRDA's GAA expansion mutation leads to deficiency of the frataxin protein, causing mitochondrial dysfunction through promoting ROS production.56 Recently, studies by Abeti et al57 revealed that FRDA mouse models show a decrease in mitochondrial membrane potential that is caused by an activity imbalance between Complex I and II in the electron transport chain. This imbalance causes ROS generation in the mitochondrial intermembrane space and the matrix, and the subsequent lipid peroxidation results in neuron degeneration. PGC‐1a, the master regulator of mitochondria biogenesis, also plays a role in FRDA development. Lin et al58 demonstrated that in diseased FRDA mouse models, both PGC‐1a and its downstream effectors are significantly reduced compared to healthy controls. This impairment occurred early in the mitochondrial biogenesis pathway and is considered a potential therapeutic target for FRDA treatment.
4.5. Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis is a progressive disease that affects motor neurons in spinal cord and brain. Over 90% of ALS patients are sporadic. The ALS patients have movement abnormalities as well as a progressive loss of intellectual function. Mitophagy, the process of eliminating damaged mitochondria, plays a significant role in the ALS disease mechanisms. The neuromuscular junction (NMJ) of SOD1G39A mice, an ALS disease model, contains significantly fewer phagosomes than do in wild‐type mice, indicating a disruption in mitophagy. The mitophagy‐related proteins PINK1 and Parkin also are downregulated. In PINK1‐Parkin double‐knockout models, mice experience exacerbated NMJ degeneration and axon swelling which corresponds to ALS symptoms.
Furthermore, double‐knockout mice show an increased amount of ATP synthase beta subunit. This suggests that the increasing quantity of mitochondria at the junction resulted from dysfunctional mitophagy.59
5. THE ELIMINATION OF ABNORMAL MITOCHONDRIA
5.1. Mitophagy maintains the quality of mitochondria in cells
Mitophagy is part of the mitochondrial quality control system and is regulated by mitochondria fission‐ and fusion‐promoting proteins. In these processes, impaired mitochondria are engulfed into an autophagosome, which then fuses with a lysosome for degradation by lysosomal enzymes (Figure 2). Mitochondrial dynamics is a crucial process in maintaining proper mitochondrial morphology and in regulating mitochondrial function, responses to apoptotic stimuli, and monitoring mitochondrial quality.60, 61
The PINK1/Parkin interaction is crucial in regulating mitophagy in mammalian cells. PTEN‐induced putative kinase 1 (PINK1) is a serine/threonine protein kinase present in the cytosol but also targeted to the outer mitochondrial membrane. Mitochondria with positive mitochondrial membrane potential import and degrade PINK1, preventing its accumulation on the outer mitochondrial membrane. PINK1 accumulates on the impaired mitochondria with a decreased Δψm. Parkin, a component of a multi‐protein E3 ubiquitin ligase complex, binds with PINK1 accumulated on the impaired mitochondria and tags the damaged mitochondria with ubiquitin for degradation through mitophagy.62, 63
In addition, increasing evidence supports, that mitophagy also can occur in a Parkin‐independent way. Without the participation of Parkin, some proteins, such as NIX, FUNDC1, or BNIP3, and cardiolipin directly interact with LC3 protein and engulf the dysfunctional mitochondria into autophagosome.64, 65 Meanwhile, other E3 ubiquitin ligases such as SMURF1 and MUL1 also can ubiquitinate the damaged mitochondria and promote mitophagy.67
Endosomes also play a role in mitochondrial elimination. Dysfunctional mitochondria are marked with ubiquitin by parkin, sequestered into Rab5‐positive early endosomes, and ultimately delivered into lysosomes for degradation. In certain cell types, disruption of the endosomal pathway through loss of Rab5 function increases the likelihood of cell death due to mitochondrial stress.68
5.2. Abnormal mitochondrial function affects lysosomal activity
Maintaining mitochondrial homeostasis requires the cooperation of mitochondrial biogenesis, mitochondrial fusion, fission, and mitophagy. The lysosome is the main degradation and recycling organelle. Mitophagy tags and delivers the dysfunctional mitochondria to the lysosome for degradation. Meanwhile, mitochondrial dysfunction also affects the structure and function of lysosomes.69 Mfn2 promotes lysosomal autophagocytosis. Mfn2 depletion in cardiomyocytes retards the fusion of autophagosomes with lysosomes.70 Phagocytosis also can be impaired by the depletion of proteins AIF, OPA1, or PINK1, and by chemical inhibition of the electron transport chain, causing the enlarged lysosomal vacuoles. CCCP increases mitochondrial ROS levels via membrane depolarization and specifically activates the lysosomal TRPML1 channels, causing lysosomal Ca2+ release. This activation triggers nuclear translocation of transcription factor EB (TFEB). This calcineurin‐dependent transcription factor can activate the autophagy/lysosome pathway by regulating the biogenesis of autophagy/lysosome organelles.71
6. METHODS TO ENHANCE MITOCHONDRIAL FUNCTION
6.1. Pharmacological methods
There are several pharmacological strategies aimed at triggering mitochondria biogenesis to treat neurodegenerative diseases. Rosiglitazone and bezafibrate can activate the PPAR‐PGC‐1α axis, and the SIRT1 agonists quercetin and resveratrol can activate sirtuins and AMPK.72, 73 Bezafibrate increase the mitochondrial proteins and mitochondrial ATP generating capacity; as the result, it had a neuroprotective effect in this mouse model of mitochondrial encephalopathy.76 Rosiglitazone can increase mitochondrial mass and attenuate mitochondrial dysfunction in mutant Htt‐expressing cells.77
Several antioxidants such as creatine, coenzyme Q10, and mitochondrially‐targeted antioxidants/peptides seems to improve the patient's ankle mobility in PD clinical trials.78 However, most of the antioxidants, such as dimebon, seem to have beneficial effects in the AD preclinical research, but failed in the AD clinical trials.79, 80 Since mitochondrial dysfunction occurs at the early stage of diseases, the method of pharmacological interventions should be considered.
Besides enhancing mitochondria biogenesis and the scavenging of ROS, inducing mitophagy would be another strategy to maintain mitochondria homeostasis. Urolithin A, a metabolite of ellagitannins produced by the human gut microbiota, induces mitophagy, prevents the accumulation of dysfunctional mitochondria with age, and prolongs lifespan in Caenorhabditis elegans and increases muscle function in rodents.81 Urolithin A can go through the blood‐brain barrier, which may have neuroprotective effects against neurodegenerative diseases. Spermidine is produced from putrescine and could be a precursor of spermine generation. Spermidine acts as acetylase inhibitor and induces autophagy in a SIRT1‐independent manner.82 Therefore, spermidine promotes basal autophagic flux and stimulates mitophagy repairing mitochondrial activity in aged cardiomyocytes.83 Spermidine feeding protects from age‐induced memory impairment in an autophagy‐dependent manner.84 Spermidine induces the formation of mitophagosomes and decreases the aggregation of dysfunctional mitochondrial through the PINK1/Parkin pathway.85
6.2. Caloric restriction
Caloric restriction (CR) is to decrease the calories intake but maintain all the essential nutrients and without malnutrition. CR decreases the production of ROS and reduces oxidative DNA damage, slows down the transcriptional changes associated with aging.86
Sirtuins are considered to have an important role in mediating the beneficial effects of CR on longevity.87, 88 Similar with CR, SIRT1 overexpression is helpful to extend the life span and decrease the disease syndromes of neurodegenerative diseases. CR induced the expressions of sirtuins, such as SIRT1, SIRT3, SIRT5, and SIRT7.89 SIRT1 knockout mice cannot live longer even with the CR diet.90 CR delayed the onset of prion disease mice but failed to delay the onset in the SIRT1 knockout strain.91 Knocking out the mitochondrial SIRT3 prevents the protective effect of CR against hearing loss.92
Caloric restriction enhances the number of mitochondrial cristae as well as the number of mitochondria in per cell.93 CR also prevents excitotoxic conditions through the indirect decrease in mitochondrial permeability and calcium retention. These are mediated through CR‐activated SIRT3 deacetylation and inhibition of cyclophilin D, a peptidylprolyl isomerase.94
Additionally, CR inhibits the PI3K/AKT pathway, induces autophagy, which may increase mitophagy and maintain mitochondria homeostasis.
6.3. Physical exercise
Some research results show that exercise might be helpful in retarding the progress of neurodegenerative diseases,95, 96 which may be associated with the recovery of mitochondrial function by exercise. Exercise stimulates brain mitochondrial activity. Exercise not only increases resistance against rotenone, an inhibitor of complex I activity, but also increases mRNA expression of TFAM and Ndufa6, subunits of mitochondrial complex I.97 At the same time, exercise increases mtDNA repair capacity in the mouse hippocampus and activates mitochondrial uncoupling proteins (UCP) which can regulate mitochondrial proliferation98 and control the production of mitochondrial‐derived ROS. Exercise upregulates UCP2 levels in the hippocampus, lowers cellular oxidative stress99 and can activate autophagy, which is helpful in maintaining muscle mass.100, 101 We have found that exercise could ameliorate the detrimental effect of chloroquine on skeletal muscles through restoring autophagic flux104 and activating the autophagy/lysosomal pathway through AMPK pathway in cerebral cortex and striatum.105 The enhanced mitophagy lessens the level of dysfunctional mitochondria to maintain a high quality of mitochondria in cells.
7. CONCLUDING REMARKS
Mitochondrial dysfunction, the downstream oxidative stress and impaired autophagy/lysosomal activity are the main factors involved in neurodegeneration. Thus, drugs that improve mitochondria function, scavenge the excessive ROS, or enhance the autophagic flux may have the potential to treat neurodegenerative diseases. However, pharmacological agents enhancing mitochondria integrity to treat neurodegenerative diseases remain to be developed. At this point, both CR and exercise, which can enhance mitochondria biogenesis and the autophagy/lysosome pathway (including mitophagy), maybe helpful in retarding the onset and progression of neurodegenerative diseases.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Grant number No. 81571252, 2016), Natural Science Foundation of Jiangsu Province (Grant number BK20151233), and Suzhou Applied Basic Research Program (SYS201509).
Wang Y, Xu E, Musich PR, Lin F. Mitochondrial dysfunction in neurodegenerative diseases and the potential countermeasure. CNS Neurosci Ther. 2019;25:816–824. 10.1111/cns.13116
REFERENCES
- 1. Westermann B. Molecular machinery of mitochondrial fusion and fission. J Biol Chem. 2008;283(20):13501‐13505. [DOI] [PubMed] [Google Scholar]
- 2. Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014;24(12):761‐770. [DOI] [PubMed] [Google Scholar]
- 3. Escriva H, Rodriguez‐Pena A, Vallejo CG. Expression of mitochondrial genes and of the transcription factors involved in the biogenesis of mitochondria Tfam, NRF‐1 and NRF‐2, in rat liver, testis and brain. Biochimie. 1999;81(10):965‐971. [DOI] [PubMed] [Google Scholar]
- 4. Picca A, Lezza AM. Regulation of mitochondrial biogenesis through TFAM‐mitochondrial DNA interactions: useful insights from aging and calorie restriction studies. Mitochondrion. 2015;25:67‐75. [DOI] [PubMed] [Google Scholar]
- 5. Scarpulla RC. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene. 2002;286(1):81‐89. [DOI] [PubMed] [Google Scholar]
- 6. Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC‐1. Cell. 1999;98(1):115‐124. [DOI] [PubMed] [Google Scholar]
- 7. Wareski P, Vaarmann A, Choubey V, et al. PGC‐1{alpha} and PGC‐1{beta} regulate mitochondrial density in neurons. J Biol Chem. 2009;284(32):21379‐21385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin J, Wu PH, Tarr PT, et al. Defects in adaptive energy metabolism with CNS‐linked hyperactivity in PGC‐1alpha null mice. Cell. 2004;119(1):121‐135. [DOI] [PubMed] [Google Scholar]
- 9. Tsunemi T, La Spada AR. PGC‐1alpha at the intersection of bioenergetics regulation and neuron function: from Huntington's disease to Parkinson's disease and beyond. Prog Neurogibol. 2012;97(2):142‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gleyzer N, Scarpulla RC. Concerted action of PGC‐1‐related coactivator (PRC) and c‐MYC in the stress response to mitochondrial dysfunction. J Biol Chem. 2016;291(49):25529‐25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vercauteren K, Gleyzer N, Scarpulla RC. Short hairpin RNA‐mediated silencing of PRC (PGC‐1‐related coactivator) results in a severe respiratory chain deficiency associated with the proliferation of aberrant mitochondria. J Biol Chem. 2009;284(4):2307‐2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanfi Y, Naiman S, Amir G, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483(7388):218‐221. [DOI] [PubMed] [Google Scholar]
- 13. Satoh A, Brace CS, Rensing N, et al. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18(3):416‐430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chakrabarti P, English T, Karki S, et al. SIRT1 controls lipolysis in adipocytes via FOXO1‐mediated expression of ATGL. J Lipid Res. 2011;52(9):1693‐1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25(3):138‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gerhart‐Hines Z, Rodgers JT, Bare O, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC‐1alpha. EMBO J. 2007;26(7):1913‐1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang BL. Sirt1 and the mitochondria. Mol Cells. 2016;39(2):87‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kong X, Wang R, Xue Y, et al. Sirtuin 3, a new target of PGC‐1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE. 2010;5(7):e11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu L, Peritore C, Ginsberg J, Kayhan M, Donmez G. SIRT3 attenuates MPTP‐induced nigrostriatal degeneration via enhancing mitochondrial antioxidant capacity. Neurochem Res. 2015;40(3):600‐608. [DOI] [PubMed] [Google Scholar]
- 20. Kim SH, Lu HF, Alano CC. Neuronal Sirt3 protects against excitotoxic injury in mouse cortical neuron culture. PLoS ONE. 2011;6(3):e14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeon SM. Regulation and function of AMPK in physiology and diseases. Exp Mol Med. 2016;48(7):e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Won JS, Im YB, Kim J, Singh AK, Singh I. Involvement of AMP‐activated‐protein‐kinase (AMPK) in neuronal amyloidogenesis. Biochem Biophys Res Commun. 2010;399(4):487‐491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amigo I, da Cunha FM, Forni MF, et al. Mitochondrial form, function and signalling in aging. Biochem J. 2016;473(20):3421‐3449. [DOI] [PubMed] [Google Scholar]
- 24. Chistiakov DA, Sobenin IA, Revin VV, Orekhov AN, Bobryshev YV. Mitochondrial aging and age‐related dysfunction of mitochondria. Biomed Res Int. 2014;2014:238463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guarente L. Aging research‐where do we stand and where are we going? Cell. 2014;159(1):15‐19. [DOI] [PubMed] [Google Scholar]
- 26. Wang AL, Lukas TJ, Yuan M, Neufeld AH. Age‐related increase in mitochondrial DNA damage and loss of DNA repair capacity in the neural retina. Neurobiol Aging. 2010;31(11):2002‐2010. [DOI] [PubMed] [Google Scholar]
- 27. Kujoth GC, Hiona A, Pugh TD, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481‐484. [DOI] [PubMed] [Google Scholar]
- 28. Kauppila T, Bratic A, Jensen MB, et al. Mutations of mitochondrial DNA are not major contributors to aging of fruit flies. Proc Natl Acad Sci U S A. 2018;115(41):E9620‐E9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst). 2007;6(4):470‐480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Szczepanowska K, Trifunovic A. Different faces of mitochondrial DNA mutators. Biochim Biophys Acta. 2015;1847(11):1362‐1372. [DOI] [PubMed] [Google Scholar]
- 31. Pinto M, Moraes CT. Mechanisms linking mtDNA damage and aging. Free Radic Biol Med. 2015;85:250‐258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ameur A, Stewart JB, Freyer C, et al. Ultra‐deep sequencing of mouse mitochondrial DNA: mutational patterns and their origins. PLoS Genet. 2011;7(3):e1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146(5):682‐695. [DOI] [PubMed] [Google Scholar]
- 34. Shaik A, Schiavi A, Ventura N. Mitochondrial autophagy promotes healthy aging. Cell Cycle. 2016;15(14):1805‐1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamamoto T, Takabatake Y, Kimura T, et al. Time‐dependent dysregulation of autophagy: implications in aging and mitochondrial homeostasis in the kidney proximal tubule. Autophagy. 2016;12(5):801‐813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85(2):257‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Menzies FM, Fleming A, Caricasole A, et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93(5):1015‐1034. [DOI] [PubMed] [Google Scholar]
- 38. Branch SY, Chen C, Sharma R, Lechleiter JD, Li S, Beckstead MJ. Dopaminergic neurons exhibit an age‐dependent decline in electrophysiological parameters in the mitopark mouse model of Parkinson's disease. J Neurosci. 2016;36(14):4026‐4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Appelqvist H, Waster P, Kagedal K, Ollinger K. The lysosome: from waste bag to potential therapeutic target. J Mol Cell Biol. 2013;5(4):214‐226. [DOI] [PubMed] [Google Scholar]
- 40. Esposito L, Raber J, Kekonius L, et al. Reduction in mitochondrial superoxide dismutase modulates Alzheimer's disease‐like pathology and accelerates the onset of behavioral changes in human amyloid precursor protein transgenic mice. J Neurosci. 2006;26(19):5167‐5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen L, Na R, Boldt E, Ran Q. NLRP3 inflammasome activation by mitochondrial reactive oxygen species plays a key role in long‐term cognitive impairment induced by paraquat exposure. Neurobiol Aging. 2015;36(9):2533‐2543. [DOI] [PubMed] [Google Scholar]
- 42. Gwon AR, Park JS, Arumugam TV, et al. Oxidative lipid modification of nicastrin enhances amyloidogenic gamma‐secretase activity in Alzheimer's disease. Aging Cell. 2012;11(4):559‐568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mattson MP, Fu W, Waeg G, Uchida K. 4‐Hydroxynonenal, a product of lipid peroxidation, inhibits dephosphorylation of the microtubule‐associated protein tau. NeuroReport. 1997;8(9–10):2275‐2281. [DOI] [PubMed] [Google Scholar]
- 44. Hoglinger GU, Lannuzel A, Khondiker ME, et al. The mitochondrial complex I inhibitor rotenone triggers a cerebral tauopathy. J Neurochem. 2005;95(4):930‐939. [DOI] [PubMed] [Google Scholar]
- 45. Melov S, Adlard PA, Morten K, et al. Mitochondrial oxidative stress causes hyperphosphorylation of tau. PLoS ONE. 2007;2(6):e536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sorrentino V, Romani M, Mouchiroud L, et al. Enhancing mitochondrial proteostasis reduces amyloid‐beta proteotoxicity. Nature. 2017;552(7684):187‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Orr Al, Li S, Wang C‐E, et al. N‐terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28(11):2783‐2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reddy PH, Shirendeb UP. Mutant huntingtin, abnormal mitochondrial dynamics, defective axonal transport of mitochondria, and selective synaptic degeneration in Huntington's disease. Biochim Biophys Acta. 2012;1822(2):101‐110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yano H, Baranov SV, Baranova OV, et al. Inhibition of mitochondrial protein import by mutant huntingtin. Nat Neurosci. 2014;17(6):822‐831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium‐induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13(14):1407‐1420. [DOI] [PubMed] [Google Scholar]
- 51. Gil‐Mohapel J, Brocardo PS, Christie BR. The role of oxidative stress in Huntington's disease: are antioxidants good therapeutic candidates? Curr Drug Targets. 2014;15(4):454‐468. [DOI] [PubMed] [Google Scholar]
- 52. Galpern WR, Matthews RT, Beal MF, Isacson O. NGF attenuates 3‐nitrotyrosine formation in a 3‐NP model of Huntington's disease. NeuroReport. 1996;7(15‐17):2639‐2642. [DOI] [PubMed] [Google Scholar]
- 53. Tariq M, Khan HA, Elfaki I, Al Deeb S, Al Moutaery MK. Neuroprotective effect of nicotine against 3‐nitropropionic acid (3‐NP)‐induced experimental Huntington's disease in rats. Brain Res Bull. 2005;67(1–2):161‐168. [DOI] [PubMed] [Google Scholar]
- 54. Mao Z, Choo YS, Lesort M. Cystamine and cysteamine prevent 3‐NP‐induced mitochondrial depolarization of Huntington's disease knock‐in striatal cells. Eur J Neurosci. 2006;23(7):1701‐1710. [DOI] [PubMed] [Google Scholar]
- 55. Hwang S, Disatnik MH, Mochly‐Rosen D. Impaired GAPDH‐induced mitophagy contributes to the pathology of Huntington's disease. EMBO Mol Med. 2015;7(10):1307‐1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pandolfo M. Friedreich ataxia: the clinical picture. J Neurol. 2009;256(suppl 1):3‐8. [DOI] [PubMed] [Google Scholar]
- 57. Abeti R, Parkinson MH, Hargreaves IP, et al. Mitochondrial energy imbalance and lipid peroxidation cause cell death in Friedreich's ataxia. Cell Death Dis. 2016;7:e2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin H, Magrane J, Rattelle A, et al. Early cerebellar deficits in mitochondrial biogenesis and respiratory chain complexes in the KIKO mouse model of Friedreich ataxia. Dis Model Mech. 2017;10(11):1343‐1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rogers RS, Tungtur S, Tanaka T, et al. Impaired mitophagy plays a role in denervation of neuromuscular junctions in ALS mice. Front Neurosci. 2017;11:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Twig G, Elorza A, Molina AJA, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27(2):433‐446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liesa M, Palacin M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89(3):799‐845. [DOI] [PubMed] [Google Scholar]
- 62. Durcan TM, Fon EA. The three 'P's of mitophagy: PARKIN, PINK1, and post‐translational modifications. Genes Dev. 2015;29(10):989‐999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jin SM, Youle RJ. PINK1‐ and Parkin‐mediated mitophagy at a glance. J Cell Sci. 2012;125(Pt 4):795‐799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chu CT, Ji J, Dagda RK, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15(10):1197‐1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11(1):45‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12(1):9‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Orvedahl A, Jr RS, Xiao G, et al. Image‐based genome‐wide siRNA screen identifies selective autophagy factors. Nature. 2011;480(7375):113‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hammerling BC, Najor RH, Cortez MQ, et al. A Rab5 endosomal pathway mediates Parkin‐dependent mitochondrial clearance. Nat Commun. 2017;8:14050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Demers‐Lamarche J, Guillebaud G, Tlili M, et al. Loss of mitochondrial function impairs lysosomes. J Biol Chem. 2016;291(19):10263‐10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhao T, Huang X, Han L, et al. Central role of mitofusin 2 in autophagosome‐lysosome fusion in cardiomyocytes. J Biol Chem. 2012;287(28):23615‐23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Redmann M, Darley‐Usmar V, Zhang J. The role of autophagy, mitophagy and lysosomal functions in modulating bioenergetics and survival in the context of redox and proteotoxic damage: implications for neurodegenerative diseases. Aging Dis. 2016;7(2):150‐162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Menzies KJ, Singh K, Saleem A, Hood DA. Sirtuin 1‐mediated effects of exercise and resveratrol on mitochondrial biogenesis. J Biol Chem. 2013;288(10):6968‐6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Naia L, Rosenstock TR, Oliveira AM, et al. Comparative mitochondrial‐based protective effects of resveratrol and nicotinamide in huntington's disease models. Mol Neurobiol. 2017;54(7):5385‐5399. [DOI] [PubMed] [Google Scholar]
- 74. Valero T. Mitochondrial biogenesis: pharmacological approaches. Curr Pharm Des. 2014;20(35):5507‐5509. [DOI] [PubMed] [Google Scholar]
- 75. Augustyniak J, Lenart J, Gaj P, et al. Bezafibrate upregulates mitochondrial biogenesis and influence neural differentiation of human‐induced pluripotent stem cells. Mol Neurobiol. 2018;816‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Noe N, Dillon L, Lellek V, et al. RETRACTED: bezafibrate improves mitochondrial function in the CNS of a mouse model of mitochondrial encephalopathy. Mitochondrion. 2013;13(5):417‐426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Quintanilla RA, Jin YN, Fuenzalida K, Bronfman M, Johnson GV. Rosiglitazone treatment prevents mitochondrial dysfunction in mutant huntingtin‐expressing cells: possible role of peroxisome proliferator‐activated receptor‐gamma (PPARgamma) in the pathogenesis of Huntington disease. J Biol Chem. 2008;283(37):25628‐25637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Procaccio V, Bris C, Chao de la Barca JM, et al. Perspectives of drug‐based neuroprotection targeting mitochondria. Rev Neurol (Paris). 2014;170(5):390‐400. [DOI] [PubMed] [Google Scholar]
- 79. Reddy PH. Role of mitochondria in neurodegenerative diseases: mitochondria as a therapeutic target in Alzheimer's disease. CNS Spectr. 2009;14(8 suppl 7):8‐13; discussion 16‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Peters OM, Shelkovnikova T, Tarasova T, et al. Chronic administration of Dimebon does not ameliorate amyloid‐beta pathology in 5xFAD transgenic mice. J Alzheimers Dis. 2013;36(3):589‐596. [DOI] [PubMed] [Google Scholar]
- 81. Ryu D, Mouchiroud L, Andreux PA, et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med. 2016;22(8):879‐888. [DOI] [PubMed] [Google Scholar]
- 82. Morselli E, Mariño G, Bennetzen MV, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192(4):615‐629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Eisenberg T, Abdellatif M, Schroeder S, et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med. 2016;22(12):1428‐1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gupta VK, Scheunemann L, Eisenberg T, et al. Restoring polyamines protects from age‐induced memory impairment in an autophagy‐dependent manner. Nat Neurosci. 2013;16(10):1453‐1460. [DOI] [PubMed] [Google Scholar]
- 85. Qi Y, Qiu Q, Gu X, Tian Y, Zhang Y. ATM mediates spermidine‐induced mitophagy via PINK1 and Parkin regulation in human fibroblasts. Sci Rep. 2016;6:24700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mercken EM, Crosby SD, Lamming DW, et al. Calorie restriction in humans inhibits the PI3K/AKT pathway and induces a younger transcription profile. Aging Cell. 2013;12(4):645‐651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cunha‐Santos J, Duarte‐Neves J, Carmona V, Guarente L, Pereira de Almeida L, Cavadas C. Caloric restriction blocks neuropathology and motor deficits in Machado‐Joseph disease mouse models through SIRT1 pathway. Nat Commun. 2016;7:11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Guarente L. Calorie restriction and sirtuins revisited. Genes Dev. 2013;27(19):2072‐2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Canto C, Auwerx J. Caloric restriction, SIRT1 and longevity. Trends Endocrinol Metab. 2009;20(7):325‐331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Boily G, Seifert EL, Bevilacqua L, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3(3):e1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chen D, Steele AD, Hutter G, et al. The role of calorie restriction and SIRT1 in prion‐mediated neurodegeneration. Exp Gerontol. 2008;43(12):1086‐1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Someya S, Yu W, Hallows WC, et al. Sirt3 mediates reduction of oxidative damage and prevention of age‐related hearing loss under caloric restriction. Cell. 2010;143(5):802‐812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Khraiwesh H, López‐Domínguez JA, Fernández del Río L, et al. Mitochondrial ultrastructure and markers of dynamics in hepatocytes from aged, calorie restricted mice fed with different dietary fats. Exp Gerontol. 2014;56:77‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Amigo I, Menezes‐Filho SL, Luevano‐Martinez LA, Chausse B, Kowaltowski AJ. Caloric restriction increases brain mitochondrial calcium retention capacity and protects against excitotoxicity. Aging Cell. 2016;16:73‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lima LO, Scianni A, Rodrigues‐de‐Paula F. Progressive resistance exercise improves strength and physical performance in people with mild to moderate Parkinson's disease: a systematic review. J Physiother. 2013;59(1):7‐13. [DOI] [PubMed] [Google Scholar]
- 96. Wang Z, Guo Y, Myers KG, et al. Exercise alters resting‐state functional connectivity of motor circuits in parkinsonian rats. Neurobiol Aging. 2015;36(1):536‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Aguiar AS, Stragier E, da Luz Scheffer D, et al. Effects of exercise on mitochondrial function, neuroplasticity and anxio‐depressive behavior of mice. Neuroscience. 2014;271:56‐63. [DOI] [PubMed] [Google Scholar]
- 98. Andrews ZB, Diano S, Horvath TL. Mitochondrial uncoupling proteins in the CNS: in support of function and survival. Nat Rev Neurosci. 2005;6(11):829‐840. [DOI] [PubMed] [Google Scholar]
- 99. Vaynman S, Ying Z, Wu A, Gomez‐Pinilla F. Coupling energy metabolism with a mechanism to support brain‐derived neurotrophic factor‐mediated synaptic plasticity. Neuroscience. 2006;139(4):1221‐1234. [DOI] [PubMed] [Google Scholar]
- 100. Grumati P, Coletto L, Schiavinato A, et al. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI‐deficient muscles. Autophagy. 2011;7(12):1415‐1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Grumati P, Coletto L, Sandri M, Bonaldo P. Autophagy induction rescues muscular dystrophy. Autophagy. 2011;7(4):426‐428. [DOI] [PubMed] [Google Scholar]
- 102. He C, Sumpter R Jr., Levine B. Exercise induces autophagy in peripheral tissues and in the brain. Autophagy. 2012;8(10):1548‐1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. He C, Bassik MC, Moresi V, et al. Exercise‐induced BCL2‐regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481(7382):511‐515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Jiang D, Chen K, Lu X, Gao HJ, Qin ZH, Lin F. Exercise ameliorates the detrimental effect of chloroquine on skeletal muscles in mice via restoring autophagy flux. Acta Pharmacol Sin. 2014;35(1):135‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Jun H, Xue W, Yi Z, et al. Exercise activates lysosomal function in the brain through AMPK‐SIRT1‐TFEB pathway. CNS Neurosci Ther. 2019;816–12. [DOI] [PMC free article] [PubMed] [Google Scholar]


