Abstract
Background: The incidence of Type 2 Diabetes Mellitus (T2DM) in Egypt is considered one of the highest in the world. Metformin and Sulfonylureas are usually prescribed together due to their efficacy and their relatively low cost. Organic cation transport 1, encoded by SLC22A1 gene, is the main transporter of metformin into hepatocytes, which is considered metformin site of action. Sulfonylureas enhance insulin release from pancreatic B-cells through binding to sulfonylurea receptor 1, encoded by ABCC8 gene. Single nucleotide polymorphisms in the SLC22A1 and ABCC8 genes might affect the response of each drug.
Aims: To investigate the influence of SLC22A1 rs622342 (A>C) and ABCC8 rs757110 (A>C) genetic variants on the efficacy of metformin and glimepiride combination therapy in Egyptian T2DM patients.
Methods: Observational cross-sectional study in which patients receiving metformin and glimepiride combination therapy for at least 6 months were included for genotyping and classified into either responders or non-responders, based on their HbA1C level.
Results: A total of 127 patients were included and genotyped. They were divided into 93 responders (HbA1C<7%) and 34 non-responders (HbA1C≥7%). Minor allele frequencies for rs622342 and rs757110 were 0.189 and 0.271, respectively. Only SLC22A1 rs622342 variant was found to be associated with the response of combination therapy, in which AA alleles carriers were 2.7-times more responsive to metformin than C allele carriers (Recessive model, odds ratio = 2.718, p = 0.025, 95% CI = 1.112–6.385).
Conclusion: Genotyping of rs622342 can be useful in predicting the response to metformin in combination therapy in Egyptian T2DM patients.
Keywords: Metformin, sulfonylurea, pharmacogenetics, glimepiride, T2DM, SNP, genotyping
Introduction
Diabetes Mellitus is a worldwide health problem. It was reported that, by year 2040, the number of patients with diabetes is expected to rise to 642 million patients1. Type 2 Diabetes Mellitus (T2DM) accounts for almost 90% of all diabetes mellitus cases around the globe2. Currently, Egypt is ranked first, with the largest number of patients with diabetes in the Middle East and North Africa region3. By 2045, the number of diabetes patients in Egypt is predicted to be doubled from 8.2 million to 16.7 million4. Many factors, including obesity, unhealthy lifestyles, and family history could be the reasons behind the increased prevalence of DM in Egypt5.
The pivotal goal for T2DM management is to reduce the risk of long-term complications and mortality1. There are many available treatment options for T2DM, many guidelines recommended initiating treatment as monotherapy; however, this might be insufficient in many cases, and dual therapy or combination therapy should be prescribed5–7. Many studies supported the superiority of combination therapy over monotherapy8,9. Owing to their acceptable efficacy and low cost7, metformin and sulfonylureas are usually prescribed together. Both drugs decrease blood glucose by different mechanisms, metformin decreases the hepatic glucose production, and acts as insulin sensitizer, while sulfonylureas lower blood glucose by increasing insulin secretion10. There is a great variation in response to many oral hypoglycemics, interestingly up to 40% of inter-individual variabilities could be explained by genetic factors11,12.
Hepatocytes uptake of metformin is essential to elucidate its pharmacological actions13. Organic cation transporter 1 (OCT1), encoded by SLC22A1 gene, is the main hepatic transporter of metformin14,15. Metformin inhibits hepatic gluconeogenesis, but the exact mechanism of how it enhances insulin sensitivity is still unknown16. SLC22A1 gene is highly polymorphic, and there are numerous single nucleotide polymorphisms (SNP) that has been reported to be associated with its activity. This might explain some of the inter-individual variabilities in metformin efficacy17,18. Rs622342 (A changes to C) is a SLC22A1 genetic variant which was previously studied in Caucasians and was found to be associated with metformin response19. Rs622342 genetic variant is located on chromosome 6 (position: chr6:160151834), in which the C minor allele was suggested to have a reduced or altered OCT1 uptake function20,21. Therefore, genotyping of rs622342 could help practitioners to determine whether current diabetic patients on metformin therapy will benefit from the medication or should be switched to another oral hypoglycemic class.
On the other hand, sulfonylureas bind to the Sulfonylurea receptor 1 (SUR1), encoded by gene ABCC8, and enhance insulin release through inhibiting the K+ channel activity in the pancreatic B-cells14,22,23. Glimepiride is a third-generation sulfonylurea with a long half-life and low incidence of hypoglycemia. Unlike the older generation, Glimepiride was found to preserve B-cell functions24. ABCC8 rs757110 (A changes to C) gene variant is a polymorphism in which alanine is replaced by serine at position 1369 (S1369A) of SUR1 located in exon 33 (position: chr11:17396930), resulting in changing the sensitivity of the sulfonylureas to bind to the A-site of the SUR117,25,26. Rs757110 genetic variant affects only the A-site of SUR1, and sulfonylureas interact differently with SUR1, as some interact with A-site only, while others can interact with both A and B sites27. Therefore, only glimepiride was included for this study.
The ethnicity and genetic build-up of the Egyptians are not well defined nor studied enough. Many studies have regarded Egyptians as Caucasians; however, other studies have found discrepancies between allele frequencies of some genetic variants between Egyptians and Caucasians and concluded that Egyptians might have admixed ancestry28–30.
The aim of the present study was to investigate and evaluate the association between SLC22A1 rs622342 and ABCC8 rs757110 genetic variants, and the response to metformin and glimepiride combination therapy in Egyptian T2DM patients. Moreover, to determine allele frequencies of these variants in an Egyptian population. In our hypothesis, we postulated that, in combination therapy, any effect due to rs622342 genetic variant is due to the direct influence on metformin because sulfonylureas are not substrates to the OCT1 receptors, while the rs757110 genetic variant might influence sulfonylureas efficacy but not metformin.
Patients and methods
Study design
This observational cross-sectional non-interventional study took place between August 2017 and July 2018 at the outpatient clinics of the National Institute of Diabetes and Endocrinology (NIDE) at Kasr El-Ainy, Cairo, Egypt. The study was approved by both the Institutional Review Board of the NIDE, and the Ethical Committee of the Faculty of Pharmacy Helwan University. Annual updates of the American Diabetes Association (ADA) diagnostic and classification criteria for diabetes diagnosis are adopted by NIDE.
Patients selection and classification
Patients previously diagnosed with T2DM were included in the study if they were (1) unrelated Egyptians, (2) adults above 18 years old, and (3) started and received metformin and glimepiride at the same time as combination therapy for at least 6 months, not longer than 4 years, to ensure that the patient reached maximum glycated hemoglobin (HbA1C) reduction was achieved after 6 months31,32. Patients were excluded if (1) they received triple therapy or insulin therapy within the 6 months preceding the study, or if they had (2) renal impairment (eGFR < 45 mL/minute/m2), (3) hepatic impairments such as chronic hepatitis or cirrhosis, (4) severe/unstable congested heart failure that required hospitalization, (5) endocrine disorders that affects glucose metabolism such as thyroid dysfunctions, (6) chronic diabetes complications such as diabetic retinopathy or amputations, (7) they were pregnant or breastfeeding, (8) they refused/failure to obtain informed consent, or (9) patients received any medications that interact with the OCT1 receptor, such as, diltiazem, verapamil, proton pump inhibitors, spironolactone, ketoconazole, or clopidogrel33,34.
Data collection
All needed patients’ data and demographics, such as full medical and medication history, treatment duration, Body Mass Index (BMI), metformin and glimepiride daily doses for the past 6 months, family history, and blood pressure, were collected from either patient profile or patient interviewing during monthly appointment checkup at NIDE. Estimated average glucose (eAG) was derived from HbA1C measured during patient appointment and calculated based on the equation (eAG = 28.7 × HbA1C – 46.7)35. For compliance assessment, patients were asked about their therapy regimen and instruction of the dosage for their prescribed medication.
Metformin and glimepiride treatment protocol
According to the treatment protocol of NIDE, patients were prescribed oral hypoglycemics from the date of diabetes diagnosis as follows: metformin was prescribed as 500 mg daily, and up to a maximum dose of 2,000 mg/day, and glimepiride was prescribed with doses ranging from 4–6 mg daily, and up to a maximum dose of 8 mg/day. Doses of both agents were titrated based on tolerability and on glycemic control during each monthly visit. All patients received vitamins B12/B6 as a supplement treatment. Daily doses of metformin and glimepiride were defined as the average doses per day of each drug during the last 6 months.
Patients stratification and response definition
HbA1C was used to determine the therapeutic and glycemic goals for diabetic patients, and a cut-off of 7% was recommended by ADA36. Therefore, patients were divided according to the HbA1C level obtained at the patient clinic appointment and classified into responders or non-responders. Responders were defined as patients who received metformin and glimepiride combination therapy for at least 6 months, and their HbA1C was less than 7%. Non-responders were considered to be on combination therapy for at least 6 months, and their HbA1C was equal or higher than 7%.
Blood sampling, FBGL, and HbA1C measurement
From each subject, 5 mL of venous blood were collected during appointment, in which, 1 mL was collected in fluoride vacutainer for fasting blood glucose level (FBGL). The remaining 4 mL were collected in EDTA vacutainer for the measurement of HbA1C and DNA extraction. Blood was stored at –20 °C until DNA extraction. FBGL was measured using automatic ARCHITE CT8000 chemistry analyzer, according to manufacturer’s instructions (US, supplied by Abbott, Alkamal Company Cairo, Egypt). HbA1C was measured by immunoturbidimetry technique using the auto-analyzer system BT3000 (Biotechnica, Rome, Italy) and according to manufacturer’s instructions.
DNA extraction and genotyping
DNA was extracted using the GeneJet Whole Blood Genomic DNA purification Mini Kit (Thermo Fisher Scientific, Waltham, MA). DNA was stored at –80 °C until genotyping. Concentration and purity of the extracted DNA were assessed using Quawell Q5000 nanodrop (Quawell Technology, Inc., San Jose, CA). DNA purity of A260/280 ranging from 1.7–1.9 were considered pure enough for genotyping. Genotyping of the selected genetic variants, rs622342 (assay ID = C____928527_20, assay type = functionally tested) and rs757110 (assay ID = C____600632_20, assay type = drug metabolizing enzyme), was carried out using TaqMan SNP Genotyping Assay (Applied Biosystems, Foster City, CA) and Sensifast Probe NO-ROX Kit master mix (Bioline, London, UK), on Rotor-Gene Q real time thermocycler instrument (Qiagen, Hilden, Germany). Each reaction mix was composed of 10 µL master mix, 1 µL DNA sample, 1 µL SNP assay, and DNase Free water to a final volume of 20 µL. To test for genotyping accuracy, randomly selected samples were retested. The concordance rates were 100% for the re-tested sample.
Statistical analysis
Patients’ demographics were summarized using descriptive statistics. Percentages were used to describe categorical data, whereas continuous data was reported as means ± standard deviations. T-test and Chi-square (χ2) were used to compare demographics between responder and non-responder groups. Deviation from Hardy-Weinberg Equilibrium (HWE) was used to assess the distribution of genotypes. Recessive model (AA vs C allele carriers) was adapted to account for the small number of homozygote minor allele carriers. To test the association between the genetic variants and response, the Chi-square test was used first to determine whether there was a statistically significant difference in the distribution of genotypes between the responder and non-responder groups. If a statistically significant distribution was found, logistic regression analysis was used to assess the association between the patients’ covariates, including genetic variants, and the response. Odds ratios (OR) were used to represent the association between genetic variants and response, with confidence intervals (CI) at 95%. A p-value <0.05 was considered statistically significant. All statistical tests were performed using SPSS (Version 22, IBM Corp., Armonk, NY).
Results
Patients characteristics
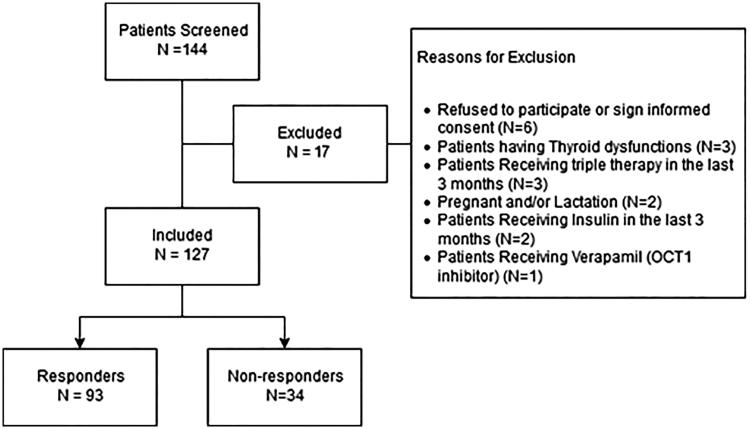
A total of 144 T2DM patients were screened, and only 17 patients were excluded. A schematic diagram for inclusion and exclusion is represented in Figure 1. One patient was excluded due to the use of verapamil, which has an inhibitory effect on OCT1. Informed consent was obtained from all 127 patients included in the study. The response rate to combination therapy was found to be 73.23%, by which 93 patients had their HbA1C below 7%. More than two thirds of patients (n = 94, 74%) had a positive family history for T2DM. Moreover, half of the patients (n = 64, 50.4%) had hypertension and, interestingly, 17.3% of the patients had high blood pressure and were not diagnosed with hypertension nor received any medication for elevated blood pressure.
Figure 1.
Patients inclusion and exclusion criteria.
All included patients showed good knowledge and compliance to their therapy regimen and medication dosing. The mean age for the responders and non-responders was 51.2 years and 49.6 years, respectively. The non-responders had a higher duration of diabetes, with an average of 2.6 years, compared to the 2.3 years of the responders group. All included patients, regardless of their group, were found to be obese, with a mean BMI of 39 Kg/m2.
Both groups were receiving an average daily dose of 4.5 mg for glimepiride. The non-responders group were receiving a slightly higher metformin daily dose of 1,221 mg compared to the responder group, who received 1,154 mg. Patients characteristics and demographics were all found to be statistically insignificant between the two groups, except for glycemic parameters. FBGL, HbA1C, and eAG, which were lower in the responders group compared to the non-responder group. The non-responders group had a higher mean FBGL (197.3 mg/dL) vs the responders group (123.6 mg/dL) (p ≤ 0.001). HbA1C was higher as well in the non-responders group, with an average of 8.3% compared to 6.0% in the responders group (p ≤ 0.001). Finally, the eAG calculated for non-responders was higher (192.9 mg/dL) than that for the responders group (123.7 mg/dL). All patients’ demographics are summarized in Table 1.
Table 1.
Patients characteristics.
| Responders, n = 93 (73.2%) | Non-responders, n = 34 (26.8%) | p-valuea | |
|---|---|---|---|
| Gender | |||
| Male (25.2%) | 27 | 5 | 0.100b |
| Female (74.8%) | 66 | 29 | |
| Age (years) | 51.2 (±7.7) | 49.6 (±9.2) | 0.327 |
| Treatment duration (years) | 2.3 (±1.1) | 2.6 (±1.2) | 0.239 |
| Metformin daily dose (mg/day) | 1,154.2 (±483) | 1,220.6 (±521) | 0.501 |
| Glimepiride daily dose (mg/day) | 4.5 (±1.8) | 4.5 (±1.7) | 0.895 |
| FBGL (mg/dL) | 123.6 (±21.4) | 197.3 (±35.7) | <0.001* |
| HbA1C (%) | 6.0 (±0.54) | 8.3 (±1.09) | <0.001* |
| eAG (mg/dL)c | 123.7 (±13.8) | 192.9 (±28.7) | <0.001* |
| BMI (Kg/m2) | 38.9 (±8.4) | 39 (±7.2) | 0.990 |
| Systole (mmHg) | 130.2 (±23.7) | 124.3 (±38.2) | |
| Diastole (mmHg) | 84.1 (±9.4) | 86.2 (±8.3) | |
| Concomitant medication | |||
| Lisinopril | 35 | 16 | |
| Aspirin | 24 | 9 | |
| Thioctic acid | 6 | 2 |
Abbreviations. BMI, body mass index; eAG, estimated average glucose; FBGL, fasting blood glucose level; HbA1C, glycated hemoglobin.
Mean ± standard deviation was used to express continuous variables.
Unpaired Student t-test was used for continuous variables.
Chi-squared test was used.
Calculated using the following equation: eAG = 28.7 × HbA1C – 46.7.
*Statistically significant at p ≤ 0.05 level.
Association between rs622342 and rs757110 and response
Both genetic variants were in Hardy–Weinberg equilibrium (p = 0.396 for rs622342, and p = 0.867 for rs757110). The minor allele frequencies were 18.9% for the C allele of rs622342, and 27.1% for the C allele of rs757110. The distribution of the two genetic variants among sample is presented in Table 2.
Table 2.
SLC22A1 rs622342 and ABCC8 rs757110 variants distribution among study groups.
| Genotype | Total sample (n = 127), n (%) | Responders (n = 93), n (%) | Non-responders (n = 34), n (%) | MAF | p-valuea | |
|---|---|---|---|---|---|---|
| Rs622342 A>C |
AA | 85 (66.9) | 67 (72) | 18 (52.9) | 0.189 | 0.043* |
| AC | 36 (28.3) | 26 (28)b | 16 (47.1)b | |||
| CC | 6 (4.8) | |||||
| Rs757110 A>C |
AA | 67 (52.8) | 47 (50.5) | 20 (58.8) | 0.271 | 0.408 |
| AC | 51 (40.2) | 46 (49.5)b | 14 (41.2)b | |||
| CC | 9 (7) |
Abbreviations. MAF, minor allele frequency.
Chi-square test was used to test the distribution of each genetic variant (AA alleles carriers and C allele carriers) between responder and non-responder groups.
Represents C-allele carriers (AA and AC).
*Statistically significant at p ≤ 0.05 level.
More than half of the responders were found to be carriers of AA alleles for SLC22A1 rs622342 variant with 72%. On the other hand, the non-responders were approximately equally distributed between AA alleles carriers (52.9%) and C allele carriers (47.1%). A statistically significant distribution between rs622342 allele groups and response groups was found (χ2 test, p = 0.043, Table 2). Logistic regression analysis was used, and none of the patients’ covariates were significantly associated with response, except for rs622342 genetic variant. AA alleles carriers of rs622342 variants were more responsive to metformin in combination therapy than C allele carriers (base model, OR = 2.291, p = 0.045, 95% CI = 1.017–5.157). The final model was adjusted for age, gender, BMI, treatment duration, metformin daily dose, and glimepiride daily dose, and the OR increased to 2.7 (OR = 2.718, p = 0.025, 95% CI = 1.112–6.385).
For the ABCC8 rs757110 genetic variant, the distribution of AA alleles carriers and C allele carriers was almost the same in both responders and non-responders groups (Table 2). The responders were found to be of approximately equally distributed between AA alleles carriers (50.5%) and C allele carriers (49.5%). ABCC8 rs757110 genetic variant did not show any association with response to combination therapy (p = 0.408).
Discussion
Diabetes is a global health problem, and its nature is multifactorial18. In some situations, such as in the case of a patient admitted with high HbA1c ≥ 9% or having an unhealthy and bad lifestyle, combination therapy would be recommended. Sulfonylureas are recommended among the treatment options available for combination therapy with metformin7,37. Several guidelines, including ADA, recommended achieving a range of 6.5–7% for good glycemic control and minimal risks for the associated complications38. Pharmacogenetic studies could help in investigating and identifying those variabilities seen in practice between patients receiving oral hypoglycemic agents11,12.
A total of 127 T2DM patients were included in the study, and the response rate to combination therapy was 73.2%. More than half of the patients were found to be hypertensive, with either already having a known diagnosis (50.4%) or relatively high blood pressure without a known diagnosis (17.3%); therefore, routine hypertension screening should be incorporated for patients with T2DM. Interestingly, all patients, regardless to their response to therapy, were found to have high BMI, and considered obese. This might be correlated to the unhealthy lifestyle of the Egyptians5, moreover, this might be one of the reasons that most practitioners prescribe combination therapy.
In previous studies, eAG was shown to be higher than FBGL39,40. The FBGL and eAG were expected to show a difference; however, they did not show any statistically significant difference. The insignificant difference between FBGL and eAG could result from patient incompliance to the FBGL test, which is a mandatory procedure in order to dispense monthly medications at the NIDE. Therefore, healthcare providers must ensure that all patients are compliant to the test and they had enough fasting.
Adherence and compliance to therapy is important in type 2 diabetic patients by which 50% of patients might fail to achieve glycemic goals due to their low adherence and compliance41. Poor patient compliance and adherence is very common in diabetic patients, and is associated with poor clinical outcomes42. In our sample, all patients (100%) showed good compliance when asked about their medication regimen, doses, and instructions. A study on Egyptian T2DM patients found that up to 84% of patients showed fair to good drug adherence43. The high compliance rate may arise from the weak method of measuring patient’s compliance, as no structured approach was adopted to capture the level of compliance and adherence.
SLC22A1, the encoding gene for OCT1, is highly polymorphic in different populations, and many polymorphisms have shown altered hepatic transporter function13,18. Therefore, OCT1 polymorphisms can affect metformin pharmacodynamics, but not its pharmacokinetics44,45. SLC22A1 rs622342 genetic variant is an intronic non-synonymous mutation that might be in linkage disequilibrium with another variant that results in a protein with less effective transporting capacity, or reduced OCT1 transcription and expression on hepatocytes, and eventually will decrease influx of metformin into the hepatocytes21,46.
To the best of our knowledge, this was the first study to investigate the pharmacogenetics of metformin in an Egyptian population. Additionally, the first to explore the minor allele frequency of rs622342 SLC22A1 genetic variant in Egyptian patients. The MAF in our sample (0.189) was considered lower than previously reported MAFs in Caucasians, which was 0.3719, and 0.253447 and 0.245 in Indian populations48, however it was almost similar to MAF (0.18) in a Cape Admixed population of South Africa49.
The influence of rs622342 on the response to metformin is controversial. Most of the studies include naive patients receiving metformin monotherapy, while in the current study we investigated the influence of such variants in diabetic patients already receiving metformin in combination with glimepiride. In the current study we found that AA alleles carriers of SLC22A1 rs622342 will be more responsive than C allele carriers; therefore, it was suggested that the C allele of rs622342 variant might have a defective transporter function. Our finding supported other studies in which they reported that C allele carriers had a reduced glucose lowering effect for metformin monotherapy18,19.
A study in South India was consistent with our finding in which AA alleles carriers were reported to have 3.56-times chance to respond to metformin monotherapy (recessive model, OR = 3.56, 95% CI = 0.83–15.26)18. On the other hand, two studies reported that rs622342 had no impact on metformin blood glucose lowering effects47,50.
The ABCC8 rs757110 genetic variant has a significant influence on the sensitivity and function of SUR1. Additionally, it was suggested that mutation in the ABCC8 gene could be a factor in developing T2DM51,52. Our study was not the first to explore MAF of ABCC8 rs757110 in an Egyptian population; however, Ghanem et al.23 found that the MAF of rs757110 was 0.245, which is slightly lower than our reported MAF (MAF = 0.271). In Caucasians, rs757110 was reported to have a MAF between 0.352 and 0.38826.
The effect of ABCC8 polymorphism on Sulfonylureas response has been debatable as well. Many studies reported that ABCC8, especially rs757110, has no influence on sulfonylureas response22,53. These studies supported our finding, despite that they had not unified the used sulfonylureas agent among their samples. In a Korean population, rs757110 was reported to have no effect on the efficacy of glimepiride24.
The rs757110 variant in ABCC8 gene results in a change of amino acid in position 1369, which is in close proximity to A-site of SUR154,55. Hence, only Sulfonylureas which interact with SUR1 through the A-site are expected to be affected by the ABCC8 rs757110 polymorphism. Glimepiride binds to both A and B sites of SUR127,54,55, therefore it should be least affected by such mutation, and our finding was consistent with such postulation. On the contrary, gliclazide binds only to the A-site of SUR1, and therefore its response would be affected by any alteration in that site, as in the case of the ABCC8 rs757110 polymorphism27,54,55, and this was supported by a Chinese study which investigated the influence of rs757110 on gliclazide only56.
Differences between MAF observed for the studied genetic variants in our Egyptian population and Caucasians could suggest that Egyptians might have admixed genetic build-up and should not be regarded as Caucasians. This was previously proposed by pharmacogenetic studies of warfarin and clopidogrel on Egyptian populations29,30.
A limitation to the current study is the variation in the treatment duration among the included patients. Although treatment duration was not statistically significantly different between the two groups, the difference in drug exposure can affect clinical outcomes. Another limitation is the lack of lifestyle information and the weak compliance and adherence assessment of the patients, which could affect the glycemic control and response to hypoglycemic agents.
Conclusion
SLC22A1 rs622342 genotyping can be a good predictor for the response of metformin in combination therapy in patients with type 2 diabetes. Moreover, the current study could help in increasing the awareness of the importance of pharmacogenetics in clinical practice in Egypt. Finally, pharmacogenetic studies help in identifying variabilities of drug response between patients, and aid in selecting the most appropriate drug for each patient, and ultimately will save costs. We recommend having more pharmacogenetics studies with larger sample sizes and a wider genetic panel for the available T2DM treatments in Egyptian populations.
Transparency
Declaration of funding
There is no funding to declare for this research.
Declaration of financial/other relationships
The authors and JDA peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Acknowledgements
The authors would like to thank Ms Shaimaa Mohamed Ewis, Department of Biochemistry and Molecular Biology, Helwan University, for her guidance and assistance in conducting the genotyping.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Upadhyay J, Polyzos SA, Perakakis N, et al. Pharmacotherapy of type 2 diabetes: An update. Metabolism. 2018;78:13–42. [DOI] [PubMed] [Google Scholar]
- 2.Zheng Y, Ley SH, Hu FB. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol. 2017;14:88–98. [DOI] [PubMed] [Google Scholar]
- 3.Majeed A, El-Sayed AA, Khoja T, et al. Diabetes in the Middle-East and North Africa: An update. Diabetes Res Clin Pract. 2014;103:218–222. [DOI] [PubMed] [Google Scholar]
- 4.Assaad Khalil SH, Megallaa MH, Rohoma KH, et al. Prevalence of type 2 diabetes mellitus in a sample of the adult population of Alexandria, Egypt. Diabetes Res Clin Pract. 2018;144:63–73. [DOI] [PubMed] [Google Scholar]
- 5.Shelbaya S, Rakha S. Effectiveness and safety of vildagliptin and vildagliptin add-on to metformin in real-world settings in Egypt - results from the GUARD study. Curr Med Res Opin. 2017;33:797–801. [DOI] [PubMed] [Google Scholar]
- 6.Cai X, Gao X, Yang W, et al. Efficacy and safety of initial combination therapy in Treatment-Naïve type 2 diabetes patients: a systematic review and Meta-analysis. Diabetes Ther. 2018;9:1995–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.American Diabetes Association 8. Pharmacologic approaches to glycemic treatment: Standards of Medical Care in Diabetes—2018. Diabetes Care [Internet]. 2018;41:S73 LP-S85. [DOI] [PubMed] [Google Scholar]
- 8.Cersosimo E, Johnson EL, Chovanes C, et al. Initiating therapy in patients newly diagnosed with type 2 diabetes: Combination therapy vs a stepwise approach. Diabetes Obes Metab. 2018;20:497–507. [DOI] [PubMed] [Google Scholar]
- 9.Phung OJ, Sobieraj DM, Engel SS, et al. Early combination therapy for the treatment of type 2 diabetes mellitus: systematic review and meta-analysis. Diabetes Obes Metab. 2014;16:410–417. [DOI] [PubMed] [Google Scholar]
- 10.Hassan MH, Abd-Allah GM. Effects of metformin plus gliclazide versus metformin plus glimepiride on cardiovascular risk factors in patients with type 2 diabetes mellitus. Pak J Pharm Sci. 2015;28:1723–1730. [PubMed] [Google Scholar]
- 11.Staiger H, Schaeffeler E, Schwab M, et al. Pharmacogenetics: Implications for modern type 2 diabetes therapy. Rev Diabet Stud. 2015;12:363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ordelheide A, Hrabě de Angelis M, Häring H-U, et al. Pharmacogenetics of oral antidiabetic therapy. Pharmacogenomics. 2018;19:577–587. [DOI] [PubMed] [Google Scholar]
- 13.Sundelin EIO, Gormsen LC, Jensen JB, et al. Genetic polymorphisms in organic cation transporter 1 attenuates hepatic metformin exposure in humans. Clin Pharmacol Ther. 2017;102:841–848. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez-Ibarra HE, Reyes-Cortes LM, Jiang X-L, et al. Genotypic and phenotypic factors influencing drug response in mexican patients with Type 2 diabetes mellitus. Front Pharmacol. 2018;9:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dujic T, Zhou K, Yee SW, et al. Variants in pharmacokinetic transporters and glycemic response to metformin: a metgen Meta-Analysis. Clin Pharmacol Ther. 2017;101:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pawlyk AC, Giacomini KM, McKeon C, et al. Metformin pharmacogenomics: current status and future directions. Diabetes. 2014;63:2590–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Florez JC. The pharmacogenetics of metformin. Diabetologia. 2017;60:1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umamaheswaran G, Praveen RG, Damodaran SE, et al. Influence of SLC22A1 rs622342 genetic polymorphism on metformin response in South Indian type 2 diabetes mellitus patients. Clin Exp Med. 2015;15:511–517. [DOI] [PubMed] [Google Scholar]
- 19.Becker ML, Visser LE, van Schaik RHN, et al. Genetic variation in the organic cation transporter 1 is associated with metformin response in patients with diabetes mellitus. Pharmacogenomics J. 2009;9:242–247. [DOI] [PubMed] [Google Scholar]
- 20.rs622342 RefSNP Report - dbSNP - NCBI [Internet]. [cited 2019. Apr 20]. Available from: https://www.ncbi.nlm.nih.gov/snp/rs622342#publications.
- 21.Becker ML, Visser LE, van Schaik RHN, et al. OCT1 polymorphism is associated with response and survival time in anti-Parkinsonian drug users. Neurogenetics. 2011;12:79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klen J, Dolžan V, Janež A. CYP2C9, KCNJ11 and ABCC8 polymorphisms and the response to sulphonylurea treatment in type 2 diabetes patients. Eur J Clin Pharmacol. 2014;70:421–428. [DOI] [PubMed] [Google Scholar]
- 23.Ghanem AI, Rushdy S, Mokhtar M. Association between KCNJ11 & ABCC8 Genetic Polymorphism and Type 2 Diabetes in Egyptian Patients Med J Cairo Univ. 2016;84:1501–1510. [Google Scholar]
- 24.Cho H, Lee S, Kim Y-G, et al. Effect of genetic polymorphisms on the pharmacokinetics and efficacy of glimepiride in a Korean population. Clin Chim Acta. 2011;412:1831–1834. [DOI] [PubMed] [Google Scholar]
- 25.van Leeuwen N, Swen JJ, Guchelaar H-J, et al. The role of pharmacogenetics in drug disposition and response of oral glucose-lowering drugs. Clin Pharmacokinet. 2013;52:833–854. [DOI] [PubMed] [Google Scholar]
- 26.rs757110 RefSNP Report - dbSNP - NCBI [Internet]. [cited 2019. Apr 20]. Available from: https://www.ncbi.nlm.nih.gov/snp/rs757110.
- 27.Sato R, Watanabe H, Genma R, et al. ABCC8 polymorphism (Ser1369Ala): influence on severe hypoglycemia due to sulfonylureas. Pharmacogenomics. 2010;11:1743–1750. [DOI] [PubMed] [Google Scholar]
- 28.Knapp S, Zakaria Z, Hashem M, et al. Influence of IFNL3.rs12979860 and IFNL4.ss469415590 polymorphism on clearance of hepatitis C virus infection among Egyptians. Hepatol Int. 2015;9:251–257. [DOI] [PubMed] [Google Scholar]
- 29.Khalil B, Shahin M, Solayman M, et al. Genetic and nongenetic factors affecting clopidogrel response in the Egyptian Population. Clin Transl Sci. 2016;9:23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shahin MHA, Khalifa SI, Gong Y, et al. Genetic and nongenetic factors associated with warfarin dose requirements in Egyptian patients. Pharmacogenet Genomics. 2011;21:130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipscombe L, Booth G, Butalia S, et al. Diabetes Canada 2018 clinical practice guidelines for the prevention and management of diabetes in canada: pharmacologic glycemic management of Type 2 diabetes in adults. Can J Diabetes. 2018;42:S88–S103. [DOI] [PubMed] [Google Scholar]
- 32.Sherifali D, Nerenberg K, Pullenayegum E, et al. The effect of oral antidiabetic agents on A1C levels: a systematic review and meta-analysis. Diabetes Care. 2010;33:1859–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dujic T, Zhou K, Donnelly LA, et al. Association of organic cation transporter 1 with intolerance to metformin in type 2 diabetes: A GoDARTS study. Diabetes. 2015;64:1786–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahlin G, Chen L, Lazorova L, et al. Genotype-dependent effects of inhibitors of the organic cation transporter, OCT1: predictions of metformin interactions. Pharmacogenomics J. 2011;11:400–411. [DOI] [PubMed] [Google Scholar]
- 35.eAG/A1C Conversion Calculator | American Diabetes Association [Internet]. [cited 2018 Sep 20]. Available from: https://professional.diabetes.org/diapro/glucose_calc.
- 36.American Diabetes Association 6. Glycemic targets: Standards of Medical Care in Diabetes—2018. Diabetes Care. 2018;41:S55–S64. [DOI] [PubMed] [Google Scholar]
- 37.Ren Q, Xiao D, Han X, et al. Genetic and clinical predictive factors of sulfonylurea failure in patients with Type 2 diabetes. Diabetes Technol Ther. 2016;18:586–593. [DOI] [PubMed] [Google Scholar]
- 38.Qaseem A, Wilt TJ, Kansagara D, et al. Hemoglobin A1c targets for glycemic control with pharmacologic therapy for nonpregnant adults with type 2 diabetes mellitus: a guidance statement update from the American College of Physicians. Ann Intern Med. 2018;168:569–576. [DOI] [PubMed] [Google Scholar]
- 39.Kim HY, Lee SY, Suh S, et al. The relationship between estimated average glucose and fasting plasma glucose. Clin Chem Lab Med. 2013;51:2195–2200. [DOI] [PubMed] [Google Scholar]
- 40.Bozkaya G, Ozgu E, Karaca B. The association between estimated average glucose levels and fasting plasma glucose levels. Clinics. 2010;65:1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polonsky WH, Henry RR. Poor medication adherence in type 2 diabetes: Recognizing the scope of the problem and its key contributors. PPA. 2016;10:1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pattnaik S, Ausvi S, Salgar A, et al. Treatment compliance among previously diagnosed type 2 diabetics in a rural area in Southern India. J Family Med Prim Care. 2019;8:919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shams MEE, Barakat E. Measuring the rate of therapeutic adherence among outpatients with T2DM in Egypt. Saudi Pharm J. 2010;18:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zamek-Gliszczynski MJ, Giacomini KM, Zhang L. Emerging clinical importance of hepatic organic cation transporter 1 (OCT1) in drug pharmacokinetics, dynamics, pharmacogenetic variability, and drug interactions. Clin Pharmacol Ther. 2017;0:1–3. [DOI] [PubMed] [Google Scholar]
- 45.Christensen MMH, Højlund K, Hother-Nielsen O, et al. Steady-state pharmacokinetics of metformin is independent of the OCT1 genotype in healthy volunteers. Eur J Clin Pharmacol. 2015;71:691–697. [DOI] [PubMed] [Google Scholar]
- 46.Becker ML, Visser LE, van Schaik RHN, et al. Interaction between polymorphisms in the OCT1 and MATE1 transporter and metformin response. Pharmacogenet Genomics. 2010;20:38–44. [DOI] [PubMed] [Google Scholar]
- 47.Tkáč I, Klimčáková L, Javorský M, et al. Pharmacogenomic association between a variant in SLC47A1 gene and therapeutic response to metformin in type 2 diabetes. Diabetes Obes Metab. 2013;15:189–191. [DOI] [PubMed] [Google Scholar]
- 48.Umamaheswaran G, Praveen RG, Arunkumar AS, et al. Genetic analysis of OCT1 gene polymorphisms in an Indian population. Indian J Hum Genet. 2011;17:164–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du Plessis M, Pearce B, Jacobs C, et al. Genetic polymorphisms of the organic cation transporter 1 gene (SLC22A1) within the Cape Admixed population of South Africa. Mol Biol Rep. 2015;42:665–672. [DOI] [PubMed] [Google Scholar]
- 50.Jablonski KA, McAteer JB, de Bakker PIW, et al. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59:2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fatehi M, Carter CRJ, Youssef N, et al. Molecular determinants of ATP-sensitive potassium channel MgATPase activity: diabetes risk variants and diazoxide sensitivity. Biosci Rep. 2015;35:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rasheed MA, Kantoush N, Abd El-Ghaffar N, et al. Expression of JAZF1, ABCC8, KCNJ11and Notch2 genes and vitamin D receptor polymorphisms in type 2 diabetes, and their association with microvascular complications. Ther Adv Endocrinol Metab. 2017;8:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holstein JD, Kovacs P, Patzer O, et al. The Ser1369Ala variant of ABCC8 and the risk for severe sulfonylurea-induced hypoglycemia in German patients with Type 2 diabetes. Pharmacogenomics. 2012;13:5–7. [DOI] [PubMed] [Google Scholar]
- 54.Hamming KSC, Soliman D, Matemisz LC, et al. Coexpression of the type 2 diabetes susceptibility gene variants KCNJ11 E23K and ABCC8 S1369A alter the ATP and sulfonylurea sensitivities of the ATP-sensitive K(+) channel. Diabetes. 2009;58:2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lang VY, Fatehi M, Light PE. Pharmacogenomic analysis of ATP-sensitive potassium channels coexpressing the common type 2 diabetes risk variants E23K and S1369A. Pharmacogenet Genomics. 2012;22:206–214. [DOI] [PubMed] [Google Scholar]
- 56.Feng Y, Mao G, Ren X, et al. Ser 1369Ala variant in sulfonylurea receptor gene ABCC8 Is associated with antidiabetic efficacy of gliclazide in Chinese Type 2 diabetic patients. Diabetes Care. 2008;31:1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.