Short abstract
Objective
MicroRNA (miR)-142-3p may function as a tumor suppressor in the development of various cancers. In this study, we measured serum levels of miR-142-3p in patients with colorectal cancer (CRC) to evaluate the diagnostic and prognostic value of miR-142-3p.
Methods
Serum samples from 363 consecutive CRC patients and 156 healthy controls were retrospectively collected. Serum miR-142-3p levels were measured using real-time quantitative reverse transcription polymerase chain reaction. All patients were followed up regularly after tumor resection. The correlation between serum miR-142-3p level and survival outcomes was analyzed.
Results
Serum levels of miR-142-3p were significantly lower in CRC patients than in healthy volunteers. A low serum miR-142-3p level was significantly associated with advanced cancer. Survival analysis demonstrated that patients with a low serum miR-142-3p had a lower 5-year overall survival rate than patients with a high serum miR-142-3p level (67.4% vs. 76.9%). Serum miR-142-3p level was also shown to be an independent risk factor for CRC in multivariate analysis (hazard ratio, 2.68; 95% confidence interval: 1.21–7.95).
Conclusions
Serum miR-142-3p might serve as a useful diagnostic and prognostic marker for CRC.
Keywords: Serum miR-142-3p, prognosis, colorectal cancer, biomarker, tumor suppression, microRNA
Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed and lethal gastrointestinal cancers in China and worldwide.1,2 Although considerable advancements in diagnostic and therapeutic modalities have improved early detection and decreased mortality over the previous decades, the prognosis of CRC remains poor, especially for patients who are diagnosed at advanced stages.3–5 Recurrence after treatment is one of the main contributors to unsatisfactory long-term prognosis for localized and operable CRC.6,7 Moreover, in clinical practice, CRC patients present with heterogeneous prognosis and treatment responses. Therefore, efficient and effective risk stratification of patients is required for clinical management of CRC.8 Previous studies have established several classical methods for the risk stratification of CRC patients and biomarkers to guide administration of adjuvant treatments, such as TNM classification and histological stage.9,10 However, these biomarkers cannot be obtained until the postoperative histological evaluation of the carcinoma tissue has been completed, which is not conducive to determining preoperative neo-adjuvant treatments.11–13 Therefore, more feasible and effective biomarkers that can be obtained before treatment are needed for CRC prognosis and risk stratification.
The microRNAs (miRNAs) are a subset of small noncoding RNAs consisting of approximately 18 to 22 nucleotides that can inhibit gene expression by specifically binding the 3′ untranslated regions of their target messenger RNAs (mRNAs), resulting in translation suppression of specific protein-coding genes.14 Dysregulation of miRNAs plays a crucial role in the development of various malignancies.15–20 Previous studies have demonstrated that miRNAs are remarkably stable in serum or plasma samples at measurable concentrations.21–24 Serum miRNAs can bind to specific proteins or be packaged into apoptotic bodies or exosomes, rendering them resistant to endogenous ribonuclease activity.25,26 Thus, expression of different serum miRNAs is a valuable biomarker for cancer.27–29
MicroRNA-142-3p is a tumor suppressor miRNA that targets several oncogenes and is strongly downregulated in various types of cancer.30,31 The aberrant miR-142-3p expression not only has diagnostic value but also predicts prognosis for cancer patients.32 Overexpression of miR-142-3p can inhibit the proliferation, invasion, and migration of gastric cancer cells.33 However, no reports have established the significance of serum miR-142-3p levels in patients with CRC. Therefore, in the current study, we evaluated serum levels of miRNA-142-3p in CRC patients to determine the prognostic value.
Methods
Patients
This study was conducted according to the relevant global and local guidelines or regulations. All subjects provided written informed consent before enrollment. Our study was also approved by the institutional review boards of the Second Affiliated Hospital of Zhejiang Chinese Medical University.
Data of primary colorectal cancer patients who underwent curative surgery between June 1, 2013, and June 1, 2017, were retrospectively collected. A number of cases were excluded from the study because of the following exclusion criteria: histology other than adenocarcinoma, preoperative acute and severe comorbidity, distant metastasis, preoperative neo-adjuvant chemotherapy, unavailable clinical and histopathological data, and life expectancy less than 24 weeks. All patients underwent R0 resection and postoperative adjuvant radiotherapy or chemotherapy, or both, according to 2015 NCCN Colorectal Cancer Practice Guidelines,34 and all of the tumor specimens were pathologically evaluated as colorectal adenocarcinoma. The control group consisted of age-matched healthy subjects without history of cancer and in good health status based on self-report.
Clinical and pathological data of all patients were collected and checked, including sex, age, tumor site and size, tumor invasion depth, lymph node involvement, TNM stage, histopathological differentiation, and surgery records. Tumor stages were evaluated based on the Union for International Cancer Control (UICC) classification system. All patients were regularly followed up through clinical visits or telephone. Clinical follow-up lasted from the surgery day to either death or June 2018. The primary outcome of interest was 5-year overall survival (OS) rate; OS was defined as the duration from surgery day to death.
Sample preparation and RNA isolation
Five milliliters of sterile peripheral venous blood was collected from each patient on the day before surgery and from healthy controls in the corresponding period. Serum was extracted from blood samples by centrifugation and then transferred to RNase/DNase-free tubes and immediately stored at −80 °C until further processing. Total RNA was extracted from serum by use of a miRNEasy Serum/Plasma Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The RNA concentration and integrity were quantified by using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies LLC, Wilmington, DE, USA).
Quantification of miRNA by qRT-PCR
Total RNA from study participants was applied to reverse transcribe miRNAs to cDNA using a miScript Reverse Transcription Kit (Qiagen). Amplifications were performed using a miScript SYBR Green PCR kit (Qiagen) and real-time quantitative (qRT)PCR was run on an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The sequence of the primer used in reverse transcription for miR-142-3p was GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCCATAA. The expression levels of each miRNA were normalized against miR-16 expression, and threshold cycle (Ct) values ≥40 were considered undetectable. The relative expression of serum miR-142-3p was quantitatively analyzed by the 2–ΔΔCT method. Each sample was analyzed in triplicate.
Statistical analysis
All statistical analyses in this study were performed using IBM SPSS Statistics for Windows, Version 20.0 (IBM Corp., Armonk, NY, USA). The results were considered statistically significant when p < 0.05 (two sided). Continuous variables expressed as means ± standard deviations were compared by using analysis of variance, and comparisons of categorical variables were conducted using χ2 or Fisher’s exact test and presented as frequencies and percentages. Receiver operating characteristic (ROC) curve analysis was conducted to assess the feasibility of serum miRNA-142-3p levels as a diagnostic indicator for CRC detection. The cut-off value for the serum miRNA-142-3p levels predicting survival was also evaluated by ROC analysis. Kaplan–Meier survival curves were analyzed by log-rank test in the univariate analysis. A multivariate Cox hazard regression model was used to confirm the independent prognostic factors for CRC.
Results
Patients
Data of 363 primary colorectal cancer patients who underwent curative surgery were retrospectively collected. The control group consisted of 156 age-matched healthy individuals.
Serum miR-142-3p expression profiling in patients with CRC
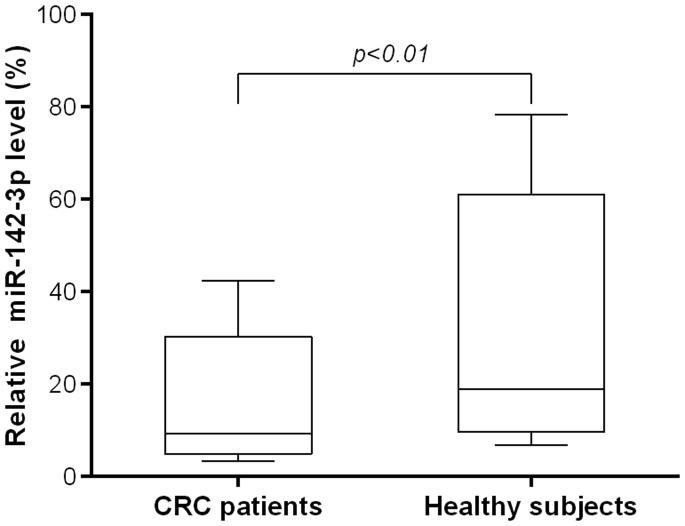
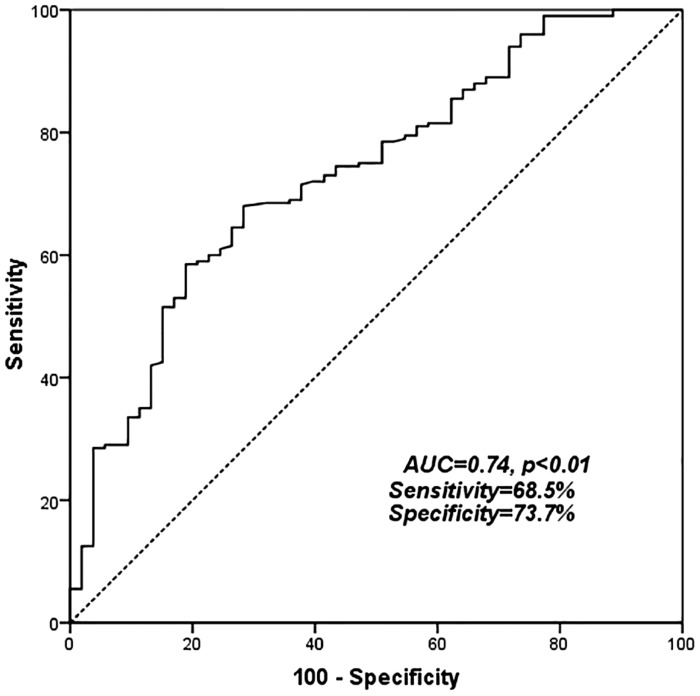
Serum miR-142-3p was detected in all blood samples from 363 patients with CRC and 156 controls. The serum levels of miR-142-3p were significantly lower in CRC patients than in healthy control subjects (p < 0.01; Figure 1). Furthermore, ROC curve analysis showed that serum miR-142-3p level could serve as a diagnostic biomarker, distinguishing patients CRC and healthy individuals with a sensitivity of 68.5% and a specificity of 73.7%. The area under the curve was 0.74 [95% confidence interval (CI): 0.68–0.86, p < 0.01; Figure 2].
Figure 1.
Serum microRNA (miR)-142-3p level in patients with colorectal cancer (CRC) and healthy controls. The serum miR-142-3p levels of 363 patients with CRC was significantly lower than that of 156 age-matched healthy volunteers (p < 0.01). The box indicates the interquartile range, the line indicates the median, and the whiskers indicate the standard deviation.
Figure 2.
Receiver operator characteristic (ROC) curve for colorectal cancer detection. ROC analysis showed an area under the curve (AUC) of 0.74 for microRNA (miR)-142-3p with a 95% confidence interval of 0.68–0.86 (p < 0.01).
Relationship between serum miR-142-3p and clinicopathological variables
In this study, we analyzed associations between serum miR-142-3p levels and clinicopathological characteristics by using the χ2 test. The results showed that a low serum miR-142-3p levels were significantly associated with advanced T stage (p < 0.01) and TNM stage (p < 0.01). We detected no significant differences between high and low serum miR-142-3p groups for other clinicopathological characteristics, including sex, age, tumor site and size, or pathological differentiation and N stage (Table 1).
Table 1.
Correlation between serum microRNA (miR)-142-3p level and clinical variables of patients with colorectal cancer.
| Characteristic | Serum miR-142-3p |
p-value | |
|---|---|---|---|
| High (n = 127) | Low (n = 236) | ||
| Age (years) | 0.38 | ||
| ≥60 | 61 | 102 | |
| <60 | 66 | 134 | |
| Sex | 0.70 | ||
| Male | 79 | 142 | |
| Female | 48 | 94 | |
| Tumor site | 0.07 | ||
| Colon | 56 | 81 | |
| Rectum | 71 | 155 | |
| Tumor size (cm) | 0.92 | ||
| ≥5 | 48 | 88 | |
| <5 | 79 | 148 | |
| T stage | <0.01 | ||
| T1 + T2 | 84 | 107 | |
| T3 | 43 | 129 | |
| Node involvement | 0.80 | ||
| N0 | 51 | 98 | |
| N1 | 76 | 138 | |
| Clinical stage (TNM) | <0.01 | ||
| I + II | 79 | 81 | |
| III | 48 | 155 | |
| Pathological differentiation | 0.24 | ||
| Good/moderate | 71 | 147 | |
| Poor | 56 | 89 | |
OS, overall survival; CI, confidence interval; HR, hazard ratio; miR-142-3p, microRNA-142-3p.
Prognostic value of serum miR-142-3p level in patients with CRC
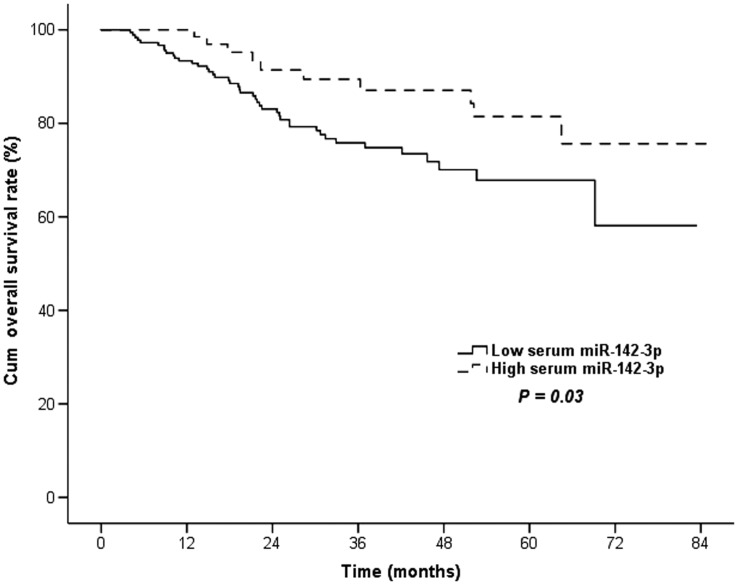
The median follow-up period was 35.4 months (range, 6.1–85.1 months). During follow-up, there were 79 (21.8%) tumor-specific deaths in the group of patients with CRC. In the univariate survival analysis, we found that patients with low expression of serum miR-142-3p had a significantly worse 5-year OS rate compared with those with high expression of miR-142-3p (67.4% vs. 76.9%, p = 0.03; Table 2, Figure 3).
Table 2.
Prognostic characteristics of patients with colorectal cancer.
| Characteristic | Univariate |
Multivariate |
||||
|---|---|---|---|---|---|---|
| n | 5-year OS rate (%) | p-value | HR | 95% CI | p-value | |
| Age (years) | 0.82 | |||||
| ≥60 | 163 | 67.2 | ||||
| <60 | 200 | 72.1 | ||||
| Sex | 0.12 | |||||
| Male | 221 | 72.5 | ||||
| Female | 142 | 71.7 | ||||
| Tumor site | 0.37 | |||||
| Colon | 137 | 70.4 | ||||
| Rectum | 226 | 69.6 | ||||
| Tumor size (cm) | 0.16 | |||||
| ≥5 | 136 | 69.3 | ||||
| <5 | 227 | 72.8 | ||||
| Tumor invasion depth | 0.02 | |||||
| T1 + T2 | 191 | 75.3 | ||||
| T3 + T4 | 172 | 69.2 | ||||
| Lymph node involvement | 0.01 | |||||
| N0 | 149 | 74.2 | ||||
| N1 | 214 | 67.6 | ||||
| Clinical stage (TNM) | 0.04 | 3.32 | 1.98–8.62 | <0.01 | ||
| I + II | 160 | 75.3 | ||||
| III | 213 | 6412 | ||||
| Pathological differentiation | 0.01 | 1.27 | 1.02–3.67 | 0.01 | ||
| Good/moderate | 218 | 75.1 | ||||
| Poor | 145 | 64.2 | ||||
| Serum miR-142-3p | 0.03 | 2.68 | 1.21–7.95 | <0.01 | ||
| Low | 236 | 67.4 | ||||
| High | 127 | 76.9 | ||||
OS, overall survival; HR, hazard ratio; CI, confidence interval; miR-142-3p, microRNA-142-3p.
Figure 3.
Lower plasma microRNA (miR)-142-3p level was associated with a worse prognosis for colorectal cancer. Prognostic analysis revealed that a low serum miR-142-3p level was significantly associated with a worse cumulative overall survival rate (p = 0.03).
The multivariate analysis included age and sex of patients, tumor size, lymph node involvement, clinical stage, histological differentiation type, and preoperative miR-142-3p level in a Cox regression model to determine independent prognostic biomarkers for patients with CRC. After adjustment for potential confounders, a low serum miR-142-3p level (p < 0.01; hazard ratio, 2.68; 95% CI: 1.21–7.95) was identified as an independent predictive risk factor for survival outcome of CRC patients, independent of classical prognostic biomarkers such as TNM stage (p < 0.01; hazard ratio, 3.32; 95% CI: 1.98–8.62) and pathological differentiation (p = 0.01; hazard ratio, 1.27; 95% CI: 1.02–3.67) (Table 2).
Discussion
In the current study, we found that the serum level of the tumor suppressor miR-142-3p was significantly downregulated in patients with CRC compared with healthy controls. Moreover, serum miR-142-3p was evaluated and determined to be a good indicator to discriminate CRC patients from healthy individuals. We also evaluated the potential value of serum miR-142-3p obtained before surgery as a candidate indicator to predict postoperative prognosis of CRC. We showed that low serum miR-142-3p was significantly related to poor survival of CRC patients. Low serum miR-142-3p levels were significantly correlated with unfavorable clinicopathological characteristics in CRC patients. According to these results, we propose that miR-142-3p could be used for diagnosis and optimal risk stratification of CRC patients, and that serum miR-142-3p is a promising biomarker for postoperative prognosis of CRC patients.
MiR-142-3p is generally expressed in human tissues and organs, and it has critical roles in biological processes of cancer cleavage and inhibiting translation.35 This microRNA has been confirmed as a tumor suppressor that is downregulated in colon cancer.36 It has been reported that miR-142-3p binds to the 3′-untranslated regions and coding sequences (thus inhibiting gene expression) of three risk genes associated with poor outcomes in colon cancer: CD133, ABCG2 (ATP binding cassette G2), and Lgr5 (leucine-rich-repeat-containing G-protein coupled receptor 5).37 Levels of miR-142-3p were shown to be decreased in color cancer samples and negatively correlated with expression of these three genes.37 Moreover, miR-142-3p may be involved in the regulation of cancer cell proliferation and metastasis in colorectal cancer by targeting transcription factor 7 (TCF-7), fatty acid synthase (FASN), and MYC oncogene.38 In the current study, we showed that miR-142-3p is highly downregulated and correlated with poor prognosis in CRC patients, which is consistent with results from the earlier studies.
The function, mechanism, and origin of circulating miR-142-3p in cancer patients have not yet been fully elucidated. Several mechanisms for the release of circulating miRNAs have been suggested, including passive leakage from cells due to injury, chronic inflammation, or necrosis; active secretion via membrane vesicles such as exosomes; and active secretion by complex formation with lipoproteins or RNA binding proteins. Furthermore, circulating miRNAs secreted from cancer cells can induce tumorigenesis in recipient cells.23,39,40 Wu et al.41 reported that miR-142-3p directly and negatively regulated RAC1 in hepatocellular carcinoma cells. Wang et al.33 reported that miR-142-3p acts as a tumor suppressor in gastric cancer carcinogenesis partly by downregulating CCNT2. Increased expression of certain miRNAs is significantly associated with advanced clinical stages of cancer.42 Our results were consistent with these previous studies. Therefore, serum miR-142-3p level could be a novel treatment target for patients CRC.
This study is the first to report, in a large group of patients, that miR-142-3p level is depleted in the serum of CRC patients and that miR-142-3p can serve as both a serum biomarker and a novel therapeutic target for CRC. However, the present study has several limitations, one of which is that the study was conducted at a single center and used a retrospective design. A large-scale, multicenter prospective study with a longer follow-up is needed to confirm our results. Furthermore, the roles of miR-142-3p in development of colorectal cancer and the underlying mechanisms are not yet fully understood. Further experiments must be conducted to elucidate the mechanisms of miR-142-3p in carcinogenesis.
Conclusions
We demonstrated that serum miR-142-3p levels were downregulated in patients with CRC. Moreover, low serum miR-142-3p levels were positively correlated with poor prognosis of CRC, suggesting that miR-142-3p may function as a tumor suppressor gene in CRC. Serum miR-142-3p might serve not only as a diagnostic and prognostic indicator for operable CRC but also as a potential novel treatment target.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017; 67: 177–193. DOI: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66: 115–132. DOI: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Barton MK. Primary tumor location found to impact prognosis and response to therapy in patients with metastatic colorectal cancer. CA Cancer J Clin 2017; 67: 259–260. DOI: 10.3322/caac.21372. [DOI] [PubMed] [Google Scholar]
- 4.Feng W, Cui G, Tang CW, et al. Role of glucose metabolism related gene GLUT1 in the occurrence and prognosis of colorectal cancer. Oncotarget 2017; 8: 56850–56857. DOI: 10.18632/oncotarget.18090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y, Gaedcke J, Emons G, et al. Colorectal cancer susceptibility loci as predictive markers of rectal cancer prognosis after surgery. Genes Chromosomes Cancer 2018; 57: 140–149. DOI: 10.1002/gcc.22512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song XM, Yang ZL, Wang L, et al. [Clinicopathological characteristics and prognosis of patients with recurrent colorectal cancer]. Zhonghua wei chang wai ke za zhi 2006; 9: 492–494[in Chinese, English Abstract]. [PubMed] [Google Scholar]
- 7.Moon A, Do SI, Kim HS, et al. Downregulation of osteoprotegerin expression in metastatic colorectal carcinoma predicts recurrent metastasis and poor prognosis. Oncotarget 2016; 7: 79319–79326. DOI: 10.18632/oncotarget.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagtegaal ID, Quirke P, Schmoll HJ. Has the new TNM classification for colorectal cancer improved care? Nat Rev Clin Oncol 2011; 9: 119–123. DOI: 10.1038/nrclinonc.2011.157. [DOI] [PubMed] [Google Scholar]
- 9.Han J, Zhang X, Zhang AD, et al. [Impact of primary tumor site on the prognosis in different stage colorectal cancer patients after radical resection]. Zhonghua wai ke za zhi 2018; 56: 68–73[in Chinese, English Abstract]. DOI: 10.3760/cma.j.issn.0529-5815.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Zhang G, Wang HL, et al. Analysis of expression of cyclin E, p27kip1 and Ki67 protein in colorectal cancer tissues and its value for diagnosis, treatment and prognosis of disease. Eur Rev Med Pharmacol Sci 2016; 20: 4874–4879. [PubMed] [Google Scholar]
- 11.Van Schaeybroeck S, Allen WL, Turkington RC, et al. Implementing prognostic and predictive biomarkers in CRC clinical trials. Nat Rev Clin Oncol 2011; 8: 222–232. DOI: 10.1038/nrclinonc.2011.15. [DOI] [PubMed] [Google Scholar]
- 12.Quirke P, Williams GT, Ectors N, et al. The future of the TNM staging system in colorectal cancer: time for a debate? Lancet Oncol 2007; 8: 651–657. DOI: 10.1016/S1470-2045(07)70205-X. [DOI] [PubMed] [Google Scholar]
- 13.Lorenc Z, Waniczek D, Lorenc-Podgorska K, et al. Profile of expression of genes encoding Matrix Metallopeptidase 9 (MMP9), Matrix Metallopeptidase 28 (MMP28) and TIMP Metallopeptidase Inhibitor 1 (TIMP1) in colorectal cancer: assessment of the role in diagnosis and prognostication. Med Sci Monit 2017; 23: 1305–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis 2015; 35: 3–11. DOI: 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Guo L, Li Y, et al. MicroRNA-494 promotes cancer progression and targets adenomatous polyposis coli in colorectal cancer. Mol Cancer 2018; 17: 1. DOI: 10.1186/s12943-017-0753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zabaglia LM, Bartolomeu NC, Dos Santos MP, et al. Decreased MicroRNA miR-181c expression associated with gastric cancer. J Gastrointest Cancer 2018; 49: 97–101. DOI: 10.1007/s12029-017-0042-7. [DOI] [PubMed] [Google Scholar]
- 17.Polasik A, Tzschaschel M, Schochter F, et al. Circulating tumour cells, circulating tumour DNA and circulating MicroRNA in metastatic breast carcinoma - what is the role of liquid biopsy in breast cancer? Geburtshilfe Frauenheilkd 2017; 77: 1291–1298. DOI: 10.1055/s-0043-122884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin C, Zhao Y, Gong C, et al. MicroRNA-154/ADAM9 axis inhibits the proliferation, migration and invasion of breast cancer cells. Oncol Lett 2017; 14: 6969–6975. DOI: 10.3892/ol.2017.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Jiang X, Niu X. Long Non-Coding RNA Reprogramming (ROR) promotes cell proliferation in colorectal cancer via affecting P53. Med Sci Monit 2017; 23: 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun X, Yuan W, Hao F, et al. Promoter methylation of RASSF1A indicates prognosis for patients with Stage II and III colorectal cancer treated with oxaliplatin-based chemotherapy. Med Sci Monit 2017; 23: 5389–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: 857–866. DOI: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008; 105: 10513–10518. DOI: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18: 997–1006. DOI: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 24.Ichikawa D, Komatsu S, Konishi H, et al. Circulating microRNA in digestive tract cancers. Gastroenterology 2012; 142: 1074–1078.e1. DOI: 10.1053/j.gastro.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011; 108: 5003–5008. DOI: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011; 13: 423–433. DOI: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010; 101: 2087–2092. DOI: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Wang T, Zhang Y, et al. Upregulation of serum miR-494 predicts poor prognosis in non-small cell lung cancer patients. Cancer Biomark 2018; 21: 763–768. DOI: 10.3233/CBM-170337. [DOI] [PubMed] [Google Scholar]
- 29.Shi M, Jiang Y, Yang L, et al. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem 2018: 119: 4711–4716. DOI: 10.1002/jcb.26650. [DOI] [PubMed] [Google Scholar]
- 30.Cao XC, Yu Y, Hou LK, et al. miR-142-3p inhibits cancer cell proliferation by targeting CDC25C. Cell Prolif 2016; 49: 58–68. DOI: 10.1111/cpr.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua S, Liu C, Liu L, et al. miR-142-3p inhibits aerobic glycolysis and cell proliferation in hepatocellular carcinoma via targeting LDHA. Biochem Biophys Res Commun 2018; 496: 947–954. DOI: 10.1016/j.bbrc.2018.01.112. [DOI] [PubMed] [Google Scholar]
- 32.Jia XP, Meng LL, Fang JC, et al. Aberrant expression of miR-142-3p and its target gene HMGA1 and FZD7 in breast cancer and its clinical significance. Clin Lab 2018; 64: 915–921. DOI: 10.7754/Clin.Lab.2017.171114. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Cao Z, Wang L, et al. Downregulation of microRNA-142-3p and its tumor suppressor role in gastric cancer. Oncol Lett 2018; 15: 8172–8180. DOI: 10.3892/ol.2018.8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ransohoff DF, Sox HC. Clinical practice guidelines for colorectal cancer screening: new recommendations and new challenges. JAMA 2016; 315: 2529–2531. DOI: 10.1001/jama.2016.7990. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y, Gao J, Zhang S, et al. miR-142-3p regulates autophagy by targeting ATG16L1 in thymic-derived regulatory T cell (tTreg). Cell Death Dis 2018; 9: 290. DOI: 10.1038/s41419-018-0298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghanbari R, Mosakhani N, Asadi J, et al. Downregulation of plasma MiR-142-3p and MiR-26a-5p in patients with colorectal carcinoma. Iran J Cancer Prev 2015; 8: e2329. DOI: 10.17795/ijcp2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen WW, Zeng Z, Zhu WX, et al. MiR-142-3p functions as a tumor suppressor by targeting CD133, ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl) 2013; 91: 989–1000. DOI: 10.1007/s00109-013-1037-x. [DOI] [PubMed] [Google Scholar]
- 38.Zaytseva YY, Rychahou PG, Gulhati P, et al. Inhibition of fatty acid synthase attenuates CD44-associated signaling and reduces metastasis in colorectal cancer. Cancer Res 2012; 72: 1504–1517. DOI: 10.1158/0008-5472.can-11-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zernecke A, Bidzhekov K, Noels H, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal 2009; 2: ra81. DOI: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 40.Wang K, Zhang S, Weber J, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res 2010; 38: 7248–7259. DOI: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu L, Cai C, Wang X, et al. MicroRNA-142-3p, a new regulator of RAC1, suppresses the migration and invasion of hepatocellular carcinoma cells. FEBS Lett 2011; 585: 1322–1330. DOI: 10.1016/j.febslet.2011.03.067. [DOI] [PubMed] [Google Scholar]
- 42.Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev 2015; 81: 75–93. DOI: 10.1016/j.addr.2014.09.001. [DOI] [PubMed] [Google Scholar]