Abstract
MicroRNAs (miRNAs) are emerging as important regulators of various physiological and pathological processes and may serve key roles in the maintenance of bone homeostasis via effects on osteoblast differentiation. The aim of the present study was to define the role of miR-877-3p in osteoblast differentiation using MC3T3-E1 cells, an osteoblast precursor cell line. It was demonstrated using RT-qPCR analysis that miR-877-3p was gradually increased in MC3T3-E1 cells during the osteoblastic differentiation induced by transforming growth factor (TGF)-β1. Gain-of-function and loss-of-function experiments revealed that the overexpression of miR-877-3p promoted the osteoblastic differentiation of MC3T3-E1 cells, whereas depletion of miR-877-3p inhibited this process in vitro and in vivo. Bioinformatics analysis and validation experiments demonstrated that Smad7, which acts as a negative regulator of osteogenesis, was a target of miR-877-3p. Furthermore, the overexpression of Smad7 partially reversed the osteoblastic differentiation of MC3T3-E1 cells induced by miR-877-3p. In conclusion, the results of the present study suggest that the miR-877-3p/Smad7 axis is associated with the osteoblastic differentiation of MC3T3-E1 cells and may indicate a potential therapeutic approach for osteogenesis disorders.
Keywords: miR-877-3p, osteoblast differentiation, Smad7, transforming growth factor-β1
Introduction
Bone homeostasis mainly relies on the balance between bone-forming cells (osteoblasts) and bone-resorptive cells (osteoclasts) (1). The imbalance between bone formation and resorption leads to skeletal diseases associated with progressive bone degeneration, such as osteoporosis. Osteoporosis is a disease characterized by lower than normal bone mass and greater than normal bone loss, and is associated with increased risk of bone weakness and fracture (2). Studies on osteoblastic differentiation may help elucidate the pathogenesis of osteoporosis.
Transforming growth factor (TGF)-β1 is a polypeptide member of the TGF family that is involved in cellular functions, including the control of cell growth, proliferation and differentiation (3). TGF-β1 can maintain postnatal bone mass by coupling bone resorption and bone formation (4). TGF-β1 favors osteoclast differentiation by binding to a receptor on osteoclasts, and thereby activating Smad2/3, which is directly associated with the TRAF6-TAB1-TAK1 complex (5).
The possible mechanisms of osteoblastic differentiation and bone formation include the coordination of cell signals and the activity of post-transcriptional factors. MicroRNAs (miRNAs/miRs) serve an important role in osteoblastic differentiation. miRNAs are small non-coding single-stranded RNAs that inhibit the translation or promote the degradation of target mRNAs through binding to their 3′-untranslated region (UTR), and thereby attenuate protein synthesis (6). Numerous miRNAs have been characterized in diverse biological and pathological processes, including cell proliferation, apoptosis, differentiation and tumorigenesis (7–10). There is also evidence that aberrant miRNA expression is strongly associated with osteoblast differentiation through various mechanisms (11). miR-877-3p has been demonstrated to promote cell proliferation and differentiation in lung and bladder cancer (12,13). In addition, miR-877-3p has been revealed to inhibit the myofibroblast differentiation of lung-resident mesenchymal stem cells and attenuate the lung fibrosis induced by bleomycin (12). However, it is unclear whether miR-877-3p is one of the key miRNAs that regulate the proliferation and differentiation of osteoblasts.
The aim of the present study was to define the role of miR-877-3p in osteoblast differentiation using MC3T3-E1 cells, an osteoblast precursor cell line derived from mouse calvaria that has been widely used as a model in bone biology research (14). Results demonstrated that miR-877-3p was upregulated during TGF-β1-mediated osteoblastic differentiation. Bioinformatics analysis identified Smad7 as a target of miR-877-3p. Osteoblastic differentiation of MC3T3-E1 cells was induced by miR-877-3p overexpression whilst Smad7 overexpression partially reversed this effect. Therefore, miR-877-3p/Smad7 may represent a potential therapeutic target for bone regeneration-associated diseases.
Materials and methods
Cell culture and osteoblastic differentiation
The MC3T3-E1 cell line was obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China) and cultured in α-minimal essential medium (α-MEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing penicillin (50 U/ml), 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and streptomycin (50 µg/ml) at 37°C in a humidified atmosphere with 5% CO2. For TGF-β1 treatment, recombinant human TGF-β1 (cat. no. CSB-AP003861HU; CUSABIO Technology LLC, Wuhan, China) with final concentration of 4 ng/ml was added to cell culture for 7 or 14 days. To induce osteoblastic differentiation, cells were seeded (1×105 cells per well) in 6-well plates and treated with osteogenic medium (10 ng/ml β-glycerophosphate; 10 mmol/l dexamethasone and 50 µg/ml vitamin C; all from Sigma-Aldrich; Merck KGaA) for 7 and 14 days when they reached 70% confluence (15).
Cell transfection
pcDNA3.1-Smad negative control (NC) and pcDNA3.1-Smad7 were synthesized by GenePharma Co., Ltd. (Jiangsu, China). miR-877-3p mimics, inhibitor and their corresponding negative controls (NCs), miR-877-3p agomir, miR-877-3p antagomir and their corresponding controls (mimics NC: miR01101, RiboBio Co., Ltd., and inhibitor NC; miR02101; RiboBio Co., Ltd.) were synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The sequences were as follows: miR-877-3p mimics, 5′-UGUCCUCUUCUCCCUCCUCCCA-3′; miR-877-3p inhibitor, 5′-UGGGAGGAGGGAGAAGAGGACA-3′; miR-877-3p agomir, 5′-UGUCCUCUUCUCCCUCCUCCCA-3′ and miR-877-3p antagomir, 5′-UGGGAGGAGGGAGAAGAGGACA-3′. MC3T3-E1 cells were seeded in 6-well plates at a density of 30,000 cells/well and cultured for 24 h in antibiotic-free fresh medium. Subsequently, the cells were transfected with miRNA or vector (100 nM) using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After 48 h incubation, the cells were used for further experiments.
RNA isolation and RT-qPCR
Total RNA was extracted from cells or tissue with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA was quantified by Nanodrop 2000 spectrophotometry (Thermo Fisher Scientific, Inc.). First-strand cDNA synthesis was then performed using a Superscript RT kit (Biouniquer Technology, Nanjing, China) following the manufacturer's instructions. The reaction was performed under the following conditions: 37°C for 15 min and 98°C for 5 min. qPCR was conducted with SYBR Green I (Biouniquer Technology) using the LightCycler 480 II System (Roche Diagnostics, Basel, Switzerland). Primer sequences were as follows: miR-877-3p-F, 5′-GCGTGTATTTGACAAGCTGAGTTG-3′; collagen type I α1 chain (COL1A1) forward, 5′-TCGTGGTGAGACTGGTCCTG-3′ and reverse, 5′-TGTCACCTTGTTCGCCTGTC-3′; runt-related transcription factor 2 (RUNX2) forward, 5′-CGGACGAGGCAAGAGTTTC-3′ and reverse, 5′-TGGCTCAGATAGGAGGGGTAA-3′; osterix (OSX) forward, 5′-AGCAGCAGCAGCAACAGAAG-3′ and reverse, 5′-CTCAGGCCAGCTCACTCTTG-3′; Smad7 forward, 5′-GCTATTCCAGAAGATGCTGTTC-3′ and reverse, 5′-GTTGCTGAGCTGTTCTGATTTG-3′; β-actin forward 5′-GTCCCTCACCCTCCCAAAAG-3′ and reverse, 5′-GCTGCCTCAACACCTCAACCC-3′. The thermocycling conditions were as follows: 95°C for 5 min, followed by 35 cycles at 95°C for 10 sec then 60°C for 30 sec. The 2−ΔΔCq method was used for data analysis with normalization to β-actin (16). U6 (cat. no. B532461-0001; Sangon Biotech Co., Ltd., Shanghai, China) was used as endogenous control for the analysis of miR-877-3p expression.
Measurement of alkaline phosphatase (ALP) activity
Total proteins were extracted from cells using a radioimmunoprecipitation assay (RIPA) lysis buffer (Biouniquer Technology) and quantified by bicinchoninic acid assay according to the manufacturer's protocol. The cell lysate was added to 96-well plates and incubated with ALP staining solution (Sigma-Aldrich; Merck KGaA) for 60 min at 37°C. Following the addition of the stop solution, ALP activity was measured spectrophotometrically at 405 nm, and normalized to the total protein concentration.
Mineralization assay
The MC3T3-E1 cells were cultured with TGF-β1 (4 ng/ml) and osteoblastic differentiation medium for 7 days in 24-well plates at a density of 2×104 cells per well, and the mineralization of the MC3T3-E1 cells was observed via Alizarin Red S (ARS) and ALP staining with an optical light microscope (ARS, Solarbio, China; ALP staining kit, Beyotime Institute of Biotechnology, Haimen, China).
In vivo transplantation
A total of 24 female NOD/SCID mice (age, 4 weeks; weight, 17–19 g) were purchased from the Provincial Animal Center (Guangdong, China) and housed for 1 week to adapt them to the laboratory environment (24°C, 12-h light/dark cycles and humidity 60±10%) with free access to water and food. The mice were randomly divided into four groups (each n=6). MC3T3-E1 cells transfected with miR-877-3p mimics, miR-877-3p mimics inhibitor or their corresponding controls were induced with osteogenic medium for 14 days in vitro. Then, 5×106 cells were loaded onto 20 mg hydroxyapatite-tricalcium phosphate (Sigma-Aldrich; Merck KGaA) and subcutaneously implanted in the dorsal region of the NOD/SCID mice. When the xenografts reached 50 mm3 in volume, miR-877-3p agomir or antagomir or their corresponding controls (5 nmol), respectively, was injected into the tumor every other day. Four weeks later, the xenografts were removed, fixed with 4% paraformaldehyde for 3 days and then decalcified in 10% EDTA (pH 6.0) for 7 days. The xenografts were then embedded in paraffin, sectioned (4 µm) and stained with hematoxylin and eosin (H&E), or Masson's trichrome stain (Sigma-Aldrich; Merck KGaA) according to the manufacturer's protocols. All animal procedures were approved by the Animal Care Committee of Southern Medical University (L2016113).
Western blotting
The cells were harvested and lysed with RIPA buffer (Beyotime Institute of Biotechnology) containing phenylmethane sulfonyl fluoride and protease inhibitors. Equal amounts of protein (30 µg per lane) were loaded on a 10% sodium dodecyl sulfate-polyacrylamide gel, separated by electrophoresis, and transferred onto a 0.4-µm polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). The membranes were sealed with 5% non-fat milk for 1 h at room temperature, then incubated with primary antibodies overnight at 4°C. The primary antibodies were anti-Smad7 (1:100; cat. no. sc-365846, Santa Cruz Biotechnology, Ltd., Dallas, TX, USA), anti-OSX (1:200; cat. no. sc-393325, Santa Cruz Biotechnology, Ltd.), anti-COL1A1 (1:1,000; cat. no. ab90395, Abcam, Cambridge, UK), anti-RUNX2 (1:1,000; cat. no. 12556; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-phospho-Smad2 (1:1,000; cat. no. 18338; Cell Signaling Technology, Inc.), anti-Smad2 (1:1,000; cat. no. 5339; Cell Signaling Technology, Inc.), anti-phospho-Smad3 (1:1,000; cat. no. ab52903; Abcam, Cambridge, UK), Smad3 (1:1,000; cat. no. 9523, Cell Signaling Technology, Inc.) and anti-β-actin antibody (1:1,000; cat. no. 4970; Cell Signaling Technology, Inc.). The membranes were probed with horseradish peroxidase-conjugated anti-rabbit secondary antibody for 1 h at room temperature (1:5,000; cat. no. 7074; Cell Signaling Technology, Inc.) and the protein signals were obtained with ECL Plus substrate (EMD Millipore).
Luciferase reporter assay
The wild-type and mutant putative miR-877-3p binding sites in the 3′UTR of Smad7 were synthesized and cloned into a pmirGLO luciferase vector (Shanghai Genechem Co., Ltd., Shanghai, China). The plasmids, miRNA (miR-877-3p and miR-NC) and Renilla luciferase plasmid (Shanghai Genechem Co., Ltd.) were co-transfected into cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). Firefly and Renilla luciferase activities were then measured using the Dual Luciferase Reporter Assay system (Promega Corporation, Madison, WI, USA) at 48 h after transfection. All experiments were repeated three times.
Bioinformatics target prediction
The candidate target genes of miR-877-3p were predicted using TargetScan online software (http://www.targetscan.org/).
Statistical analysis
Data are expressed as mean ± standard deviation and were analyzed using SPSS 22.0 software (IBM Corp., Armonk, NY, USA). All experiments were repeated at least three times with comparable results unless indicated otherwise. Statistical evaluation of the data was performed using the unpaired Student's t-test for comparisons between two groups and one-way analysis of variance followed by Bonferroni correction post hoc test for comparisons amongst multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
TGF-β1 promotes the osteoblastic differentiation of MC3T3-E1 cells
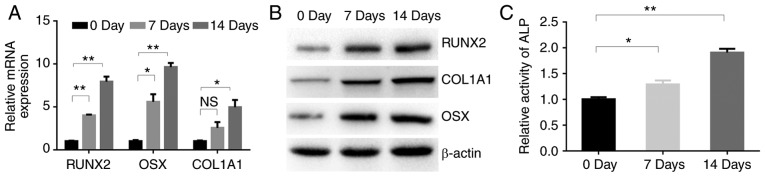
To induce osteoblastic differentiation, the MC3T3-E1 cells were treated with TGF-β1. The mRNA expression levels of three osteogenic genes, namely RUNX2, OSX and COL1A1, were then determined in the MC3T3-E1 cells at 0, 7 and 14 days after TGF-β1 treatment. The expression levels of RUNX2, OSX and COL1A1 mRNA in the MC3T3-E1 cells were increased at days 7 and 14 compared with the respective levels at day 0, with the highest levels being observed at day 14 (Fig. 1A). Consistently, the corresponding protein levels in the MC3T3-E1 cells appeared to increase in a time-dependent manner (Fig. 1B). Furthermore, ALP activity gradually increased during treatment with TGF-β1 (Fig. 1C), indicating the key role of TGF-β1 in osteoblastic differentiation.
Figure 1.
TGF-β1 promotes the osteoblastic differentiation of MC3T3-E1 cells. (A) RT-qPCR analysis of the osteogenic-related genes RUNX2, OSX and COL1A1 in MC3T3-E1 cells after treatment with TGF-β1 (4 ng/ml) for 0, 7 and 14 days. (B) Western blotting of RUNX2, COL1A1-and OSX protein in MC3T3-E1 cells cultured with TGF-β1 (4 ng/ml) for 0, 7 and 14 days. (C) ALP activity in MC3T3-E1 cells after treatment with TGF-β1 (4 ng/ml) for 0, 7 and 14 days. NS, no significance change; *P<0.05, **P<0.01 as indicated. TGF, transforming growth factor; RUNX2, runt-related transcription factor 2; OSX, osterix; COL1A1 collagen type I α1 chain; ALP, alkaline phosphatase.
Expression of miR-877-3p is significantly altered during osteoblast induction and promotes the osteoblastic differentiation of MC3T3-E1 cells
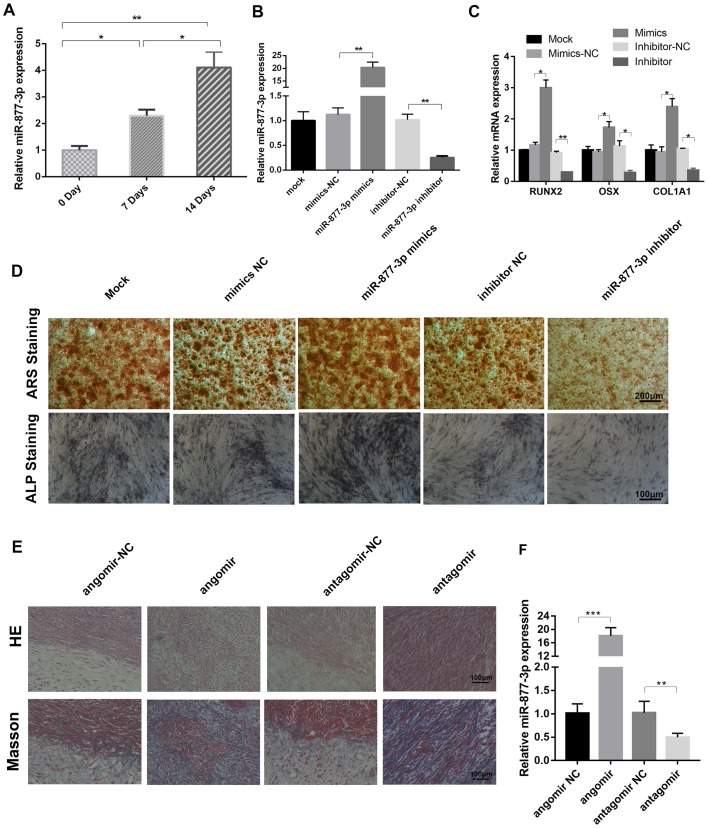
The role of miR-877-3p during osteoblast differentiation is unclear. Therefore, the aim of the present study was to investigate whether miR-877-3p is associated with osteogenic differentiation. Following the treatment of MC3T3-E1 cells with TGF-β1, the expression of miR-877-3p was detected using RT-qPCR analysis, and a time-dependent increase in the level of miR-877-3p was observed (Fig. 2A). To further elucidate the functional role of miR-877-3p during osteoblastic differentiation, miR-877-3p was overexpressed and knocked down in MC3T3-E1 cells via transfection with miR-877-3p mimics and miR-877-3p inhibitor, respectively, and corresponding controls were established for comparison (Fig. 2B). The mRNA levels of the osteoblast-associated markers RUNX2, OSX and COL1A1 were found to be significantly elevated in miR-877-3p-overexpressing MC3T3-E1 cells, and significantly reduced in cells with miR-877-3p knockdown (Fig. 2C). Consistently, ARS and ALP staining results demonstrated that the staining intensity of mineralization nodes was markedly increased in the miR-877-3p-overexpressing cells, and decreased in the miR-877-3p-depleted cells (Fig. 2D). The effect of miR-877-3p on MC3T3-E1 osteogenesis was further evaluated in vivo. The H&E and Masson's trichrome staining results revealed that osteoid formation was increased in the xenografts of mice treated with miR-877-3p agomir, and decreased in those of the mice treated with miR-877-3p antagomir (Fig. 2E). RT-qPCR analysis confirmed that the expression of miR-877-3p was elevated in the agomir-treated group and reduced in antagomir-treated group, compared with the respective controls (Fig. 2F). These results demonstrate that miR-877-3p promoted the osteogenic differentiation of MC3T3-E1 cells.
Figure 2.
miR-877-3p promotes the osteoblastic differentiation of MC3T3-E1 cells. (A) miR-877-3p expression in MC3T3-E1 cells after treatment with TGF-β1 (4 ng/ml) for 0, 7 and 14 days. (B) Expression of miR-877-3p was validated in MC3T3-E1 cells transfected with miR-877-3p mimics or inhibitor and their corresponding controls by RT-qPCR. (C) RUNX2, OSX and COL1A1 mRNA expression in MC3T3-E1 cells. (D) MC3T3-E1 cells transfected with miR-877-3p mimics or inhibitor and their corresponding controls were cultured in osteogenic medium. At day 14, the mineralization of differentiated MC3T3-E1 cells was detected with ARS and ALP staining. (E) Transfected MC3T3-E1 cells were cultured in osteogenic medium for 14 days and then mixed with tricalcium phosphate/hydroxyapatite and transplanted into the dorsal region of nude mice for 4-weeks. The results were then evaluated by H&E and Masson's trichrome staining. (F) (F) Transfected MC3T3-E1 cells were cultured in osteogenic medium for 14 days and then mixed with tricalcium phosphate/hydroxyapatite and transplanted into the dorsal region of nude mice for 4-weeks. The Expression expression of miR-877-3p in the xenografts from each group was detected by RT-qPCR analysis. *P<0.05, **P<0.01 and ***P<0.001, as indicated. miR, microRNA; TGF, transforming growth factor; RUNX2, runt-related transcription factor 2; OSX, osterix; COL1A1 collagen type I α1 chain; NC, negative control; ARS, Alizarin Red S; ALP, alkaline phosphatase; H&E, hematoxylin and eosin.
MiR-877-3p inhibits the expression of Smad7
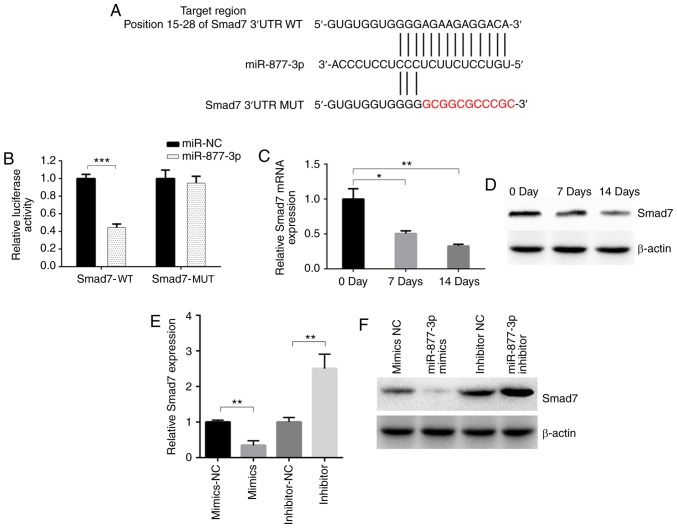
miR-877-3p has been demonstrated to promote the TGF-β1-induced osteoblastic differentiation of MC3T3-E1 cells; however, its downstream target has not yet been determined. Using the online bioinformatics tool TargetScan, Smad7 was predicted to be a downstream target of miR-877-3p with a potential binding site (Fig. 3A). Fragments of the 3′UTR of Smad7 containing a putative miR-877-5p binding site or mutant fragment were synthesized and inserted into plasmids. The predicted binding site of miR-877-3p in Smad7 was confirmed using a luciferase reporter assay (Fig. 3B). The mRNA and protein levels of Smad7 were decreased after TGF-β1 induction (Fig. 3C and D). Whether miR-877-3p regulated osteogenic differentiation by suppressing Smad7 expression was then investigated. The results revealed a notable downregulation of Smad7 expression following the transfection of miR-877-3p mimic into MC3T3-E1 cells, whereas Smad7 was upregulated in cells transfected with miR-877-3p inhibitor, as confirmed by RT-qPCR and western blot assays (Fig. 3E and F). These results support the hypothesis that miR-877-3p directly binds to Smad7.
Figure 3.
miR-877-3p inhibits the expression of Smad7. (A) Design of luciferase reporters with the WT Smad7 3′UTR or the site-directed MUT Smad7 3′UTR. (B) Effect of miR-877-3p and miR-NC on luciferase activity in MC3T3-E1 cells transfected with the WT or MUT Smad7 3′UTR. Smad7 expression detected by (C) RT-qPCR and (D) western blotting in MC3T3-E1 cells after culture with TGF-β1 (4 ng/ml) for 0, 7 and 14 days. Smad7 (E) mRNA and (F) protein expression in MC3T3-E1 cells transfected with miR-877-3p mimics or inhibitors and their corresponding controls. *P<0.05, **P<0.01 as indicated. miR, microRNA; WT, wild type; MUT, mutant; UTR, untranslated region; NC, negative control; TGF, transforming growth factor.
Smad7 may be a negative target of miR-877-3p involved in osteoblast differentiation
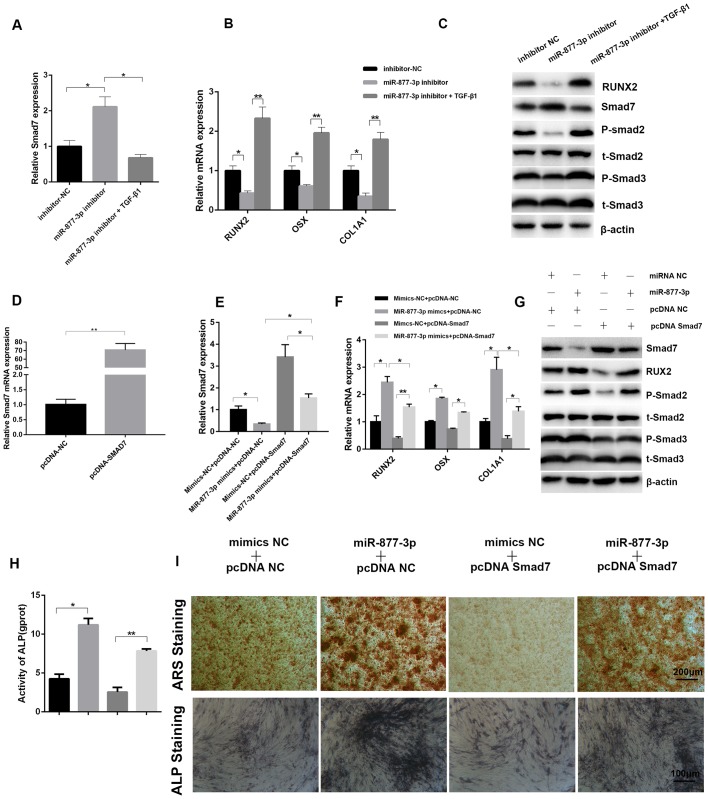
Although Smad7 expression has been found to be affected by miR-877-3p, the interaction between Smad7 and miR-877-3p required further evaluation. To confirm the role of the miR-877-3p/Smad7 axis in the regulation of osteoblastic differentiation, MC3T3-E1 cells transfected with miR-877-3p inhibitor or inhibitor-NC were cultured with or without TGF-β1 for 7 days. Compared with the control cells transfected with inhibitor-NC, the expression of Smad7 was upregulated when cells were transfected with miR-877-3p inhibitor alone, while TGF-β1 reversed the expression of Smad7 induced by the miR-877-3p inhibitor, as confirmed by RT-qPCR (Fig. 4A). By contrast, the osteogenesis-associated genes RUNX2, OSX and COL1A1 exhibited the opposite tendency (Fig. 4B). The protein levels of RUNX2 and Smad7 were also verified by western blotting, and were consistent with the mRNA results. Western blotting also revealed that the levels of phospho-Smad2 and phospho-Smad3 were decreased by the miR-877-3p inhibitor, and this effect was reversed by TGF-β1 (Fig. 4C). Furthermore, Smad7-overexpressing and control MC3T3-E1 cells were established by transfection with a pcDNA3.1-Smad7 plasmid (pcDNA-Smad7) and control plasmid (pcDNA-NC), and then co-transfected with miR-877-3p mimics or mimics-NC, and the success of transfection was verified using RT-qPCR (Fig. 4D and E). The promoting effects of miR-877-3p mimics on osteogenesis-related genes, phospho-Smad2 and phospho-Smad3 were markedly inhibited by pcDNA-Smad7 (Fig. 4F and G). ALP activity, ARS and ALP staining exhibited the same tendency (Fig. 4H and I). Collectively, these results confirmed that Smad7 inhibits the osteoblastic differentiation of MC3T3-E1 cells and is able to reverse the osteoblastic differentiation mediated by miR-877-3p.
Figure 4.
Smad7 is identified as a downstream target of miR-877-3p during the osteoblastic differentiation of MC3T3-E1 cells. mRNA expression of (A) Smad7 and (B) osteoblast differentiation-associated genes in MC3T3-E1 cells transfected with inhibitor-NC, miR-877-3p inhibitor or miR-877-3p inhibitor plus TGF-β1. (C) Expression of RUNX2, Smad7, p-Smad2, t-Smad2, p-Smad3 and t-Smad3 protein in MC3T3-E1 cells transfected with inhibitor-NC, miR-877-3p inhibitor or miR-877-3p inhibitor plus TGF-β1. (D) The mRNA expression of Smad7 was validated in MC3T3-E1 cells transfected with pcDNA-NC or pcDNA-Smad7. The mRNA expression of (E) Smad7 and (F) osteoblast differentiation-associated genes in MC3T3-E1 cells transfected with miR-877-3p mimics and/or pcDNA-Smad7. (G) Protein levels of Smad7, RUNX2, p-Smad2, t-Smad2, p-Smad3 and t-Smad3 in MC3T3-E1 cells transfected with miR-877-3p mimics and/or pcDNA-Smad7. (H) ALP activity of MC3T3-E1 cells transfected with miR-877-3p mimics and/or pcDNA-Smad7 after culture with osteogenic medium for 7 days. (I) MC3T3-E1 cells transfected with miR-877-3p mimics and/or pcDNA-Smad7 were induced with osteogenic medium. At day 14, the mineralization of differentiated MC3T3-E1 cells was detected with ARS and ALP staining. *P<0.05, **P<0.01 as indicated. miR, microRNA; NC, negative control; TGF, transforming growth factor; RUNX2, runt-related transcription factor 2; p, phospho; t, total; ALP, alkaline phosphatase; ARS, Alizarin Red S.
Discussion
Bone formation and mineralization are triggered by the differentiation of mesenchymal precursor cells into osteoblasts. Osteoblast differentiation is significantly affected by a key transcriptional factor, RUNX2, that regulates the expression of several osteogenic genes, including ALP, OSX and COL1A1 (17–19). These osteogenic genes are associated with the expression of bone matrix genes such as type 1 collagen, osteopontin, osteocalcin and bone sialoprotein (20–23). In addition, several signaling pathways, including Wnt/β-catenin, BMP/TGF-β, JAK/STAT and MAPK, have been reported to promote osteoblast differentiation in vivo and in vitro (23–25). TGF-β1 is a polypeptide member of the TGF-β superfamily of cytokines that regulate several cellular functions, including cell proliferation, survival and differentiation (26–28). The combined actions of these biological processes rely on the effect of TGF-β1 on bone formation (29).
MC3T3-E1 cells, which are mouse pre-osteoblast cells considered to be good models for the study of in vitro osteoblast differentiation (14), were used in the present study. These cells were treated with TGF-β1 to induce osteoblastic differentiation, which was evaluated by the expression of the aforementioned osteoblast gene markers. It must be acknowledged that a limitation of the present study is that only one cell line was investigated; different cell lines, such as primary osteoblasts, should be included in further studies. However, the experiments in the present study were, to the best of our knowledge, the first to provide evidence that miR-877-3p is upregulated during the osteoblastic differentiation of MC3T3-E1 cells in a time-dependent manner. Consistent with this observation, miR-877-3p has also been found to be highly upregulated during myofibroblast differentiation (12). Subsequently, the effect of miR-877-3p on TGF-β1-induced osteoblast differentiation and mineralization was investigated, by overexpression and knockdown experiments performed in the osteoblast cell line. The results clearly demonstrated that miR-877-3p promotes osteoblast differentiation.
miRNAs have been identified as post-transcriptional inhibitors that exert their biological effects via the downregulation of target gene expression. Bioinformatics analysis in the present study identified Smad7 as a putative target of miR-877-3p. By transient transfection with miR-877-3p mimics or inhibitor, it was demonstrated that miR-877-3p suppressed Smad7 mRNA and protein expression in MC3T3-E1 cells. Smad7 is a well-defined negative regulator of the TGF-β1 signaling pathway and bone metabolism (30). As regards to bone formation initiated by osteoblasts, decreasing expression of Smad7 during osteoblast differentiation of MC3T3-E1 cells was observed in the present study, which is consistent with previously reported findings (31). The present study also indicated that the expression of Smad7 increased in MC3T3-E1 cells with miR-877-3p downregulation, while this effect was eliminated in the presence of TGF-β1. Therefore, it was hypothesized that Smad7 is a negative target of miR-877-3p and TGF-β1, and that one of the important mechanisms by which miR-877-3p promotes the osteoblastic differentiation induced by TGF-β1 may be through suppressing the expression of the Smad7 gene. This hypothesis was confirmed in the co-transfection experiment. The results suggested that overexpression of Smad7 partially inhibited the miR-877-3p-mediated osteoblastic differentiation of MC3T3-E1 cells. However, evidence is required to demonstrate that microRNAs-877-3p regulate osteoblast differentiation by interacting with Smad7. In addition, the role of miRNAs in osteoblast differentiation is complex; the bypass or crosstalk of microRNAs-877-3p in this process was not excluded, and requires further study.
In summary, the results of the present study revealed the mechanism underlying the regulatory effect of miR-877-3p on the osteoblastic differentiation of MC3T3-E1 cells. Through this mechanism, miR-877-3p promoted osteoblastic differentiation of MC3T3-E1 cells induced by TGF-β1 via silencing Smad7. These findings provided novel insights into the effects of miR-877-3p on osteoblast differentiation and bone formation, which may provide a new therapeutic approach for degenerative bone diseases.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
DH conceived and designed the experiments, conducted the data analysis, contributed reagents/materials/tools and wrote the manuscript. GH and JC performed the experiments. All authors read and approved the final manuscript. All authors contributed to the data analysis, drafting and critical revision of the paper, and agree to be accountable for all aspects of the work.
Ethics approval and consent to participate
All animal procedures were approved by the Animal Care Committee of Southern Medical University (L2016113).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Takayanagi H. Osteoimmunology: Shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7:292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- 2.Manolagas SC, Jilka RL. Bone marrow, cytokines, and bone remodeling. Emerging insights into the pathophysiology of osteoporosis. N Engl J Med. 1995;332:305–311. doi: 10.1056/NEJM199502023320506. [DOI] [PubMed] [Google Scholar]
- 3.Bernabeu C, Lopez-Novoa JM, Quintanilla M. The emerging role of TGF-beta superfamily coreceptors in cancer. Biochim Biophys Acta. 2009;1792:954–973. doi: 10.1016/j.bbadis.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Crane JL, Xian L, Cao X. Role of TGF-β signaling in coupling bone remodeling. Methods Mol Biol. 2016;1344:287–300. doi: 10.1007/978-1-4939-2966-5_18. [DOI] [PubMed] [Google Scholar]
- 5.Yasui T, Kadono Y, Nakamura M, Oshima Y, Matsumoto T, Masuda H, Hirose J, Omata Y, Yasuda H, Imamura T, et al. Regulation of RANKL-induced osteoclastogenesis by TGF-β through molecular interaction between Smad3 and Traf6. J Bone Miner Res. 2011;26:1447–1456. doi: 10.1002/jbmr.357. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiang LY, Ou HH, Liu XC, Chen ZJ, Li XH, Huang Y, Yang DH. Loss of tumor suppressor miR-126 contributes to the development of hepatitis B virus-related hepatocellular carcinoma metastasis through the upregulation of ADAM9. Tumour Biol. 2017;39:1010428317709128. doi: 10.1177/1010428317709128. [DOI] [PubMed] [Google Scholar]
- 9.Cao M, Hou D, Liang H, Gong F, Wang Y, Yan X, Jiang X, Wang C, Zhang J, Zen K, et al. miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signalling inhibitor 1. Eur J Cancer. 2014;50:1013–1024. doi: 10.1016/j.ejca.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 10.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taipaleenmäki H, Bjerre Hokland L, Chen L, Kauppinen S, Kassem M. Mechanisms in endocrinology: micro-RNAs: Targets for enhancing osteoblast differentiation and bone formation. Eur J Endocrinol. 2012;166:359–371. doi: 10.1530/EJE-11-0646. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Gu S, Cao H, Li Z, Xiang Z, Hu K, Han X. miR-877-3p targets Smad7 and is associated with myofibroblast differentiation and bleomycin-induced lung fibrosis. Sci Rep. 2016;6:30122. doi: 10.1038/srep30122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Zhu Y, Liang Z, Wang X, Meng S, Xu X, Xu X, Wu J, Ji A, Hu Z, et al. Up-regulation of p16 by miR-877-3p inhibits proliferation of bladder cancer. Oncotarget. 2016;7:51773–51783. doi: 10.18632/oncotarget.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu YB, Kong XH, Li YH, Fan L, Pan YL, Li CR, Wu XL, Lu TL, Mei QB. Radix Dipsaci total saponins stimulate MC3T3-E1 cell differentiation via the bone morphogenetic protein-2/MAPK/Smad-dependent Runx2 pathway. Mol Med Rep. 2015;11:4468–4472. doi: 10.3892/mmr.2015.3249. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Wu F, Song Y, Li X, Wu Q, Duan Y, Jin Z. Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death Dis. 2016;7:e2327. doi: 10.1038/cddis.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Dacic S, Kalajzic I, Visnjic D, Lichtler AC, Rowe DW. Col1a1-driven transgenic markers of osteoblast lineage progression. J Bone Miner Res. 2001;16:1228–1236. doi: 10.1359/jbmr.2001.16.7.1228. [DOI] [PubMed] [Google Scholar]
- 18.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/S0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 19.Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi JY, Komori T, Stein JL, Lian JB, et al. Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- 20.Baek WY, Kim JE. Transcriptional regulation of bone formation. Front Biosci (Schol Ed) 2011;3:126–135. doi: 10.2741/s138. [DOI] [PubMed] [Google Scholar]
- 21.Bruderer M, Richards RG, Alini M, Stoddart MJ. Role and regulation of RUNX2 in osteogenesis. Eur Cell Mater. 2014;28:269–286. doi: 10.22203/eCM.v028a19. [DOI] [PubMed] [Google Scholar]
- 22.Ohata Y, Ozono K. Bone and stem cells. The mechanism of osteogenic differentiation from mesenchymal stem cell. Clin Calcium. 2014;24:501–508. (In Japanese) [PubMed] [Google Scholar]
- 23.Bodine PV, Komm BS. Wnt signaling and osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:33–39. doi: 10.1007/s11154-006-9002-4. [DOI] [PubMed] [Google Scholar]
- 24.Levy JB, Schindler C, Raz R, Levy DE, Baron R, Horowitz MC. Activation of the JAK-STAT signal transduction pathway by oncostatin-M cultured human and mouse osteoblastic cells. Endocrinology. 1996;137:1159–1165. doi: 10.1210/endo.137.4.8625884. [DOI] [PubMed] [Google Scholar]
- 25.Beederman M, Lamplot JD, Nan G, Wang J, Liu X, Yin L, Li R, Shui W, Zhang H, Kim SH, et al. BMP signaling in mesenchymal stem cell differentiation and bone formation. J Biomed Sci Eng. 2013;6:32–52. doi: 10.4236/jbise.2013.68A1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massagué J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 27.Park JS, Chu JS, Tsou AD, Diop R, Tang Z, Wang A, Li S. The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-β. Biomaterials. 2011;32:3921–3930. doi: 10.1016/j.biomaterials.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegel PM, Massagué J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 29.Janssens K, ten Dijke P, Janssens S, Van Hul W. Transforming growth factor-beta1 to the bone. Endocr Rev. 2005;26:743–774. doi: 10.1210/er.2004-0001. [DOI] [PubMed] [Google Scholar]
- 30.Zhang F, Ren CC, Liu L, Chen YN, Yang L, Zhang XA. HOXC6 gene silencing inhibits epithelial-mesenchymal transition and cell viability through the TGF-β/smad signaling pathway in cervical carcinoma cell. Cancer Cell Int. 2018;18:204. doi: 10.1186/s12935-018-0680-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yano M, Inoue Y, Tobimatsu T, Hendy G, Canaff L, Sugimoto T, Seino S, Kaji H. Smad7 inhibits differentiation and mineralization of mouse osteoblastic cells. Endocr J. 2012;59:653–662. doi: 10.1507/endocrj.EJ12-0022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are available from the corresponding author upon reasonable request.