Abstract
The Gli family is known to be required for the activation of hedgehog signalling, which participates in the pathogenesis of liver fibrosis. The aim of the present study was to identify the association between microRNA (miR)-152 and GLI family zinc finger 3 (Gli3) and their roles in liver fibrosis. In a carbon tetrachloride (CCl4)-treated rat model, fibrogenesis-associated indexes, including hydroxyproline content, collagen deposition, and α-smooth muscle actin (α-SMA) and albumin expression, were examined in in vivo and in vitro models. The expression of miR-152 and Gli3 in cells and tissues was determined by reverse transcription quantitative polymerase chain reaction and western blot analysis. The interaction of Gli3 and miR-152 was evaluated by bioinformatical analysis and a dual-luciferase reporter assay. The results demonstrated that miR-152 was significantly downregulated in serum samples from clinical patients, liver tissues from CCl4-treated rats and activated LX2 cells. Furthermore, at the cellular level, the mRNA and protein expression levels of α-SMA and albumin were increased and decreased, respectively, in LX2 cells. Nevertheless, following transfection with an miR-152 mimic, the expression levels of α-SMA and albumin were reversed, and Gli3 expression was notably decreased in LX2 cells. Additionally, the target interaction between miR-152 and Gli3 was demonstrated. Finally, an miR-152 mimic was introduced into the rat model and additionally demonstrated that the changes in α-SMA, albumin and Gli3 expression levels were similar to the expression pattern in LX2 cells following miR-152 mimic transfection. These data provided insight into the potential function of miR-152 as an anti-fibrotic therapy through the modulation of Gli3.
Keywords: liver fibrosis, microRNA-152, GLI family zinc finger 3
Introduction
Liver fibrosis is a common pathological consequence of continued damage to the liver tissue due to infection [primarily hepatitis B virus (HBV) and hepatitis C virus (HCV)], toxic/drug-induced injury, or metabolic or autoimmune factors, and the associated chronic activation of the wound healing reaction (1). With ongoing liver damage, fibrosis may progress to cirrhosis, which is characterized by a distortion of the liver vasculature and architecture and is the major determinant of morbidity and mortality in patients with liver disease, predisposing them to liver failure and primary liver cancer (2,3). At present, the limited available curative treatment options primarily include antiviral therapy for chronic HBV and HCV infection, weight loss and exercise for non-alcoholic steatohepatitis or liver transplantation (4). However, certain patients with liver fibrosis are either not sensitive to these causal drug treatments or are diagnosed at late end-stages, when satisfactory therapeutic strategies are not available, ultimately resulting in mortality (5). In addition, liver transplantation is considered to be the only therapy to significantly improve lifespan, but the inadequate availability of organs, increasing numbers of patients requiring transplants, and issues of compatibility and comorbidity factors mean that not all patients are eligible for transplantation (6). Therefore, the development of novel effective and safe therapeutic regimens for liver fibrosis are urgently required.
MicroRNAs (miRNAs/miR), a group of endogenous, small (18–23 nucleotides in length), non-coding RNAs, have been identified in a variety of eukaryotic organisms and post-transcriptionally regulate gene expression by interacting with the 3′-untranslated region (3′-UTR) of target gene mRNAs to repress translation or increase mRNA cleavage (7). A growing body of evidence has revealed that miRNAs may regulate a large number of biological processes, including cell proliferation, differentiation, and apoptosis (8) and that aberrant miRNA expression was closely associated with the development of multiple diseases, including liver fibrosis (9). For example, miR-130a-3p inhibited transforming growth factor-β (TGF-β)/Mothers against decapentaplegic (Smad) signalling by directly targeting TGF-β receptors 1 and 2, which may contribute to the pathogenesis of hepatic fibrosis and provide a potential novel drug target for the treatment of non-alcoholic steatohepatitis (10). In addition, restoration of miR-9 expression inhibited the activation of hepatic stellate cells (HSCs), the primary extracellular matrix (ECM)-producing cells in the fibrotic liver, by targeting multidrug resistance-associated protein 1; therefore, miR-9 may serve a suppressive role in liver fibrosis (11); members of the miR-34 family (miR-34a, miR-34b and miR-34c) were identified to be the most upregulated compared with other present miRs and may be involved in lipid/fatty acid metabolism by targeting acyl-CoA synthetase long-chain family member 1 in the progression of hepatic fibrosis. These studies indicated that dysregulated miRNAs exert an important role in the fibrotic process, and miRNA gene therapies have also been proposed as a promising therapeutic approach for the treatment of liver fibrosis (12).
Previously, miR-152 was suggested to be a regulator in certain fibrotic diseases (13,14). For example, miR-152 levels were significantly downregulated in a rat model of peritoneal fibrosis, indicating that miR-152 may be associated with the pathogenesis of this disease (15). Furthermore, it was also identified that miR-152 contributed to DNA methyltransferase 1 downregulation and epigenetically regulated Patched1, resulting in the inhibition of epithelial-mesenchymal transition (EMT) in liver fibrosis (13). Hedgehog (Hh) signalling is critically important in hepatic fibrogenesis, and GLI family zinc finger 3 (Gli3) may function as an Hh signalling-independent transcriptional activator (16,17). Nevertheless, the interaction and underlying mechanisms between miR-152 and Gli3 in the progression of liver fibrosis remain unclear. Therefore, the present study examined the expression of miR-152 in clinical samples, and in in vivo animal and in vitro cell models, verified the interaction between miR-152 and Gli3 and additionally explored the role of miR-152 in the process of liver fibrosis.
Materials and methods
Study population and serum sample preparation
Clinical samples were collected from two independent cohorts recruited from the First People's Hospital of Kunming City (Kunming, China) between January 2015 and June 2016. Cohort 1 comprised 25 patients with liver fibrosis, whereas cohort 2 comprised 25 healthy people. All patients were diagnosed on the basis of history, clinical and pathological examination, by at least two experienced clinicians. Following collection of the liver samples via resection, tissues were partially embedded with paraffin and preserved in liquid nitrogen. Diagnoses of the samples were confirmed by pathological examination. The presence of liver fibrosis in a sample was the first inclusion criterion. In addition, patients with liver cancer, autoimmune hepatitis, drug-induced injury or alcohol abuse were excluded. Informed written consent was obtained from all participants prior to enrolment in the study, and the study was approved by the Ethical Committee of the First People's Hospital of Kunming City.
Fasting venous blood samples were collected by trained laboratory technicians. Peripheral blood samples (5 ml) were incubated at 4°C for 12 h, and then the sera in the upper layers were aspirated and centrifuged at 400 × g for 10 min at 4°C. Sera were aliquoted and stored at −80°C until examination.
Animal grouping and model preparation
Male Sprague-Dawley (SD) rats (n=30; 150–200 g) were purchased from Shanghai Laboratory Animal Centre (Shanghai, China) and housed with 5 animals per cage under specific pathogen-free conditions. All animal experiments were approved by the Animal Care and Use Committee of the First People's Hospital of Kunming City, in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (18). Animals were housed in a temperature-controlled environment (20–22°C) at 75±2% relatively humidity with a 12 h light/dark cycle and free access to food and purified water. Rats were acclimated for 1 week prior to the experimentation. Then, a liver fibrosis model was generated, and rats were randomly separated into the following six treatment groups (n=5 animals per group): i) Model-0 week; ii) Model-2 week; iii) Model-4 week; iv) Model-8 week; v) Model + Negative control (NC; injected with NC plasmid); vi) Model + miR-152 (injected with miR-152 mimic).
All groups, excluding the Model-0 week group, received a subcutaneous injection of 10 ml/kg carbon tetrachloride (CCl4) dissolved in olive oil (25%, v/v). The Model-2 week, Model-4 week and Model-8 week groups received CCl4 twice a week for 2, 4 and 8 weeks, respectively as described previously (18,19). However, the Model + negative control (NC) and Model + miR-152 groups were injected intraperitoneally with an NC plasmid and miR-152 mimic, respectively, during the period of CCl4 treatment twice a week for 8 weeks.
At the end of the treatments, all animals were anaesthetized with ketamine hydrochloride (50 mg/kg) and sodium pentobarbital (30 mg/kg, iv). At the end of the study period, animals were sacrificed via an overdose of pentobarbital. Blood samples were immediately collected into tubes and then centrifuged at 400 × g for 10 min at 4°C for serum preparation. Specimens were removed from the liver and washed immediately with ice-cold PBS to remove blood. Then, one-half of each specimen was fixed in a 10% formalin solution at 4°C overnight for histopathological analysis, and one-half was stored at −80°C for reverse transcription quantitative polymerase chain reaction (RT-qPCR) and western blot (WB) analysis.
Cell culture and treatments
Human normal hepatocytes, including AML12 and L02 cell lines, and the HSC LX2 cell line were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in Petri dishes (Corning, Inc., Corning, NY, China) containing Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin, 0.25 g/l glutamine and 10% foetal bovine serum (FBS; Hyclone GE Healthcare Life Sciences, Logan, UT, USA) at 37°C in the presence of 95% air and 5% CO2. Culture medium was changed every 2 or 3 days.
THP-1 cells purchased from the ATCC were cultivated in RPMI-1640 medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% FBS, 1% penicillin/streptomycin and maintained in a humidified incubator at 37°C with 5% CO2. THP-1 cells were differentiated into macrophages over 48 h in RPMI-1640 medium containing 5–25 ng/ml phorbol 12-myristate 13-acetate, as described previously (20). Then, LX2 cells with the phenotype of activated HSCs were pre-cocultured with the treated THP-1 cells and challenged with LPS (1 µg/ml) as described by Prestigiacomo et al (21). After 0, 6, 12, 24 and 48 h, the cells were collected for subsequent analysis.
293T cells obtained from the ATCC were grown in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS, L-glutamine and 1% penicillin/streptomycin at 37°C in a humidified incubator with 5% CO2 for the dual luciferase assay.
Determination of hepatic hydroxyproline content
Hydroxyproline is an amino acid that stabilizes collagen deposited in the liver and is exclusively associated with collagenous connective tissue; therefore, it is a good surrogate for the quantification of collagen deposition (22). Briefly, liver samples were weighed and hydrolysed in 2.5 ml 6N HCl at 110°C for 18 h in Teflon-coated tubes. The hydrolysate was centrifuged at 300 × g at room temperature for 10 min, and then the pH of the resulting supernatant was adjusted to pH 7.4. Finally, the optical density was measured at an absorbance wavelength of 558 nm by a microplate reader (Tecan GENios 2032218; Tecan Group Ltd., Männedorf, Switzerland). Total hydroxyproline content was calculated against a standard curve prepared with a trans4hydroxyLproline (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) preparation and expressed per mg of wet tissue weight.
Liver histology and morphometric collagen determination
The liver tissues were fixed in 4% paraformaldehyde at 4°C overnight and embedded in paraffin, sectioned at 4 µm and stained with haematoxylin-eosin (H&E) at room temperature for 10 min and Masson's trichrome at room temperature for 30 min according to the manufacture's protocol (C0105; Beyotime Institiute of Biotechnology, Haimen, China). The extent of fibrosis was evaluated on slides by a member of the Department of Pathology from the First People's Hospital of Kunming City blinded to the experimental conditions. Fibrosis was determined histologically by observing the intensity of fibrosis in 5 digital images by a light microscopy (magnification, ×200) captured from slides from each tissue sample stained with Masson's trichrome for visual assessment.
Immunohistochemistry
To detect the immunohistochemical localization of α-smooth muscle actin (α-SMA), sections from 4% formalin-fixed at room temperature for 10 min, paraffin-embedded specimens were deparaffinized and rehydrated in decreasing concentrations (90, 50, 10 and 0%) of ethyl alcohol at room temperature. All tissue sections were treated with fresh 3% hydrogen peroxide (H2O2) for 20 min at room temperature to eliminate endogenous peroxidase activity and then washed with PBS. The sections were then sequentially incubated with 2% bovine serum albumin (BSA) for 30 min, then rabbit anti-rat α-SMA monoclonal antibody (1:300 dilution; ab5694; Abcam, Cambridge, MA, USA) for 2 h, and then with the appropriate horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:200 dilution; Wuhan Boster Biological Technology, Ltd., Wuhan, China) for 40 min, followed by incubation with 3,3′-diaminobenzidine (DAB) as a substrate for 10 min. All the incubation steps were performed at room temperature, and three washes with PBS were applied between each step. In the negative control experiments, the primary antibodies were replaced with PBS. Finally, the sections were counterstained with haematoxylin at room temperature for 6 min, covered with a glycerine gel and observed under light microscopy (magnification, ×200) by a pathologist blinded to the experimental conditions. The readout was recorded visually and was not quantitatively analyzed.
RNA extraction and RT-qPCR
TRIzol® reagent (Thermo Fisher Scientific, Inc.) was utilized to extract total RNA from clinical samples, rat tissues and cell lines. RNA concentration and purity were determined using an Agilent 2100 Bioanalyzer and RNA 6000 Nano/Pico LabChip (Agilent Technologies GmbH, Waldbronn, Germany). The expression levels of miR-152, α-SMA, albumin and Gli3 were quantified by RT-qPCR. For miRNA analysis, a total of 100 ng RNA was reverse transcribed using an miScript RT-II kit (Qiagen GmbH, Hilden Germany) in a reaction volume of 20 µl and subjected to 60 min incubation at room temperature, followed by 5 min incubation at 95°C. miR-152 expression was assessed with an miScript miRNA PCR Array (Qiagen GbmH) in accordance with the manufacturer's protocol in 96-well plates using an ABI PRISM® 7500 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Initial denaturation at 30 min at 95°C, followed by 40 cycles of denaturation for 15 sec at 94°C, annealing for 30 sec at 55°C, elongation for 30 sec at 70°C and final extension at 70°C for 5 min.
For mRNA analysis, 2 µg total RNA was reverse transcribed to cDNA using an Moloney-Murine Leukaemia Virus reverse transcriptase kit (Promega Corporation, Madison, WI, USA), and then PCR was performed using a specific primer set to examine the expression levels of mRNAs with a SYBR Green qPCR SuperMix kit (Invitrogen; Thermo Fisher Scientific, Inc.) on an ABI PRISM® 7500 Sequence Detection System. The PCR thermocycler conditions were 95°C for 5 min, then 40 cycles of denaturation at 94°C for 2 min and annealing and extension at 62°C for 30 sec, followed by extension at 72°C for 30 sec. The gene-specific primer pairs used for RT-qPCR in the present study, listed in Table I, were designed according to the published GenBank database (https://www.uniprot.org/database/DB-0028) using Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA, USA). The 2−ΔΔCq method was performed using U6 and β-actin as the internal reference for miRNA and mRNA, respectively, and the following formula: ΔΔCq=ΔCq experimental group-ΔCq control group, where ΔCq=Cq detected gene-Cq internal reference (22). All experiments were performed in triplicate.
Table I.
Sequences of primers used for the RT-quantitative polymerase chain reaction assays.
| miRNA/genes | Primer sequences |
|---|---|
| miR-152 | RT: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCAAGTTC-3′ |
| Forward: 5′-ACACTCCAGCTGGGTCAGTGCATGACAGAACT-3′ | |
| Reverse: 5′-CTCAACTGGTGTCGTGGA-3′ | |
| U6 | RT: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAAATATGG-3′ |
| Forward: 5′-CTCGCTTCGGCAGCACA-3′ | |
| Reverse: 5′-AACGCTTCACGAATTTGCGT-3′ | |
| α-SMA (Human) | Forward: 5′-GTTCCAGCCATCCTTCATCGG-3′ |
| Reverse: 5′-CCTTCTGCATTCGGTCGGCAA-3′ | |
| Albumin (Human) | Forward: 5′-CAGAATGCGCTATTAGTTCG-3′ |
| Reverse: 5′-CTGGCGTTTTCTCATGCAA-3′ | |
| Gli3 (Human) | Forward: 5′-TTTTCCCCTTTAATCTTGCCAT-3′ |
| Reverse: 5′-CCAGTGGCAAATCAACCTCC-3′ | |
| β-actin (Human) | Forward: 5′-CTCCATCCTGGCCTCGCTGT-3′ |
| Reverse: 5′-GCTGTCACCTTCACCGTTCC-3′ | |
| α-SMA (Rat) | Forward: 5′-GCGTGACTCACAACGTGCCTA-3′ |
| Reverse: 5′-CCCATCAGGCAGTTCGTAGCTCT-3 | |
| Albumin (Rat) | Forward: 5′-GATCTGCCCTCAATAGCTG-3′ |
| Reverse: 5′-TGGCTTCATATTTCTTAGCAA-3′ | |
| Gli3 (Rat) | Forward: 5′-CTCGACCATTTCCACGGCAAC-3′ |
| Reverse: 5′-TCAGCACAGTGAAGTCTACACC-3′ | |
| β-actin (Rat) | Forward: 5′-TCAGGTCATCACTATCGGCAAT-3′ |
| Reverse: 5′-AAAGAAAGGGTGTAAAACGCA-3′ |
RT, reverse transcription; miR, microRNA; α-SMA, α-smooth muscle actin; Gli3, GLI family zinc finger 3.
WB analysis
A total of 3 independent samples from each group were harvested, and proteins were extracted using radioimmunoprecipitation assay lysis buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.1% SDS, 1% NP-40) supplemented with protease inhibitor cocktail on ice for 30 min. Cell lysates were centrifuged at 1,200 × g for 10 min at 4°C. The supernatants were retained, and protein concentrations were determined by a BCA protein assay (Promega Corporation). Protein extracts (30 µg) were resuspended and subjected to 10–12% SDS-PAGE and then blotted onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA) at 200 mA for 1.5 h by wet electrophoretic transfer. Following blocking with 5% non-fat milk in TBS with 0.1% Tween-20 (TBST) for 1 h at room temperature, membranes were immunoblotted with primary antibodies against GAPDH (1:1,000; mAbM20028; Abmart, Berkeley Heights, NJ, USA), α-SMA (1:1,000; ab5694; Abcam), albumin (1:1,500; ab106582; Abcam) and Gli3 (1:2,000; ab69838; Abcam) overnight at 4°C, and then additionally incubated with secondary horseradish peroxidase-conjugated antibodies (1:12,000; M21002; Abmart) at room temperature for 2 h. Finally, protein bands were detected by developing the blots with an enhanced chemiluminescence WB detection kit (Beyotime Institute of Biotechnology, Haimen, China). The band intensity was analysed by Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville, MD, USA).
Luciferase reporter assay
The sequences of the 3′-UTR of wild-type (WT) and mutant Gli3 mRNA containing the putative miR-152 binding sites were synthesized by Sangon Biotech Co., Ltd., (Shanghai, China) and cloned downstream of the luciferase gene in a psiCHECK-2 reporter vector (Promega Corporation) to generate the vectors psiCHECK-2-Gli3-3′-UTR-WT and psiCHECK-2-Gli3-3′-UTR-Mutant. The aforementioned 293T cells were seeded into 96-well plates (1,000 cells/well) 24 h prior to transfection and then co-transfected with 50 ng WT or mutant luciferase vector containing Gli3 3′-UTR and 20 µM miR-152 mimics (5′-AGGUUCUGUGAUACACUCCGACU-3′), miR-152 inhibitor (5-AGUCGGAGUGUAUCACAGAACCU-3′) or their respective controls (5′UUCUCCGAACGUGUCACGUTT-3′; Guangzhou RiboBio Co., Ltd., Guangzhou, China). Cell transfection was using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The cells were harvested 48 h after co-transfection and then the luciferase activity was measured with a Dual-Luciferase reporter assay system (Promega Corporation) following the protocol of the manufacturer. The fluorescence intensity was normalized to Renilla intensity.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). Data are presented as the mean ± standard deviation from triplicate experiments. A student's t-test was used for the comparison of two groups, and one-way analysis of variance followed by a Bonferroni post-hoc test was used for the comparison of >two groups. P<0.05 was considered to indicate a statistically significant difference. Mean values were obtained using SPSS v. 19.0 software (IBM Corp., Armonk, NY, USA).
Results
Expression patterns of miR-152 in patients with liver fibrosis and cell lines
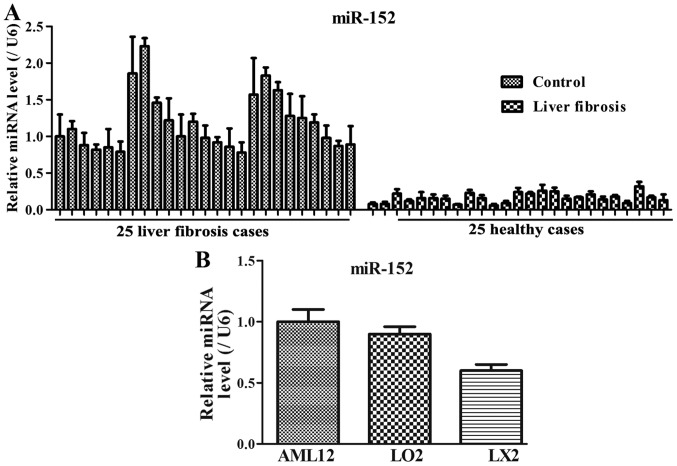
To investigate the miR-152 expression levels in serum samples of patients with liver fibrosis and cellular samples, RT-qPCR was performed in vivo and in vitro. Among the 25 patients with liver fibrosis, the miR-152 expression level was decreased compared with that in the healthy controls (Fig. 1A). In addition, compared with that in the normal immortalized human liver AML12 and L02 cells, miR-152 expression was markedly decreased in LX2 cells (Fig. 1B). These results suggested that decreased miR-152 expression may be closely associated with the progression of liver fibrosis.
Figure 1.
miR-152 expression is downregulated in serum samples of (A) patients with liver fibrosis and (B) cellular samples. Data are presented as the mean ± standard deviation from triplicate experiments. U6 was used as an internal loading control. miR, microRNA.
Evaluation of the animal model
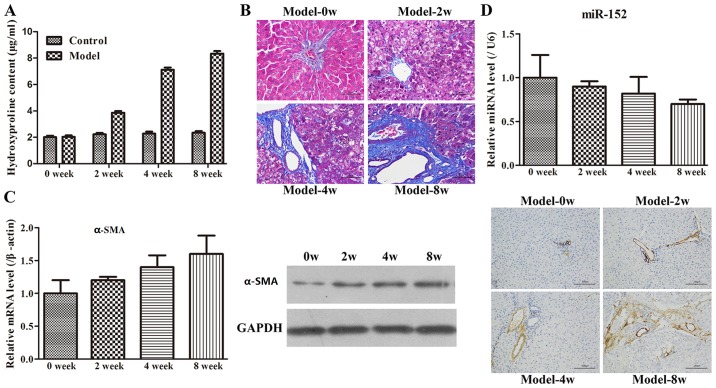
Administration of CCl4 is frequently used to construct liver fibrosis models (18), and hydroxyproline content is commonly measured to assess liver fibrosis (23). The hydroxyproline contents of the Model-0 week, Model-2 week, Model-4 week and Model-8 week groups were 2.04±0.49, 3.86±0.52, 7.12±0.63 and 8.34±0.72 µg/mg, respectively (Fig. 2A). The hydroxyproline level in the CCl4-treated groups was increased compared with that in the control group. In addition, the increase in hydroxyproline following CCl4 treatment was observed to occur in a time-dependent manner. Subsequently, H&E or Masson's trichrome staining was performed to detect collagen fibres. The results indicated that the rats in the Model-0 week group exhibited normal, clear and complete liver tissue structures with large and round nuclei and abundant cytoplasm, and with limited collagen deposition at the venous walls and bile duct walls in the portal area (Fig. 2B). However, the rats in the other three groups demonstrated increased levels of hyperplasia of fibrous connective tissue, fatty degeneration, steatosis, cell necrosis, infiltration of inflammatory cells and a larger number of collagen fibres, which were primarily deposited in the portal area and interlobular septa in comparison with the Model-0 group. In addition, longer modelling time intervals exhibited more marked changes compared with the shorter modelling time intervals. Finally, to examine the rat model of liver fibrosis in more detail, the expression of α-SMA at the mRNA and protein levels was examined by RT-qPCR, WB and immunohistochemistry methods. As demonstrated in Fig. 2C, the α-SMA expression was significantly increased with increases in the modelling time intervals. Furthermore, the immunohistochemistry result also revealed that limited α-SMA-positive tissues were detected at the vascular walls of the liver tissues in the Model-0 week group, whereas the expression of α-SMA was not only identified in the vascular walls but also widely spread throughout the portal area, fibrous septum and the adjacent hepatic sinusoids in the other three groups. Therefore, these results indicated that the rat model of liver fibrosis was successfully established.
Figure 2.
Rat model of liver fibrosis is constructed by administrating carbon tetrachloride. (A) Liver hydroxyproline content was determined to estimate collagen content and expressed as µg hydroxyproline per mg liver tissue (wet weight). The values are expressed as the mean ± SD. (B) Changes in the liver tissues of rats at different time points following modelling were visualized using haematoxylin-eosin and Masson staining (magnification, ×200). (C) RT-qPCR examination, western blot analysis and immunohistochemistry staining of liver tissues were performed with α-SMA specific-primers and antibodies. Data in the bar graphs are presented as the mean ± SD. (D) Hepatic expression level of miR-152 was measured by RT-qPCR. The relative levels of miR-152 were normalized against U6 in the same samples. SD, standard deviation; RT-qPCR, reverse transcription quantitative polymerase chain reaction; miR, microRNA; α-SMA, α-smooth muscle actin.
miR-152 changes in the rat model of fibrosis
Based on the miR-152 results in the clinical samples, the expression level of miR-152 in the rat model of fibrosis was examined using RT-qPCR. It was identified that miR-152 expression gradually decreased with increasing time intervals (Fig. 2D). This result implied that the dynamic change in miR-152 expression may be involved in the development of liver fibrosis.
miR-152 and fibrosis-associated gene expression in stimulated LX2 cells
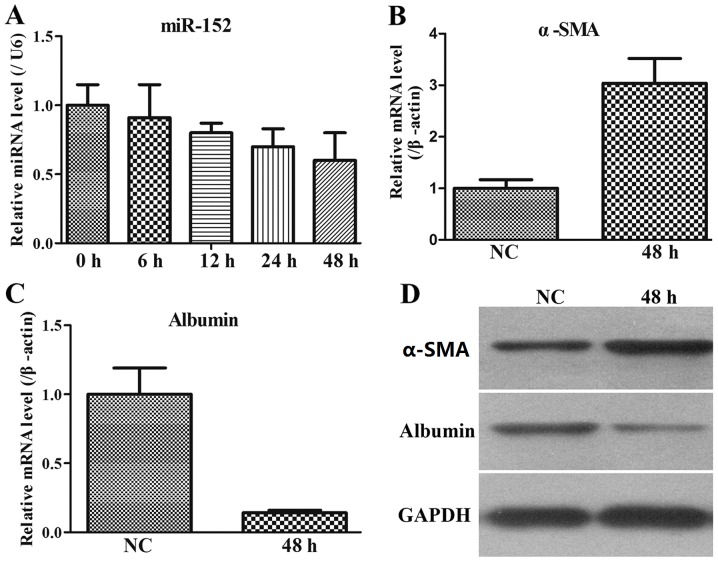
The LX-2 human HSC line has been widely characterized and maintains key features of hepatic stellate cytokine signalling, retinoid metabolism and fibrogenesis, making it a suitable model of human hepatic fibrosis. Therefore, the miR-152 expression was additionally assessed by RT-qPCR in stimulated LX2 cells. The results indicated that in the co-culture system of LX2 and THP-1 cells, miR-152 expression was gradually decreased with increasing time intervals (Fig. 3A).
Figure 3.
LX2 cells are activated in a co-cultured system with THP-1 cells. (A) miR-152 expression in the LX2 cells during culture activation was detected by RT-qPCR. Data are presented as the mean ± standard deviation. (B) α-SMA mRNA expression in LX2 cells was analysed with RT-qPCR. Each value represents the mean ± SD of 3 experiments. (C) Albumin mRNA expression in LX2 cells was examined using RT-qPCR. Each value represents the mean ± SD of 3 experiments. (D) Western blot analysis of α-SMA and albumin in stimulated LX2 cells. The housekeeping protein GAPDH was used as loading controls for the western blot analysis and protein normalization. miR, microRNA; SD, standard deviation; RT-qPCR, reverse transcription quantitative polymerase chain reaction; α-SMA, α-smooth muscle actin; NC, negative control.
As α-SMA is the most well-established marker for activated LX2 cells (24), the levels of α-SMA in stimulated LX2 cells at 48 h were monitored. It was demonstrated that α-SMA expression at the mRNA and protein levels was upregulated in stimulated LX2 cells compared with that in the NC group (Fig. 3B and C). In addition, albumin level is also a biochemical marker during liver fibrosis, and therefore, its expression pattern was measured. The results indicated that albumin expression at the mRNA and protein levels was downregulated in stimulated LX2 cells compared with that in the NC group (Fig. 3C and D). Therefore, these data from the co-culture system of activated LX2 cells and THP-1 cells indicated that miR-152 served an important role in the process of liver fibrosis.
Effects of miR-152 on activated LX2 cells
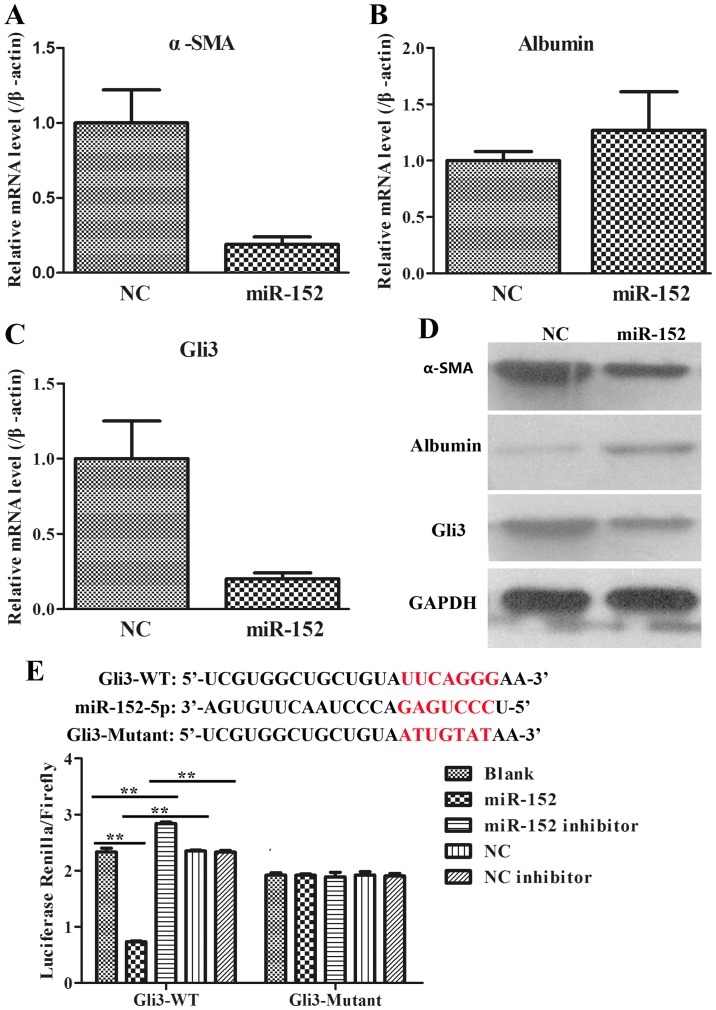
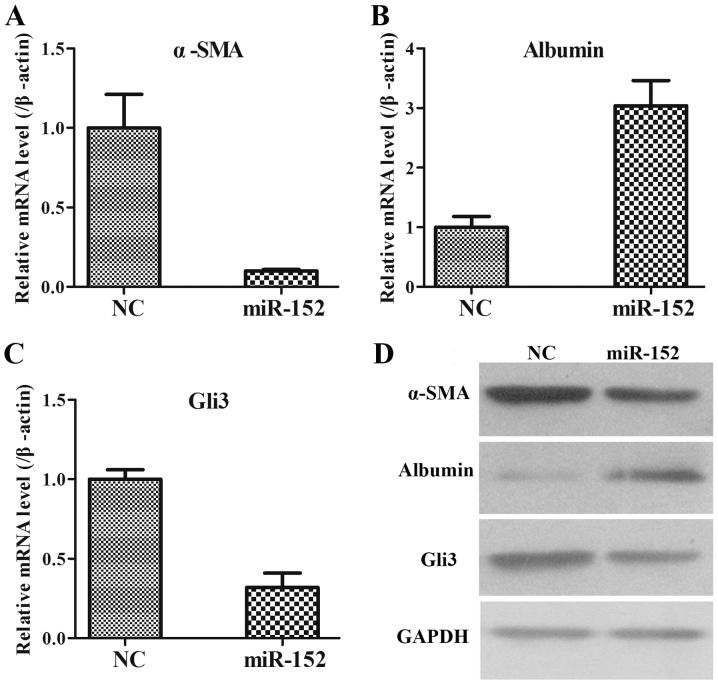
To additionally explore the role of miR-152 in the formation of liver fibrosis, fibrosis-associated genes, including α-SMA, albumin and Gli3, were analysed using RT-qPCR and WB in LX2 cells transfected with an NC plasmid and miR-152 mimic. It was observed that when compared with those in NC group, the mRNA expression of α-SMA was significantly decreased (Fig. 4A); the mRNA expression of Albumin was increased (Fig. 4B); and the mRNA expression of Gli3 was significantly reduced in miR-152 mimic transfected cells (Fig. 4C). The protein expression levels of these genes were consistent with the mRNA expression levels (Fig. 4D). Therefore, these data demonstrated that miR-152 may inhibit α-SMA and Gli3 expression and promote albumin expression.
Figure 4.
Interaction between miR-152 and fibrosis-associated genes. (A) The downregulation of α-SMA mRNA expression in LX2 cells transfected with an miR-152 mimic was determined by RT-qPCR. (B) The upregulation of albumin mRNA expression in LX2 cells transfected with an miR-152 mimic was examined by RT-qPCR. (C) The downregulation of Gli3 mRNA expression in LX2 cells transfected with an miR-152 mimic was measured by RT-qPCR. (D) α-SMA, albumin and Gli3 protein expression in LX2 cells transfected with miR-152 mimics were analysed via western blotting with GAPDH as an internal control. (E) Relative luciferase activities of luciferase reporters bearing WT or mutant Gli3 were analysed 48 h following transfection with the indicated miR-152 mimics/inhibitor or NC mimic/inhibitor. **P<0.01. miR, microRNA; α-SMA, α-smooth muscle actin; NC, negative control; Gli3, GLI family zinc finger 3; WT, wild type.
miR-152 is predicted to target the 3′-UTR of Gli3
Due to the opposite expression patterns of miR-152 and Gli3, bioinformatic analysis was used to predict the potential target interaction between miR-152 and Gli3 (Targetscan Human 7.2, http://www.targetscan.org/). It was confirmed by luciferase assays that miR-152 decreased the relative activity of luciferase by directly binding to the 3′-UTR of Gli3 in 293T cells (Fig. 4E). A combined plasmid with mutations in the predicted binding site was generated and co-transfected with different groups of miR-152 in luciferase assays, and no significant differences in luciferase activity levels among the different groups were observed. These data suggested that miR-152 may directly target Gli3.
Role of miR-152 in the rat model of liver fibrosis
Subsequent to demonstrating the effects of miR-152 on activated LX2 cells, the role of miR-152 in the rat liver fibrosis model was eventually confirmed. As indicated in Fig. 5, the changing expression patterns of α-SMA, albumin and Gli3 at the mRNA and protein levels were notably coincident with LX2 cells. Taken together, these data suggested that miR-152 may suppress α-SMA and Gli3 expression and facilitate albumin expression in vivo and in vitro.
Figure 5.
Role of miR-152 in the rat model of liver fibrosis. (A) The mRNA levels of α-SMA were measured by RT-qPCR. Each value is the mean ± SD of 3 experiments. (B) The mRNA levels of albumin were assessed by RT-qPCR. Each value is the mean ± SD of 3 experiments. (C) The mRNA levels of Gli3 were assessed by RT-qPCR. Each value is the mean ± SD of 3 experiments. (D) The protein levels of α-SMA, albumin and Gli3 were examined by western blot analysis and compared with GAPDH. Data are presented as the mean ± SD. miR, microRNA; α-SMA, α-smooth muscle actin; NC, negative control; RT-qPCR, reverse transcription quantitative polymerase chain reaction; Gli3, GLI family zinc finger 3; SD, standard deviation.
Discussion
Liver fibrosis is a scarring response to liver damage (1). It is a common pathological process for a number of liver disorders (25). A small number of patients will progress to cirrhosis and/or hepatocellular carcinoma (26). Presently, a considerable number of studies have focused on the roles of miRNAs in the pathophysiology of liver fibrosis in view of their regulatory effects on fibrogenesis-associated genes (27). For example, the downregulation of miR-145 may contribute to liver fibrosis in biliary atresia by targeting gamma-adducin (28); and miR-101 suppressed liver fibrosis by targeting the TGF-β signalling pathway (29). Zheng et al (14) demonstrated that the long non-coding RNA Pvt1 oncogene epigenetically downregulated protein patched homolog 1 (PTC1) expression via competitively binding miR-152, contributing to the EMT process in liver fibrosis. Yu et al (13) revealed that salvianolic acid B-induced miRNA-152 inhibited liver fibrosis by attenuating DNA (cytosine-5)-methyltransferase 1-mediated PTC1 methylation. In the present study, it was identified that miR-152 was significantly decreased in serum samples from clinical patients, liver tissues from CCl4-treated rats and activated LX2 cells, suggesting that downregulated miR-152 may serve an important role in liver fibrosis.
Subsequently, fibrogenesis-associated indexes, including hydroxyproline content, collagen deposition, and α-SMA and albumin expression levels, were examined in in vivo and in vitro models. The hydroxyproline content was increased in the livers of CCl4-treated rats compared with in the livers of the control rats, and H&E and Masson staining revealed deposition of excessive collagen fibres in CCl4-treated livers over time. In addition, the expression level of α-SMA in liver tissues was markedly increased. Therefore, these results indicated that CCl4-treated rats exhibited the apparent features of liver fibrosis. Additionally, it was identified that α-SMA and albumin expression levels were notably upregulated and downregulated, respectively, in LX2 cells co-cultured with treated THP-1 cells. During fibrosis, HSCs were hypothesized to serve a crucial role, as they are responsible for the proliferation and migration of HSCs and excessive deposition of ECM to effectively amplify the fibrotic response (26). Furthermore, activation of HSCs is regulated by multiple signalling pathways, including the TGF-β/Smad, phosphatidylinositol 3-kinase/RAC-alpha serine/threonine-protein kinase and the Hh signalling pathways (30). Among these pathways, Hh pathway components are usually expressed at relatively low levels in normal liver tissues; however, they progressively increase during the process of liver injury, and Gli3 is a vital transcription factor in the Hh pathway (31). The present study primarily focused on whether miR-152 was involved in the regulation of Gli3, which may trigger the Hh pathway, in human liver fibrosis samples, stimulated LX2 cells, a common type of HSCs, and a rat model for verification. The results demonstrated that overexpression of miR-152 inhibited α-SMA and Gli3 expression and facilitated albumin expression. In addition, it was identified that Gli3 was a directly target gene of miR-152, as indicated by bioinformatical analysis and a dual-luciferase reporter assay. Finally, in concordance with the α-SMA, albumin and Gli3 expression patterns exhibited in activated LX2 cells following administration of an miR-152 mimic to CCl4-treated rats, it was additionally identified that these genes presented a similar trend of expression in the liver tissues, implying that miR-152 may suppress activation of LX2 cells and liver fibrosis by regulating Gli3.
In conclusion, the present study demonstrated that miR-152 was markedly decreased during the progression of liver fibrosis in vivo and in vitro. It was also confirmed that the interaction target of miR-152 is Gli3. In addition, the overexpression of miR-152 in a CCl4-induced liver fibrosis rat model and activated LX2 cells decreased pro-fibrotic gene expression and increased anti-fibrotic gene expression. Taken together, the identification of miR-152 and its target gene provides useful insights into the mechanisms underlying liver fibrosis.
Acknowledgements
Not applicable.
Funding
The present study was supported by a Research Fund from the Yunnan Provincial Department of Education (grant. no., 2013C239) and a Postdoctoral Supporting Fund from Kunming Human Resources and Social Security Bureau.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LL designed the current study and drafted the manuscript. LZ and XZ performed the animal experiments. JC and JL collected clinical and cellular data. GC analyzed the data.
Ethics approval and consent to participate
Informed written consent was obtained from all participants prior to enrolment in the study, and the study was approved by the Ethical Committee of the First People's Hospital of Kunming City. All animal experiments were approved by the Animal Care and Use Committee of the First People's Hospital of Kunming City, in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (18).
Patient consent for publication
Informed written consent was obtained from all participants prior to enrolment in the study.
Competing interests
The authors declare that they have no competing interest.
References
- 1.Lee UE, Friedman SL. Mechanisms of hepatic fibrogenesis. Best Pract Res Clin Gastroenterol. 2011;25:195–206. doi: 10.1016/j.bpg.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elpek GÖ. Cellular and molecular mechanisms in the pathogenesis of liver fibrosis: An update. World J Gastroenterol. 2014;20:7260–7276. doi: 10.3748/wjg.v20.i23.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seki E, Brenner DA. Recent advancement of molecular mechanisms of liver fibrosis. J Hepatobiliary Pancreat Sci. 2015;22:512–518. doi: 10.1002/jhbp.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trautwein C, Friedman SL, Schuppan D, Pinzani M. Hepatic fibrosis: Concept to treatment. J Hepatolo 62 (1 Suppl) 2015:S15–S24. doi: 10.1016/j.jhep.2015.02.039. [DOI] [PubMed] [Google Scholar]
- 5.Friedman SL. Mechanisms of disease: Mechanisms of hepatic fibrosis and therapeutic implications. Nat Clin Pract Gastroenterol Hepatol. 2004;1:98–105. doi: 10.1038/ncpgasthep0055. [DOI] [PubMed] [Google Scholar]
- 6.Coletta M, Nicolini D, Benedetti Cacciaguerra A, Mazzocato S, Rossi R, Vivarelli M. Bridging patients with hepatocellular cancer waiting for liver transplant: All the patients are the same? Transl Gastroenterol Hepatol. 2017;2:78. doi: 10.21037/tgh.2017.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Treiber T, Treiber N, Plessmann U, Harlander S, Daiß JL, Eichner N, Lehmann G, Schall K, Urlaub H, Meister G. A Compendium of RNA-binding proteins that regulate MicroRNA biogenesis. Mol Cell. 2017;66:270–284.e13. doi: 10.1016/j.molcel.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Fortin K, Mourelatos Z. MicroRNAs: Biogenesis and molecular functions. Brain Pathol. 2008;18:113–121. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami Y, Kawada N. MicroRNAs in hepatic pathophysiology. Hepatol Res. 2017;47:60–69. doi: 10.1111/hepr.12730. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Du J, Niu X, Fu N, Wang R, Zhang Y, Zhao S, Sun D, Nan Y. MiR-130a-3p attenuates activation and induces apoptosis of hepatic stellate cells in nonalcoholic fibrosing steatohepatitis by directly targeting TGFBR1 and TGFBR2. Cell Death Dis. 2017;8:e2792. doi: 10.1038/cddis.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun J, Zhang H, Li L, Yu L, Fu L. MicroRNA-9 limits hepatic fibrosis by suppressing the activation and proliferation of hepatic stellate cells by directly targeting MRP1/ABCC1. Oncol Rep. 2017;37:1698–1706. doi: 10.3892/or.2017.5382. [DOI] [PubMed] [Google Scholar]
- 12.Li WQ, Chen C, Xu MD, Guo J, Li YM, Xia QM, Liu HM, He J, Yu HY, Zhu L. The rno-miR-34 family is upregulated and targets ACSL1 in dimethylnitrosamine-induced hepatic fibrosis in rats. FEBS J. 2011;278:1522–1532. doi: 10.1111/j.1742-4658.2011.08075.x. [DOI] [PubMed] [Google Scholar]
- 13.Yu F, Lu Z, Chen B, Wu X, Dong P, Zheng J. Salvianolic acid B-induced microRNA-152 inhibits liver fibrosis by attenuating DNMT1-mediated Patched1 methylation. J Cell Mol Med. 2015;19:2617–2632. doi: 10.1111/jcmm.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng J, Yu F, Dong P, Wu L, Zhang Y, Hu Y, Zheng L. Long non-coding RNA PVT1 activates hepatic stellate cells through competitively binding microRNA-152. Oncotarget. 2016;7:62886–62897. doi: 10.18632/oncotarget.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin F, Wu X, Zhang H, You X, Zhang Z, Shao R, Huang C. A microrna screen to identify regulators of peritoneal fibrosis in a rat model of peritoneal dialysis. BMC Nephrol. 2015;16:48. doi: 10.1186/s12882-015-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renault MA, Roncalli J, Tongers J, Misener S, Thorne T, Jujo K, Ito A, Clarke T, Fung C, Millay M, et al. The Hedgehog transcription factor Gli3 modulates angiogenesis. Circ Res. 2009;105:818–826. doi: 10.1161/CIRCRESAHA.109.206706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Syn WK, Jung Y, Omenetti A, Abdelmalek M, Guy CD, Yang L, Wang J, Witek RP, Fearing CM, Pereira TA, et al. Hedgehog-mediated epithelial-to-mesenchymal transition and fibrogenic repair in nonalcoholic fatty liver disease. Gastroenterology. 2009;137:1478–1488.e8. doi: 10.1053/j.gastro.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X, Wang X, Lv Y, Xu L, Lin J, Diao Y. Protection effect of kallistatin on carbon tetrachloride-induced liver fibrosis in rats via antioxidative stress. PLoS One. 2014;9:e88498. doi: 10.1371/journal.pone.0088498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahreen S, Khan MR, Khan RA. Evaluation of Rumex hastatus leaves against hepatic fibrosis: A rat model. BMC Complement Altern Med. 2017;17:435. doi: 10.1186/s12906-017-1943-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park EK, Jung HS, Yang HI, Yoo MC, Kim C, Kim KS. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm Res. 2007;56:45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 21.Prestigiacomo V, Weston A, Messner S, Lampart F, Suter-Dick L. Pro-fibrotic compounds induce stellate cell activation, ECM-remodelling and Nrf2 activation in a human 3D-multicellular model of liver fibrosis. PLoS One. 2017;12:e0179995. doi: 10.1371/journal.pone.0179995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Knight V, Lourensz D, Tchongue J, Correia J, Tipping P, Sievert W. Cytoplasmic domain of tissue factor promotes liver fibrosis in mice. World J Gastroenterol. 2017;23:5692–5699. doi: 10.3748/wjg.v23.i31.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buniatian G, Hamprecht B, Gebhardt R. Glial fibrillary acidic protein as a marker of perisinusoidal stellate cells that can distinguish between the normal and myofibroblast-like phenotypes. Biol Cell. 1996;87:65–73. doi: 10.1111/j.1768-322X.1996.tb00967.x. [DOI] [PubMed] [Google Scholar]
- 25.Ismail MH, Pinzani M. Reversal of hepatic fibrosis: Pathophysiological basis of antifibrotic therapies. Hepat Med. 2011;3:69–80. doi: 10.2147/HMER.S9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang CY, Yuan WG, He P, Lei JH, Wang CX. Liver fibrosis and hepatic stellate cells: Etiology, pathological hallmarks and therapeutic targets. World J Gastroenterol. 2016;22:10512–10522. doi: 10.3748/wjg.v22.i48.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang XP, Ai WB, Wan LY, Zhang YQ, Wu JF. The roles of microRNA families in hepatic fibrosis. Cell Biosci. 2017;7:34. doi: 10.1186/s13578-017-0161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye Y, Li Z, Feng Q, Chen Z, Wu Z, Wang J, Ye X, Zhang D, Liu L, Gao W, et al. Downregulation of microRNA-145 may contribute to liver fibrosis in biliary atresia by targeting ADD3. PLoS One. 2017;12:e0180896. doi: 10.1371/journal.pone.0180896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu X, Zhang H, Zhang J, Zhao S, Zheng X, Zhang Z, Zhu J, Chen J, Dong L, Zang Y, Zhang J. MicroRNA-101 suppresses liver fibrosis by targeting the TGFβ signalling pathway. J Pathol. 2014;234:46–59. doi: 10.1002/path.4373. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Q, Qin CY, Zhao ZH, Fan YC, Wang K. Epigenetic modifications in hepatic stellate cells contribute to liver fibrosis. Tohoku J Exp Med. 2013;229:35–43. doi: 10.1620/tjem.229.35. [DOI] [PubMed] [Google Scholar]
- 31.Yang JJ, Tao H, Li J. Hedgehog signaling pathway as key player in liver fibrosis: New insights and perspectives. Expert Opin Ther Targets. 2014;18:1011–1021. doi: 10.1517/14728222.2014.927443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.