Abstract
Glioma is one of the most common types of malignant cancer and the significance of microRNAs (miRNAs) in cancer therapy has been demonstrated. In the current study, miR-490-3p expression was significantly downregulated in glioma tissue and cell lines. Overexpression of miR-490-3p inhibited glioma cell proliferation and migration in vitro. In addition, the high-mobility group AT-hook 2 (HMGA2) was identified as a candidate target gene of miR-490-3p. The current study demonstrated that miR-490-3p mimic could inhibit HMGA2 protein expression in glioma cells. In addition, correlation analysis demonstrated that miR-490-3p and HMGA2 expression was inversely correlated in glioma tissues. Furthermore, the inhibitory effect of miR-490-3p mimic on cell proliferation and migration was partially reversed by the overexpression of HMGA2. Taken together, these results suggest that miR-490-3p may have a tumor suppressive role in glioma and therefore miR-490-3p may be a new target for the treatment of glioma.
Keywords: microRNA-490-3p, high-mobility group AT-hook 2, glioma, tumor suppressor
Introduction
Approximately 250,000 novel central nervous system (CNS) tumors are reported to occur each year in Europe (1). Glioma is one of the most common types of malignant cancer and accounts for the majority of CNS tumor-related mortality (2). Due to the limited treatment strategies available, the overall 5-year survival rate for patients with glioma remains poor (3). Therefore, considerable efforts are required in order to investigate the underlying mechanisms of glioma progression in order to facilitate the development of novel therapeutic approaches in malignant glioma.
microRNAs (miRNAs) are endogenous RNAs that can bind to the 3′-untranslated region (3′-UTR) of target genes (4). Emerging evidence suggests that miRNAs may serve vital roles in regulating cellular behavior (5,6). Human cancer progression is often associated with abnormal cellular behavior, which includes cell proliferation, apoptosis, migration and invasion (7). Therefore, it may be important to investigate the roles of miRNAs in several types of human cancer, including glioma (8,9). miR-490-3p, a newly identified miRNA, has been previously shown to function as a tumor suppressor in several types of cancer (10–13). Liu et al (10) revealed that miR-490-3p regulates colorectal cancer cell proliferation, metastasis and apoptosis by targeting the voltage dependent anion channel 1. Tian et al (11) observed that miR-490-3p regulates the sensitivity of ovarian cancer cells to cisplatin by directly targeting ABCC2. Previous studies also demonstrated that miR-490-3p may be a potential therapeutic target for osteosarcoma (12,13). However, the clinical significance of miR-490-3p in glioma remains unknown.
High-mobility group AT-hook 2 (HMGA2) belongs to the high-motility group (HMG) protein family and is reported to function as an oncogene in several types of human cancer (14–16). A previous study demonstrated that HMGA2 overexpression was correlated with poor prognosis in patients with lung cancer (15). In addition, HMGA2 overexpression was revealed to be closely associated with aggressive cell behaviors of glioma (16). Furthermore, HMGA2 is inversely regulated by miRNAs in several types of human cancer including squamous cell lung carcinoma and clear cell renal carcinoma (17,18). However, whether miRNAs regulate HMGA2 expression in glioma remains unknown.
In the current study, miR-490-3p expression was significantly downregulated in glioma tissues and cell lines. Subsequently, the current study demonstrated that miR-490-3p may regulate glioma cell proliferation and migration in vitro. HMGA2 was identified as a target gene of miR-490-3p in glioma. Furthermore, the inhibitory effect of miR-490-3p on glioma cell proliferation and migration may be partially regulated by HMGA2 expression. Therefore miR-490-3p may be a potential novel target for the treatment of glioma.
Materials and methods
Clinical samples
Glioma tissue and adjacent noncancerous tissue samples (>3 cm from cancer tissues) were collected from 24 patients (12 male and 12 female; average age, 59.1±6.3 years; age range, 42–79 years) with glioma at The First Affiliated Hospital of Guangzhou University of Traditional Chinese Medicine between April 2014 and May 2016. Tumor stage was classified based on the World Health Organization (WHO) stage and grading system (19). The Karnofsky performance scale (KPS) ranking runs from 100 to 0, where 100 is ‘perfect health’ and 0 is death. Its purpose is to evaluate a patient's ability to survive chemotherapy treatment (20). All patients involved in the current study did no receive anti-cancer treatment prior to the surgery. All tissue samples were stored at −80°C for further analyses. The study protocol was approved by the Ethics Committee of The First Affiliated Hospital of Guangzhou University of Traditional Chinese Medicine (Guangzhou, China). Written informed consent was obtained from all patients prior to the study.
Cell culture
Human glioma cell lines A172, T98 and U87-MG (cat. no. TCHu138, glioblastoma of unknown origin) were purchased from Cell Bank of the Chinese Academy of Science (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Thermo Fisher Scientific, Inc.). Normal human astrocytes (NHAs) were purchased from Lonza Group, Ltd. and cultured in AGM™ Astrocyte Growth Medium (Lonza Group, Ltd.) supplemented with 10% FBS (Thermo Fisher Scientific, Inc.). Cells were maintained at 37°C in a humidified atmosphere containing 5% of CO2.
Cell transfection
miR-490-3p mimic (5′-CAACCUGGAGGACUCCAUGCUG-3′), inhibitor (5′-CAGCAUGGAGUCCUCCAGGUUG-3′) and negative control miRNA (NC-miRNA; 5′-ACCGCUAAUCAUACGAAUACAC-3′) were purchased from Guangzhou RiboBio Co., Ltd. HMGA2 overexpression vector based on pcDNA3.1 and NC plasmid (pcDNA3.1) was purchased from GenScript Co., Ltd. (Nanjing, China). Cells (U87-MG and T98) were seeded into 6-well plates at the density of 5×105 cells/well. Synthetic miRNAs (100 pmol) or HMGA2 plasmid (2 µg) were mixed with Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol and used to transfect cells for 48 h.
Reverse transcription-quantitative (RT-q) PCR
Total RNA was extracted from tissue samples and cells (U87-MG, A172 and T98) using TRIzol® reagent (Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into first-strand cDNA using the PrimeScript™ RT Reagent kit (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. qPCR was subsequently performed using an ABI7500 CPR machine (Applied Biosystems; Thermo Fisher Scientific, Inc.) and SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd.). The following primer pairs were used for the qPCR: miR-490-3p, forward, 5′-TGCGGTTCAAGTAATTCAGGA-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′; and U6 small nuclear (sn)RNA, forward, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′. The following thermocycling conditions were used: 10 min at 95°C; 40 cycles of 1 min at 95°C; 2 min at 63°C; and 1 min at 72°C. Relative miR-490-3p levels were quantified using the 2−ΔΔCq method and normalized to U6 snRNA, the internal control (21).
Western blotting
Total protein was extracted from tissue samples and cells (U87-MG and T98) using radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology) and quantified using bicinchoninic acid kit (Beyotime Institute of Biotechnology). Total protein (50 µg per lane) was separated by SDS-PAGE on a 12% gel. The separated proteins were subsequently transferred onto polyvinylidene difluoride membranes (Thermo Fisher Scientific, Inc.) and blocked with 5% fat-free milk at 4°C for 3 h. The membranes were incubated with primary antibodies against HMGA2 (1:1,000; ab109329), matrix metallopeptidase-9 (MMP-9; 1:1,000; ab73734) and GAPDH (ab181602; 1:1,000; all Abcam) at 4°C for overnight. Membranes were washed three times with Tris-buffered saline and Polysorbate 20 (TBST). Following primary incubation, membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000; ab97051; Abcam) at room temperature for 4 h. Protein bands were visualized using an BeyoEnhanced Chemiluminescence kit (Beyotime Institute of Biotechnology) and protein expression was quantified using ImageJ 1.48 software (National Institutes of Health).
Cell proliferation assay
Cell proliferation rate was analyzed using the cell counting kit-8 (CCK-8; Beyotime Institute of Biotechnology). Cells (U87-MG and T98) were seeded into 96-well plates at the density of 5×103 cells/well and incubated for 0, 24, 48 and 72 h prior at 37°C to the addition of 10 ml CCK-8 reagent. Cells were incubated at 37°C for a further 2 h. Optical density was measured at a wavelength of 450 nm using microplate reader (Bio-Rad Laboratories, Inc.). Each sample was performed in triplicate.
Cell invasion assay
Cell invasion was examined using a Transwell invasion assay. A total of 1×105 cells (U87-MG and T98) were plated in the upper chamber (8 mm; Corning Inc.) that pre-coated with Matrigel at 37°C for 30 min (BD Biosciences, San Jose, CA, USA) in DMEM. DMEM supplemented with 10% FBS (Thermo Fisher Scientific, Inc.) was plated in the lower chambers. Following incubation for 24 h, migratory cells in the lower membrane were fixed with methanol and stained with 0.5% crystal violet solution at 37°C for 4 h. Stained cells were counted from five independent fields using an inverted fluorescent microscope (magnification, ×200; Olympus IX53; Olympus Corporation).
Dual-luciferase assay
Bioinformatic analysis was performed using TargetScan (www.targetscan.org/vert_72) to predict potential targets of miR-490-3p. The wild-type (wt) 3′-UTR of HMGA2 was cloned using the following primers: HMGA2-wt forward, 5′-CCGTCTAGACGGGGGGCGCCAACGTTCGATTTCT-3′ and reverse, 5′-CTGTTTTGACCAAACTTTATTAATAGTTTAAGATCTATTCTTAT-3′. The cloned 3′-UTR fragment of HMGA1 was subsequently inserted into the XbaI/XbaI sites of pGL3 vector (Promega Corporation). A site-directed mutagenesis kit (Takara Biotechnology Co., Ltd.) was used to construct the mutant (mut) 3′-UTR of HMGA2 using the following primers: HMGA2-mut forward, 5′-AAAAAAGGGGGGGGCAATCTCTCGGTCCAATTTCTCTCTCTCTCTTCCTC-3′ and reverse, 5′-GAGGAAGAGAGAGAGAGAAATTGGACCGAGAGATTGCCCCCCCCTTTTTT-3′. For luciferase activity assays, cells (U87-MG and T98) were co-transfected with wt or mut 3′-UTR HMGA2 and miR-490-3p mimic or NC-miRNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Luciferase activity was measured 48 h post-transfection using a Dual-Luciferase® (DLR™) Reporter Assay System (Promega Corporation) with Renilla luciferase activity as the internal control.
Statistical analysis
Data were analyzed with SPSS 19.0 (IBM Corp.) and presented as the mean ± standard deviation. Differences between groups were analyzed using Wilcoxon signed-rank test or one-way analysis of variance followed by Tukey's post hoc test. Association between miR-490-3p expression and clinicopathological parameters was analyzed by χ2 test. Correlation between miR-490-3p and HMGA2 was conducted using Pearson's correlation co-efficient. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-490-3p expression is downregulated in glioma tissues and cell lines
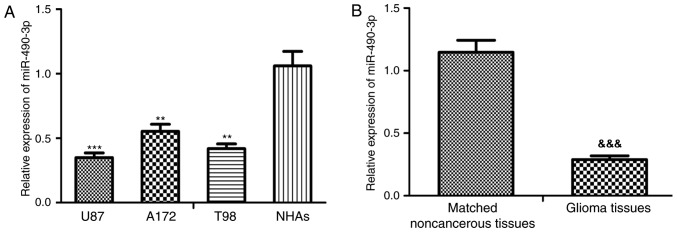
The relative miR-490-3p expression level was significantly downregulated in all glioma cell lines, U87-MG, A172 and T98, compared with the NHAs (P<0.01 and P<0.001; Fig. 1A). In addition, the U87-MG and T98 cells lines demonstrated the greatest decrease and therefore the U87-MG and T98 cells lines were selected for subsequent experimentation. Furthermore, the relative miR-490-3p expression level was significantly downregulated in glioma tissue compared with adjacent noncancerous tissue samples (P<0.001; Fig. 1B). Patients with glioma were classified into a high (n=9) or low (n=15) miR-490-3p expression group based on the relative miR-490-3p levels (cut-off value: 0.27). The current study demonstrated that low miR-490-3p expression was closely associated with WHO grade (P=0.031) and KPS (P=0.014) in patients with glioma, however, there was no significant association observed between miR-490-3p expression with age or sex (Table I).
Figure 1.
miR-490-3p expression is significantly downregulated in glioma. (A) The relative expression level of miR-490-3p was determined by RT-qPCR in glioma cell lines, U87-MG, A172, and T98 as well as in normal human astrocytes. (B) The relative expression level of miR-490-3p was determined by RT-qPCR in glioma tissue and adjacent noncancerous tissue samples. **P<0.01; ***P<0.001 vs. NHAs; &&&P<0.001 vs. noncancerous tissues. miR-490-3p, microRNA-490-3p; RT-qPCR, quantitative real-time PCR; NHAs, normal human astrocytes.
Table I.
Association between miR-490-3p expression and clinicopathological features of patients with glioma.
| miR-490-3p expression | ||||
|---|---|---|---|---|
| Clinicopathological features | No. of cases | Low (n=15) | High (n=9) | P-value |
| Age, years | ||||
| >60 | 12 | 9 | 3 | 0.083 |
| <60 | 12 | 6 | 6 | |
| Sex | ||||
| Male | 12 | 8 | 4 | 0.558 |
| Female | 12 | 7 | 5 | |
| KPS | ||||
| >90 | 7 | 5 | 2 | 0.014 |
| <90 | 17 | 10 | 7 | |
| WHO grade | ||||
| I–II | 8 | 4 | 4 | 0.031 |
| III | 16 | 11 | 5 | |
miR-490-3p, microRNA-490-3p; KPS, Karnofsky performance scale; WHO, world health organization.
miR-490-3p directly regulates of HMGA2 expression
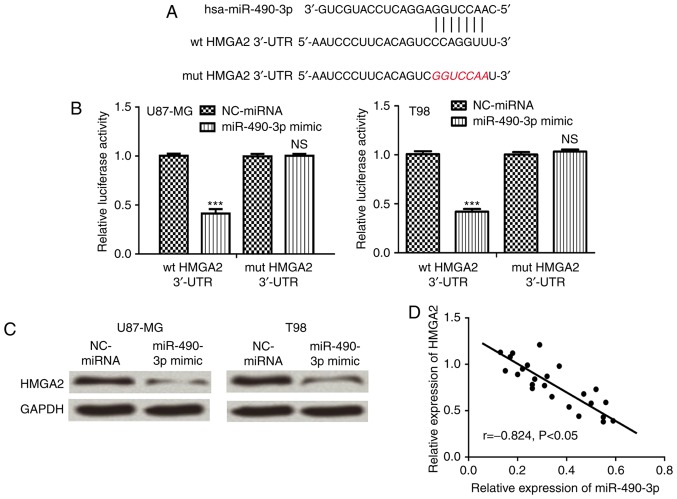
Bioinformatics analysis was performed to predict target genes of miR-490-3p. HMGA2 was identified as a putative target gene of miR-490-3p and bioinformatics analysis was used to predict the miR-490-3p target sequence in the 3′-UTR of HMGA2 (Fig. 2A). Luciferase reporter assays demonstrated that miR-490-3p overexpression significantly reduced wt HMGA2 luciferase activity compared with mut HMGA2 3′-UTR (P<0.001), which did not affect luciferase activity (Fig. 2B). Furthermore, HMGA2 protein expression was analyzed in cells following transfection with miR-490-3p mimic or NC-miRNA. The relative HMGA2 protein expression level was markedly reduced in cells following transfection with miR-490-3p mimic compared with NC-miRNA (Fig. 2C). Furthermore, correlation analysis demonstrated that there was a negative correlation between miR-490-3p and HMGA2 expression in glioma tissues (Fig. 2D).
Figure 2.
HMGA2 is a direct target gene of miR-490-3p in glioma. (A) Bioinformatics analysis was used to predict the miR-490-3p target sequence in the 3′-UTR of HMGA2. (B) Luciferase reporter assays were performed in cells following co-transfection with wt or mut 3′-UTR HMGA2 and miR-490-3p mimic or NC-miRNA. (C) The relative HMGA2 protein expression was determined by western blot analysis in cells following transfection with miR-490-3p mimic or NC-miRNA. (D) Correlation analysis of miR-490-3p and HMGA2 in glioma. ***P<0.001 vs. NC-miRNA. ns, not significant; miR-490-3p, microRNA-490-3p; wt, wild-type; mut, mutant; NC, negative control; UTR, untranslated region; HMGA2, high-mobility group AT-hook 2.
miR-490-3p suppresses glioma cell proliferation and invasion
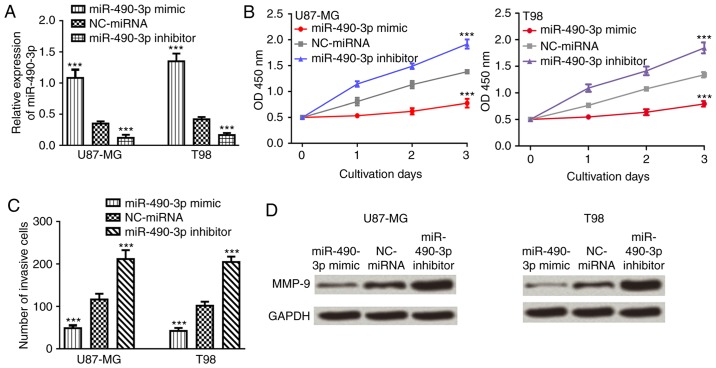
To investigate the role of miR-490-3p in glioma, CCK-8 and Transwell invasion assays were used to examine the effect of miR-490-3p on glioma cell proliferation and invasion following transfection with miR-490-3p mimic, NC-miRNA or miR-490-3p inhibitor. The relative miR-490-3p expression level was significantly increased in glioma cell lines following transfection with miR-490-3p mimic compared with NC-miRNA (P<0.001; Fig. 3A). By contrast, the relative miR-490-3p expression level was significantly decreased in glioma cell lines following transfection with miR-490-3p inhibitor (P<0.001; Fig. 3A). Cell proliferation and invasion were significantly decreased in glioma cells following transfection with miR-490-3p mimic compared with NC-miRNA (P<0.001; Fig. 3B and C). Furthermore, the expression level of MMP-9, an invasion marker (22), was examined in glioma cell lines following transfection with miR-490-3p mimic, NC-miRNA or miR-490-3p inhibitor. The relative protein expression level of MMP-9 was markedly decreased following transfection with miR-490-3p mimic, while relative protein expression level of MMP-9 was markedly increased following transfection with miR-490-3p inhibitor compared with NC-miRNA (Fig. 3D).
Figure 3.
miR-490-3p overexpression suppresses in vitro cell proliferation and invasion. (A) The relative expression level of miR-490-3p expression, (B) cell proliferation and (C) cell invasion was determined in cells following transfection with miR-490-3p mimic, miR-490-3p inhibitor or NC-miRNA. (D) The relative MMP-9 protein expression was determined by western blot analysis in cells following transfection with miR-490-3p mimic, miR-490-3p inhibitor or NC-miRNA. ***P<0.001 vs. NC-miRNA. miR-490-3p, microRNA-490-3p; miRNA, microRNA; NC, negative control; MMP-9, matrix metallopeptidase-9.
Overexpression of HMGA2 partially reverses the inhibitory effect of miR-490-3p on glioma cell proliferation and invasion
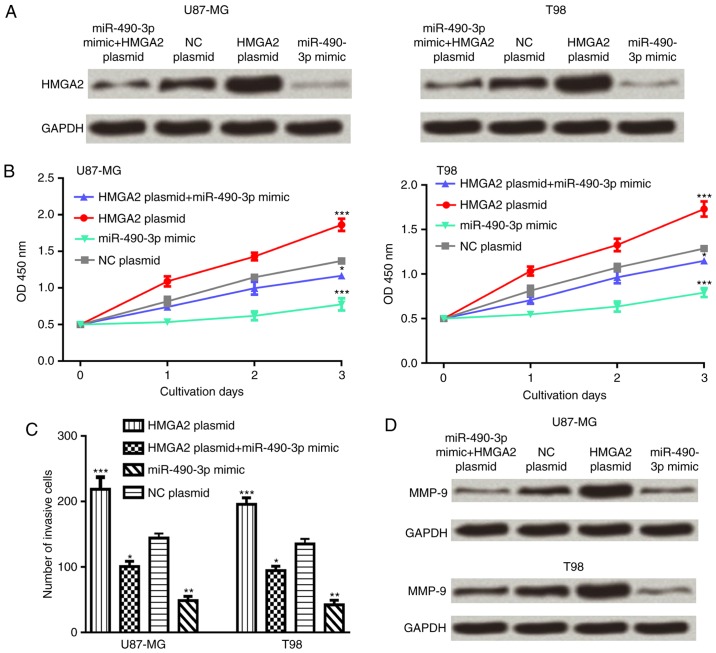
To investigate whether HMGA2 was an effector for miR-490-3p, glioma cell lines were co-transfected with the HMGA2 expression plasmid and miR-490-3p mimic. The results demonstrated that overexpression of HMGA2 partially reversed the inhibitory effect of miR-490-3p on the HMGA2 protein expression level in glioma cells (Fig. 4A). In addition, overexpression of HMGA2 partially reversed the inhibitory effect of miR-490-3p on glioma cell proliferation and invasion (Fig. 4B and C). Furthermore, the protein expression level of MMP-9 was increased following co-transfection with the HMGA2 expression plasmid and miR-490-3p mimic compared with the miR-490-3p mimic alone (Fig. 4D).
Figure 4.
HMGA2 is a functional target of miR-490-3p in glioma. (A) The relative HMGA2 protein expression, (B) cell proliferation and (C) cell invasion was determined in cells following transfection with HMGA2 expression plasmid and/or miR-490-3p mimic. (D) The relative MMP-9 protein expression was determined in cells following transfection with HMGA2 expression plasmid and/or miR-490-3p mimic. *P<0.05; **P<0.01; ***P<0.001 vs. NC plasmid. miR-490-3p, microRNA-490-3p; NC, negative control; MMP-9, matrix metallopeptidase-9; HMGA2, high-mobility group AT-hook 2.
Discussion
Approximately 60% of human genes are thought to be regulated by miRNAs, which suggests that miRNAs may be involved in multiple cellular processes (23). Numerous miRNAs have been identified to be critical players in the progression of glioma (9,24,25). For example, miR-30b-5p overexpression significantly inhibited glioma cell proliferation in vitro by downregulating the expression of metadherin (24). Ding et al (25) demonstrated that miR-122 expression was downregulated in glioma tissues. In addition, miR-122 may function as tumor suppressor through targeting runt-related transcription factors (25).
The current study demonstrated that miR-490-3p expression was significantly downregulated in glioma tissue compared with adjacent noncancerous tissue samples. Furthermore, the current study demonstrated that low miR-490-3p expression was closely associated with advanced tumor grade and high KPS scores in patients with glioma. Taken together, these results suggest that miR-490-3p may act as a tumor suppressor in glioma and decreased miR-490-3p expression may be associated with tumor progression.
In the current study, the biological role of miR-490-3p in glioma was further investigated. The results demonstrated that ectopic expression of miR-490-3p suppresses glioma cell proliferation and invasion in vitro, while the opposite effects were observed following downregulation of miR-490-3p. Furthermore, the protein expression level of MMP-9 in glioma cell lines was increased following transfection with miR-490-3p inhibitor, while the protein expression level of MMP-9 in glioma cell lines was decreased following transfection with miR-490-3p mimic. Previous studies demonstrated that miR-490-3p has different targets in several types of cancer (10,11).
To examine the underlying mechanism of miR-490-3p in glioma cell proliferation and invasion, bioinformatics analysis was used to identify target genes of miR-490-3p. In the current study, luciferase reporter assays and western blot analysis confirmed HMGA2 as a direct target of miR-490-3p in glioma. HMGA2 was previously reported to be involved in the regulation of cellular behavior in several types of human cancer (15–18). In addition, HMGA2 was recently identified to be dispensable for pancreatic cancer progression, metastasis and therapy resistance (26). Hawsawi et al (27) revealed that HMGA2 may promote epithelial-mesenchymal transition of prostate cancer via the mitogen-activated protein kinase pathway. In the current study, functional assays demonstrated that HMGA2 overexpression partially reversed the inhibitory effect of miR-490-3p on the malignant phenotype of glioma cells, which suggests that miR-490-3p may function as a tumor suppressor in glioma by targeting HMGA2. Several studies demonstrated that HMGA2 overexpression promotes stemness, invasion and tumorigenicity in glioma (16,28). A previous study revealed that the miRNA let-7a may target HMGA2 expression to regulate glioma cell proliferation, invasion and metastasis via the transforming growth factor-β/Smad3 signaling pathway (29). Furthermore, the expression levels of let-7a and HMGA2 were correlated with glioma grades (29). These results suggest that targeting HMGA2 may have therapeutic benefits in glioma therapy. A previous study revealed an inverse correlation between miR-490-3p and HMGA2 expression and miR-490-3p may regulate cell proliferation and apoptosis in osteosarcoma (13). In the current study, an inverse correlation between miR-490-3p and HMGA2 expression was observed in glioma, which confirmed the results from previous studies. Furthermore, the current study revealed the inhibitory effect of miR-490-3p on cell proliferation and invasion. Taken together, these results demonstrate the importance of the miR-490-3p/HMGA2 axis, which may be valuable in understanding the roles of miR-490-3p and HMGA2 in different types of human cancer.
In conclusion, the current study demonstrated that miR-490-3p serves an important role in regulating glioma cell proliferation and invasion. In addition, a correlation between miR-490-3p and HMGA2 expression in glioma was identified, which may be valuable in understanding the underlying mechanisms of glioma progression. Taken together, these results suggest that the miR-490-3p/HMGA2 axis may be a potential therapeutic target for glioma.
Acknowledgements
Not applicable.
Funding
This work was supported by the Foundation of the Science and Technology Program of Guangzhou, People's Republic of China (grant no. 201607010365).
Availability of data and materials
The datasets generated/analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FZ, AW, YW and JL designed the study, performed the experiments, analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The current study was approved by the Ethics Committee of The First Affiliated Hospital of Guangzhou University of Traditional Chinese Medicine (Guangzhou, China). All patients provided written informed consent prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Crocetti E, Trama A, Stiller C, Caldarella A, Soffietti R, Jaal J, Weber DC, Ricardi U, Slowinski J, Brandes A, RARECARE working group Epidemiology of glial and non-glial brain tumours in Europe. Eur J Cancer. 2012;48:1532–1542. doi: 10.1016/j.ejca.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 2.Chhabda S, Carney O, D'Arco F, Jacques TS, Mankad K. The 2016 world health organization classification of tumours of the central nervous system: What the paediatric neuroradiologist needs to know. Quant Imaging Med Surg. 2016;6:486–489. doi: 10.21037/qims.2016.10.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mittal S, Pradhan S, Srivastava T. Recent advances in targeted therapy for glioblastoma. Expert Rev Neurother. 2015;15:935–946. doi: 10.1586/14737175.2015.1061934. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Banelli B, Forlani A, Allemanni G, Morabito A, Pistillo MP, Romani M. MicroRNA in glioblastoma: An overview. Int J Genomics. 2017;2017:7639084. doi: 10.1155/2017/7639084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, He B, Xu T, Pan Y, Hu X, Chen X, Wang S. MiR-490-3p functions as a tumor suppressor by inhibiting oncogene VDAC1 expression in colorectal cancer. J Cancer. 2018;9:1218–1230. doi: 10.7150/jca.23662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian J, Xu YY, Li L, Hao Q. MiR-490-3p sensitizes ovarian cancer cells to cisplatin by directly targeting ABCC2. Am J Transl Res. 2017;9:1127–1138. [PMC free article] [PubMed] [Google Scholar]
- 12.Tang B, Liu C, Zhang QM, Ni M. Decreased expression of miR-490-3p in osteosarcoma and its clinical significance. Eur Rev Med Pharmacol Sci. 2017;21:246–251. [PubMed] [Google Scholar]
- 13.Liu W, Xu G, Liu H, Li T. MicroRNA-490-3p regulates cell proliferation and apoptosis by targeting HMGA2 in osteosarcoma. FEBS Lett. 2015;589:3148–3153. doi: 10.1016/j.febslet.2015.08.034. [DOI] [PubMed] [Google Scholar]
- 14.Hammond SM, Sharpless NE. HMGA2, microRNAs and stem cell aging. Cell. 2008;135:1013–1016. doi: 10.1016/j.cell.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarhadi VK, Wikman H, Salmenkivi K, Kuosma E, Sioris T, Salo J, Karjalainen A, Knuutila S, Anttila S. Increased expression of high mobility group A proteins in lung cancer. J Pathol. 2006;209:206–212. doi: 10.1002/path.1960. [DOI] [PubMed] [Google Scholar]
- 16.Liu B, Pang B, Hou X, Fan H, Liang N, Zheng S, Feng B, Liu W, Guo H, Xu S, Pang Q. Expression of high mobility group AT-hook protein 2 and its prognostic significance in malignant gliomas. Hum Pathol. 2014;45:1752–1758. doi: 10.1016/j.humpath.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Du B, Lu QJ, Fan XW, Tang K, Yang L, Liao WL. miR-541 suppresses proliferation and invasion of squamous cell lung carcinoma cell lines via directly targeting high-mobility group AT-hook 2. Cancer Med. 2018;7:2581–2591. doi: 10.1002/cam4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H, Zhao H, Xia X, Liu X. MicroRNA-599 targets high-mobility group AT-hook 2 to inhibit cell proliferation and invasion in clear cell renal carcinoma. Mol Med Rep. 2018;17:7451–7459. doi: 10.3892/mmr.2018.8755. [DOI] [PubMed] [Google Scholar]
- 19.Komori T. The 2016 WHO classification of tumours of the central nervous system: The major points of revision. Neurol Med Chir (Tokyo) 2017;57:301–311. doi: 10.2176/nmc.ra.2017-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karnofsky DA, Burchenal JH. Columbia University Press; New York, NY: 1949. Evaluation of chemotherapeutic agents. In: The Clinical Evaluation of Chemotherapeutic Agents in Cancer. MacLeod CM (ed) pp. 191–205. [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 23.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 24.Zhang D, Liu Z, Zheng N, Wu H, Zhang Z, Xu J. MiR-30b-5p modulates glioma cell proliferation by direct targeting MTDH. Saudi J Biol Sci. 2018;25:947–952. doi: 10.1016/j.sjbs.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding CQ, Deng WS, Yin XF, Ding XD. MiR-122 inhibits cell proliferation and induces apoptosis by targeting runt-related transcription factors 2 in human glioma. Eur Rev Med Pharmacol Sci. 2018;22:4925–4933. doi: 10.26355/eurrev_201808_15631. [DOI] [PubMed] [Google Scholar]
- 26.Chiou SH, Dorsch M, Kusch E, Naranjo S, Kozak MM, Koong AC, Winslow MM, Grüner BM. Hmga2 is dispensable for pancreatic cancer development, metastasis, and therapy resistance. Sci Rep. 2018;8:14008. doi: 10.1038/s41598-018-32159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hawsawi O, Henderson V, Burton LJ, Dougan J, Nagappan P, Odero-Marah V. High mobility group A2 (HMGA2) promotes EMT via MAPK pathway in prostate cancer. Biochem Biophys Res Commun. 2018;504:196–202. doi: 10.1016/j.bbrc.2018.08.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaur H, Ali SZ, Huey L, Hütt-Cabezas M, Taylor I, Mao XG, Weingart M, Chu Q, Rodriguez FJ, Eberhart CG, Raabe EH. The transcriptional modulator HMGA2 promotes stemness and tumorigenicity in glioblastoma. Cancer Lett. 2016;377:55–64. doi: 10.1016/j.canlet.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Zhang X, Chen D, Ma C. Let-7a suppresses glioma cell proliferation and invasion through TGF-β/Smad3 signaling pathway by targeting HMGA2. Tumour Biol. 2016;37:8107–8119. doi: 10.1007/s13277-015-4674-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated/analyzed during the current study are available from the corresponding author on reasonable request.