Abstract
Developing molecular tools to visualize and control Rho GTPase signaling in living cells has been instrumental in elucidating the mechanisms of cytoskeletal reorganization and causal relationships between activation events in cell function. An indispensable part of such studies is the quantitative characterization of the spatiotemporal GTPase activity. Here we present a computational pipeline, EdgeProps, designed for comparative/correlative analysis of cell dynamics (edge velocity) and near-edge protein activity (intensity of a fluorescent signal). The tool offers a user-friendly interface with three functional modules for processing, visualization, and statistical characterization of single-cell imaging data.
Keywords: GTPase activity, Image Analysis, Morphodynamics, Protrusive Activity, Correlation
1. Introduction
Molecular imaging tools such as fluorescence resonance energy transfer (FRET)-based biosensors offer a unique opportunity for observing intracellular localization of the key signaling proteins specifically in their activated states [1–3]. These observations allow researchers to relate protein activity to signaling and/or biomechanical events in living cells [4,5]. Because, in general, signaling events are localized and transient, understanding cell dynamics and function requires characterization of causal relationships between its regulatory components in different compartments and in real time. In particular, quantitative analysis of Rho GTPase activity helps in elucidating the mechanisms of cytoskeletal reorganization that drives various cellular processes, including cell motility, cell polarity, axon guidance, vesicle trafficking, and cell cycle progression[6–8]. Here we describe a MATLAB-based image analysis pipeline, EdgeProps, for accurate measurement of the correlation between the velocity of the cell edge and the local activity of a protein of interest. This tool serves two purposes: (1) providing several alternative options for visualization of the fluorescent signal distribution in a moving cell, and (2) providing statistical measures of a correlation between the intensity of the signal and the protrusive activity of the cell. The pipeline includes a segmentation module MovThresh (to define the cell edge) [9], an optional cell-body detection module BisectoGraph (to filter out filopodia, retraction fibers, or any other fine processes at the cell edge) [9], and the main quantification module EdgeProps2 [10]. The quantification module processes imaging data (a stack of cell images from a time-lapse record) to extract 2D histograms for intensity vs. velocity, intensity vs. orientation, and velocity vs. orientation values, kymographs for intensity and velocity, and the kymograph correlation. EdgeProps2 also includes the capability to perform the correlation analysis at a user-defined distance from the edge, which might be critical for avoiding artifacts associated with a high signal noise at thin parts of the lamellipodia where the protein abundance is too low.
2. Materials
The EdgeProps is a collection of MATLAB scripts divided into three folders corresponding to the three modules of the pipeline. Each folder includes the main file of the program, supporting functions, and an example of data that can be used for getting acquainted with the functionality of the tool. Each module represents a graphical user interface (GUI) with an intuitive user-friendly design. The basic installation of a MATLAB (MathWorks®) version R2014b or later is required for running the GUIs. However, no knowledge of computer programming and no experience with MATLAB is needed for the data analysis with EdgeProps.
3. Methods
3.1. Cell segmentation
The purpose of the correlative analysis using the EdgeProps pipeline is to quantify the relationship between the protrusive activity and the activity of a protein of interest, such as a Rho family GTPase, at the cell edge. Thus, accurate detection of the cell edge is the absolutely critical first step in the pipeline. There are multiple algorithms and tools for image segmentation and any of them can be used here as long as the output of the processing is an image stack with all background pixels having the intensity value 0 and all pixels of the cell of interest having positive values (> 0). The EdgeProps pipeline includes the MovThresh module that might be particularly useful when the threshold values for segmentation vary significantly over the course of the data record.
Open the MovThresh module, i.e. run MovThesh.m file in MATLAB. Accept “Change Folder” if prompted. All the GUI controls will be disabled until a data file is imported.
Import a TIFF file with a single-channel grayscale multi-frame (stack) image of a cell by clicking File -> Import (.tif) in the menu at the top-left corner of the GUI window. An image with multi-channel or colored data requires preprocessing before the import (see Note 1). The threshold for each time frame will be determined automatically and displayed in two ways: (1) the cell edge defined by the threshold will be highlighted in yellow on the cell image in the left panel, and (2) the threshold values will be shown in the right panel in blue as a function of time. The vertical red line in the right panel indicates the current time frame. Use Options -> Colormap and Options -> Boundary Line in the menu, to change the colormap of the image (grayscale by default) and the color of the edge line (yellow by default) if needed .
Use the time slider under the cell image to visually assess the quality of the segmentation for all time points. Use the toolbar icons below the menu (“zoom in”, “zoom out”, and “pan”) to explore the location of the detected edge more closely. Click Options -> Algorithm in the menu, to switch between Histogram [11] and Otsu [12] segmentation methods and to test which one works better for the imported data. If the automatic segmentation is satisfactory, click File -> Save As Masks in the menu and close the GUI window. Otherwise, try one of the following options:
If the automatically found threshold values have a reasonable trend but jump too much up and down from frame to frame, switch to “Smoothed Curve” and click the “Re-threshold” button. The updated segmentation is based now on the values shown in the right panel as a smooth red curve. The degree of smoothness is regulated by the value of the “Smoothing Window”, which is the width of the Gaussian filter used for the smoothing. The whole smoothed curve can be moved up and down using the vertical slider on the right. The range of this movement is regulated by the max and min values above and below the slider, respectively. If these adjustments had improved the segmentation, save the results and close the GUI window. Otherwise, try the manual option:
Switch to “Custom Curve” and adjust the threshold value for any time frame, for which the automatically found threshold did not work well. Click the “Threshold range” button to see the edge line interactively as you change the value of the threshold using the vertical slider. Click the “Re-Threshold” button to see the result of the thresholding (based on the new values indicated by the black curve). Every time the threshold is adjusted for a given time point, a red pivot is added to the black curve. To remove the pivot from the current time frame, click “Remove pivot”. After manual adjustment of the threshold values, save the results and close the GUI window.
3.2. Filtering out filopodia
This step can be skipped if the edge of the cell is relatively smooth, i.e. no high-curvature processes, such as filopodia, are present. Otherwise, consider using the BisectoGraph module for automated cell body detection (i.e. filtering out thin protrusions). This step is not necessary and the rest of the processing can be performed for the cells with filopodia. However, if the edge analysis is intended for the study of the effects of GTPase activity on the large-scale protrusive dynamics and the overall cell morphology, filtering out small-scale processes at the cell edge might improve the accuracy of the analysis.
Open the BisectoGraph module, i.e. run BisectoGraph.m file in MATLAB. Accept “Change Folder” if prompted. All the GUI controls will be disabled until a data file is imported.
By clicking File ->Import Masks (.tif) in the menu at the top-left corner of the GUI, import a TIFF file with the cell masks generated by MovThresh or any other segmentation tool used in the previous step of the processing pipeline.
Click the “Graph All Frames” button to construct a tree graph (Voronoi diagram inside a polygon) for the cell outline at each time frame, as displayed in the left-side panel.
Click the “Profile” button and the “Show Cell Body” checkbox. The automatically detected cell body will be highlighted in green on the left panel.
Use the vertical slider in the bottom-right corner of the GUI to interactively adjust the parameter CrR for all frames at once or on the frame-to-frame basis if needed. Visually inspect the detected cell body at all time points using the horizontal slider under the cell image. If no further adjustments of the CrR are needed, save the cell body masks as a tif file using File -> Save Results (.tif) in the menu.
3.3. Edge Analysis
Open the EdgeProps2 module, i.e. run EdgeProps2.m file in MATLAB. Accept “Change Folder” if prompted. All the GUI controls will be disabled until a data file is imported.
Import a TIFF file with a multi-frame mask of a cell of interest (see Section 3.1) by clicking File -> Import in the menu at the top-left corner of the GUI window. Import the original TIFF file with a single-channel grayscale multi-frame (stack) image of a cell. An image with multi-channel or colored data requires preprocessing before importing (see Note 1). The size and the number of frames in the mask and the data files must be the same, otherwise, the corresponding error message will be displayed. If the original data file has all background pixels set to 0, this file will work as a mask too (simply import this same file twice). The cell of interest will appear in the top-left panel of the GUI. The file name will appear above the panel. The cell image will be overlaid with the outline of the cell at a later time frame depending on the time lag (the default value is 1). Specifically, the cell at time frame T will be overlaid with the cell at the time frame (T + TL), where TL is the time lag value shown in the top-right corner of the GUI window. The rationale for exploring different values of the time lag is provided in Note 2. Use the slider under the panel to display any time frame up to (TM − TL), where TM is the total number of the frames in the imported data file.
Choose a value for the averaging radius in the top-right corner of the GUI (the default value is 5). This value defines the radius of the disk with the center at a pixel on the cell edge. The intensity at the edge will be calculated as the average intensity of the cell pixels within the disk, the center of which runs point by point along the cell outline (see Note 3). If no averaging is required, set the averaging radius to the value 0.
Press the “Perform analysis” button in the “Edge Analysis” button group. The program will calculate the edge velocity, local intensity, and orientation with respect to the cell centroid for each point of the cell outline at each time frame (see Note 4). The resulting values will be displayed as the colormaps in the bottom panels on the left and the right side of GUI, respectively. These graphs should help in visual comparisons of the velocity and intensity at each time frame without moving the time slider back and forth. To take a closer look at a specific region of the cell, use the toolbar icons below the menu (“zoom in”, “zoom out”, and “pan”). Click the “=” button in the “Figure” group, to synchronize the views in the three plotting panels. To return to the original views, click the “Reset” button.
The cumulative statistics of the measures for all time frames is displayed as the colormap of the two-dimensional histogram at the top-right panel. The white curve shows the average values of the columns of this colormap, representing the correlation between all intensity and velocity values of the record. The maximal and minimal values of the measures are shown in the top-right corner of the GUI and can be adjusted within the original [min, max] range. If any of these values are changed, the 2D histogram will be re-calculated so that the number of the bins is always the same: 100×100. This is different from zooming in or out, which would not change the bin size. The program also calculates the angular orientation of the outline points with respect to the cell centroid. Therefore, in addition to intensity/velocity correlation, one can display intensity/orientation or velocity/orientation correlations. To do this, choose the measures of interest from the drop-down lists at the top-right corner of the GUI.
3.4. Interior Analysis
In some cases, the intensity of the signal at the very edge of the cell is too noisy (the number of fluorescing molecules is too low) because the thickness of the cell is too small, which might lead to quantification artifacts. One solution is to analyze the intensity at some distance from the edge (a pixel or two might be enough). However, the correlative analysis of such off-edge intensity with the velocity requires interpolation of the edge velocity to the regions inside the cell. EdgeProps2 provides this option with the “Interior Analysis” button group.
Clicking the “Current” button initiates the velocity interpolation only for the current time frame (see Note 5). Alternatively, all time frames can be processed in one run if the “All” button is clicked. The second option will take some time. Thus, it is advised first to explore the results for different values of the Hill coefficient and the offset in a single frame before applying these parameters to the analysis of the entire time record.
After the completion of the interpolation step, click the “Graph” button to display the correlation on the measures of interest (intensity, velocity, or orientation) as a new figure.
Click “Save” button to save the resulting correlation plot as a PNG image.
3.5. Kymographs
The last type of data visualization and analysis is a kymograph representation of the edge velocity and intensity. The kymographs show each of these measurements as a single spatiotemporal graph with each horizontal line being a colormap of the values along the original or the offset outline (black) at a given time frame. Thus, the vertical axis is time and the horizontal axis is the arc length along the outline. The program also calculates the correlation between the two kymographs (see Note 6).
Once the edge analysis is performed, click the “Perform Analysis in the “Kymographs” button group. The velocity and intensity kymographs will be displayed in the bottom panels. The correlation graph will be displayed in the top-right panel. The left-most (right-most) columns of the kymographs correspond to the location of the cell edge “cut” (zero of the arc length) highlighted by a black marker on the cell image in the top-left plotting panel.
If the scaled kymographs are preferred, check the box named “scale lines”. This scaling is performed for each line (i.e. each time frame) so that the maximal value becomes 1 and the minimal value becomes −1.
All graphs (scaled and not scaled) can be displayed as separate MATLAB figures for any additional adjustments of the graphics before saving. In no adjustments needed, the graphs can be also saved automatically as TIFF files by clicking the “Save” button.
4. Notes
If data is saved as a stack of images of RGB type (colored), please use the standard FIJI (ImageJ) program to resave the data as a grayscale image stack. To do this, open the file in FIJI, then go to Image -> Type -> 16-bit and then to File -> Save As -> Tiff…. If data is saved as a multi-channel stack of images, open the file in FIJI, then go to Image ->Color -> Split Channels,and then save the channel of interest as a separate Tiff file.
In order to get a good representation of cellular morphodynamics, the frame rate of the recording should be high enough, so that cell shape changes in small increments from frame to frame. Such recording allows for accurate association of the cell outlines in the consecutive time points for velocity calculation. However, if the frame rate is too high and the cell shape changes on a scale of 1 pixel from frame to frame, the velocity measurements might become very noisy. One solution is to resave the data only keeping every nth frame. This way the number of frames for the analysis will be significantly reduced. EdgeProps2 offers an alternative solution by keeping all time frames while calculating the velocity based of the frames with a time lag. For example, if the time lag is 5, the velocity for frame 1 will be calculated based on the outlines at frames 1 and 6, for frame 2 – between 2 and 7, frame 3 – between 3 and 8, and so on.
The cell outline is defined as a middle-height contour of the cell mask smoothed by a Gaussian filter. This approach generates a smooth outline passing the interface between the pixels of the cell and the pixels of the background. In other words, the outline represent the cell edge but doesn’t include sharp corners resulting from the pixelated nature of the image. In addition, the program distributes a large number of points on the outline equidistantly, so that this number is consistent between the outlines from all time frames and the distance between two neighboring points is <= 1 pixel size.
The velocity for a point of the cell outline at time T is calculated based on the minimal distance from this point to the outline at time (T + TL). If the point at a time T is located within the area confined by the outline at time (T + TL), the velocity has a positive value (protrusion), otherwise it is negative (retraction).
- Velocity interpolation is performed according to the formula
where di(x,y) is the distance from a point i of the cell outline to the point (x,y) inside the cell and vi is the value of the edge velocity at the point i of the outline. This form of interpolation is chosen so that the largest influence on the values of the velocity V(x,y) is provided by the nearest points of the outline. Directly on the outline V(xi,yi)=vi. The bigger the value of the Hill coefficient, n, the stronger the influence of the outline points on the near-edge areas of the cell, with a sharp change further from the edge as compared to a smoother (global) influence of the edge when n is small. - The kymograph correlation is calculated by shifting one kymograph, K2, with respect to the other K1, in the horizontal direction by mx pixels and the vertical direction by my pixels. The overlapping parts of the kymographs are used for calculating the correlation coefficient:
where and are the average values over all pixels of the overlapping parts of the kymographs. The shift includes both positive and negative values: mx∈[−max(y),+max(y)] and my ∈[−max(y),+max(y)]. Such correlation plot is useful for identifying statistically significant time delays in the onset of protrusion or retraction events upon the protein activation.
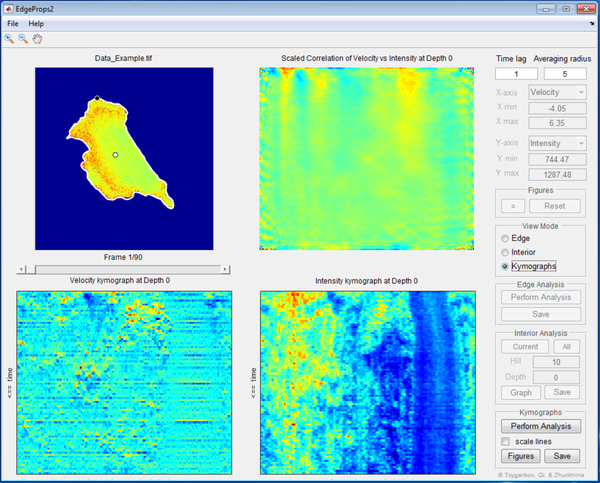
Figure 1.
A screenshot of the EdgeProps2 GUI layout in one of the quantification mode, showing velocity and protein activity kymographs and the correlation between them as a two-dimensional plot.
Acknowledgement
This work was supported by U.S. Army Research Office (ARO) Grant W911NF-17–1-0395 to DT and U.S. Army Research Office (ARO) Grant W911NF-15-1-0631 to TE and KH.
5. References
- 1.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G (2009) Coordination of Rho GTPase activities during cell protrusion. Nature 461 (7260):99–103. doi: 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson HJ, Campbell RE (2009) Genetically encoded FRET-based biosensors for multiparameter fluorescence imaging. Current opinion in biotechnology 20 (1):19–27. doi: 10.1016/j.copbio.2009.01.003 [DOI] [PubMed] [Google Scholar]
- 3.Hochreiter B, Garcia AP, Schmid JA (2015) Fluorescent proteins as genetically encoded FRET biosensors in life sciences. Sensors 15 (10):26281–26314. doi: 10.3390/s151026281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karginov AV, Tsygankov D, Berginski M, Chu PH, Trudeau ED, Yi JJ, Gomez S, Elston TC, Hahn KM (2014) Dissecting motility signaling through activation of specific Src-effector complexes. Nature chemical biology 10 (4):286–290. doi: 10.1038/nchembio.1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu PH, Tsygankov D, Berginski ME, Dagliyan O, Gomez SM, Elston TC, Karginov AV, Hahn KM (2014) Engineered kinase activation reveals unique morphodynamic phenotypes and associated trafficking for Src family isoforms. Proceedings of the National Academy of Sciences of the United States of America 111 (34):12420–12425. doi: 10.1073/pnas.1404487111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420 (6916):629–635. doi: 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- 7.Hodge RG, Ridley AJ (2016) Regulating Rho GTPases and their regulators. Nature reviews Molecular cell biology 17 (8):496–510. doi: 10.1038/nrm.2016.67 [DOI] [PubMed] [Google Scholar]
- 8.Wennerberg K, Der CJ (2004) Rho-family GTPases: it’s not only Rac and Rho (and I like it). Journal of cell science 117 (Pt 8):1301–1312. doi: 10.1242/jcs.01118 [DOI] [PubMed] [Google Scholar]
- 9.Tsygankov D, Bilancia CG, Vitriol EA, Hahn KM, Peifer M, Elston TC (2014) CellGeo: a computational platform for the analysis of shape changes in cells with complex geometries. The Journal of cell biology 204 (3):443–460. doi: 10.1083/jcb.201306067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacNevin CJ, Toutchkine A, Marston DJ, Hsu CW, Tsygankov D, Li L, Liu B, Qi T, Nguyen DV, Hahn KM (2016) Ratiometric Imaging Using a Single Dye Enables Simultaneous Visualization of Rac1 and Cdc42 Activation. Journal of the American Chemical Society 138 (8):2571–2575. doi: 10.1021/jacs.5b09764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prewitt JMS, Mendelsohn ML (1966) Analysis of Cell Images. Ann Ny Acad Sci 128 (A3):1035.-+ [DOI] [PubMed] [Google Scholar]
- 12.Otsu N (1979) Threshold Selection Method from Gray-Level Histograms. Ieee T Syst Man Cyb 9 (1):62–66 [Google Scholar]