Abstract
Modifications of lysine contribute to the amount of dietary advanced glycation end-products reaching the colon. However, little is known about the ability of intestinal bacteria to metabolize dietary N-ε-carboxymethyllysine (CML). Successive transfers of fecal microbiota in growth media containing CML were used to identify and isolate species able to metabolize CML under anaerobic conditions. From our study, only donors exposed to processed foods degraded CML, and anaerobic bacteria enrichments from two of them used 77 and 100% of CML. Oscillibacter and Cloacibacillus evryensis increased in the two donors after the second transfer, highlighting that the bacteria from these taxa could be candidates for anaerobic CML degradation. A tentative identification of CML metabolites produced by a pure culture of Cloacibacillus evryensis was performed by mass spectrometry: carboxymethylated biogenic amines and carboxylic acids were identified as CML degradation products. The study confirmed the ability of intestinal bacteria to metabolize CML under anoxic conditions.
Keywords: microbiota, Maillard reaction, N-ε-carboxymethyllysine, intestinal metabolism, dietary advanced glycation end-products
Introduction
Thermal treatment of foods is well-known to result in the formation of Maillard reaction products, also named dietary advanced glycation end-products (d-AGEs).1 Free amino groups of proteins and amino acids react with reducing carbohydrates, forming compounds that are poorly bioavailable and cannot be used for de novo protein synthesis by the human body. In some foods, Maillard reaction (MR) is limited to the initial stages, and the main detectable products are Amadori and Heyns compounds.2 In severe thermally treated foods, reaction products such as hydroimidazolones, pyrraline, hydroxymethylfurfural, and N-ε-carboxymethyllysine (CML) are predominantly found.3 CML is a particularly stable compound present in the range of 0.01–5.09 mg/100 g in many different foods, such as dairy products, bread crust, cookies.4 Because of its chemical nature and ubiquity in foods and in vivo, CML has been largely used as a biomarker of the dietary intake of thermally treated foods, and it is probably the most investigated d-AGE.4
The nutritional and health concerns of thermally treated foods are related to the decrease of available lysine and arginine and to the metabolism of the d-AGEs.5,6 A substantial percentage of lysine can react with carbohydrate through MR, and since most of the d-AGE-containing proteins are not bioavailable, they become part of unabsorbed digesta that reach the colon every day.7 This is not only due to the direct lysine blockage but also a consequence of the fact that glycation and oxidation promote significant protein cross-linking, decreasing the overall digestibility and funneling more material to the gut.8 As a result, these d-AGEs serve as substrate for intestinal bacteria. The adoption of a diet with plenty of cooked and processed foods has surely been beneficial to increase energy intake.9 The ability of microbiota to use d-AGEs may provide an additional advantage to individuals harboring suitable species. Nevertheless, animal studies have shown that some of the d-AGEs are potentially toxic, and their degradation may contribute to the general health of the consumer.10
The ability of the intestinal microbiota to metabolize glycated proteins has become of general interest as part of the larger discussion on the metabolism of dietary protein by colonic bacteria.11 Mounting evidence suggests that a diet rich in glycated proteins profoundly changes the intestinal microbiota composition. Recently, it was reported that one of the intermediates of the MR, N-ε-fructosyllysine, could no longer be detected after 4 h of incubation in fecal material from healthy volunteers who consumed a Western diet.12Intestinimonas butyriciproducens was isolated from stool of a healthy individual, and this strain is able to convert N-ε-fructosyllysine into butyrate, highlighting microbiota responses to the dietary component.13,14
Our present insights in the capability of human microbiota to metabolize CML under anaerobic conditions are very limited. The first pivotal work estimated that 10% of the orally ingested AGEs was absorbed, and only 30% of that was eliminated in the urine.15 It was also observed that CML and N-ε-fructosyllysine are available for absorption during simulated gastrointestinal digestion, while lysinoalanine and other cross-link products are not.16 Both d-AGEs appear as small peptides similar to proteinogenic amino acids and have similar biochemical characteristics. Interestingly, it was found that dietary CML (d-CML) is partially bioavailable, and a significant amount is accumulated in various organs.17 It is known that d-CML does not bind to the receptor of AGEs (RAGE) as is the case for the endogenously formed CML, which is formed by the lysine present in collagen and other tissue protein. Hence, the mechanisms responsible for the inflammatory action of d-CML are not known, and no scientific consensus has been reached on the physiological relevance of CML so far.18 Besides accumulation in the organ and urinary excretion, metabolism of d-CML showed that the large majority is recovered in the feces: the consumption of the high d-AGE diet led to a higher CML input associated with a higher fecal excretion in a two period crossover trial with 11–14 year old adolescent males.19 Recent data showed that d-CML is partially metabolized by the gut microbiota.12 Notably, it has been shown that under aerobic and low-amino-acid conditions, in the presence of CML dipeptide and CML, five different strains of Escherichia coli were able to linearly produce three CML bacterial metabolites, up to 8.4% of initial CML dose: N-carboxymethylcadaverine (CM-CAD), N-carboxymethylaminopentanoic acid (CM-APA), and the N-carboxymethyl-Δ1-piperideinium ion.20 However, the involved anaerobic intestinal microbes have been neither identified nor isolated or characterized.
In this work, we aimed at studying the capability of intestinal bacteria to degrade dietary CML. A strategy of successive transfers of fecal microbiota in growth media containing CML as the major carbon source was adopted to enrich, identify, and isolate the anaerobes able to metabolize d-AGEs and corroborate previous findings on CML degradation pathways.
Material and Methods
Chemicals and Reagents
Acetonitrile, methanol, and water for mass spectrometry analyses were obtained from Merck (Darmstadt, Germany). Analytical standards N-ε-carboxymethyllysine (CML) and Nε-(carboxy[2H4]methyl)-l-lysine (d4-CML) were obtained from TRC Chemicals (Toronto, Canada), while ammonium hydroxide solution (28% in water w/v), ammonium acetate, formic acid, lysine, and l-lysine-4,4,5,5-d4 hydrochloride (d4-Lys) were purchased from Sigma (St. Louis, MO).
Enrichment and Growth Media
The study involved six subjects that donated fecal samples (Table 1). Informed consent was obtained from the mothers for their approximately 6 month old infants and the Asian adult, while the two African samples were derived from a previously reported intervention study and preserved in 25% glycerol (kind gift from Prof O’Keefe, Pittsburgh, USA).21
Table 1. Volunteer Information.
| subject | origin | code | inoculum |
|---|---|---|---|
| formula-fed infant | Netherlands | F1 | fresh |
| breast-fed infant | Netherlands | M1 | fresh |
| breast-fed infant | Netherlands | M2 | fresh |
| adult | Asian | AS1 | fresh |
| adult | African | AF1A | frozen |
| adult | African | AF4A | frozen |
Fresh fecal samples were collected in 15 mL Falcon tubes containing anaerobic phosphate buffer (pH 7.0) and later stored in 25% glycerol in 5 mL anaerobic bottles kept at −80 °C. Aliquots (0.5 mL) of these fecal slurries were transferred to 10 mL anaerobic bicarbonate-buffered mineral salt medium (CP medium, see below) containing CML as the energy and carbon source for the first enrichment. CML concentration at time 0 is reported in Table 2. These anaerobic bottles were filled with a headspace of CO2/N2 (1:4) at 1.5 atm and incubated at 37 °C. Subsequently, 0.5 mL of the fully grown first enrichment was transferred to 10 mL of fresh CP medium containing CML as in a 10 mL bottle with a head space and incubation regimen as before.
Table 2. CML Degradation in the First Enrichments from Six Volunteersa.
| samples |
||||||
|---|---|---|---|---|---|---|
| CML concentration (mM) | F1* | AF4A* | M1* | AF1A | AS1 | M2 |
| T0 | 6.30 | 7.90 | 4.84 | 4.50 | 2.19 | 2.14 |
| after 3w*/5w | 5.13 | 7.47 | 3.84 | 1.00 | 0.00 | 2.11 |
| consumed (mM) | 1.17 | 0.43 | 1.00 | 3.50 | 2.19 | 0.03 |
| consumed (%) | 18.62 | 5.46 | 20.65 | 77.69 | 100.00 | 1.39 |
The measurement was performed after 3 weeks of incubation (*) or 5 weeks of incubation.
The growth experiments on the CML degradation capacity of the newly isolated C. evryensis strain AS3 were also performed in CP medium (see below) containing either 5 mM CML alone or 5 mM CML with a combination of 10 mM lysine, 10 mM serine, 10 mM histidine, and 10 mM arginine. Lysine, serine, histidine, and arginine were individually added to CP medium from 0.5 M sterile anoxic stock solutions upon inoculation, while CML was added to the medium as powder and filter sterilized afterward. The cultures were sampled at regular intervals for CML analysis by liquid chromatography tandem mass spectrometry (LC–MS/MS).
Enrichments were performed in anaerobic bicarbonate-buffered mineral salt medium (CP medium) consisting of 0.53 g/L Na2HPO4·2H2O, 0.41 g/L KH2PO4, 0.3 g/L NH4Cl, 0.11 g/L CaCl2·2H2O, 0.10 g/L MgCl2·6H2O, 0.3 g NaCl, 4.0 g/L NaHCO3, and 0.48 g/L Na2S·9H2O as well as alkaline and acid trace elements (each 0.1% v/v) and vitamins (0.2% v/v).22 The alkaline trace element solution contained the following (mM): 0.1 Na2SeO3, 0.1 Na2WO4, 0.1 Na2MoO4, and 10 NaOH. The acid trace element solution was composed of the following (mM): 7.5 FeCl2, 1 H3BO4, 0.5 ZnCl2, 0.1 CuCl2, 0.5 MnCl2, 0.5 CoCl2, 0.1 NiCl2, and 50 HCl. The vitamin solution had the following composition (g/L): 0.02 biotin, 0.2 niacin, 0.5 pyridoxine, 0.1 riboflavin, 0.2 thiamine, 0.1 cyanocobalamin, 0.1 p-aminobenzoic acid, and 0.1 pantothenic acid.
Isolation of anaerobes was further performed by streaking on plates of Reinforced Clostridium Medium (BD) solidified with 1.5% agar that were incubated in anaerobic jars filled with CO2/N2 (1:4) at 1.5 atm. The picking and streaking were performed in an anaerobic tent filled with N2/H2 (95/5 v/v).
Volatile Fatty Acids Detection
The formation of volatile fatty acids and alcohols was measured on a Thermo Scientific Spectra HPLC system equipped with a P2000 pump, an AS3000 autosampler, and an Agilent Metacarb 67H 300 × 6.5 mm column kept at 45 °C and running with H2SO4 as an eluent. The detector was an RI-150 refractive index detector. The eluent flow was 0.8 mL/min. The bacterial cultures were centrifuged at 14 000g for 5 min at room temperature to obtain the supernatants, of which 400 μL was used to analyze in the HPLC. The measurement was performed at 45 °C for separation of volatile fatty acids and alcohols.23
CML and Free Lysine Detection by Liquid Chromatography Tandem Mass Spectrometry
Free lysine and CML were analyzed in order to combine target analysis and CML degradation product investigation; 0.5 mL cultures of the enrichments were collected at different time intervals of up to 5–6 weeks. Upon the sampling, the supernatants were collected by centrifugation at 21 000g for 10 min at 4 °C and subsequently diluted 500× in acetonitrile: water (50:50, v/v), and 5 μL was used for injection. Hydrophilic interaction liquid chromatography (HILIC) separation of CML, lysine, d4-lysine, and d4-CML was achieved on a thermostated (40 °C) Luna amino column (3.0 μm, 100 × 2.0 mm, Phenomenex, Torrance, CA) equipped with an amino security guard (4.0 × 2.0 mm). The following mobile phases were used: A, acetonitrile, and B, 20 mM ammonium acetate in water with ammonium hydroxide added to adjust alkalinity (pH 9.0), according to the procedure that was optimized previously.24 The compounds were eluted at 300 μL/min through the following gradient of solvent B (t in [min]/[%B]): (0/2), (2.5/2), (7/90), (9/90). Positive electrospray ionization was used for detection, and the source parameters were selected as follows: spray voltage = 5.5 kV; capillary temperature = 400 °C; dwell time = 100 ms. The chromatographic profile was recorded in multiple reaction monitoring mode (MRM) by using an API 3000 triple quadrupole (ABSciex, Carlsbad, CA). Target analytes and their labeled internal standards were analyzed using the mass transitions given in parentheses, and in bold is the transition used for the quantitation in the case of target analytes and for qualification in the case of internal standards experiments: lysine (m/z 147 → 84, 130), d4-lysine (m/z 151 → 88, 134), CML (m/z 205 → 84, 130), d4-CML (m/z 209 → 88, 134). Analytical performances such as robustness, sensitivity, reproducibility, repeatability, linearity, accuracy, carryover, and matrix effects were evaluated by following the procedures previously reported.25 Typical retention times were 4.5 and 5.3 min for lysine and CML, respectively. Quantitation of CML and lysine was performed by using the internal standard technique, and results are reported as mmol/L.
Identification of CML Degradation Metabolites
An internally generated database of CML degradation metabolites was constructed following lysine metabolite entries from the publicly available online metabolite database KEGG (www.genome.jp/kegg/) and by adding a carboxymethyl group (−CH2COOH) to the lysine amino side chain. Tentative identification of CML degradation products was achieved by high-resolution mass spectrometry (HRMS). Optimal conditions for chromatographic separation of CML metabolites were obtained after two consecutive trials by using the amino column with mobile phases as described above for tandem mass spectrometry quantitation of CML and by means of a silica sulfobetaine zwitterionic modified HILIC column (50 × 2.1 mm, 3.0 μm, Thermo Fisher, Bremen, Germany) at 35 °C. Mobile phases for the zwitterionic column were 0.1% formic acid in acetonitrile (solvent A) and 0.1% formic acid in water (solvent B). Samples were diluted 10× in a mixture of acetonitrile/water, 50:50 (v/v), and the following gradient of solvent B (minutes/%B): (0/10), (0.80/10), (3.5/5), (5.5/5) was used. The flow rate was set to 300 μL/min, and the injection volume was 5 μL. The U-HPLC system (Accela 1250, Thermo Fisher, Bremen, Germany) was interfaced to an Exactive Orbitrap HRMS (Thermo Fisher, Bremen, Germany), equipped with an heated electrospray source (HESI-II) working in polarity switching mode. The current ion of potential candidates was scanned in the m/z range of 50–350 by using a scan time of 1 s. The interface parameters were spray voltage = 4.2 kV, capillary voltage = 35.0 V, capillary temperature = 350 °C, heater temperature = 300 °C, sheath gas flow = 42 arbitrary units, and auxiliary gas flow = 12 arbitrary units, while for negative acquisition mode, the interface parameters were spray voltage = −3.8 kV and capillary voltage = −25.0 kV. HRMS conditions were optimized by infusing a mixture of CML, d4-CML, lysine, and d4-lysine (20 μg/mL) at a flow rate of 3 μL/min. Mass tolerance was fixed at 5 ppm, and the exact mass 83.06037 [M2+H]+ of acetonitrile was used as a recalibrating agent (lock mass) to improve selectivity and signal stability. Analyte concentrations were monitored by external standard technique with CML as standard and the intensities, signal-to-noise ratio (S/N) stability, and reproducibility among different batches of samples were used to select between the amino and zwitterionic column. Metabolites concentration was reported as μmol/L
Bacterial Community Analysis
The cell biomass was harvested from an aliquot (1–2 mL) of well-homogenized liquid culture by centrifuging at 13 000g for 10 min. DNA was extracted from the pellet by using a MasterPure Gram Positive DNA Purification Kit (Epicenter, Madison, United States) according to the manufacturer’s instructions. The PCR was performed using 27F and 1492R to amplify the complete 16S rRNA genes of the bacteria using the program starting at 94 °C for 5 min and continuing with 35 cycles consisting of 94 °C for 90 s, 52 °C for 30 s, 72 °C for 90 s, and finally 72 °C for 10 min.26 The PCR products were subsequently purified by a PCR purification kit (Qiagen, Hilden, Germany) and used to generate a clone library of full-length 16S rRNA gene sequences using pGEM Easy Vector Systems (Promega, Madison, United States). All steps mentioned above were done following the manufacturers’ instructions. A total of 24 to 60 clones were selected for Sanger sequencing at GATC (Biotech, Konstanz, Germany) using SP6 (5′-ATTTAGGTGACACTATAGAA-3′) as the sequencing primer. The sequences were trimmed with DNASTAR to remove vector contamination and manually checked. Later, they were aligned with the multiple sequence aligner SINA and merged with the Silva SSU ref database (release 111). Phylogenetic trees were constructed in the ARB software package (v. 6) by the same algorithm.27
Results and Discussion
CML Enrichments from Stool
Fecal samples from three infant and three adult volunteers (Table 1) were enriched directly in bicarbonate-buffered medium containing CML as the only carbon and energy source without any intermediate step or substrate, under anaerobic conditions. The first enrichments showed that the fecal microbiota of all volunteers were able to degrade CML, however, at very different levels (Table 2). While the microbiota of adult donors AF1A and AS1 degraded this substrate the best after 5 weeks of incubation, those of M2 and AF4A had the lowest CML degradation capabilities. The degradation was rather slow, and this could be attributed to a nonoptimized medium for growth of potential CML degraders. The microbiota of the infants (both breast- and formula-fed) showed very limited CML degradation capacity. Among the three adults, two showed a good degradation capacity, and one did not. This is in line with the possibility that dietary exposure to CML is connected to the ability of microbiota to metabolize it.
Interestingly, CML degradation activities of the samples from donors F1, AF4A, M1, and M2 were not detected in the second transfer, while those of samples AF1A and AS1 were somewhat reduced (see below). This may indicate that either some growth factors were absent or microbial partners involved in CML degradation were missing at the second transfer.
To get further insights in the bacteria that were able to degrade CML, we focused on the incubations of the samples from donors AF1A and AS1, as these were two samples with the highest degradation activity (77 and 100% of the added CML, respectively). The first enrichments were transferred a second time, and the bacterial composition in the two CML enrichments was determined using Sanger sequencing of clone libraries of 16S rRNA amplicons. Sequences of approximately 900 bp obtained were subsequently used to construct phylogenetic trees and assign taxonomic groups.
Microbial Composition Analysis Indicates Uncultured Oscillibacter spp. as Potential CML Degrader
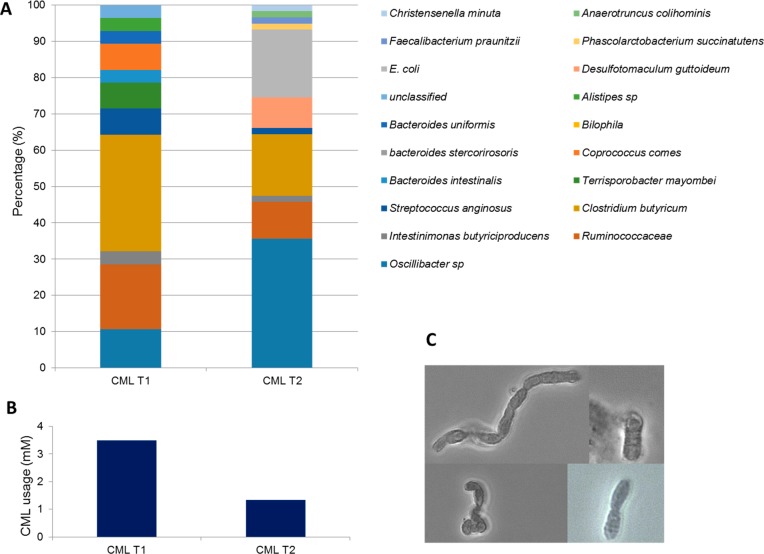
The microbial composition of the first and second enrichments was determined to investigate the microbes that were involved in anaerobic CML degradation in the fecal sample of volunteer AF1A (Figure 1). After 5 weeks of incubation, 3.5 mM CML was degraded in the first enrichment, while 1.2 mM CML was utilized in the second transfer. In the first enrichment, Clostridium butyricum, Ruminococaceae, and Oscillibacter were the most dominant, while after the second transfer, the majority of the enrichment belonged to Oscillibacter, Clostridium butyricum, Clostridium sphenoides, and Escherichia coli with Oscillibacter spp. as the most prevalent taxon (Figure 1). On the basis of the clone library abundance, the sequence of Oscillibacter spp. made up 10.7% of the total microbial community in the first enrichment and increased to 35.6% after the second transfer. The typical morphology of Oscillibacter was also observed in the first and second enrichment (Figure 1C).28 Up to now, there is no report about anaerobic bacteria that are able to degrade CML in anoxic conditions. However, an increase in abundance of Oscillibacter spp. in the second transfer indicated a possible role of this bacterium in anaerobic CML degradation. So far, efforts to isolate this bacterium from this enrichment using different media have not been successful, which is in line with the reported difficulties in culturing members of the genus.29
Figure 1.
Microbial composition of the first and second enrichments in CML (A); CML usage in the first and second enrichment from sample AF1A (B) and morphology of Oscillibacter spp. observed in the enrichment (C). The incubation time was 5 weeks at 37 °C under anoxic conditions.
Complete Degradation of CML by an Enrichment of Bacteria Related to Cloacibacillus evryensis
The microbiota in the fecal sample from volunteer AS1 showed the capacity to completely degrade CML in the first enrichment (Table 2). While more than 2 mM CML was completely degraded in 5 weeks in the first enrichment, only 0.8 mM CML was degraded in the second enrichment (Figure 2B). As with the enrichment of the fecal samples of volunteer AF1A, this suggested that some growth factors present in the fecal sample could be missing in the second enrichment. Interestingly, the microbial community analysis of the the second enrichment from the fecal sample of volunteer AS1 revealed an up to 80% enriched population of a single taxonomic group, Cloacibacillus evryensis (Supplementary Figure 1 and Figure 2A). This species has been known as an active amino-acid degrader in a mesophilic anaerobic digester.30,31 However, C. evryensis has not been isolated yet from a human specimen, although its presence has been suggested in stools of human and other animals. The CML degradation results in the second transfer of AS1 enrichment also indicated that bacteria related to C. evryensis could well be responsible for the anaerobic CML degradation.
Figure 2.
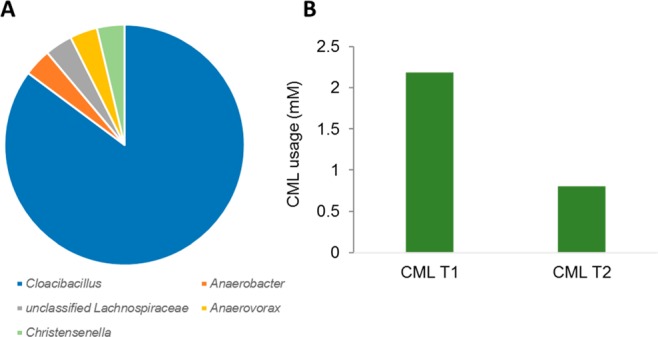
Microbial composition of the second transfer of CML enrichment from AS1 (A) and the CML usage in the first and second transfer (B). The incubation time was 5 weeks at 37 °C under anoxic conditions.
Cloacibacillus evryensis Isolation with CML Degradation Capacity
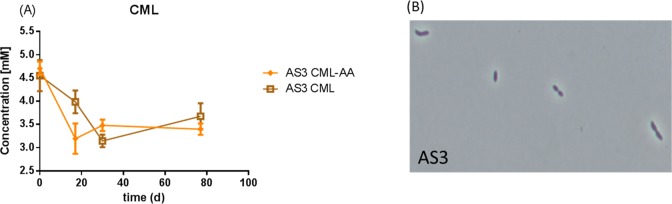
We made several attempts to isolate the enriched bacteria related to C. evryensis by plating the enrichment on Reinforced Clostridium agar. After picking and streaking colonies several times, we got an axenic culture, termed C. evryensis strain AS3, since a 16S rRNA gene sequence of the pure culture showed >99% similarity to that of the type strain of C. evryensis strain DSM 19522T.30 Moreover, this sequence was identical to that detected in the clone library (Figure 2A) indicating that C. evryensis strain AS3 was representative of the enriched cells. The cells of C. evryensis strain AS3 were short rods and often grown in duplococci (Figure 3B). Interestingly C. evryensis strain AS3 was not able to grow in glucose but could grow on a mixture of amino acids including lysine, histidine, serine, and arginine producing propionate and acetate as major end-products. As C. evryensis strain AS3 was able to ferment lysine, histidine, serine, and arginine like other Cloacibacillus spp., a CML degradation test was performed in two conditions with and without these amino acids under anaerobic conditions (Figure 3A). Without lysine, histidine, serine, and arginine, C. evryensis strain AS3 was able to slowly degrade CML, but in the presence of the amino-acid mixture, CML degradation was somewhat faster, with acetate and propionate as main end-products under both conditions. This result indicated that the pure culture of C. evryensis AS3 was able to degrade CML anaerobically. The slow CML degradation might be explained by a number of factors, including lack of growth factors, potential syntrophic partners, or the formation of toxic CML degradation compounds that inhibit the growth/activity of the bacteria. Of note, we tested the growth of C. evryensis strain AS3 in several rich media , but it never grew to a high OD600 of over 1 even in an optimized medium.30 A recent study on CML degradation showed that E. coli was able to degrade only a limited part of an initial low concentration (250 μM) of CML at aerobic conditions while producing three different metabolites.20 In our study, much higher CML concentrations were used, suggesting the possibility that C. evryensis strain AS3 may also degrade initial low concentrations of CML.
Figure 3.
CML degradation by strain AS3 (A) and morphology of strain AS3 (B). AA_CML indicates a condition in which an amino-acid mixture was added along with CML, while the other only contained CML as the sole carbon and energy source.
Identification of Potential Markers of CML Degradation Products
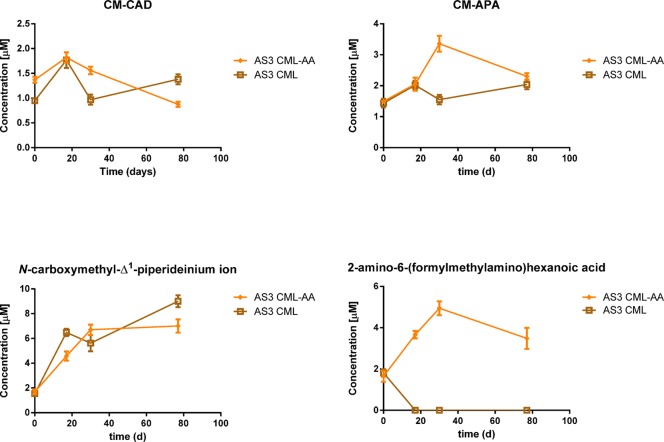
The formation of CML degradation products was investigated by zwitterionic HILIC and HRMS in bacterial cultures showing CML degradation capacity. Chromatographic conditions were optimized by means of acetonitrile and water in an acidic environment (0.1% formic acid) in order to improve ionization in positive ion mode as well as retention and separation of metabolites from their precursor CML. Several compounds were considered as potential candidates of CML degradation and included in the in-house database according to KEEG pathway. Full-scan analysis in high-resolution polarity switching mode (m/z range 50–350, resolving power set to 50 000 full width at half-maximum (fwhm), m/z 200) was considered as the optimal strategy to combine the chemical nature of the potential candidates to their behavior in HILIC chromatography.32 A total of 4 out of 25 candidates were tentatively identified as potential markers of CML metabolism and included N-carboxymethylcadaverine (CM-CAD), N-carboxymethylaminopentanoic acid (CM-APA), 2-amino-6-(formylmethylamino)hexanoic acid, and the N-carboxymethyl-Δ1-piperideinium ion. The analytical characteristics of these indicator compounds were determined (Table 3). The use of HILIC-HRMS allowed the separation of CML metabolites from their precursor, based on the typical retention time and partition coefficients: three out of four compounds exhibited a logP below 0, resulting in a capacity factor higher than 2 in the chromatographic conditions tested.
Table 3. Zwitterionic HILIC High-Resolution Mass Spectrometry (HRMS) Analysisa.
| compound name | RT | EC | m/z T | m/z E | Δ ppm |
|---|---|---|---|---|---|
| CM-CAD | 2.8 | C7H16N2O2 | 161.12845 | 161.12822 | –1.4 |
| CM-APA | 3.1 | C7H13NO4 | 176.09173 | 176.09201 | 1.6 |
| 2-amino-6-(formylmethylamino)hexanoic acid | 3.5 | C8H16N2O3 | 189.09955 | 189.09899 | –3.0 |
| N-carboxymethyl-Δ1-piperideinium ion | 3.6 | C7H12NO2+ | 142.08626 | 142.08599 | –1.9 |
Error (Δ ppm) was calculated as the ratio between the difference of the theoretical mass minus the experimental mass and the theoretical mass, multiplied per 1 000 000. Theoretical mass (m/zT), experimental mass (m/zE), retention time (RT, min), elemental composition (EC). N-Carboxymethylcadaverine (CM-CAD), N-carboxymethylaminopentanoic acid (CM-APA).
To test the usefulness of these four indicator compounds, we analyzed their presence during the degradation by the C. evryensis strain AS3 of CML (Figure 4). Data indicated an overall accumulation trend for the four metabolites during the time course of incubation with AS3 strains, in particular up to the first 5 weeks. The sum of the four metabolites explained only 1.5% of the CML degradation, suggesting that other metabolites not included in target HRMS analysis experiments could be formed as well as a dose-dependent relationship between metabolites and the initial concentration of CML. Besides the multitude of compounds arisen from lysine degradation mediated by Cloacibacillus metabolism, the presence of a carboxymethyl group on the epsilon side chain of lysine and its steric hindrance limited the number of potential candidates included in our in-house database. The concentration of CM-CAD remained constant during incubation in the presence of lysine, arginine, serine, and histidine, suggesting that long incubation may promote reaction of the primary amino group, leading to carboxymethylation of cadaverine, arising from lysine metabolism. However, when the microbial culture enriched in C. evryensis strain AS3 was incubated in the presence of CML, the concentration of CM-CAD increased up to 86% during the first 17 days toward initial concentration. Similar trends were observed for CM-APA: in the case of CML incubation with another amino-acid source, the formation of the metabolite was characterized by an increase up to 3.36 ± 0.26 μM after 30 days, and then, it constantly decreased at the end of the incubation. In the case of incubation with CML alone, the formation of CM-APA was characterized by a decrease after the first 30 days, suggesting that the presence of another amino-acid source had a key role for the yield of CM-APA. Formation of N-carboxymethyl-Δ1-piperideinium ion was substrate specific: only CML degradation led to its formation, while the degradation of other amino acids did not have an influence on the formation of this ion. Indeed N-carboxymethyl-Δ1-piperideinium ion formation exhibited a similar trend in the presence of cofactor, suggesting that its formation was linked only to the presence of CML as a carbon source able to generate piperidein ions. The 2-amino-6-(formylmethylamino)hexanoic acid is the only metabolite having a completely different trend in the presence of other amino acids with respect to CML alone. When other amino acids are present, its concentration sharply increased during the first month, while in the presence of CML alone, the concentration suddenly dropped during the first days. This figure suggests a potential interconversion in the catabolic pathways of different amino acids.
Figure 4.
Formation of CML degradation products during the anaerobic incubation of CML by C. evryensis strain AS3. CM-CAD (carboxymethylcadaverine); CM-APA (carboxymethylaminopentanoic acid). The label AA indicates the presence of another carbon source along with CML.
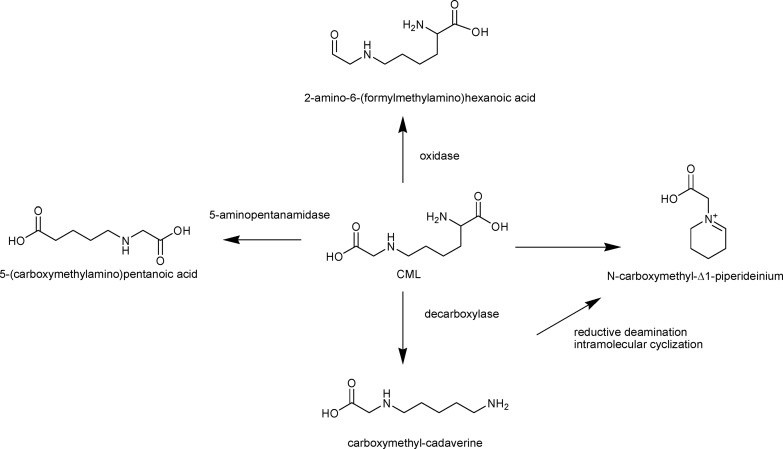
The lysine degradation pathway and anaerobic conditions were used as reference to tentatively explain CML metabolism. As depicted in Figure 5, decarboxylation of the alpha carboxylic group mediated by lysine decarboxylase is one of the possible routes that led to the formation of CM-CAD. The formation of N-carboxymethyl-Δ1-piperideinium ion could include CM-CAD as precursors: in this respect, a reductive deamination followed by an intramolecular cyclization of the epsilon amino group was the key step for piperidine derived compound formation. On the opposite, the formation of CM-APA required a preliminary amide formation followed by oxidation into the corresponding carboxylic acid. This mechanism can be catalyzed by 5-aminopentanamidase similarly to lysine metabolism. The formation of 2-amino-6-(formylmethylamino)hexanoic acid requires the presence of a carboxylic acid oxidase for the formation of the aldehyde. Once aldehyde is formed, there is a possibility of a reduction of the aldehyde into the corresponding alcohol through the Ehrlich pathway.33
Figure 5.
Hypothesized pathways for CML degradation according to KEGG and lysine metabolism of AS3 strains.
Post-translational modifications of amino acids and the consequent formation of Amadori compounds, Heyns compounds, and d-AGEs contribute to the variety of compounds metabolized by the intestinal microbiota.34 The identification of markers of d-AGE metabolism is as intricate as the Maillard cascade, and multiple factors need to be considered before depicting the appropriate metabolic pathway. Even if aerobic conditions favored a faster CML degradation by E. coli strains, anaerobic environment is not a limitation, and similar compounds can be obtained as end-products (or intermediate) during incubation with fecal microbiota. For the four compounds identified, the presence of other amino acids along with CML was not determinant, and no significant difference in the yields were obtained except for 2-amino-6-(formylmethylamino) hexanoic acid and CM-APA after 5 weeks. In agreement with previous results, the appropriate substrate can be relevant to determine the optimal conditions for CML metabolism;20 in the present paper, we demonstrated that successive transfer of fecal microbiota are able to generate CML metabolites following similar pathways of lysine degradation. As lysine is a preferred substrate of colonic bacteria, and different pathways are responsible of its catabolism, when hypothesizing parallel pathways between lysine and CML, multiple conditions need to be taken into account, such as time of incubation, pH changes, and presence of cofactors.35 Finally, our results in anaerobic conditions are in line with aerobic incubation of CML in the presence of E. coli strains; the concentration of CM-CAD was close to 10 μM (around 4% of 250 μM of CML) detected by Hellwig and co-workers, while CM-APA was 1 order of magnitude higher. meaning that its formation prevailed in anaerobic conditions, while a key role can be attributed to N-carboxymethyl-Δ1-piperideinium ion in anaerobic conditions, as its concentrations throughout the incubation were 3 and 4 times higher than those of CM-APA and CM-CAD, respectively.20
In conclusion, our results indicated that intestinal bacteria from adults are able to degrade CML under anaerobic conditions. This study not only provides a better understanding of CML degradation by gut bacteria but also enables further studies on identifying enzymes that are responsible for this degradation and factors that might have an influence on the degradation. Even if studies on larger population are required, these results paved the way to introduce a closer view on the relationship between diet and CML metabolic fate, revealing that also human microbiota is able to generate metabolites with a potential impact on pathophysiological outcomes connected to d-AGEs.
Acknowledgments
The authors acknowledge Geert Meijer for the technical assistance in CML analysis. This work was partially supported by The SIAM Gravitation Grant 024.002.002 and Spinoza Award of The Netherlands Organization for Scientific Research to W.M.d.V.
Glossary
Abbreviations Used
- dAGEs
dietary advanced glycation end-products
- CML
N-ε-carboxymethyllysine
- d-CML
dietary N-ε-carboxymethyllysine
- CM-CAD
N-carboxymethylcadaverine
- CM-APA
N-carboxymethylaminopentanoic acid
- HILIC
hydrophilic interaction liquid chromatography
- LC–MS/MS
liquid chromatography tandem mass spectrometry
- HRMS
high-resolution mass spectrometry
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jafc.9b02208.
Phylogenetic tree based on 16S rRNA sequences obtained from the second transfer of CML enrichment from AS1 and closely related species (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Delgado-Andrade C.; Fogliano V. Dietary Advanced Glycosylation End-Products (DAGEs) and Melanoidins Formed through the Maillard Reaction: Physiological Consequences of Their Intake. Annu. Rev. Food Sci. Technol. 2018, 9, 271–291. 10.1146/annurev-food-030117-012441. [DOI] [PubMed] [Google Scholar]
- Mossine V. V.; Mawhinney T. P. 1-Amino-1-Deoxy-d-Fructose (“Fructosamine”) and Its Derivatives. In Adv. Carb. Chem. Biochem. 2010, 64, 291–402. 10.1016/S0065-2318(10)64006-1. [DOI] [PubMed] [Google Scholar]
- Hellwig M.; Henle T. Baking Ageing, Diabetes: A Short History of the Maillard Reaction. Angew. Chem., Int. Ed. 2014, 53 (39), 10316–10329. 10.1002/anie.201308808. [DOI] [PubMed] [Google Scholar]
- Scheijen J. L. J. M.; Clevers E.; Engelen L.; Dagnelie P. C.; Brouns F.; Stehouwer C. D. A.; Schalkwijk C. G. Analysis of Advanced Glycation Endproducts in Selected Food Items by Ultra-Performance Liquid Chromatography Tandem Mass Spectrometry: Presentation of a Dietary AGE Database. Food Chem. 2016, 190, 1145–1150. 10.1016/j.foodchem.2015.06.049. [DOI] [PubMed] [Google Scholar]
- Pischetsrieder M.; Henle T. Glycation Products in Infant Formulas: Chemical, Analytical and Physiological Aspects. Amino Acids 2012, 42 (4), 1111–1118. 10.1007/s00726-010-0775-0. [DOI] [PubMed] [Google Scholar]
- Erbersdobler H. F.; Somoza V. Forty Years of Furosine - Forty Years of Using Maillard Reaction Products as Indicators of the Nutritional Quality of Foods. Mol. Nutr. Food Res. 2007, 51 (4), 423–430. 10.1002/mnfr.200600154. [DOI] [PubMed] [Google Scholar]
- Tuohy K. M.; Hinton D. J. S.; Davies S. J.; Crabbe M. J. C.; Gibson G. R.; Ames J. M. Metabolism of Maillard Reaction Products by the Human Gut Microbiota - Implications for Health. Mol. Nutr. Food Res. 2006, 50 (9), 847–857. 10.1002/mnfr.200500126. [DOI] [PubMed] [Google Scholar]
- Erbersdobler H. F.; Faist V. Metabolic Transit of Amadori Products. Nahrung 2001, 45 (3), 177–181. . [DOI] [PubMed] [Google Scholar]
- Carmody R. N.; Weintraub G. S.; Wrangham R. W. Energetic Consequences of Thermal and Nonthermal Food Processing. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (48), 19199–19203. 10.1073/pnas.1112128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassara H.; Striker G. E. AGE Restriction in Diabetes Mellitus: A Paradigm Shift. Nat. Rev. Endocrinol. 2011, 7 (9), 526. 10.1038/nrendo.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portune K. J.; Beaumont M.; Davila A. M.; Tomé D.; Blachier F.; Sanz Y. Gut Microbiota Role in Dietary Protein Metabolism and Health-Related Outcomes: The Two Sides of the Coin. Trends Food Sci. Technol. 2016, 57, 213–232. 10.1016/j.tifs.2016.08.011. [DOI] [Google Scholar]
- Hellwig M.; Bunzel D.; Huch M.; Franz C. M. A. P.; Kulling S. E.; Henle T. Stability of Individual Maillard Reaction Products in the Presence of the Human Colonic Microbiota. J. Agric. Food Chem. 2015, 63 (30), 6723–6730. 10.1021/acs.jafc.5b01391. [DOI] [PubMed] [Google Scholar]
- Bui T. P. N.; Ritari J.; Boeren S.; De Waard P.; Plugge C. M.; De Vos W. M. Production of Butyrate from Lysine and the Amadori Product Fructoselysine by a Human Gut Commensal. Nat. Commun. 2015, 6, 10062. 10.1038/ncomms10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui T. P. N.; Shetty S. A.; Lagkouvardos I.; Ritari J.; Chamlagain B.; Douillard F. P.; Paulin L.; Piironen V.; Clavel T.; Plugge C. M.; et al. Comparative Genomics and Physiology of the Butyrate-Producing Bacterium Intestinimonas Butyriciproducens. Environ. Microbiol. Rep. 2016, 8 (6), 1024–1037. 10.1111/1758-2229.12483. [DOI] [PubMed] [Google Scholar]
- Koschinsky T.; He C. J.; Mitsuhashi T.; Bucala R.; Liu C.; Buenting C.; Heitmann K.; Vlassara H. Orally Absorbed Reactive Glycation Products (Glycotoxins): An Environmental Risk Factor in Diabetic Nephropathy. Proc. Natl. Acad. Sci. U. S. A. 1997, 94 (12), 6474–6479. 10.1073/pnas.94.12.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig M.; Matthes R.; Peto A.; Löbner J.; Henle T. N-ε-Fructosyllysine and N-ε-Carboxymethyllysine, but Not Lysinoalanine, Are Available for Absorption after Simulated Gastrointestinal Digestion. Amino Acids 2014, 46 (2), 289–299. 10.1007/s00726-013-1501-5. [DOI] [PubMed] [Google Scholar]
- Tessier F. J.; Niquet-Léridon C.; Jacolot P.; Jouquand C.; Genin M.; Schmidt A. M.; Grossin N.; Boulanger E. Quantitative Assessment of Organ Distribution of Dietary Protein-Bound 13C-Labeled Nε-Carboxymethyllysine after a Chronic Oral Exposure in Mice. Mol. Nutr. Food Res. 2016, 60 (11), 2446–2456. 10.1002/mnfr.201600140. [DOI] [PubMed] [Google Scholar]
- Buetler T.; Henle T.; Vlassara H. The Effects of AGEing on Diet. Am. J. Pathol. 2009, 174 (1), 351–353. 10.2353/ajpath.2009.080805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiquer I.; Rubio L. A.; Peinado M. J.; Delgado-Andrade C.; Navarro M. P. Maillard Reaction Products Modulate Gut Microbiota Composition in Adolescents. Mol. Nutr. Food Res. 2014, 58 (7), 1552–1560. 10.1002/mnfr.201300847. [DOI] [PubMed] [Google Scholar]
- Hellwig M.; Auerbach C.; Müller N.; Samuel P.; Kammann S.; Beer F.; Gunzer F.; Henle T. Metabolization of the Advanced Glycation End Product N-ϵ-Carboxymethyllysine (CML) by Different Probiotic E. Coli Strains. J. Agric. Food Chem. 2019, 67 (7), 1963–1972. 10.1021/acs.jafc.8b06748. [DOI] [PubMed] [Google Scholar]
- O’Keefe S. J. D.; Li J. V.; Lahti L.; Ou J.; Carbonero F.; Mohammed K.; Posma J. M.; Kinross J.; Wahl E.; Ruder E.; et al. Fat, Fibre and Cancer Risk in African Americans and Rural Africans. Nat. Commun. 2015, 6, 6342. 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stams A. J. M.; Van Dijk J. B.; Dijkema C.; Plugge C. M. Growth of Syntrophic Propionate-Oxidizing Bacteria with Fumarate in the Absence of Methanogenic Bacteria. Appl. Environ. Microbiol. 1993, 59 (4), 1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelder A. H.; Aydin R.; Alves M. M.; Stams A. J. M. 1,3-Propanediol Production From Glycerol by a Newly Isolated Trichococcus strain. Microb. Biotechnol. 2012, 5 (4), 573–578. 10.1111/j.1751-7915.2011.00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajad S. U.; Lu W.; Kimball E. H.; Yuan J.; Peterson C.; Rabinowitz J. D. Separation and Quantitation of Water Soluble Cellular Metabolites by Hydrophilic Interaction Chromatography-tandem Mass Spectrometry. J. Chromatogr. A 2006, 1125 (1), 76–88. 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Troise A. D.; Fiore A.; Wiltafsky M.; Fogliano V. Quantification of Nε-(2-Furoylmethyl)-L-Lysine (Furosine), Nε-(Carboxymethyl)-L-Lysine (CML), Nε-(Carboxyethyl)-L-Lysine (CEL) and Total Lysine through Stable Isotope Dilution Assay and Tandem Mass Spectrometry. Food Chem. 2015, 188, 357–364. 10.1016/j.foodchem.2015.04.137. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G.; Barns S. M.; Pelletier D. A.; Lane D. J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173 (2), 697–703. 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C.; Pruesse E.; Yilmaz P.; Gerken J.; Schweer T.; Yarza P.; Peplies J.; Glöckner F. O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41 (D1), D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie R. I.; Aminov R. I.; Hu W.; Klieve A. V.; Ouwerkerk D.; Sundset M. A.; Kamagata Y. Ecology of Uncultivated Oscillospira Species in the Rumen of Cattle, Sheep, and Reindeer as Assessed by Microscopy and Molecular Approaches. Appl. Environ. Microbiol. 2003, 69 (11), 6808–6815. 10.1128/AEM.69.11.6808-6815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.; Kim J.; Hatzenpichler R.; Karymov M. A.; Hubert N.; Hanan I. M.; Chang E. B.; Ismagilov R. F. Gene-Targeted Microfluidic Cultivation Validated by Isolation of a Gut Bacterium Listed in Human Microbiome Project’s Most Wanted Taxa. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (27), 9768–9773. 10.1073/pnas.1404753111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan A.; Chaussonnerie S.; Tarrade A.; Dauga C.; Bouchez T.; Pelletier E.; Le Paslier D.; Sghir A. Cloacibacillus Evryensis Gen. Nov., Sp. Nov., a Novel Asaccharolytic, Mesophilic, Amino-Acid-Degrading Bacterium within the Phylum “Synergistetes”, Isolated from an Anaerobic Sludge Digester. Int. J. Syst. Evol. Microbiol. 2008, 58 (9), 2003–2012. 10.1099/ijs.0.65645-0. [DOI] [PubMed] [Google Scholar]
- Marchandin H.; Damay A.; Roudière L.; Teyssier C.; Zorgniotti I.; Dechaud H.; Jean-Pierre H.; Jumas-Bilak E. Phylogeny, Diversity and Host Specialization in the Phylum Synergistetes with Emphasis on Strains and Clones of Human Origin. Res. Microbiol. 2010, 161 (2), 91–100. 10.1016/j.resmic.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Creek D. J.; Jankevics A.; Breitling R.; Watson D. G.; Barrett M. P.; Burgess K. E. V. Toward Global Metabolomics Analysis with Hydrophilic Interaction Liquid Chromatography-Mass Spectrometry: Improved Metabolite Identification by Retention Time Prediction. Anal. Chem. 2011, 83 (22), 8703–8710. 10.1021/ac2021823. [DOI] [PubMed] [Google Scholar]
- Hellwig M.; Börner M.; Beer F.; van Pée K. H.; Henle T. Transformation of Free and Dipeptide-Bound Glycated Amino Acids by Two Strains of Saccharomyces Cerevisiae. ChemBioChem 2017, 18 (3), 266–275. 10.1002/cbic.201600486. [DOI] [PubMed] [Google Scholar]
- Han K.; Jin W.; Mao Z.; Dong S.; Zhang Q.; Yang Y.; Chen B.; Wu H.; Zeng M. Microbiome and Butyrate Production Are Altered in the Gut of Rats Fed a Glycated Fish Protein Diet. J. Funct. Foods 2018, 47, 423–433. 10.1016/j.jff.2018.06.007. [DOI] [Google Scholar]
- Neis E. P. J. G.; Dejong C. H. C.; Rensen S. S. The Role of Microbial Amino Acid Metabolism in Host Metabolism. Nutrients 2015, 7, 2930–2946. 10.3390/nu7042930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.