Abstract
Leaf blade width, curvature, and cuticular wax are important agronomic traits of rice. Here, we report the rice Oschr4-5 mutant characterized by pleiotropic phenotypes, including narrow and rolled leaves, enhanced cuticular wax deposition and reduced plant height and tiller number. The reduced leaf width is caused by a reduced number of longitudinal veins and increased auxin content. The cuticular wax content was significantly higher in the Oschr4-5 mutant, resulting in reduced water loss rate and enhanced drought tolerance. Molecular characterization reveals that a single-base deletion results in a frame-shift mutation from the second chromodomain of OsCHR4, a CHD3 (chromodomain helicase DNA-binding) family chromatin remodeler, in the Oschr4-5 mutant. Expressions of seven wax biosynthesis genes (GL1-4, WSL4, OsCER7, LACS2, LACS7, ROC4 and BDG) and four auxin biosynthesis genes (YUC2, YUC3, YUC5 and YUC6) was up-regulated in the Oschr4-5 mutant. Chromatin immunoprecipitation assays revealed that the transcriptionally active histone modification H3K4me3 was increased, whereas the repressive H3K27me3 was reduced in the upregulated genes in the Oschr4-5 mutant. Therefore, OsCHR4 regulates leaf morphogenesis and cuticle wax formation by epigenetic modulation of auxin and wax biosynthetic genes expression.
Keywords: auxin, chromatin remodeling factor, cuticular wax, drought tolerance, epigenetic regulation, leaf width, histone modification, narrow leaf, OsCHR4, rice
1. Introduction
In plants, leaves are the major organ used for photosynthesis and transpiration, and are considered to be the main source of dry matter accumulation in plants. Blade width and curvature are key factors of leaf morphology. As reported, blade width, particularly that of the first and second uppermost leaves, can significantly influence the rice grain yield, whereas appropriately rolled leaves can optimize canopy light transmission, increase the effective leaf area per unit of land and, consequently, the canopy photosynthesis efficiency [1].
To date, a number of narrow and/or rolled leaf genes have been reported in rice. The majority of narrow leaf genes are related to auxin signaling. For example, a mutant of Nal1 (encoding a plant-specific protein with unknown biochemical function) exhibited a characteristic phenotype of narrow leaves due to decreased number of longitudinal veins and polar auxin transport capacity [2,3]. Mutation in TDD1 (encoding a protein homologous to anthranilate synthase beta-subunit involved in the upstream of Trp-dependent auxin biosynthesis) could induce obviously narrow leaves [4]. NAL7 and its allelic gene CONSTITUTIVELY WILTED1 encode an auxin biosynthesis enzyme OsYUCCA8 and their mutants exhibit narrow leaves [5,6]. Similar to the auxin-deficient mutant, the leaves of the osarf11-1, a loss-of-function mutant of the auxin-response factor 11 (OsARF11), are also moderately narrowed [7]. Mutations in genes involved in other pathways could also produce narrow leaves such as NAL9 (encoding an ATP-dependent Clp protease proteolytic subunit) and NAL11 (encoding a heat shock DNAJ protein) [8,9]. For rolled leave, it could be divided into adaxialized and abaxialized curling. The mutant of SLL1/RL9 (encoding a SHAQKYF class MYB family transcription factor) displayed completely adaxialized leaves [10]. This gene mainly functions in the modulation of sclerenchymatous cells formation on the abaxial side during leaf development [11]. The mutant of ADL1 (a plant-specific calpain-like cysteine proteinase) exhibited abaxially rolled leaves due to abnormal bulliform-like cells distribution [12]. Meanwhile, overexpression of ACL1 (encodes a protein with unknown functional domains) and its homolog ACL2 caused abaxial leaf curling [13], while overexpression of OsAGO7 was correlated with adaxial leaf curling [14]. SRL1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates leaf adaxial rolling by regulating the formation of bulliform cells [15]. Interestingly, several genes were associated with both narrow and rolled leaves. The mutants of NRL1 (encoding cellulose synthase-like protein D4), NRL2 and NRL3 (both encoding a novel protein with unknown biochemical function) could induce semi-rolled leaves with reduced blade width [16,17,18,19].
The surface of rice leaf is covered by cuticular wax which provides self-protection against potential external stresses, such as non-stomatal water loss, pathogen infection and ultraviolet (UV) radiation [20]. The main composition of cuticular wax is the very-long-chain fatty acids (VLCFAs) as well as their following derivatives: aldehydes, primary and secondary alcohols, alkanes, ketones and wax esters. Through years of study, many genes encoding enzymes and transcription factors involved in biosynthesis of cuticular wax in rice have been uncovered by bioinformatics and genetic approaches. The Glossy1 (GL1) gene family, reported first in maize, encodes the fatty aldehyde decarbonylase required for the synthesis of cuticular wax. Through homologous sequence alignment, 11 OsGL1 family genes were found in rice [21]. Subsequent studies validated that OsGL1-1/WSL2, OsGL1-2, OsGL1-3, OsGL1-5/Wda1 and OsGL1-6 affect wax synthesis on various parts of rice plant [21,22,23,24,25]. AP2/ERF family transcription factors OsWR1 and OsWR2, homologs of the Arabidopsis WIN1/SHN1 gene, promote wax production via activating wax biosynthesis gene [26,27]. Homeodomain-leucine zipper class IV family transcription factor ROC4 positively regulates cuticular wax biosynthesis via directly binding to the conserved L1 box cis-element in the promoters of Os-BDG, which encodes an extracellular synthase responsible for the formation of cuticle [28]. DROUGHT HYPERSENSITIVE (DHS), a RING-type E3 ligase, interacts with ROC4 and promotes its degradation via the ubiquitin/26S proteasome [28]. After synthesized in the endoplasmic reticulum, wax is transferred to the plant surface via adenosine triphosphate binding cassette transporters as well as lipid transfer proteins [29].
Proper regulation of gene expression during development is dependent on the dynamic modifications of chromatin, during which the accessibility of specific DNA regions to the transcription machinery may be altered. The chromodomain helicase DNA-binding (CHD) proteins are one family of chromatin remodeling factors, which are defined by a plant homeodomain (PHD) finger, two chromodomains, one SNF2 (sucrose non-fermenting)-related ATPase/helicase domain, and one DNA binding motif [30,31]. CHD chromatin remodelers directly change the chromatin structures and are important regulators of gene expression. Mutations of CHD protein could always induce pleiotropic phenotypes in plants. PICKLE is a typical CHD protein in Arabidopsis. The pickle mutant was initially characterized by pickle roots with green tuber due to failed repression of seed-specific genes [32]. This protein was then found to function in repression of ectopic stipules and meristems in leaf tissue and inhibition of meristematic genes in carpel tissue [33,34]. In rice, the closest homolog with PICKLE is OsCHR702, while both T-DNA insertion mutation and RNAi of OsCHR702 did not produce any morphological phenotypes [35,36]. OsCHR4 is another CHD member characterized in rice. The Oschr4 mutant shows defective chloroplasts in the adaxial mesophyll cells due to blocked proplastid growth and thylakoid membrane formation [37]. As an allelic gene of OsCHR4, OsCHR729 was revealed to encode a bifunctional chromatin regulator associated with gene transcription regulation through modulation of histone H3K4 and H3K27 methylation during plant development [36]. Meanwhile, the Oschr729 mutant induced by T-DNA insertion shows pleiotropic phenotypes, including small and rolled leaves, reduced stem elongation and chlorophyll contents, and absence of secondary panicle branches [36]. One study reported that CHR729 regulates seedling development by affecting contents of gibberellin acid (GA) in rice, and the transcript level of GA biosynthesis genes was altered in the corresponding mutant line t483 [38]. In addition, CRL6, an allelic gene of CHR729, was speculated to regulate crown root development via auxin signaling pathway due to decreased expression of most OsIAA genes in crl6 compared with the wild type (WT) [39]. Recently, rfs, another allelic mutant of OsCHR4/OsCHR729, was characterized by the chlorotic and cell death phenotypes due to accumulation of reactive oxygen species (ROS). Both transcript and H3K4me3 levels of five ROS-related genes declined in the rfs mutant [40].
In this study, three additional allelic mutants of OsCHR4 were reported, namely Oschr4-5, Oschr4-6, and Oschr4-7, which exhibited pleiotropic phenotypes such as narrow and rolled leaf, reduced plant height, tiller number and seed size at mature stage. Anatomical investigations revealed that the reduced leaf width was due to a decreased number of major and minor veins while the cell size remained unchanged. The auxin content was significantly higher in Oschr4-5, which may explain the altered venation pattern and leaf size at physiological level. Interestingly, more cuticular wax was deposited on the surface of the Oschr4-5 leaves, which reduced the water-loss rate and enhanced the drought tolerance. Expression levels of several auxin and wax biosynthesis genes were up-regulated in the Oschr4-5 mutant. Consistent with the role of CHD3 protein in chromatin remodeling, histone modifications H3K4me3 and H3K27me3 in the upregulated genes were increased and reduced in Oschr4-5, respectively. Our data reveal novel functions of OsCHR4 in the epigenetic regulation of auxin and wax biosynthesis-related genes, leaf development and cuticle wax formation in rice.
2. Results
2.1. Pleiotropic Phenotype of the Oschr4-5 Mutant
A rice mutant characterized by narrow and rolled leaves was isolated from the gamma ray mutagenized Indica rice cultivar Indica 9, which was named as Oschr4-5 based on the gene identity revealed in the following study. At the seedling stage, slight dwarfism and albino on the adaxial side of leaves were observed in Oschr4-5 (Figure 1A,B). At the mature stage, the plant height and tiller number of Oschr4-5 was significantly reduced by 45% and 76% when compared with that of WT (p < 0.01) (Figure 1C, Table S1). The length and width of the leaves in Oschr4-5 was significantly reduced by 38% and 50% when compared with that of the WT (p < 0.01) (Figure 1D,F, Table S1). Meanwhile, smaller seeds were also observed in Oschr4-5 (Figure 1G). Later on, two allelic mutants, namely Oschr4-6 andOschr4-7, were isolated from the Ethyl methane sulfonate (EMS) mutagenized japonica cultivar Nipponbare, which exhibited the same pleiotropic phenotypes, including narrow and rolled leaves with albino on the adaxial side, significantly reduced leaf length, plant height, tiller number and seed size (Figure S1, Table S1).
Figure 1.
Phenotypic characterization of the Oschr4-5 mutant. (A) Phenotypes of wild type (WT) and Oschr4-5 mutant plants at seedling stage. Bar = 1 cm. (B) Adaxial side of leaf blade from wild type and Oschr4-5 mutant at seedling stage. Bar = 0.5 cm. (C) Phenotypes of wild type and Oschr4-5 mutant plants at mature stage. Bar = 15 cm. (D) Comparison of leaf length between wild type and Oschr4-5 mutant at mature stage. Bar = 5 cm. (E) Cross sections through mature leaves from wild type and Oschr4-5 mutant. Bar = 0.25 cm. (F) Comparison of leaf width between wild type and Oschr4-5 mutant at mature stage. Bar = 0.5 cm. (G) Phenotypes of seeds from wild type and Oschr4-5 mutant. Bar = 1 cm.
2.2. Anatomical and Physiological Basis of Reduced Leaf Width
To reveal the underlying cellular basis of narrow leaves in the Oschr4-5 mutant, the venation pattern of leaf was first observed. The number of large veins and small veins of Oschr4-5 was significantly reduced by 36% and 42%, respectively, of that of the WT (p < 0.01) (Figure 2A–D,G–H). Then the epidermal cell profile on the abaxial side of leaf blade was examined. Neither the number of cell column between the adjacent small vein nor the cell width at the horizontal orientation showed significant difference between Oschr4-5 and WT (Figure 2E,F,I,J). These results indicate that the decreased leaf width in Oschr4-5 may be caused by reduction in the number of veins. Since auxin is required for specification of the venation pattern in leaves [3,41] and auxin overproduction usually leads to narrow and epinastic leaves in Arabidopsis [42,43], the amount of free indole acetic acid (IAA), the major auxin, in Oschr4-5 and WT was measured. As a result, the content of IAA in Oschr4-5 was significantly higher than that of WT (86.39 ± 4.55 vs. 24.39 ± 0.76 ng/g, p < 0.01) (Figure 2L).
Figure 2.
The Oschr4-5 mutant affects leaf width. (A–D) The morphology of leaf veins from wild type and Oschr4-5mutant. Red bars represent large veins. Bar = 0.2 cm (A,B), 0.2 mm (C,D). (E,F) The abaxial epidermal cells of leaf blade from wild type and Oschr4-5 mutant. Bar = 50 μm. The columns with stoma were indicated with black arrow. Sv: Small vein; Lv: Large vein. (G–K) Comparison of the large veins (G), small veins (H), cell columns (I), cell width (J) and stomatal density (K) on abaxial epidermis of leaf blade and indole acetic acid (IAA) contents between wild type and Oschr4-5 mutant (L). Data are the means ± standard errors, n = 15 (G–J), 5 (K), 3 (L). Significance of data is tested by student’s t test. ** p < 0.01.
2.3. Increased Cuticular Wax on the Surface of Leaves with Enhanced Drought Tolerance in Oschr4-5
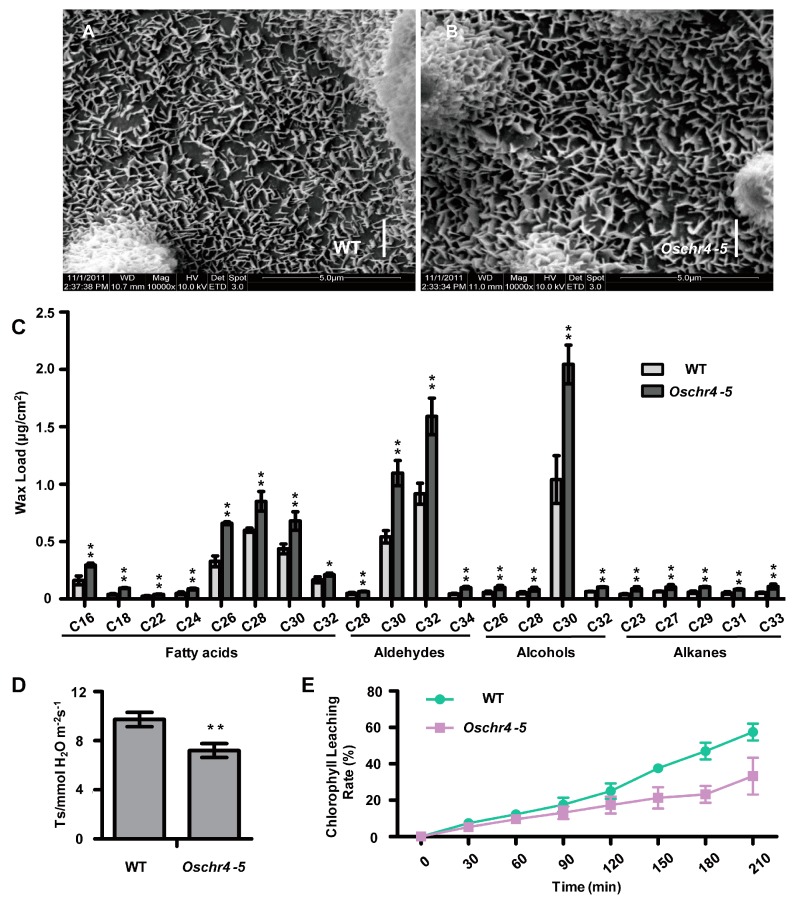
Scanning electron microscopy was used to examine the surface of leaves. Interestingly, the platelet-like wax crystals deposited on the leaves’ surface were larger and denser in Oschr4-5 compared with WT (Figure 3A,B). Then the compositions and contents of cuticular wax were measured by gas chromatography-mass spectrometry (GC-MS). As shown in Figure 3C, the wax components of fatty acids, aldehydes, alcohols and alkanes were all significantly increased in Oschr4-5 than WT (p < 0.05).
Figure 3.
More cuticular wax deposited on leaves of the Oschr4-5 mutant. (A–C) Scanning electron microscope (SEM) images of adaxially leaf surface (A,B) and cuticular wax composition and loads (C) from wild type and the Oschr4-5 mutant. Bar = 2 μm. Data are the means ± standard errors (n = 3). Significance of data is tested by student’s t test. * p < 0.05, ** p < 0.01. (D,E) The transpiration rate (D) and chlorophyll leaching rate (E) of leaf blades from wild type and Oschr4-5 mutant. Data are the means ± standard errors (n = 3). Significance of data is tested by student’s t test. ** p < 0.01.
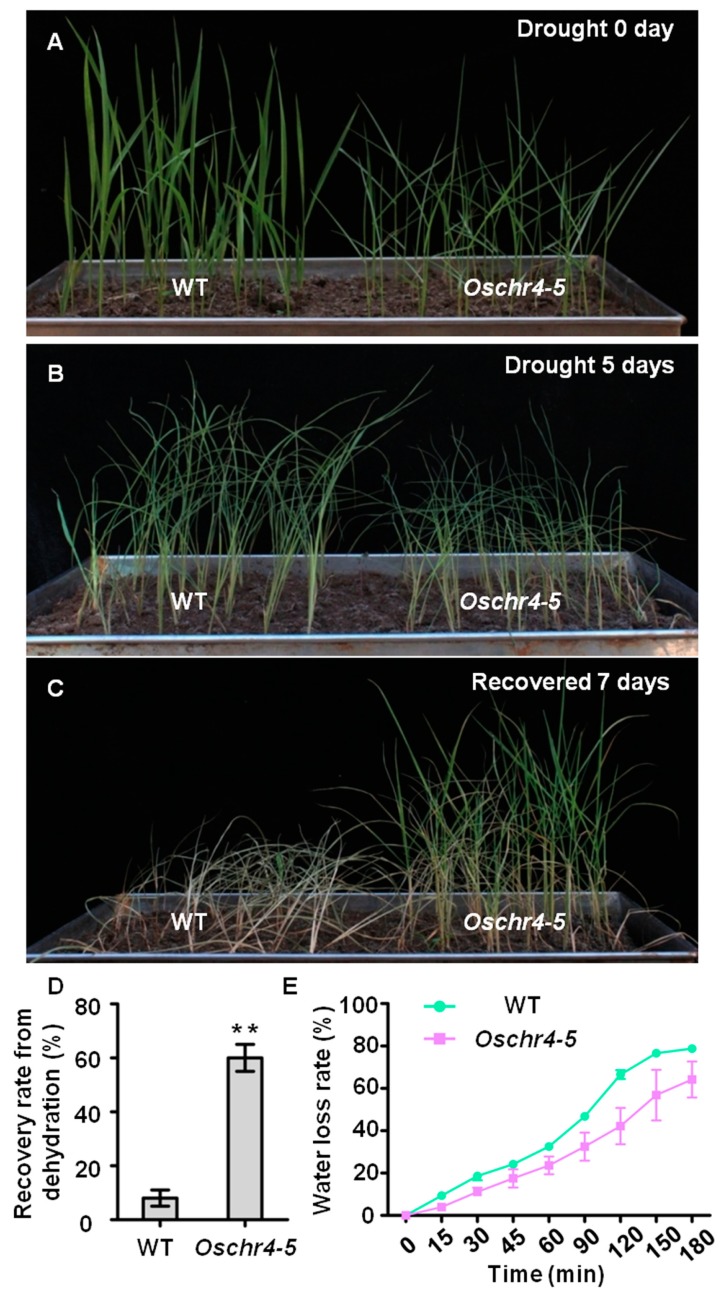
As the content and structure of cuticular wax could strongly influence cuticle permeability, its effects on transpiration rate were first evaluated. Indeed, the transpiration rate of Oschr4-5 was significantly lower than that of WT (p < 0.01) (Figure 3D). Secondly, measurement of cuticle membrane permeability as demonstrated by chlorophyll-leaching rate showed that chlorophyll leaches slower from Oschr4-5 than WT at different time points (Figure 3E). Furthermore, a dehydration-rehydartion treatment revealed that drought tolerance of Oschr4-5 was significantly enhanced. The Oschr4-5 seedlings showed significantly higher recovery rate than WT (60% vs. 8%, p < 0.01) (Figure 4A–D). The water loss rate was also measured to evaluate the water retention capacity of detached leaves. The data showed that water loss rates of Oschr4-5 were significantly slower than in WT (Figure 4E). The possible effects of stomatal density and leaf thickness on the differential water loss rate might be ruled out because no significant difference was detected in the stomatal density (Figure 2E,F,K) and leaf blade thickness (WT: 107.43 ± 7.51 μm; Oschr4-5: 108.17 ± 7.23 μm) between Oschr4-5 and WT. These results prove that Oschr4-5 exhibited lower cuticle permeability and water loss because of higher cuticular wax content on the surface of leaves compared to WT.
Figure 4.
The drought stress responses of the Oschr4-5 mutant. (A–C) Drought experiment with the 4 week-old plants for 0 day (A), 5 days (B), seedling recovered 7 days after rehydration (C). (D) Recovery rate from dehydration of wild type and Oschr4-5 mutant in (C). Data are the means ± standard errors (n = 3). Significance of data is tested by student’s t test, ** p < 0.01. (E) Water loss rate of detached leaf blades between wild type and the Oschr4-5 mutant. Data are the means ± standard errors (n = 3).
2.4. Characterization of the Molecular Lesions in the Oschr4 Mutant
To find out the molecular lesion responsible for the pleiotropic phenotypes of the Oschr4-5 mutant, three F2 populations were generated (Oschr4-5 × 9311; Oschr4-5 × Nipponbare; Oschr4-5 × 02428). Within these F2 populations, a mendelian segregation ratio of 3:1 (χ20.05 < 3.84) was calculated, which indicated the mutant phenotype was controlled by a single recessive gene. Then a large F2 population generated by Oschr4-5 × 02428 was used to map the causative locus. In primary mapping, 116 F2 mutant individuals located this gene between two Indel markers ha1 and ha10 on the long arm of chromosome 7. Fine mapping using 7 new Indel markers and 2000 F2 recessive individuals further localized this gene within 50 kb region delimited by Indel markers ha5 and ha6 (Figure 5A).
Figure 5.
Molecular characterization of mutation sites in the OsCHR4 gene. (A) Fine mapping of the gene on chromosome 7. The mutated gene was localized within 50 kb region delimited by two indel marker ha5 and ha6. (B) Predicted open reading frames (ORFs) within the fine mapping interval. Green, ORFs with known biochemical functions; Yellow, ORFs encoding expressed hypothetical proteins; Black, ORFs encoding transposons. (C) Gene structure of OsCHR4. The mutation sites in theOschr4-5, Oschr4-6 and Oschr4-7 mutant were indicated as black arrows. Blue boxes, exons; white boxes, UTR; black line, introns. (D) Protein structure of OsCHR4. Each domain is represented in different color box. Red, PHD domain; yellow, Chromo domains; green, SNF2 related helicase/ATPase domain; blue, Helicase C domain; purple, DNA-binding domain.
According to the MSU (Michigan State University) Rice Genome Annotation Release 7 (http://rice.plantbiology.msu.edu), a total of 5 predicted open reading frames (ORFs) were revealed within the 50 kb fine mapping interval, including 3 ORFs with known biochemical functions (ORF 2: phosphate-induced protein 1; ORF 4: OsCHR4/CHR729; ORF 5: peptide-Nasparagine amidase), 1 ORF encoding expressed hypothetical protein (ORF 3) and 1 transposon (ORF 1) (Figure 5B). Then genomic DNA of all these five ORFs in the Oschr4-5 mutant was sequenced and compared with that of WT. A single-base deletion at position 3688 was found in the ORF4 in Oschr4-5 (created through gamma ray radiation), whereas no mutation was found in other four ORFs. Sequence analysis of other two allelic mutants Oschr4-6 and Oschr4-7 created through EMS mutagenesis revealed a C4382T and G5190A substitution in ORF4, respectively (Figure 5C). Therefore, ORF4 encoding OsCHR4/CHR729 was found to be the target gene, which is quite large with 11 exons and 10 introns.
2.5. Alignment and Phylogenetic Analysis
OsCHR4 encodes a member of the CHD3 subfamily of chromatin remodeling factors [36,37]. OsCHR4 has 2259 amino acids in length with several putative domains, including one PHD domain, two chromodomains, one SNF2 related helicase/ATPase domain, one Helicase C and one DNA-binding domain from amino terminus to carboxyl terminus (Figure 5D). In the Oschr4-5 mutant, the single-base deletion results in a frame-shift mutation from the second chromodomain. In Oschr4-6, the amino acid substitution L830F (CTC to TTC) occurred within the SNF2 related helicase/ATPase domain. In Oschr4-7, the amino acid substitution R1099K (AGA to AAA) occurred within the Helicase C domain (Figure 5D).
Multiple sequence alignment of the OsCHR4 homologous proteins in eight species was performed. As shown in Figure S2A, the substitution mutation sites in Oschr4-6 and Oschr4-7 were highly conserved in these species, which highlights the important role of these residuals in CHR4. Then the evolutionary relationship among CHD families in rice and Arabidopsis was evaluated. As a result, CHR705 was revealed to belong to the member of subfamily I (CHD1), and the other CHR proteins belonged to subfamily II (CHD3). There were no subfamily III members found in rice or Arabidopsis. Meanwhile, PICKLE in Arabidopsis was found to be close to CHR702 in rice, and the closest gene with OsCHR4 in Arabidopsis was CHR4 (PKR1) (Figure S2B).
2.6. Expression of Several Wax and Auxin Biosynthesis Related Genes Were Upregulated in Oschr4-5
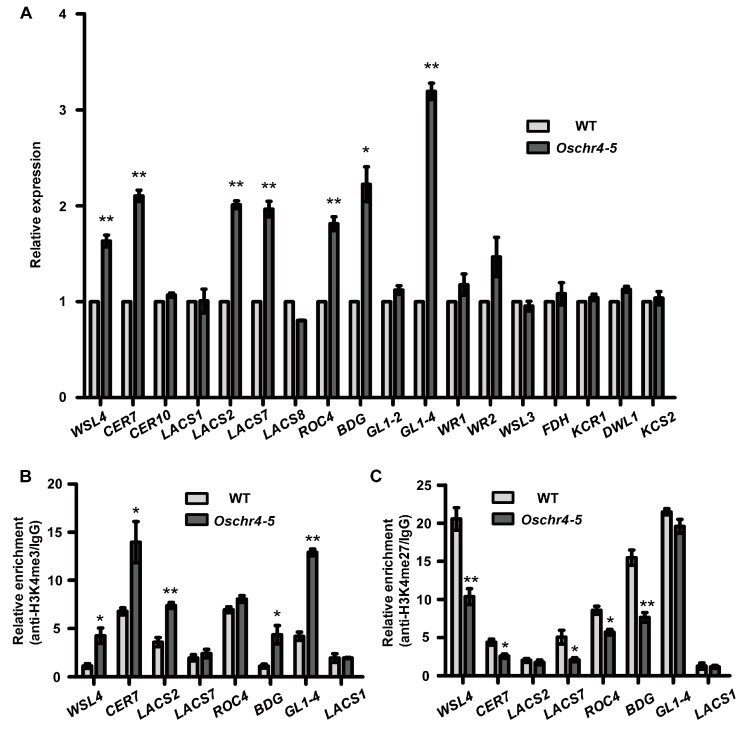
As a chromatin remodeling factor, the loss of OsCHR4 function was identified to influence the rice plant growth and development in various ways [36,37,38,39,40], but the underlying molecular mechanisms need to be further analyzed. To understand why the content of cuticular wax increased in the Oschr4-5 mutant, we examined the expression levels of 16 genes encoding enzymes involved in wax biosynthesis and three genes (ROC4, WR1, WR2) encoding transcription factors that activate the expression of wax biosynthetic genes [26,27,28,44,45]. Quantitative real-time polymerase chain reaction (RT-PCR) using RNA samples prepared from leaves of 30-day-old plants revealed that transcript levels of six wax biosynthetic genes (GL1-4, WSL4, OsCER7, LACS2, LACS7, BDG) and one transcription factor gene (ROC4) [26,27,28,44,45] were significantly upregulated in Oschr4-5, compared with WT (Figure 6A). This result suggests that loss of function of CHR4 affects the expression of important genes involved in the wax biosynthesis.
Figure 6.
OsCHR4 regulates expression of genes related to wax biosynthesis. (A) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of 19 wax genes in 30-day-old plants of WT and Oschr4-5. Data are the means ± standard errors (n = 3). Significance of data is tested by student’s t test. * p < 0.05, ** p < 0.01. (B,C) ChIP-qPCR analysis for H3K4me3 (B) and H3K27me3 (C) levels on 7 up-regulated wax genes in Oschr4-5 relative to the WT. Data are the means ± standard errors (n = 3). Significance of data is tested by student’s t test. * p < 0.05, ** p < 0.01.
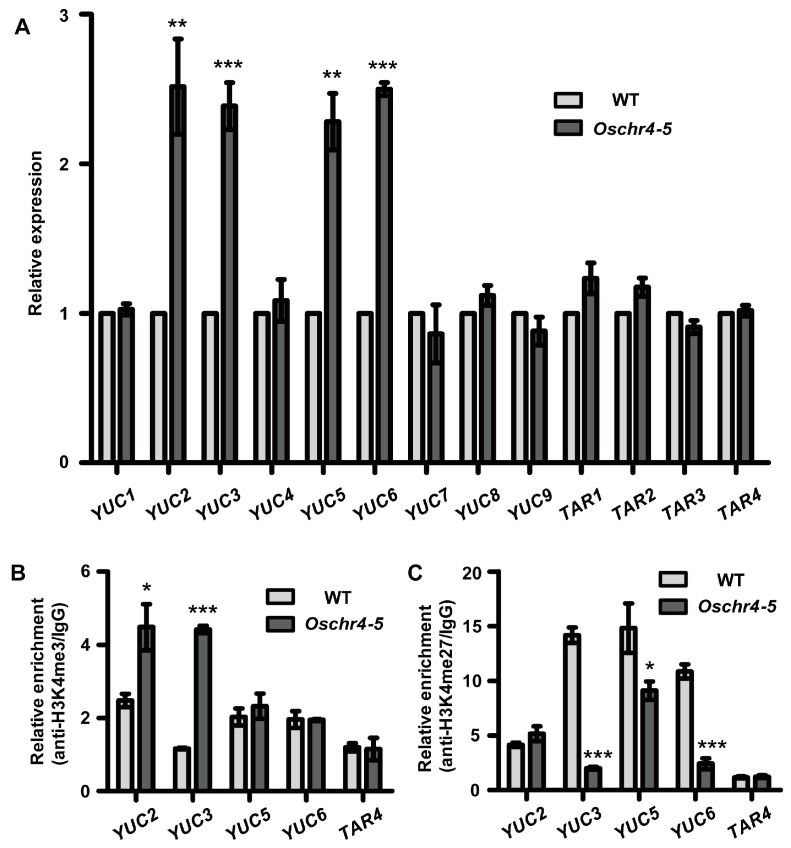
The YUCCA gene, which encodes a flavin monooxygenase-like enzyme, is known to be involved in Trp-dependent auxin biosynthesis [46]. The TAA/TAR genes, which encode putative Trp aminotransferases, have been proposed to act in the same pathway with YUCCA genes [47]. To explain the increased IAA level in the Oschr4-5 mutant, we measured the expression levels of 13 genes from the YUCCA and TAA/TAR family between three-day-old seedlings of WT and Oschr4-5. The transcript levels of four YUCCA related genes including YUC2, YUC3, YUC5 and YUC6 were upregulated significantly in the Oschr4-5 mutants (Figure 7A). However, the other 9 genes showed no significant alteration in the Oschr4-5 mutants. These results suggested that OsCHR4 may influence the expression of several wax-related and IAA-related genes, and the increased gene expressions in the Oschr4-5 mutants conduces to higher wax and IAA content in Oschr4-5 than WT.
Figure 7.
Expression and histone modification of genes involved in IAA biosynthesis in WT and Oschr4-5. (A) qRT-PCR analysis showed 4 of 13 tested IAA biosynthesis genes were up-regulated in 3-day-old seedlings of Oschr4-5 relative to the WT. Data are the means ± standard errors (n = 3). Significance of data is tested by student’s t test. ** p < 0.01, *** p < 0.001. (B,C) ChIP-qPCR analysis showed variations of H3K4me3 (B) and H3K27me3 (C) levels on 4 up-regulated IAA genes in Oschr4-5 compared to WT. Data are the means ± standard errors (n = 3). Significance of data is tested by student’s t test. * p < 0.05,*** p < 0.001.
2.7. Histone Modifications of the Wax and Auxin Biosynthesis-Related Genes Were Affected in Oschr4-5
Histone modification is recognized as an important mechanism in the epigenetic regulation of gene expression. For example, trimethylation of histone H3 lysine 4 (H3K4me3) is generally correlated with gene activation, whereas trimethylation of histone H3 lysine 27 (H3K27me3) is generally correlated with gene repression [48,49]. As reported, the rice CHD3 protein OsCHR4/CHR729 is a chromatin remodeler and can modulate H3K4me3 and H3K27me3 level to regulate the genes expression [35]. In order to test whether increased expression of wax and auxin biosynthesis related genes in Oschr4-5 could be attributed to altered histone modification, a ChIP analysis with antibody recognizing H3K4me3 and H3K27me3 was performed. Of the seven wax biosynthetic genes, three genes (WSL4, CER7, BDG) showed increased H3K4me3 and decreased H3K27me3 simultaneously, four genes showed either increased H3K4me3 (GL1-4, LACS2) or decreased H3K27me3 (LACS7, ROC4) (Figure 6B,C). Similarly, of the four auxin biosynthetic genes, one gene (YUC3) showed increased H3K4me3 and decreased H3K27me3 simultaneously, three genes showed either increased H3K4me3 (YUC2) or decreased H3K27me3 (YUC5, YUC6) (Figure 7B,C). LACS1 and TAR4, two control genes which were not differentially expressed between Oschr4-5 and WT, did not show significant changes in level of H3K4me3 and H3K27me3 (Figure 6; Figure 7). Taken together, OsCHR4 affects modifications of histone protein associated with wax and auxin biosynthesis related genes and regulates their expression epigenetically.
3. Discussion
In rice, a total of six CHD-related (CHR) genes were found in its genome (http://www.chromdb.org), including CHR705 (Os07g46590), CHR702 (PKL, Os06g08480), CHR703 (Os01g65850), CHR4/CHR729 (PKR1, Os07g31450), CHR723 (Os06g01320) and CHR744 (Os02g02050). Among these CHR proteins, CHR729 was a typical chromatin remodeling factor which belonged to subfamily II (CHD3) [38]. It has been reported that three additional allelic mutants of CHR4 displayed pleiotropic phenotypes [37,38,50]. In this study, three more allelic mutants of CHR4 (Oschr4-5, -6, -7) were isolated, which exhibited similar pleiotropic phenotypes, including slight dwarfism, narrow and rolled leaves, and reduced chlorophyll contents in adaxial cells. More importantly, we found some novel phenotypes such as reduced number of longitudinal veins, increased IAA content, enhanced cuticular wax disposition and enhanced drought tolerance in Oschr4-5, which further illustrated the multiple functions of OsCHR4 in plant development.
The closest homolog gene with OsCHR4 in Arabidopsis is considered to be AtCHR4 (PICKLE RELATED1, PKR1) (Figure S2B), while their functions in plants seem to be different. Homozygous mutants of neither PICKLE homologs PKR1 nor PKR2 exhibited significant phenotypic differences to WT plants under standard growth conditions [51]. However, OsCHR4 was revealed to be closely related with the development of rice leaves in this study. It is noteworthy that the function of OsCHR4 is more similar to CHD3-related PICKLE. PICKLE was initially isolated from the Arabidopsis pickle mutant characterized by green tuberous root phenotype [52]. It has been reported that PICKLE functions in repression of ectopic stipules and meristems in leaf tissue and represses meristematic genes in carpel tissue [51,53]. PICKLE is not only involved in organ or cell polarity, but also plays an important role in cell differentiation [52]. Loss-of-function mutations in PICKLE could always influence cell transition from embryonic to vegetative development, thereby inducing pleiotropic phenotypes in the root, leaves and even the whole plants [54,55]. These phenotypes were just consistent with those of Oschr4-5 isolated in this study and indicated a similar function between OsCHR4 and PICKLE. However, T-DNA insertion mutation and RNAi of the closest homolog gene with PICKLE in rice (OsCHR702) did not produce any morphological phenotype [36]. Therefore, we suspect that the function of CHR proteins may be diversified between Arabidopsis and rice due to specific species difference.
Cuticular waxes are complex mixtures of very-long-chain fatty acids (VLCFAs) and their derivatives and provide effective protection for plants to confront various environmental stresses [28]. In this study, we discovered that OsCHR4 negatively regulates the wax biosynthesis and more cuticle wax accumulated on the adaxial side of leaves in Oschr4-5 mutant compared to the WT. Previous reports demonstrate that excessive deposition of cuticular wax can decrease nonstomatal water loss in plants [24,56]. We also proved that increased wax content in Oschr4-5 results in reduced water loss and improved drought tolerance. As a result of increased wax deposition in Oschr4-5, the mutant Oschr4-5 exhibits less chlorophyll leaching rate compared with WT. Furthermore, increased transcript levels of genes involved in wax biosynthesis in Oschr4-5 were also in support of the regulation of OsCHR4 in wax synthesis. Among the seven up-regulated genes in Oschr4-5 (Figure 6A), WSL4 and its homolog gene OsCER7 encode β-Ketoacyl-coenzyme A synthase (KCS) which is the key enzyme in the first step of VLCFA elongation [45,57]. OsGL1-4 encodes the fatty aldehyde decarbonylase, which has been identified to positively regulate various steps of wax biosynthesis [21,22,23,24,25]. LACS2 and LACS7 encode long-chain acyl-CoA synthase that preferentially modifies VLCFAs for wax biosynthesis [58]. BDG (BODYGUARD) encodes an α/β-hydrolase fold protein important for cuticle development [28]. ROC4 encodes a HD-ZIP IV transcription factor which positively regulates wax deposition in rice by activating wax biosynthetic gene such as BDG [28]. This result suggests multiple steps of wax biosynthesis pathway are affected by the loss of function of CHR4 and is consistent with the quantification data that many wax components were significantly increased in Oschr4-5 (Figure 3C).
It has been demonstrated in previous research that OsCHR4 is essential for various aspects of plant development via influencing the gibberellin pathway and ROS homeostasis [38,40]. In this study, we found that the content of IAA in Oschr4-5 was significantly higher than that of WT. As reported, many narrow leaves genes were interrelated with IAA. Auxin overproduction is well known to lead to narrow and epinastic leaves. Ectopic overexpression of either YUCCA genes (encoding flavin monooxygenases) in Arabidopsis [46] or the Agrobacterium auxin biosynthetic gene iaaM (encoding tryptophan monooxygenase) and petunia FLOOZY gene (ortholog of YUCCA) in petunia resulted in narrow and rolled leaves with high levels of IAA [42,43]. In Arabidopsis, narrow and rolled leaves were also caused by elevated IAA levels by overexpressing cytochrome P450 CYP79B2 which converts tryptophan (Trp) to indole-3-acetaldoxime [59]. A similar case was reported in rice, where OsYUCCA8/NAL7 also affects leaf width via altered IAA content [6]. In this study, increased IAA content was also detected in the narrow leaf mutant Oschr4-5. Auxin is principally converted from tryptophan via the TAA family of aminotransfrases and the YUCCA (YUC) family of flavin-containing monooxygenases [60,61]. For further verification of the altered IAA concentration in Oschr4-5, we detected the expression of TAA and YUC family genes and found that expression levels of four YUC genes increased in Oschr4-5 mutants compared with WT. The finding is consistent with increased IAA concentration in Oschr4-5, which strengthens the evidence that OsCHR4 might regulate auxin biosynthetic process in rice. Interestingly, 31 OsAux/IAA genes involved in the auxin signaling pathway was significantly downregulated in crl6, an allelic mutant of Oschr4-5 [39]. The decreased transcript level of OsIAA genes might be due to higher IAA content as revealed in this study. As early-response genes and negative regulators of auxin signaling, Aux/IAA transcripts tend to be rapidly induced and then repressed by elevated IAA content [62].
Histone modification was an important mechanism in regulation of DNA transcription and other activities. As reported, trimethylated histone H3 lysine 27 (H3K27me3) was associated with gene silencing and repression during plant development, while trimethylated histone H3 lysine 4 (H3K4me3) is generally correlated with gene activation [63,64]. In the chr729 mutant, 56% and 23% of marked genes lost H3K27me3 and H3K4me3, while 754 and 724 genes gained ectopic H3K27me3 and H3K4me3 [36]. Meanwhile, PICKLE was revealed to be able to reduce the level of H3K27me3 on root stem cells, thereby promoting the expression of meristem marker genes [65]. To evaluate the histone modification states in the Oschr4-5 mutant, ChIP was performed on several differently expressed genes involved in wax and auxin biosynthesis in this study. As a result, the increased level of H3K4me3 and/or reduced level of H3K27me3 in the Oschr4-5 mutant were significantly correlated with increased expression of genes related to wax or IAA biosynthesis. Consistent with the genome-wide ChIP-Seq analysis showing that 724 genes were detected to have gained ectopic H3K4me3 in the chr729 mutant [36], H3K4me3 was also increased in several auxin and wax biosynthesis genes in the Oschr4-5 mutant in our study. The underlying mechanism for the increased H3K4me3 at some specific loci still remains elusive [36]. As a bifunctional chromatin regulator that recognizes and modulates both H3K4 and H3K27 methylation, the function of OsCHR4 might be determined by sequence-specific transcription factors that associates with their targets and/or by the recruitment of different histone modification enzymes that catalyze methylation or demethylation.
In conclusion, our results suggest that OsCHR4 not only regulates the accumulation of cuticle wax on leaf surface, but also plays an essential role in the modulation of IAA biosynthesis. Expression levels of several genes involved in the two pathways were increased accompanied with up-regulated H3K4me3 or down-regulated H3K27me3. Epigenetic regulation by OsCHR4 is required for multiple aspects of plant development.
4. Materials and Methods
4.1. Plant Materials and Growth Condition
The rice mutant Oschr4-5 (requested from Dale Bumpers National Rice Research Center, Stuttgart, AR, USA) was isolated from the indica cultivar Indica 9 treated by gamma ray radiation. The other two allelic mutants Oschr4-6 and Oschr4-7 were isolated from the japonica cultivar Nipponbare treated with EMS in the Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China. All rice plants were cultivated in the experimental field of Shandong Rice Research Institute (Jining, China) under natural growing conditions, and the climatic conditions during the rice growing season were listed in Table S2. The mutant phenotype inherited stably after continuous cultivation. Field data of the mutant and WT plants, including plant height, tiller number, leaf width and leaf length were measured at the heading stage. The transpiration rate of plants was detected in field using LI-6400XT Portable Photosynthesis System.
4.2. Microscopic Observation
At the heading stage, the top first leaf of the mutant and WT were cut into pieces (2 cm) in the widest part and divided into three groups. One group of samples were used for microscopic observation of leaf vein and transverse cross section. Another group of samples were air-pumped in 2% cellulase R-10 solution for 30 min and digested for at least 2 days. After washing with water, mesophyll cells were peeled and the abaxial epidermis was observed under microscope. The cell width was measured using the Image J software (https://imagej.nih.gov/ij/). Stomatal density (mm−2) was counted between two lateral veins in the center region of five individual leaves. Finally, the last group of samples were fixed in fixative solution (75% ethanol, 5% acetic acid, 5% glycerol, 5% formaldehyde and 10% deionized water) for at least one day, and then critical point dried, sputter-coated with gold and observed with a scanning electron microscope (UANTA 200).
4.3. Measurement of Chlorophyll and Indole Acetic Acid (IAA)
The top first leaf of plants at heading stage was used for chlorophyll measurement. Briefly, 0.05 g leaf samples were cut into small pieces and incubated in 80% acetone overnight in dark. Then the chlorophyll contents were measured and calculated by absorption spectrophotometry. For the measurement of IAA, 0.2 g freeze dried sample of the whole shoots of 3-day-old seedlings was harvested for each replicate. Free IAA extraction and measurement was performed as described previously [66].
4.4. Measurement of Cuticular Waxes
The content of cuticular waxes in the leaf of mutant and WT was measured as described previously [24]. Briefly, cuticular wax samples were first extracted from the top first leaf of plants at the heading stage by immersion in chloroform (60 °C, 30 s). A total of 20 µg n-tetracosane (C24, SUPELCO, Sigma, Saint Louis, USA) was added to these samples as an internal standard. Then the solutions were soaked with 100 mm3 bis-N,N-(trimethylsilyl) trifluoroacetamide (BSTFA, SUPELCO, Sigma) and 100 mm3 pyridine for 1 h at 70 °C. After evaporation and filtration (WHATMAN, PTFE, 13 mm × 0.22 μm), the compositions of cuticular waxes were analyzed using a capillary gas chromatograph equipped with an HP-1MS column (30 m length, inner diameter 0.32 mm, film thickness 0.25 µm) attached to a mass spectrometer (GCMS-QP2010, Kyoto, Japan). The GC-MS protocol included injection at 250, 50 °C for 2 min, ramped to 200 at 20 °C min−1, 2 min at 200 °C, ramped to 320 at 2 °C min−1, 14 min at 320 °C with supplied at 1.2 cm3 min−1 as the carrier gas. A flame ionization detector was used for quantitative analyses, and the quantification of each component was calculated by the equivalent ratio of mass to peak area between the component and internal standard.
4.5. Water Loss and Chlorophyll-Leaching Assays
Leaf water loss rate was measured using detached leaves of the same parts from 6 week-old seedlings cultured in climate chambers. Each leaf was kept in a box without lid, and weighed every 15 min [28]. The water loss rate was calculated by dividing the lost weight of leaf at different time points by the initial weight. In the assay of chlorophyll leaching, the leaves (0.25 g) were cut into 2 cm lengths and immersed in 25 cm3 80% ethanol. 1 cm3 chlorophyll leaching solution was taken for spectrophotometry at wave lengths of 664 and 647 nm every 30 min. 1 cm3 80% ethanol was used as control. The solution should be collected and poured back in the same tube after each measurement. The chlorophyll concentration was calculated by the formula of 7.939 A664 × 19.539 A647 [24]. The chlorophyll-leaching rate was given by dividing the concentration monitored at different time points by the chlorophyll concentration measured after 24 h of immersion. The experiment comprised three replicates.
4.6. Mapping and Sequencing
An F2 segregating population generated from a cross between Oschr4-5 and 02428 (a polymorphic japonica cultivar) was used for mapping and cloning of the target gene. For the primary mapping, a DNA pool composed of DNA from 10 individual mutant plants was used for bulked segregation analysis (BSA) [67]. Approximately 200 polymorphic indel markers distributed evenly throughout the rice genome were used for the BSA analysis. For fine mapping, new indel markers were designed by utilizing genomic sequence information from Indica and Japonica (http://rgp.dna.affrc.go.jp/) (specific primers were shown in Table S3). DNA was extracted from fresh leaves of each plant. PCR program included 5 min at 95 °C followed by 35 cycles of 30 s at 94 °C, 30 s at 55 °C, 30 s at 72 °C, and a final extension of 10 min at 72 °C. PCR products were analyzed on a polyacrylamide gel stained with silver nitrate. The genomic region of the candidate gene was divided into several overlapping 1.5-kb fragments, and amplified using high-fidelity PrimeSTAR DNA polymerase (TaKaRa Bio, DaLian, China). The PCR program included 3 min at 94 °C followed by 40 cycles at 98 °C for 30 s, 55 °C for 5 s, 72 °C for 1.5 min, and a final extension at 72 °C for 10 min. PCR products were sequenced directly.
4.7. Real-Time Polymerase Chain Reaction (RT-PCR)
Total RNA was first extracted from rice plants with Trizol method, and then first-strand cDNA was synthesized using a Superscript III Reverse Transcription Kit (Invitrogen, Carlsbad, CA, USA). Semiquantitative PCR was performed using LA Taq DNA polymerase (TaKaRa) with the rice ACTIN1 gene serving as an internal control. The PCR program included 1 min at 94 °C followed by 30 cycles at 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 10 min. Quantitative PCR was performed using a SYBR Premix Ex Taq2 kit (TaKaRa) on ABI PRISM 7900HT under the following conditions: 10 s denaturing at 95 °C, 30 s annealing at 60 °C, 40 cycles. The mRNA amount relative to ACTIN1 was finally calculated. Specific primers were shown in Table S4.
4.8. Sequence and Phylogenetic Analyses
Gene prediction was performed using online search software (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice). Exon/intron structures were identified by alignment of coding sequences (CDS) and genomic DNA sequences. Multiple sequence alignments were conducted using CLUSTALX software, and a phylogenetic tree was built using MEGA 4 software [68].
4.9. Chromatin Immunoprecipitation (ChIP) Analysis
The ChIP experiment was performed based on a previously published protocol [69]. Simply, 0.5 g of 3-day-old or four-leaf-old seedlings of Oschr4-5 and WT were first harvested and crosslinked in 1% formaldehyde under vacuum for 15 min or 30 min, respectively. Then the isolated chromatin complex was fragmented to 200–500 bp by sonication, and 1% of the fragmented chromatin from each tube was kept as input DNA. The chromatin modification states of several target genes were analyzed using antibodies recognizing H3K4me3 (ab8580, Abcam, Cambridge, UK) and H3K27me3 (ab6002, Abcam) and the pre-immune serum (02-6502, Invitrogen) as a negative control. Then the immune complexes were collected with Protein A Agarose beads (P3476, Sigma). After reversing the crosslink, the precipitated and input DNAs were detected by real-time PCR, and the enrichment of H3K4me3 and H3K27me3 of each genes was quantified by normalizing the threshold cycle (Ct) of the ChIP sample with that of the input with 2(Ct of input − Ct of sample ChIP). Specific primers used in ChIP-PCR are shown in Table S4.
4.10. Accession Number
Sequence data from this article can be found in the GenBank databases under the following accession numbers: OsCHR4 genomic DNA, MK765112; OsCHR4 cDNA, MK765113.
Acknowledgments
We thank Dale Bumpers National Rice Research Center (Stuttgart, AR, USA) for providing the rice seeds of Oschr4-5 (GSOR22) and Indica 9; Tianfu Han (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for assistance in measurement of photosynthetic and transpiration rate.
Abbreviations
| CHD3 | Chromodomain Helicase DNA-binding (CHD) protein 3 |
| ChIP | Chromatin immunoprecipitation |
| CHR4 | CHD-related protein 4 |
| NAL1 | Narrow leaf 1 |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/10/2567/s1.
Author Contributions
Conceptualization, X.L.; methodology, T.G., D.W.; validation, J.F., and J.Z.; resources, S.Y.; writing—original draft preparation, T.G. and D.W.; writing—review and editing, X.L.; supervision, X.L. and L.X.; funding acquisition, X.L.
Funding
This research was funded by National Natural Science Foundation of China, grant number 31870271; National Major Project for Developing New GM Crops, grant number 2016ZX08009-003; and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences. The APC was funded by National Natural Science Foundation of China.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lang Y., Zhang Z., Gu X., Yang J., Zhu Q. Physiological and ecological effect of crimpy leaf character in rice (Oryza sativa L.). II. Photosynthetic character, dry mass production and yield forming. Acta Agron. Sin. 2004;30:883–887. [Google Scholar]
- 2.Jiang D., Fang J., Lou L., Zhao J., Yuan S., Yin L., Sun W., Peng L., Guo B., Li X. Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS ONE. 2015;10:e0118169. doi: 10.1371/journal.pone.0118169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qi J., Qian Q., Bu Q., Li S., Chen Q., Sun J., Liang W., Zhou Y., Chu C., Li X., et al. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008;147:1947–1959. doi: 10.1104/pp.108.118778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takashi S., Noriko K., Takeshi N., Kozue O., Yutaka S., Kohei I., Yasuo N., Tomokazu K., Yoshiaki N., Motoyuki A. A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. Plant J. 2009;60:227–241. doi: 10.1111/j.1365-313X.2009.03952.x. [DOI] [PubMed] [Google Scholar]
- 5.Woo Y.M., Park H.J., Su’udi M., Yang J.I., Park J.J., Back K., Park Y.M., An G. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 2007;65:125–136. doi: 10.1007/s11103-007-9203-6. [DOI] [PubMed] [Google Scholar]
- 6.Fujino K., Matsuda Y., Ozawa K., Nishimura T., Koshiba T., Fraaije M.W., Sekiguchi H. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 2008;279:499–507. doi: 10.1007/s00438-008-0328-3. [DOI] [PubMed] [Google Scholar]
- 7.Sakamoto T., Morinaka Y., Inukai Y., Kitano H., Fujioka S. Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J. 2013;73:676–688. doi: 10.1111/tpj.12071. [DOI] [PubMed] [Google Scholar]
- 8.Li W., Wu C., Hu G., Xing L., Qian W., Si H., Sun Z., Wang X., Fu Y. Characterization and Fine Mapping of a Novel Rice Narrow Leaf Mutant nal9. J. Integr. Plant Biol. 2013;55:1016–1025. doi: 10.1111/jipb.12098. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y., Luo L., Chen L., Tao X., Huang M., Wang H., Chen Z., Xiao W. Chromosome mapping, molecular cloning and expression analysis of a novel gene response for leaf width in rice. Biochem. Biophys. Res. Commun. 2016;480:394–401. doi: 10.1016/j.bbrc.2016.10.061. [DOI] [PubMed] [Google Scholar]
- 10.Song Y., Yan C., Zeng X., Yang Y., Fang Y., Tian C., Sun Y., Cheng Z., Gu M. ROLLED LEAF 9, encoding a GARP protein, regulates the leaf abaxial cell fate in rice. Plant Mol. Biol. 2008;68:239–250. doi: 10.1007/s11103-008-9365-x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang G., Xu Q., Zhu X., Qian Q., Xue H. SHALLOT-LIKE1 Is a KANADI Transcription Factor That Modulates Rice Leaf Rolling by Regulating Leaf Abaxial Cell Development. Plant Cell. 2009;21:719–735. doi: 10.1105/tpc.108.061457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ken-Ichiro H., Mari O., Emi H., Masashi A., Tsutomu I., Hikaru S., Jun-Ichi I., Yasuo N. The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev. Biol. 2009;334:345–354. doi: 10.1016/j.ydbio.2009.07.042. [DOI] [PubMed] [Google Scholar]
- 13.Li L., Shi Z., Li L., Shen G., Wang X., An L., Zhang J. Overexpression of ACL 1 (abaxially curled leaf 1) Increased Bulliform Cells and Induced Abaxial Curling of Leaf Blades in Rice. Mol. Plant. 2010;3:807–817. doi: 10.1093/mp/ssq022. [DOI] [PubMed] [Google Scholar]
- 14.Shi Z., Wang J., Wan X., Shen G., Wang X., Zhang J. Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta. 2007;226:99–108. doi: 10.1007/s00425-006-0472-0. [DOI] [PubMed] [Google Scholar]
- 15.Xiang J., Zhang G., Qian Q., Xue H. Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 2012;159:1488–1500. doi: 10.1104/pp.112.199968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C., Fu Y., Hu G., Si H., Cheng S., Liu W. Isolation and characterization of a rice mutant with narrow and rolled leaves. Planta. 2010;232:313–324. doi: 10.1007/s00425-010-1180-3. [DOI] [PubMed] [Google Scholar]
- 17.Hu J., Zhu L., Zeng D., Gao Z., Guo L., Fang Y., Zhang G., Dong G., Yan M., Liu J. Identification and characterization of NARROW ANDROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 2010;73:283–292. doi: 10.1007/s11103-010-9614-7. [DOI] [PubMed] [Google Scholar]
- 18.Zhao S., Zhao L., Liu F., Wu Y., Zhu Z., Sun C., Tan L. NARROW AND ROLLED LEAF 2 regulates leaf shape, male fertility, and seed size in rice. J. Integr. Plant Biol. 2016;58:983–996. doi: 10.1111/jipb.12503. [DOI] [PubMed] [Google Scholar]
- 19.Chen W., Sheng Z., Cai Y., Li Q., Wei X., Xie L., Jiao G., Shao G., Tang S., Wang J., et al. Rice Morphogenesis and chlorophyll accumulation is regulated by the protein encoded by NRL3 and its interaction with NAL9. Front. Plant Sci. 2019;10:1–13. doi: 10.3389/fpls.2019.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia Y., Nikolau B.J., Schnable P.S. Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell. 1996;8:1291–1304. doi: 10.1105/tpc.8.8.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Islam M.A., Hao D., Jing N., Ye H., Xiong L. Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol. Biol. 2009;70:443–456. doi: 10.1007/s11103-009-9483-0. [DOI] [PubMed] [Google Scholar]
- 22.Qin B., Tang D., Huang J., Li M., Wu X., Lu L., Wang J., Yu X., Chen J., Gu M. Rice OsGL 1-1 is involved in leaf cuticular wax and cuticle membrane. Mol. Plant. 2011;4:985–995. doi: 10.1093/mp/ssr028. [DOI] [PubMed] [Google Scholar]
- 23.Zhou L., Ni E., Yang J., Zhou H., Liang H., Li J., Jiang D., Wang Z., Liu Z., Zhuang C. Rice OsGL1-6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS ONE. 2013;8:e65139. doi: 10.1371/journal.pone.0065139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao B., Cheng Z., Lei C., Xu F., Gao S., Ren Y., Wang J., Zhang X., Wang J., Wu F. Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta. 2012;235:39–52. doi: 10.1007/s00425-011-1481-1. [DOI] [PubMed] [Google Scholar]
- 25.Jung K., Han M., Lee D., Lee Y., Lukas S., Rochus F., Andrea F., Alexander Y., Heinz S., Kim Y., et al. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell. 2006;18:3015–3032. doi: 10.1105/tpc.106.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Wan L., Zhang L., Zhang Z., Zhang H., Quan R., Zhou S., Huang R. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 2012;78:275–288. doi: 10.1007/s11103-011-9861-2. [DOI] [PubMed] [Google Scholar]
- 27.Zhou X., Jenks M.A., Liu J., Liu A., Zhang X., Xiang J., Zou J., Peng Y., Chen X. Overexpression of Transcription Factor OsWR2 Regulates Wax and Cutin Biosynthesis in Rice and Enhances its Tolerance to Water Deficit. Plant Mol. Biol. Rep. 2014;32:719–731. doi: 10.1007/s11105-013-0687-8. [DOI] [Google Scholar]
- 28.Wang Z., Tian X., Zhao Q., Liu Z., Li X., Ren Y., Tang J., Fang J., Xu Q., Bu Q. The E3 Ligase DROUGHT HYPERSENSITIVE Negatively Regulates Cuticular Wax Biosynthesis by Promoting the Degradation of Transcription Factor ROC4 in Rice. Plant Cell. 2018;30:228–244. doi: 10.1105/tpc.17.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernard A., Joubès J. Arabidopsis cuticular waxes: Advances in synthesis, export and regulation. Prog. Lipid Res. 2013;52:110–129. doi: 10.1016/j.plipres.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Clapier C.R., Cairns B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 31.Hall J.A., Georgel P.T. CHD proteins: A diverse family with strong ties. Biochem. Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 32.Stanley D.R., Henderson J.T., Jerome R.E., Edenberg H.J., Jeanne R.S., Joe O. Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 2010;35:33–43. doi: 10.1046/j.1365-313x.2003.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hay A., Kaur H., Phillips A., Hedden P., Hake S., Tsiantis M. The Gibberellin Pathway Mediates KNOTTED1-Type Homeobox Function in Plants with Different Body Plans. Curr. Biol. 2002;12:1557–1565. doi: 10.1016/S0960-9822(02)01125-9. [DOI] [PubMed] [Google Scholar]
- 34.Franziska T., Francois R., Sara F., Marie-Laure M.M., Elodie G., Nicolas B., Séverine G., Martienssen R.A., George C., Vincent C. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:855–866. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Y., Lai Y., Zhu D. Transcription regulation by CHD proteins to control plant development. Front. Plant Sci. 2014;5:223. doi: 10.3389/fpls.2014.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y., Liu D., Zhong X., Zhang C., Zhang Q., Zhou D. CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. USA. 2012;109:5773–5778. doi: 10.1073/pnas.1203148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao C., Xu J., Chen Y., Mao C., Zhang S., Bai Y., Jiang D., Wu P. Molecular cloning and characterization of OsCHR4, a rice chromatin-remodeling factor required for early chloroplast development in adaxial mesophyll. Planta. 2012;236:1165–1176. doi: 10.1007/s00425-012-1667-1. [DOI] [PubMed] [Google Scholar]
- 38.Ma X., Ma J., Zhai H., Xin P., Chu J., Qiao Y., Han L. CHR729 Is a CHD3 Protein That Controls Seedling Development in Rice. PLoS ONE. 2015;10:e0138934. doi: 10.1371/journal.pone.0138934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Wang D., Gan T., Liu L., Long W., Wang Y., Niu M., Li X., Zheng M., Jiang L., et al. CRL6, a member of the CHD protein family, is required for crown root development in rice. Plant Physiol. Biochem. 2016;105:185–194. doi: 10.1016/j.plaphy.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 40.Cho S., Lee C., Gi E., Yim Y., Koh H., Kang K., Paek N. The Rice Rolled Fine Striped (RFS) CHD3/Mi-2 Chromatin Remodeling Factor Epigenetically Regulates Genes Involved in Oxidative Stress Responses During Leaf Development. Front. Plant Sci. 2018;9:364. doi: 10.3389/fpls.2018.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enrico S., Michalis B., Miltos T. Control of leaf and vein development by auxin. Cold Spring Harbor Perspect. Biol. 2010;2:a001511. doi: 10.1101/cshperspect.a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klee H.J., Horsch R.B., Hinchee M.A., Hein M.B., Hoffmann N.L. The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Gene Dev. 1987;1:86–96. doi: 10.1101/gad.1.1.86. [DOI] [Google Scholar]
- 43.Rafael T.S., Mattijs B., Karin L., GöRan S., Mol J.N.M., Erik S., Ronald K. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 2002;16:753. doi: 10.1101/gad.219502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu X., Xiong L. Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. Proc. Natl. Acad. Sci. USA. 2013;110:17790–17795. doi: 10.1073/pnas.1316412110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., Guan Y., Zhang D., Dong X., Tian L., Qu L. A β-ketoacyl-CoA synthase is Involved in Rice Leaf Cuticular Wax Synthesis and requires a CER2-LIKE Protein as a Cofactor. Plant Physiol. 2017;173:944–955. doi: 10.1104/pp.16.01527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 47.Abu-Zaitoon Y.M., Bennett K., Normanly J., Nonhebel H.M. A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiol. Plant. 2012;146:487–499. doi: 10.1111/j.1399-3054.2012.01649.x. [DOI] [PubMed] [Google Scholar]
- 48.Li X., Wang X., He K., Ma Y., Su N., He H., Stolc V., Tongprasit W., Jin W., Jiang J., et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell. 2008;20:259–276. doi: 10.1105/tpc.107.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Elling A., Li X., Li N., Peng Z., He G., Sun H., Qi Y., Liu X., Deng X. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell. 2009;21:1053–1069. doi: 10.1105/tpc.109.065714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang F., Tang Y., Miao R., Xu F., Lin T., He G., Sang X. Identification and gene mapping of a narrow and upper-albino leaf mutant in rice (Oryza sativa L.) Chin. Sci. Bull. 2012;57:3798–3803. doi: 10.1007/s11434-012-5154-7. [DOI] [Google Scholar]
- 51.Ernst A., Villar C.B.R., Sara F., Reyes J.C., Lars H., Claudia K.H. CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 2009;5:e1000605. doi: 10.1371/journal.pgen.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogas J., Cheng J., Sung Z., Somerville C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science. 1997;277:91–94. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- 53.Henderson J.T., Li H.-C., Rider S.D., Mordhorst A.P., Romero-Severson J., Cheng J.-C., Robey J., Sung Z.R., de Vries S.C., Ogas J. PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 2004;134:995–1005. doi: 10.1104/pp.103.030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H., King C., Henderson J.T., Stanley Dean R., Yinglin B., Heng Z., Matthew F., Jacob G., Joe O. PICKLE acts during germination to repress expression of embryonic traits. Plant J. 2005;44:1010–1022. doi: 10.1111/j.1365-313X.2005.02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogas J., Kaufmann S., Henderson J., Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou X., Li L., Xiang J., Gao G., Xu F., Liu A., Zhang X., Peng Y., Chen X., Wan X. OsGL1-3 is involved in cuticular wax biosynthesis and tolerance to water deficit in rice. PLoS ONE. 2015;10:e116676. doi: 10.1371/journal.pone.0116676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gan L., Zhu S., Zhao Z., Liu L., Wang X., Zhang Z., Zhang X., Wang J., Wang J., Guo X., et al. Wax Crystal-Sparse Leaf 4, encoding a b-ketoacyl-coenzyme A synthase 6, is involved in rice cuticular wax accumulation. Plant Cell Rep. 2017;36:1655–1666. doi: 10.1007/s00299-017-2181-5. [DOI] [PubMed] [Google Scholar]
- 58.Lco S., Song T., Kosma D.K., Parsons E.P., Rowland O., Jenks M.A. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2010;59:553–564. doi: 10.1111/j.1365-313X.2009.03892.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Y., Hull A.K., Gupta N.R., Goss K.A., José A., Ecker J.R., Jennifer N., Joanne C., Celenza J.L. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002;16:3100–3112. doi: 10.1101/gad.1035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kakei Y., Nakamura A., Yamamoto M., Ishida Y., Yamazaki C., Sato A., Narukawa-Nara M., Soeno K., Shimada Y. Biochemical and chemical biology study of rice OsTAR1 revealed that tryptophan aminotransferase is involved in auxin biosynthesis; identification of a potent OsTAR1 inhibitor, pyruvamine2031. Plant Cell Physiol. 2017;58:598–606. doi: 10.1093/pcp/pcx007. [DOI] [PubMed] [Google Scholar]
- 61.Dai X., Kiyoshi M., Chen Q., Hiroyuki K., Yuji K., Sunil O., Jennifer D.B., David B., Zhao Y. The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 2013;288:1448–1457. doi: 10.1074/jbc.M112.424077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chapman E.J., Estelle M. Mechanism of Auxin-Regulated Gene Expression in Plants. Annu. Rev. Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- 63.Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jörg F., Dmitri D., Andreas H., Ingo S. Chromosomal histone modification patterns—From conservation to diversity. Trends Plant Sci. 2006;11:199–208. doi: 10.1016/j.tplants.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 65.Ernst A., Villar C.B.R., Riccardo D.M., Sabrina S., Claudia K. The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell. 2011;23:1047–1060. doi: 10.1105/tpc.111.083352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gou J., Strauss S.H., Tsai C.J., Fang K., Chen Y., Jiang X., Busov V.B. Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell. 2010;22:623–639. doi: 10.1105/tpc.109.073239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michelmore R.W., Paran I., Kesseli R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 69.Gendrel A.V., Lippman Z., Martienssen R., Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.