Abstract
The cloning, purification, and initial characterization of the β-carbonic anhydrase (CA, EC 4.2.1.1) from the genome of the opportunistic pathogen Malassezia restricta (MreCA), which a fungus involved in dandruff and seborrheic dermatitis (SD), is reported. MreCA is a protein consisting of 230 amino acid residues and shows high catalytic activity for the hydration of CO2 into bicarbonate and protons, with the following kinetic parameters: kcat of 1.06 × 106 s−1 and kcat/KM of 1.07 × 108 M−1 s−1. It is also sensitive to inhibition by the sulfonamide acetazolamide (KI of 50.7 nM). Phylogenetically, MreCA and other CAs from various Malassezia species seem to be on a different branch, distinct from that of other β-CAs found in fungi, such as Candida spp., Saccharomyces cerevisiae, Aspergillus fumigatus, and Sordaria macrospora, with only Cryptococcus neoformans and Ustilago maydis enzymes clustering near MreCA. The further characterization of this enzyme and the identification of inhibitors that may interfere with its life cycle might constitute new strategies for fighting dandruff and SD.
Keywords: carbonic anhydrase, Malassezia restricta, cloning, enzyme inhibition, acetazolamide
1. Introduction
Carbonic anhydrases (CA, EC 4.2.1.1) catalyze the simple but physiologically crucial interconversion of carbon dioxide and water into bicarbonate and protons: CO2 + H2O ⇌ HCO3− + H+ [1,2,3,4,5]. These metalloenzymes are indispensable for maintaining the physiological equilibrium of the dissolved CO2, H2CO3, HCO3−, and CO32−, which are metabolites essential for the biosynthesis and energy metabolism of organisms [5,6,7,8,9]. Thus, the survival of a microbe will be compromised by restricting the access of a pathogen to these metabolites. Interference with pH regulation, as well as metabolic pathways connected with the inhibition of CA activity, has begun to be considered for obtaining new anti-infective agents [10,11,12,13,14], in addition to applications of such agents for antitumor therapies [14,15,16,17]—the field in which this approach was validated after more than a decade of strenuous research efforts [6,7,18,19,20,21]. Indeed, the 21st century has been affected by the spread of antibiotic resistance, and consequently, the improvement of the pharmacological arsenal against pathogens is needed [22,23,24,25,26]. Most of the existing antibiotics target a microorganism’s cellular functions, such as the synthesis of proteins, nucleic acids, cell walls, or folate [27,28,29]. Generally, the identification of novel drug targets follows criteria pointing to pathogen survival and absence from the human genome [30,31,32]. Recently, it has been demonstrated that the CA superfamily represents a valuable member of new macromolecules affecting the growth of microorganisms or making them vulnerable to host defense mechanisms [32,33,34]. Targeting CAs from pathogens belonging to various species of bacteria, fungi, and protozoans may lead to anti-infectives with new mechanisms of action different from the clinically used agents [34,35,36,37]. Among the fungal pathogens investigated in detail, Malassezia globosa (MgCA) represents a particular case [38,39,40,41], because the β-CA encoded in the genome of this fungus was shown to be a druggable target [41]. Indeed, several effective in vitro MgCA inhibitors belonging to the sulfonamide type were also shown to have significant antifungal effects in vivo, in an animal model of dandruff [42,43,44]. Thus, after considering the significant drug resistance problems with azoles and other antifungals [45,46,47,48,49], MgCA was validated as a possible antifungal target.
Recently, it has been demonstrated that, rather than one particular Malassezia sp. being involved, a complex bacterial and fungal equilibrium is involved in dandruff [50,51,52,53,54,55,56]. Another Malassezia species, Malassezia restricta is also involved in triggering the disequilibrium between the commensals Cutibacterium acnes (formerly named Propionibacterium acnes) and Staphylococcus sp., both of which contribute to dandruff and seborrheic dermatitis symptoms [50,51,52,57]. Furthermore, the genome of this pathogen was recently decoded and was published [57], making it possible to search for potential drug targets and the corresponding agents that may interfere with them. For this reason, we decided to investigate the analogous enzyme from MgCA, which should also be present in the genome of M. restricta. here, we report on the cloning, purification, and characterization of the β-CA from the pathogenic fungus, M. restricta (MreCA).
2. Results and Discussion
2.1. MreCA Features
The genome of M. restricta contains a region of 690 bp encoding for a polypeptide chain of 230 amino acid residues and is homologous to the β-CA identified in the genome of M. globosa (MgCA). To show the relevant degree of homology existing between these enzymes, MreCA was aligned with MgCA and Cryptococcus neoformans CA (Can2) [58,59], as shown in Figure 1.
Figure 1.
Multiple sequence alignment of selected β-carbonic anhydrase (β-CAs) from three fungal species. The Cryptococcus neoformans CA (Can2) numbering system was used. Zinc ligands are indicated in red, and amino acids involved in the enzyme catalytic cycle are indicated in blue. Multiple sequence alignment was performed with the program Clustal W, version 2.1. Legend: Malassezia restricta (MreCA), Malassezia globosa (MgCA), and Cryptococcus Neoformans (Can2). Conserved residues are indicated with an asterisk (*), while (:) and (.) indicate conservative and semi-conservative substitutions, respectively. The sequence accession numbers are reported in Table 1.
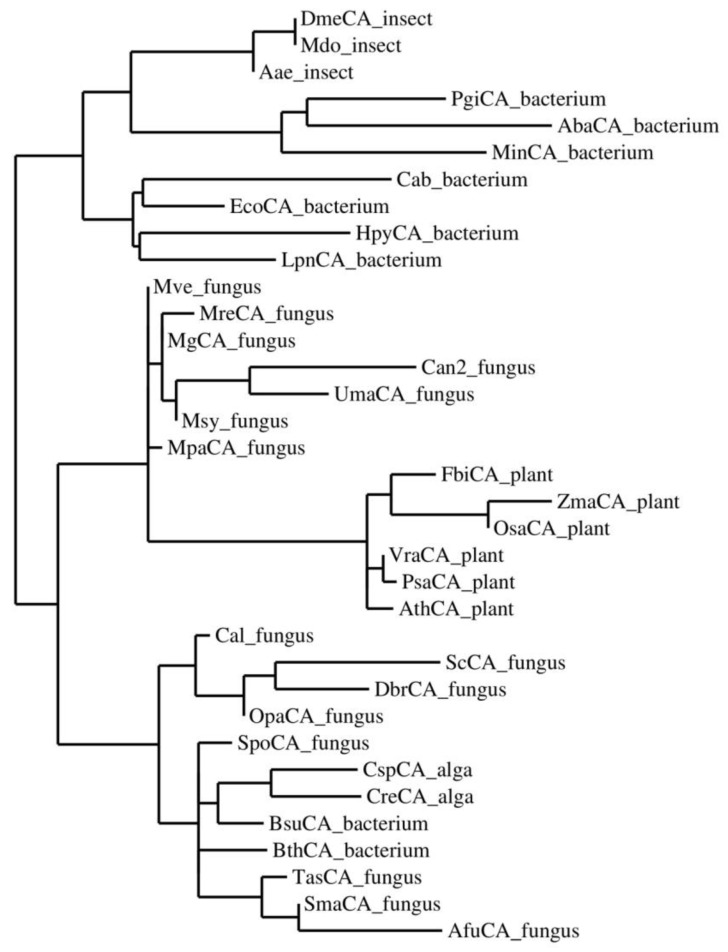
MreCA contains all the typical features of β-CAs, including the three residues that are involved in the catalytic mechanism of the enzyme, acting as zinc ligands (two cysteines and one histidine). It also contains amino acid residues, including the catalytic dyad that consists of an aspartate and an arginine, which are involved in the activation of the zinc-coordinated water molecule responsible for nucleophilic attack [38,39,40,41,42,43,44,45,58,59] (Figure 1). To better investigate the relationships between MreCA and the β-CAs identified in other species, such as insects, plants, fungi, algae, and bacteria, the most parsimonious phylogenetic tree has been constructed (Figure 2), which takes into account all the amino acid substitutions that differentiate β-CAs from various organisms indicated in Table 1.
Figure 2.
Phylogenetic tree of the β-CAs from selected prokaryotic and eukaryotic species. The tree was constructed using PhyML 3.0. For the acronyms and organism names see Table 1.
Table 1.
The accession numbers of the β-CA sequences used in the phylogenetic analysis. Groups, organism names, and acronyms are reported.
| Group | Organism Name | Acronym | Accession Number |
|---|---|---|---|
| Bacteria | |||
| Porphyromonas gingivalis | PgiCA_bacterium | YP_001929649.1 | |
| Acinetobacter baumannii | AbaCA_bacterium | YP_002326524 | |
| Myroides injenensis | MinCA_bacterium | ZP_10784819 | |
| Methanobacterium thermoautotrophicum | Cab_bacterium | GI:13786688 | |
| Helicobacter pylori | HpyCA_bacterium | BAF34127.1 | |
| Legionella pneumophila | LpnCA_bacterium | YP_003619232 | |
| Escherichia coli | EcoCa_bacterium | ACI70660 | |
| Burkholderia thailandensis | BthCA_bacterium | ZP_02386321 | |
| Brucella suis | BsuCA_bacterium | NP_699962.1 | |
| Fungi | |||
| Malassezia globosa | MgCA_fungus | XP_001730815.1 | |
| Malassezia pachydermatis | MpaCA_fungus | XP_017991749.1 | |
| Malassezia vespertilionis | Mve_fungus | PKI85431.1 | |
| Malassezia sympodialis | Msy_fungus | XP_018739548.1 | |
| Malassezia restricta | MreCA_fungus | PRJNA474956 | |
| Cryptococcus neoformans | Can2_fungus | GI:219109194 | |
| Candida albicans | Cal_fungus | XP_721792.1 | |
| Saccharomyces cerevisiae | ScCA_fungus | GAA26059 | |
| Dekkera bruxellensis | DbrCA_fungus | EFW97556 | |
| Ogataea parapolymorpha | OpaCA_fungus | EFW97556 | |
| Aspergillus fumigatus | AfuCA_fungus | XP_751704 | |
| Sordaria macrospora | SmaCA_fungus | CAT00781 | |
| Trichosporon asahii | TasCA_fungus | EKD04029 | |
| Schizosaccharomyces pombe | SpoCA_fungus | CAA21790 | |
| Ustilago maydis | UmaCA_fungus | XM_011388340.1 | |
| Algae | |||
| Coccomyxa sp. | CspCA_alga | AAC33484.1 | |
| Chlamydomonas reinhardtii | CreCA_alga | XP_001699151.1 | |
| Insect | |||
| Drosophila melanogaster | DmeCA_insect | NP_649849 | |
| Musca domestica | Mdo_insect | XP_005191496.1 | |
| Aedes aegypti | Aae_insect | XP_021707077.1 | |
| Plant | |||
| Vigna radiata | VraCA_plant | AAD27876 | |
| Pisum sativum | PsaCA_plant | AAA33652 | |
| Flaveria bidentis | FbiCA_plant | AAA86939.2 | |
| Arabidopsis thaliana | AthCA_plant | AAA50156 | |
| Zea mays | ZmaCA_plant | NP_001147846.1 | |
| Oryza sativa | OsaCA_plant | AAA86943 |
From the dendrogram reported in Figure 2, MreCA appears to be closely related to the β-CAs identified in the genome of the other Malassezia species. Intriguingly, the MreCA and MgCA clusters included the β-CA encoded by the genome of the pathogenic fungus Ustilago maydis, which is responsible for a plant disease known as common corn smut [60]. It has been reported that the genomes of M. restricta and globosa encode for many extracellular hydrolases, such as lipases, phospholipases, aspartyl proteases, and acid sphingomyelinases, which are phylogenetically close to those encoded by the U. maydis genome [60]. The dendrogram in Figure 2 also shows that the Malassezia β-CAs are neighbors to the β-CA from Cryptococcus neoformans (Can2). They are clustered distinctly away from the other β-CAs found in the genome of fungi, such as Candida albicans, Saccharomyces cerevisiae, Dekkera bruxellensis, Ogataea parapolymorpha, Aspergillus fumigatus, Sordaria macrospora, Trichosporon asahii, and Schizosaccharomyces pombe. This result may be the consequence of a gene duplication event indicating an ancestral β-CA gene in these two groups of fungi. Furthermore, Malassezia CAs are in the same cluster as the β-CAs from plants (Figure 2).
2.2. Expression, Purification, and Protonography
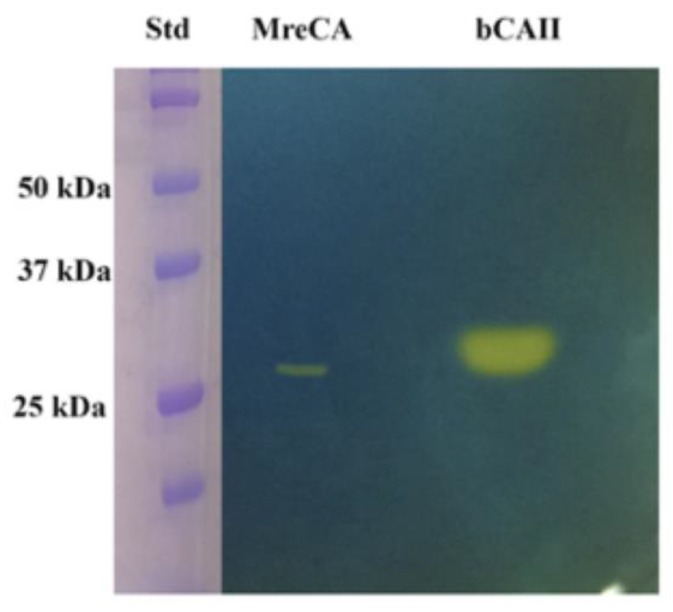
IPTG (Isopropyl β-D-1-thiogalactopyranoside) induction of Escherichia coli BL21 (DE3) cells transformed with the plasmid pET100D-Topo/MreCA resulted in the production of the recombinant MreCA as a fusion protein containing a His-tag tail at its N-terminus. After sonication and centrifugation, most of the CA activity was recovered in the soluble fraction of the E. coli cell extract. Using an affinity column (His-select HF (High Flow) nickel affinity gel), MreCA was purified to homogeneity, as shown by the appearance of the SDS-PAGE results (material not intended for publication). Samples of the purified MreCA were loaded onto the gel and subjected to protonography to investigate its hydratase activity via SDS–PAGE. Protonography is a powerful technique, which allows the detection of pH variation on polyacrylamide gel due to the CA-catalyzed conversion of CO2 into bicarbonate and protons [61,62]. The production of ions (H+) during the CO2 hydration reaction can be visualized as a yellow band on the polyacrylamide gel (Figure 3). The developed gel is called a protonogram.
Figure 3.
Protonography of MreCA with bovine CA (bCAII) as the standard enzyme.
The MreCA protonogram in Figure 3 shows a band corresponding to a monomer with an apparent molecular weight of about 27.0 kDa. The predicted molecular mass of the enzyme fused to the His-tag tail resulting from its amino acid sequence was 27.0 kDa. It is interesting to note that, as was found for other CA classes belonging to prokaryotic and eukaryotic organisms, MreCA was able to correctly refold, generating the active enzyme after removing SDS from the gel before developing the protonogram. Commercial bovine CA was used as a positive control.
2.3. Determination of the Kinetic Constants
Using stopped-flow techniques, the kinetic parameters were determined for the purified recombinant MreCA using CO2 as a substrate. The data in Table 2 demonstrate that MreCA shows a catalytic activity higher than that of MgCA, with the following kinetic parameters: kcat of 1.06 × 106 s−1 and kcat/KM of 1.07 × 108 M−1 s−1. Therefore, when compared with the high-activity human isoform hCA II, it is only slightly less effective as a catalyst for CO2 hydration. Furthermore, the activity is highly inhibited by acetazolamide, the clinically used sulfonamide inhibitor, with an inhibition constant of 50.7 nM, as seen in Table 2. Thus, the two Malassezia enzymes, MgCA and MreCA, show a net difference in their sensitivity to this sulfonamide, with the first one being quite resistant to the inhibitor and the second one being quite sensitive. It is interesting to stress the fact that β-CAs are missing in the genome of Cutibacterium acnes and Staphylococcus epidermidis, the two most abundant bacteria found on the human scalp. Thus, the synthesis of new drugs capable of interfering with MreCA and MgCA activity will not influence the catalytic mechanism of the CAs encoded by the scalp microbes, preserving not only the integrity of the human skin, but also avoiding interference with human CAs, because the human genome only encodes CAs belonging to the α-class.
Table 2.
Kinetic parameters for the newly obtained enzyme from MreCA, compared with the human hCA II (α-class) and the β-class enzyme from MgCA at 25 °C, pH 8.3 in 20 mM Tris buffer and 20 mM NaClO4, for the CO2 hydration reaction.
| Isozyme | kcat (s−1) | KM (mM) | kcat/KM (M−1 s−1) | KI (AAZ) (nM) |
|---|---|---|---|---|
| hCA II | 1.4 × 106 | 9.3 | 1.4 × 108 | 12 |
| MgCA | 9.2 × 105 | 11.1 | 8.3 × 107 | 74,000 |
| MreCA * | 1.06 × 106 | 10.1 | 1.07 × 108 | 50.7 |
* The kinetic/inhibition parameters are the mean from 3 different assays. Errors are in the range of 10% of the reported values (material not intended for publication).
3. Materials and Methods
3.1. Cloning and Purification of MreCA
The genome of M. restricta used in this study to retrieve the amino acid sequence was recently published [57]. The amino acid sequence of MreCA is shown in Figure 1. It was back translated into the nucleotide sequence and optimized for codon usage to increase its expression in E. coli cells. The synthetic M. restricta gene, as obtained from GeneArt Company (Milan, Italy), contained four base-pair sequences (CACC) necessary for directional cloning at the 5′ end of the MreCA DNA gene. The fragment was cloned into the expression vector pET100/D-TOPO (Invitrogen, Carlsbad, CA, USA), creating the plasmid pET100D-Topo/MreCA. Competent E. coli BL21 (DE3) codon plus cells (Agilent, Santa Clara, CA, USA) were transformed with the pET100D-Topo/MreCA. The level of expression of the target protein was improved by varying the time and temperature of induction and the concentration of the inducer (IPTG). After growth, the cells were harvested and disrupted by sonication at 4 °C in 20 mM of phosphate buffer, pH 8.0. Following sonication, the sample was centrifuged at 1200× g at 4 °C for 30 min. The supernatant was loaded onto a His-select HF nickel affinity column. The MreCA was eluted with 0.02 M phosphate buffer (pH 8.3) containing 250 mM imidazole and 0.5 M NaCl at a flow rate of 1.0 mL/min. Fractions were collected and dialyzed. At this stage of purification, the enzyme was at least 80% pure, and the obtained recovery was of 0.1 mg of the recombinant MreCA per liter of culture. The catalyst showed activity after undergoing the protonography test [61,62] (Figure 3) and via a kinetic, stopped-flow CO2 hydrase assay [63] (Table 2).
3.2. Protonography
Wells of 12% SDS gel were loaded with MreCA or the commercial bovine CA (Sigma, St. Louis, MO, USA), mixed with Laemmli loading buffer containing SDS (1% final concentration) but without 2-mercaptoethanol. In order to avoid protein denaturation the samples were not boiled. The gel was run at 150 V until the dye front moved off the gel. Following electrophoresis, the 12% SDS gel was subjected to protonography in order to detect the MreCA hydratase activity on the gel as described by Capasso et al. [61,62].
3.3. SDS-PAGE, Primary Structure, and Phylogenetic Analysis
Sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) was performed using 12% gels as described previously [61,62,64]. Multiple amino acid sequence alignment of the amino acid sequences of the β-CAs from the different species was performed with the program CLUSTAL W, version 2.1 [65]. The phylogenetic tree of β-CAs from the selected prokaryotic and eukaryotic species was constructed by using the program PhyML 3.0 [66].
3.4. CA Activity Measurements
An applied photophysics stopped-flow instrument was used for assaying the CA-catalyzed CO2 hydration activity [19]. Bromothymol blue (at a concentration of 0.2 mM) was used as an indicator, working at the absorbance maximum of 557 nm, with 10–20 mM TRIS (pH 8.3) as a buffer and 20 mM Na2SO4 for maintaining the ionic strength (this anion is not inhibitory and has a KI > 200 mM against this enzyme), following the initial rates of the CA-catalyzed CO2 hydration reaction for a period of 10–100 s. In order to determine the kinetic parameters and inhibition constants, the CO2 concentrations ranged from 1.7 to 17 mM. For each measurement, at least six traces of the initial 5%–10% of the reaction were used for determining the initial velocity, working with 10-fold decreasing inhibitor concentrations ranging from 1 nM to 10–100 µM (depending on the inhibitor potency, but at least 5 points at different inhibitor concentrations were employed for determining the inhibition constants). The uncatalyzed rates were determined in the same manner and then subtracted from the total observed rates. Stock solutions of inhibitor (0.1 mM) were prepared in distilled-deionized water, and dilutions up to 1 nM were done thereafter with the assay buffer. Inhibitor and enzyme solutions were pre-incubated together for 15 min at room temperature before assaying, in order to allow for the formation of the E–I complex. The inhibition constants were obtained by non-linear least-squares methods using the Cheng–Prusoff equation, and represent the mean from at least three different determinations. The human isoforms hCA I, II, and IX were assayed in the same conditions as above except that the working pH was 7.4, using HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer and phenol red as an indicator [63]. All enzymes were recombinant ones produced in our laboratory as described previously [58,59,63,67,68,69,70].
3.5. Multiple Sequence Alignment and Phylogenetic Analysis
Multiple alignment of the amino acid sequences of β-CAs identified in the genome of organisms belonging to different groups (bacteria, fungi, algae, insects, and plants) was performed with the program CLUSTAL W, version 2.1 [65]. The phylogenetic tree of β-CAs from the selected prokaryotic and eukaryotic species was constructed using the program PhyML 3.0, which searched for the tree with the highest probability [66].
4. Conclusions
We report here on the cloning, purification, and initial characterization of the β-CA (MreCA) from the genome of the pathogenic fungus M. restricta, responsible for dandruff, together with other fungi and bacteria. MreCA has a high catalytic activity for the hydration of CO2 into bicarbonate and protons, with the following kinetic parameters: kcat of 1.06 × 106 s−1 and kcat/KM of 1.07 × 108 M−1 s−1. It is also sensitive to inhibition by the classical sulfonamide inhibitor acetazolamide (KI of 50.7 nM). Phylogenetically, MreCA and other CAs from various Malassezia species seem to be on a different branch, distinct from that of other β-CAs found in fungi, such as Candida spp., Saccharomyces cerevisiae, Aspergillus fumigatus, and Sordaria macrospora, with only the Cryptococcus neoformans and Ustilago maydis enzymes being on the same branch as MreCA. As previously reported, the closest species were of plant origin. The further characterization of this enzyme and identification of inhibitors that could interfere with its life cycle might constitute new strategies for fighting dandruff and seborrheic dermatitis.
Author Contributions
S.D.P., performed the cloning, expression, and purification of the recombinant coral enzyme; D.V. performed the enzyme kinetic and all the inhibition assays; C.G., J.H., and X.M. contributed reagents and materials; C.C. (Cécile Clavaud) analyzed the data and edited the manuscript; and C.T.S. and C.C. (Clemente Capasso) wrote, edited, and supervised the manuscript.
Funding
The research was entirely funded by L’Oréal Research & Innovation.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Alterio V., Di Fiore A., D’Ambrosio K., Supuran C.T., De Simone G. Multiple binding modes of inhibitors to carbonic anhydrases: How to design specific drugs targeting 15 different isoforms? Chem. Rev. 2012;112:4421–4468. doi: 10.1021/cr200176r. [DOI] [PubMed] [Google Scholar]
- 2.Berrino E., Supuran C.T. Novel approaches for designing drugs that interfere with pH regulation. Expert Opin. Drug Discov. 2019;14:231–248. doi: 10.1080/17460441.2019.1567488. [DOI] [PubMed] [Google Scholar]
- 3.Capasso C., Supuran C.T. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: Can bacterial carbonic anhydrases shed new light on evolution of bacteria? J. Enzyme Inhib. Med. Chem. 2015;30:325–332. doi: 10.3109/14756366.2014.910202. [DOI] [PubMed] [Google Scholar]
- 4.Capasso C., Supuran C.T. Bacterial Carbonic Anhydrases, in Zinc Enzyme Inhibitors. Top. Med. Chem. 2017;1:134–152. [Google Scholar]
- 5.Capasso C., Supuran C.T. Protozoan carbonic anhydrases, in Zinc Enzyme Inhibitors. Top. Med. Chem. 2017;1:111–133. [Google Scholar]
- 6.Neri D., Supuran C.T. Interfering with pH regulation in tumours as a therapeutic strategy. Nat. Rev. Drug Discov. 2011;10:767–777. doi: 10.1038/nrd3554. [DOI] [PubMed] [Google Scholar]
- 7.Ozensoy Guler O., Capasso C., Supuran C.T. A magnificent enzyme superfamily: Carbonic anhydrases, their purification and characterization. J. Enzyme Inhib. Med. Chem. 2016;31:689–694. doi: 10.3109/14756366.2015.1059333. [DOI] [PubMed] [Google Scholar]
- 8.Supuran C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008;7:168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- 9.Supuran C.T. Carbonic anhydrases: From biomedical applications of the inhibitors and activators to biotechnological use for CO(2) capture. J. Enzyme Inhib. Med. Chem. 2013;28:229–230. doi: 10.3109/14756366.2013.761876. [DOI] [PubMed] [Google Scholar]
- 10.Supuran C.T. Structure and function of carbonic anhydrases. Biochem. J. 2016;473:2023–2032. doi: 10.1042/BCJ20160115. [DOI] [PubMed] [Google Scholar]
- 11.Supuran C.T. How many carbonic anhydrase inhibition mechanisms exist? J. Enzyme Inhib. Med. Chem. 2016;31:345–360. doi: 10.3109/14756366.2015.1122001. [DOI] [PubMed] [Google Scholar]
- 12.Supuran C.T. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin. Drug Discov. 2017;12:61–88. doi: 10.1080/17460441.2017.1253677. [DOI] [PubMed] [Google Scholar]
- 13.Supuran C.T. Carbonic Anhydrases and Metabolism. Metabolites. 2018;8:25. doi: 10.3390/metabo8020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Supuran C.T. Carbonic anhydrase inhibitors and their potential in a range of therapeutic areas. Expert Opin. Ther. Pat. 2018;28:709–712. doi: 10.1080/13543776.2018.1523897. [DOI] [PubMed] [Google Scholar]
- 15.Supuran C.T. Applications of carbonic anhydrases inhibitors in renal and central nervous system diseases. Expert Opin. Ther. Pat. 2018;28:713–721. doi: 10.1080/13543776.2018.1519023. [DOI] [PubMed] [Google Scholar]
- 16.Supuran C.T., Capasso C. Biomedical applications of prokaryotic carbonic anhydrases. Expert Opin. Ther. Pat. 2018;28:745–754. doi: 10.1080/13543776.2018.1497161. [DOI] [PubMed] [Google Scholar]
- 17.Supuran C.T., Vullo D., Manole G., Casini A., Scozzafava A. Designing of novel carbonic anhydrase inhibitors and activators. Curr. Med. Chem. Cardiovasc. Hematol. Agents. 2004;2:49–68. doi: 10.2174/1568016043477305. [DOI] [PubMed] [Google Scholar]
- 18.Supuran C.T. Carbonic Anhydrase Inhibition and the Management of Hypoxic Tumors. Metabolites. 2017;7:48. doi: 10.3390/metabo7030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Supuran C.T. Carbon-versus sulphur-based zinc binding groups for carbonic anhydrase inhibitors? J. Enzyme Inhib. Med. Chem. 2018;33:485–495. doi: 10.1080/14756366.2018.1428572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Supuran C.T. Carbonic anhydrase inhibitors as emerging agents for the treatment and imaging of hypoxic tumors. Expert Opin. Investig. Drugs. 2018;27:963–970. doi: 10.1080/13543784.2018.1548608. [DOI] [PubMed] [Google Scholar]
- 21.Nocentini A., Supuran C.T. Carbonic anhydrase inhibitors as antitumor/antimetastatic agents: A patent review (2008–2018) Expert Opin. Ther. Pat. 2018;28:729–740. doi: 10.1080/13543776.2018.1508453. [DOI] [PubMed] [Google Scholar]
- 22.Nishimori I., Minakuchi T., Maresca A., Carta F., Scozzafava A., Supuran C.T. The beta-carbonic anhydrases from Mycobacterium tuberculosis as drug targets. Curr. Pharm. Des. 2010;16:3300–3309. doi: 10.2174/138161210793429814. [DOI] [PubMed] [Google Scholar]
- 23.Nishimori I., Minakuchi T., Vullo D., Scozzafava A., Innocenti A., Supuran C.T. Carbonic anhydrase inhibitors. Cloning, characterization, and inhibition studies of a new beta-carbonic anhydrase from Mycobacterium tuberculosis. J. Med. Chem. 2009;52:3116–3120. doi: 10.1021/jm9003126. [DOI] [PubMed] [Google Scholar]
- 24.Guzel O., Maresca A., Scozzafava A., Salman A., Balaban A.T., Supuran C.T. Discovery of low nanomolar and subnanomolar inhibitors of the mycobacterial beta-carbonic anhydrases Rv1284 and Rv3273. J. Med. Chem. 2009;52:4063–4067. doi: 10.1021/jm9004016. [DOI] [PubMed] [Google Scholar]
- 25.Del Prete S., Isik S., Vullo D., De Luca V., Carginale V., Scozzafava A., Supuran C.T., Capasso C. DNA cloning, characterization, and inhibition studies of an alpha-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. J. Med. Chem. 2012;55:10742–10748. doi: 10.1021/jm301611m. [DOI] [PubMed] [Google Scholar]
- 26.Del Prete S., Vullo D., De Luca V., Carginale V., di Fonzo P., Osman S.M., AlOthman Z., Supuran C.T., Capasso C. Anion inhibition profiles of alpha-, beta- and gamma-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg. Med. Chem. 2016;24:3413–3417. doi: 10.1016/j.bmc.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 27.Del Prete S., Vullo D., De Luca V., Carginale V., Osman S.M., AlOthman Z., Supuran C.T., Capasso C. Comparison of the sulfonamide inhibition profiles of the alpha-, beta- and gamma-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg. Med. Chem. Lett. 2016;26:1941–1946. doi: 10.1016/j.bmcl.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Ferraroni M., Del Prete S., Vullo D., Capasso C., Supuran C.T. Crystal structure and kinetic studies of a tetrameric type II beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Acta Crystallogr. D Biol. Crystallogr. 2015;71:2449–2456. doi: 10.1107/S1399004715018635. [DOI] [PubMed] [Google Scholar]
- 29.Nishimori I., Onishi S., Takeuchi H., Supuran C.T. The alpha and beta classes carbonic anhydrases from Helicobacter pylori as novel drug targets. Curr. Pharm. Des. 2008;14:622–630. doi: 10.2174/138161208783877875. [DOI] [PubMed] [Google Scholar]
- 30.Capasso C., Supuran C.T. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin. Ther. Targets. 2015;19:1689–1704. doi: 10.1517/14728222.2015.1067685. [DOI] [PubMed] [Google Scholar]
- 31.Capasso C., Supuran C.T. An Overview of the Selectivity and Efficiency of the Bacterial Carbonic Anhydrase Inhibitors. Curr. Med. Chem. 2015;22:2130–2139. doi: 10.2174/0929867321666141012174921. [DOI] [PubMed] [Google Scholar]
- 32.Annunziato G., Giovati L., Angeli A., Pavone M., Del Prete S., Pieroni M., Capasso C., Bruno A., Conti S., Magliani W., et al. Discovering a new class of antifungal agents that selectively inhibits microbial carbonic anhydrases. J. Enzyme Inhib. Med. Chem. 2018;33:1537–1544. doi: 10.1080/14756366.2018.1516652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guzel-Akdemir O., Angeli A., Demir K., Supuran C.T., Akdemir A. Novel thiazolidinone-containing compounds, without the well-known sulphonamide zinc-binding group acting as human carbonic anhydrase IX inhibitors. J. Enzyme Inhib. Med. Chem. 2018;33:1299–1308. doi: 10.1080/14756366.2018.1499628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray A.B., Aggarwal M., Pinard M., Vullo D., Patrauchan M., Supuran C.T., McKenna R. Structural Mapping of Anion Inhibitors to beta-Carbonic Anhydrase psCA3 from Pseudomonas aeruginosa. ChemMedChem. 2018;13:2024–2029. doi: 10.1002/cmdc.201800375. [DOI] [PubMed] [Google Scholar]
- 35.Nocentini A., Cadoni R., Dumy P., Supuran C.T., Winum J.-Y. Carbonic anhydrases from Trypanosoma cruzi and Leishmania donovani chagasi are inhibited by benzoxaboroles. J. Enzyme Inhib. Med. Chem. 2018;33:286–289. doi: 10.1080/14756366.2017.1414808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Supuran C.T. Inhibition of carbonic anhydrase from Trypanosoma cruzi for the management of Chagas disease: An underexplored therapeutic opportunity. Future Med. Chem. 2016;8:311–324. doi: 10.4155/fmc.15.185. [DOI] [PubMed] [Google Scholar]
- 37.Vermelho A.B., da Silva Cardoso V., Ricci Junior E., Dos Santos E.P., Supuran C.T. Nanoemulsions of sulfonamide carbonic anhydrase inhibitors strongly inhibit the growth of Trypanosoma cruzi. J. Enzyme Inhib. Med. Chem. 2018;33:139–146. doi: 10.1080/14756366.2017.1405264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Prete S., De Luca V., Vullo D., Osman S.M., AlOthman Z., Carginale V., Supuran C.T., Capasso C. A new procedure for the cloning, expression and purification of the beta-carbonic anhydrase from the pathogenic yeast Malassezia globosa, an anti-dandruff drug target. J. Enzyme Inhib. Med. Chem. 2016;31:1156–1161. doi: 10.3109/14756366.2015.1102137. [DOI] [PubMed] [Google Scholar]
- 39.Nocentini A., Vullo D., Del Prete S., Osman S.M., Alasmary F.A.S., AlOthman Z., Capasso C., Carta F., Gratteri P., Supuran C.T. Inhibition of the beta-carbonic anhydrase from the dandruff-producing fungus Malassezia globosa with monothiocarbamates. J. Enzyme Inhib. Med. Chem. 2017;32:1064–1070. doi: 10.1080/14756366.2017.1355307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nocentini A., Bua S., Del Prete S., Heravi Y.E., Saboury A.A., Karioti A., Bilia A.R., Capasso C., Gratteri P., Supuran C.T. Natural Polyphenols Selectively Inhibit beta-Carbonic Anhydrase from the Dandruff-Producing Fungus Malassezia globosa: Activity and Modeling Studies. ChemMedChem. 2018;13:816–823. doi: 10.1002/cmdc.201800015. [DOI] [PubMed] [Google Scholar]
- 41.Hewitson K.S., Vullo D., Scozzafava A., Mastrolorenzo A., Supuran C.T. Molecular cloning, characterization, and inhibition studies of a beta-carbonic anhydrase from Malassezia globosa, a potential antidandruff target. J. Med. Chem. 2012;55:3513–3520. doi: 10.1021/jm300203r. [DOI] [PubMed] [Google Scholar]
- 42.Singh S., Supuran C.T. In silico modeling of beta-carbonic anhydrase inhibitors from the fungus Malassezia globosa as antidandruff agents. J. Enzyme Inhib. Med. Chem. 2016;31:417–424. doi: 10.3109/14756366.2015.1031127. [DOI] [PubMed] [Google Scholar]
- 43.Del Prete S., Vullo D., Osman S.M., AlOthman Z., Capasso C., Supuran C.T. Anion inhibition studies of the dandruff-producing fungus Malassezia globosa beta-carbonic anhydrase MgCA. Bioorg. Med. Chem. Lett. 2015;25:5194–5198. doi: 10.1016/j.bmcl.2015.09.068. [DOI] [PubMed] [Google Scholar]
- 44.Vullo D., Del Prete S., Nocentini A., Osman S.M., AlOthman Z., Capasso C., Bozdag M., Carta F., Gratteri P., Supuran C.T. Dithiocarbamates effectively inhibit the beta-carbonic anhydrase from the dandruff-producing fungus Malassezia globosa. Bioorg. Med. Chem. 2017;25:1260–1265. doi: 10.1016/j.bmc.2016.12.040. [DOI] [PubMed] [Google Scholar]
- 45.Entezari Heravi Y., Bua S., Nocentini A., Del Prete S., Saboury A.A., Sereshti H., Capasso C., Gratteri P., Supuran C.T. Inhibition of Malassezia globosa carbonic anhydrase with phenols. Bioorg. Med. Chem. 2017;25:2577–2582. doi: 10.1016/j.bmc.2017.03.026. [DOI] [PubMed] [Google Scholar]
- 46.Angiolella L., Carradori S., Maccallini C., Giusiano G., Supuran C.T. Targeting Malassezia species for Novel Synthetic and Natural Antidandruff Agents. Curr. Med. Chem. 2017;24:2392–2412. doi: 10.2174/0929867324666170404110631. [DOI] [PubMed] [Google Scholar]
- 47.Nocentini A., Cadoni R., Del Prete S., Capasso C., Dumy P., Gratteri P., Supuran C.T., Winum J.-Y. Benzoxaboroles as Efficient Inhibitors of the beta-Carbonic Anhydrases from Pathogenic Fungi: Activity and Modeling Study. ACS Med. Chem. Lett. 2017;8:1194–1198. doi: 10.1021/acsmedchemlett.7b00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bua S., Osman S.M., AlOthman Z., Supuran C.T., Nocentini A. Benzenesulfonamides incorporating nitrogenous bases show effective inhibition of beta-carbonic anhydrases from the pathogenic fungi Cryptococcus neoformans, Candida glabrata and Malassezia globosa. Bioorg. Chem. 2019;86:39–43. doi: 10.1016/j.bioorg.2019.01.030. [DOI] [PubMed] [Google Scholar]
- 49.Angeli A., Pinteala M., Maier S.S., Del Prete S., Capasso C., Simionescu B.C., Supuran C.T. Inhibition of alpha-, beta-, gamma-, delta-, zeta- and eta-class carbonic anhydrases from bacteria, fungi, algae, diatoms and protozoans with famotidine. J. Enzyme Inhib. Med. Chem. 2019;34:644–650. doi: 10.1080/14756366.2019.1571273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clavaud C., Jourdain R., Bar-Hen A., Tichit M., Bouchier C., Pouradier F., El Rawadi C., Guillot J., Menard-Szczebara F., Breton L., et al. Dandruff is associated with disequilibrium in the proportion of the major bacterial and fungal populations colonizing the scalp. PLoS ONE. 2013;8:e58203. doi: 10.1371/annotation/bcff4a59-10b7-442a-8181-12fa69209e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stalhberger T., Simenel C., Clavaud C., Eijsink V.G.H., Jourdain R., Delepierre M., Latge J.-P., Breton L., Fontaine T. Chemical organization of the cell wall polysaccharide core of Malassezia restricta. J. Biol. Chem. 2014;289:12647–12656. doi: 10.1074/jbc.M113.547034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L., Clavaud C., Bar-Hen A., Cui M., Gao J., Liu Y., Liu C., Shibagaki N., Gueniche A., Jourdain R., et al. Characterization of the major bacterial-fungal populations colonizing dandruff scalps in Shanghai, China, shows microbial disequilibrium. Exp. Dermatol. 2015;24:398–400. doi: 10.1111/exd.12684. [DOI] [PubMed] [Google Scholar]
- 53.Grice E.A., Dawson T.L.J. Host-microbe interactions: Malassezia and human skin. Curr. Opin. Microbiol. 2017;40:81–87. doi: 10.1016/j.mib.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 54.Park T., Kim H.-J., Myeong N.R., Lee H.G., Kwack I., Lee J., Kim B.J., Sul W.J., An S. Collapse of human scalp microbiome network in dandruff and seborrhoeic dermatitis. Exp. Dermatol. 2017;26:835–838. doi: 10.1111/exd.13293. [DOI] [PubMed] [Google Scholar]
- 55.Theelen B., Cafarchia C., Gaitanis G., Bassukas I.D., Boekhout T., Dawson T.L. Corrigendum: Malassezia ecology, pathophysiology, and treatment. Med. Mycol. 2019;57:e2. doi: 10.1093/mmy/myy046. [DOI] [PubMed] [Google Scholar]
- 56.Xu Z., Wang Z., Yuan C., Liu X., Yang F., Wang T., Wang J., Manabe K., Qin O., Wang X., et al. Dandruff is associated with the conjoined interactions between host and microorganisms. Sci. Rep. 2016;6:24877. doi: 10.1038/srep24877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morand S.C., Bertignac M., Iltis A., Kolder I.C.R.M., Pirovano W., Jourdain R., Clavaud C. Complete Genome Sequence of Malassezia restricta CBS 7877, an Opportunist Pathogen Involved in Dandruff and Seborrheic Dermatitis. Microbiol. Resour. Announc. 2019;8:e01543-18. doi: 10.1128/MRA.01543-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlicker C., Hall R.A., Vullo D., Middelhaufe S., Gertz M., Supuran C.T., Muhlschlegel F.A., Steegborn C. Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans. J. Mol. Biol. 2009;385:1207–1220. doi: 10.1016/j.jmb.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 59.Innocenti A., Muhlschlegel F.A., Hall R.A., Steegborn C., Scozzafava A., Supuran C.T. Carbonic anhydrase inhibitors: Inhibition of the beta-class enzymes from the fungal pathogens Candida albicans and Cryptococcus neoformans with simple anions. Bioorg. Med. Chem. Lett. 2008;18:5066–5070. doi: 10.1016/j.bmcl.2008.07.122. [DOI] [PubMed] [Google Scholar]
- 60.Xu J., Saunders C.W., Hu P., Grant R.A., Boekhout T., Kuramae E.E., Kronstad J.W., Deangelis Y.M., Reeder N.L., Johnstone K.R., et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. USA. 2007;104:18730–18735. doi: 10.1073/pnas.0706756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Luca V., Del Prete S., Supuran C.T., Capasso C. Protonography, a new technique for the analysis of carbonic anhydrase activity. J. Enzyme Inhib. Med. Chem. 2015;30:277–282. doi: 10.3109/14756366.2014.917085. [DOI] [PubMed] [Google Scholar]
- 62.Del Prete S., De Luca V., Supuran C.T., Capasso C. Protonography, a technique applicable for the analysis of eta-carbonic anhydrase activity. J. Enzyme Inhib. Med. Chem. 2015;30:920–924. doi: 10.3109/14756366.2014.990963. [DOI] [PubMed] [Google Scholar]
- 63.Khalifah R.G. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem. 1971;246:2561–2573. [PubMed] [Google Scholar]
- 64.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 65.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 66.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 67.Carta F., Scozzafava A., Supuran C.T. Sulfonamides: A patent review (2008–2012) Expert Opin. Ther. Pat. 2012;22:747–758. doi: 10.1517/13543776.2012.698264. [DOI] [PubMed] [Google Scholar]
- 68.Pastorekova S., Casini A., Scozzafava A., Vullo D., Pastorek J., Supuran C.T. Carbonic anhydrase inhibitors: The first selective, membrane-impermeant inhibitors targeting the tumor-associated isozyme IX. Bioorg. Med. Chem. Lett. 2004;14:869–873. doi: 10.1016/j.bmcl.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 69.Scozzafava A., Carta F., Supuran C.T. Secondary and tertiary sulfonamides: A patent review (2008–2012) Expert Opin. Ther. Pat. 2013;23:203–213. doi: 10.1517/13543776.2013.742065. [DOI] [PubMed] [Google Scholar]
- 70.Supuran C.T. Carbonic anhydrase inhibitors as emerging drugs for the treatment of obesity. Expert Opin. Emerg. Drugs. 2012;17:11–15. doi: 10.1517/14728214.2012.664132. [DOI] [PubMed] [Google Scholar]