Abstract
Cells utilize natural supramolecular assemblies to maintain homeostasis and biological functions. Naturally inspired modular assembly of biomaterials are now being exploited for understanding or manipulating cell biology for treatment, diagnosis, and detection of diseases. Supramolecular biomaterials, in particular peptides and oligonucleotides, can be precisely tuned to have diverse structural, mechanical, physicochemical and biological properties. These merits of oligonucleotides and peptides as building blocks have given rise to the evolution of numerous nucleic acid- and peptide-based self-assembling nanomaterials for various medical applications, including drug delivery, tissue engineering, regenerative medicine, and immunotherapy. In this review, we provide an extensive overview of the intracellular delivery approaches using supramolecular self-assembly of DNA, RNA, and peptides. Furthermore, we discuss the current challenges related to subcellular delivery and provide future perspectives of the application of supramolecular biomaterials for intracellular delivery in theranostics.
Keywords: Self-assembly, Intracellular delivery, Oligonucleotide, Peptide
1. Introduction
Nature relies on the supramolecular assembly of nucleic acids, proteins, lipids to maintain cellular homeostasis and functions 1. Supramolecular assembly is spontaneous organization of molecules to a unique structure via noncovalent interactions, such as hydrogen bonding, hydrophobic, electrostatic interactions, van der Waals forces, and π-π stacking 2. The simplicity through which complex structures can be built using fundamental molecules confers elegant architectures within the cells. These biological supramolecular assemblies are involved in a variety functions in living organisms, such as compartmentalization of environments, transport and release of molecules, and interactions and communications of cells with extracellular compartments 3. Cellular membrane, which is a supramolecular organization of phospholipids, separates the cell interior from the extracellular compartments. Actin filaments, an assembly of actin protein essential in all eukaryotic cells, are present in cytoplasm and perform various functions; their assembly and disassembly, responding to intra- and extracellular stimuli, are involved in various cellular activities including cytokinesis, endocytosis and exocytosis, maintaining mechanical stability, providing shape and motility to cells, and so on. In the past decades, supramolecular assembly of actin monomer was of high interest. The changes in pH, presence of ATP and divalent cations, and salt concentrations result in the nucleation of actin and the formation of filament 4,5.
Aspirations to acquire mechanistic understanding of these examples have inspired myriads of synthetic supramolecular assemblies that mimic some of the natural assemblies. Particularly, it enabled material scientists to develop a variety of self-assembling biomaterials, for example peptide-amphiphiles, liposomes, micelles, and dendrimers, with a precise design in a bottom-up approach. These self-assembling biomaterials can be used for diagnostic and/or therapeutic purposes. Especially, versatility of self-assembling nanostructures offers some advantages for subcellular delivery. For effective intracellular delivery, the vector should reach specific organs and enter the target cells. After that, by going through a series of endocytic pathways, the delivery system should be released into cytoplasm. Among diverse self-assembling biomaterials, oligonucleotide- and peptide-based supramolecular assemblies possess several promising features for the successful intracellular delivery. The bottom-up assembly of these nanoparticles enables to control the physicochemical properties of the final architectures. Their size, shape, and hydrophobicity can be precisely tuned to be favorable for delivery. Especially, by manipulating the hydrophobicity of the building block, loading capacity and efficiency of hydrophobic small molecules in the nanoparticles can be tailored; even their release behaviors can be programmed to be responsive to exogenous stimuli (such as temperature, or presence of specific enzyme). Another merit is the surface of nanoparticles can be decorated with functional moieties. To enhance cellular uptake of nanoparticles, antibodies or membrane-active peptides can be attached to the desired sites on surface. It is also advantageous for building blocks of self-assembling delivery systems that nucleotides and peptides are highly biocompatible and biodegradable. Nucleic acids-based self-assembly with various structures, including triangles 6, cubes 7, micelles 8, and fibers 9, have been developed for intracellular delivery. Peptides can be used as a building block to construct self-assembled nanostructures. The secondary structures of peptides allow to self-assemble into peculiar structures, such as fibers 10, ribbons 11, and tubes 12. This structural diversity of supramolecular self-assembly and the ability to precisely control its structure can be used to engage with cellular machinery with high affinity. In this regard, the physicochemical properties of self-assembled nanoparticles can be optimized to enhance their internalization into cell interior and to reach drug targets efficiently.
In this review, we recapitulated the recent advances in the supramolecular self-assemblies as it relates to subcellular delivery. We currently limit this review to DNA-, RNA-, and peptide-based supramolecular self-assemblies, focusing on their mechanisms and applications for intracellular delivery. Future direction for utilizing tools in self-assembly to overcome cellular barriers are also discussed.
2. Challenges of intracellular delivery
Ideal carriers for intracellular delivery should possess the following features. First, they should be able to not only package cargos efficiently, but protect their cargos from any degradation. Second, they reach and enter the target cells, selectively. Third, their cargos should be introduced or released into the appropriate intracellular compartments 13. To achieve these features, a diverse range of delivery systems have been developed, including viral vectors, liposomes, dendrimers, polymeric micelles, lipid nanoparticles, gold nanoparticles, carbon-based nanocarriers, et cetera. As a carrier approaches the exterior plasma membrane of cells, dynamic interactions are initiated between the carrier and the membrane, which affects the pathway of cellular entry of the carrier. These interactions act for carriers as adhesion forces including electrostatic, hydrophobic, van der Waals, receptor-ligand interactions, etc. It is also known that physicochemical properties of nanocarriers, including size, shape, surface, and stiffness, influence the internalization pathway 14,15. Engagement of the combined interactions leads carriers to the defined pathway of internalization.
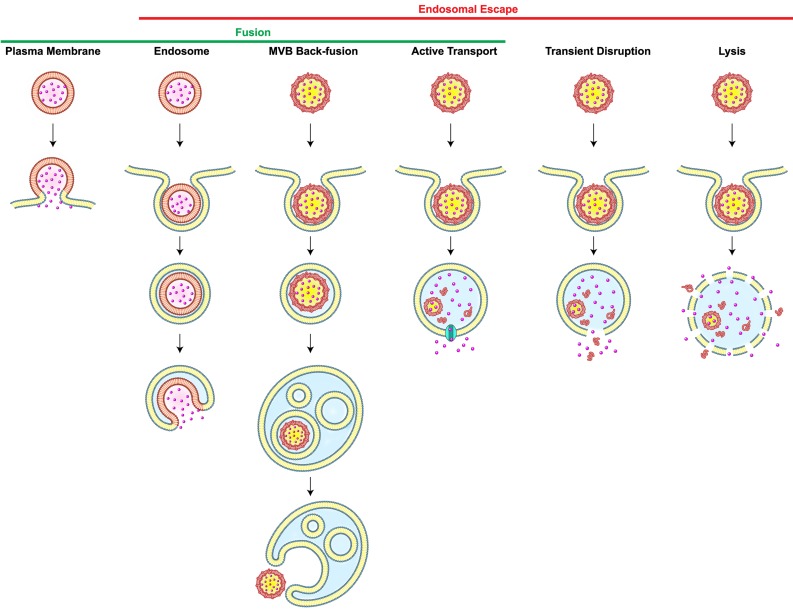
Most carriers generally enter cells via endocytosis. Once nanoparticles get internalized, they are usually transported to early endosomes which serve as a primary sorting station and direct them to their next destinations, either to late endosomes or recycling endosomes. Late endosomes are formed by the maturation of early endosomes, and they contain multiple close-packed intraluminal vesicles. These late endosomes then fuse with lysosomes which are spherical-shaped organelles. Lysosomes contain hydrolytic enzymes that degrade many kinds of macromolecules. If carriers end up in lysosomes, both cargos and carriers would be exposed to degradation processes 16. For intracellular delivery, therefore, carriers must be retrieved in the middle of the endocytic pathway, a phenomenon known as endosomal escape. However, the incidence of the endosomal escape is extremely low; it was reported that only 1-2% of nanocarriers reach the intracellular compartments owing to the entrapment in endosomes 17 or recycling pathways 18. This low incidence of endosomal escape significantly limits the efficiencies of carrier-mediated intracellular delivery, particularly in the cytosolic delivery of genes 19. Scenarios of endosomal escape includes (1) transient disruption or (2) complete lysis of endosomes; (3) active transport of small molecules; and fusion either through (4) back-fusion of MVBs with the outer limiting membrane or (5) fusion of the carrier with endosomes (Figure 1). One of the endosomal escape scenarios available to nanoparticles is based on the “proton sponge” model. The ionizable amine groups in nanoparticles are accumulated inside an endosome and they can get protonated at acidic endosomal pH. This would cause more protons and counter ions to flood the endosome. The resultant difference in osmotic pressure would lead water to enter the endosome, resulting in swelling and eventually endosome rupture. Additionally, electrostatic and hydrophobic interactions of nanoparticles with the lipids in the endosomal membranes could lead to destabilization or reorganization of the membrane, allowing the endosomal cargos to escape to the cytosol. However, understanding of endosomal escape still remains obscure because of the limited frequency of measurable escapes. Recently, it is reported that several agents and the characteristics of carriers can facilitate the endosomal escape 20,21. Lönn et al. demonstrated that the incorporation of synthetic endosomal escape domains (EEDs) enhanced the intracellular delivery of proteins in a combination with TAT peptides 22. Continuous research on the development of novel delivery systems and new tools for accurate detection of endosomal escape is much-needed to break through the current hurdles in the intracellular delivery. The process of self-assembly may be used to trigger endosomal release through precise control of architectures for cellular entry, stimuli-responsive dissociation for cargo release, and any interaction that causes cytosolic delivery.
Figure 1.
Scenarios of Carrier-Mediated Endosomal Escape and Subcellular Delivery of Cargos. Membrane fusion takes place when a membrane-bound carrier fuses with the plasma or the endosomal membrane. Alternatively, carrier containing intraluminal vesicles back-fuse with the membrane of the multi-vesicular body (MVB), thereby leading to the inadvertent endosomal escape. Dissociation of carriers inside endosomes may cause endosomal escape of cargos via active transport depending on membrane proteins (cyan), transient membrane destabilization, or complete lysis of the endosome. Redrawn from 16.
3. Supramolecular self-assembly of oligonucleotides for intracellular delivery
The ability of nucleic acids, DNA and RNA, to offer non-covalent interactions within and between themselves opens up new frontiers in biomimetic self-assembly 23. This offers the potential to construct supramolecular architectures with precise controls over morphology, physicochemical properties, surface functionalization, etc 24,25. Additionally, some unique features of self-assembling oligonucleotides distinguish them from other self-assembling systems. They include rules for complementary base pairing, universal dimensions of double helices (e.g., B-type DNA has right handed double helix with approximately 2 nm diameter, 10 base pairs for one complete turn), etc. Tuning non-covalent interactions at each nucleotide level brings controls over structural properties of nanoarchitecture 26. This section highlights the progresses in the construction of DNA- and RNA-based self-assembling nanostructures for intracellular delivery.
4. DNA self-assembly
Discoveries of DNA molecular recognition and the idea that DNA can be constructed as linear or branched structures with periodic lattices catapulted the field of biomimetic supramolecular self-assembly of DNA 27,28. These rationally designed DNA building blocks can self-assemble into much complex nanoarchitectures that led to the development of diverse applications in molecular robotics 29, chiral plasmonic structures 30, plasmonic hotspots 31, enzymatic nanoreactors 32, light harvesting systems 33, biophysical studies 34, molecular lithography 35 and intracellular delivery 36,37.
4.1. Structural features of DNA contributing self-assembly
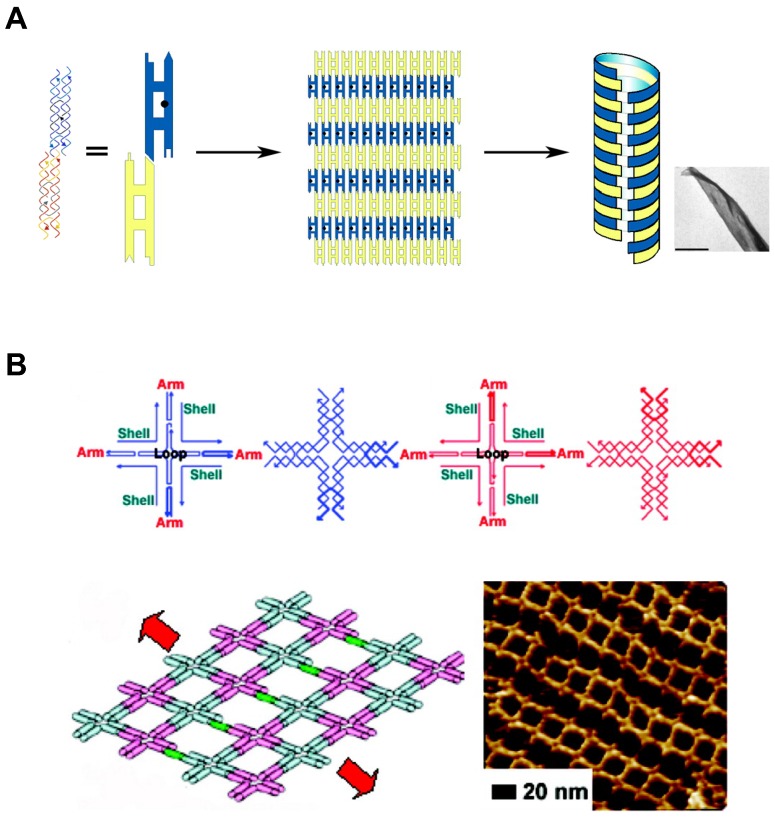
In addition to the predictable canonical base pairing (A-T, G-C), DNA is one of the well understood molecules with defined conformations and physicochemical properties, such as the structural characteristics of double helix. The basic entity of DNA self-assembly begins with the formation of “Tiles”. They are self-assembled and single-stranded DNA molecules having complementary sticky ends which can form higher-order arrays with a well-defined conformation and topology (Figure 2A, B). The tailor-made design of sticky ends dictates specific assembly patterns of the tiles in the desired way, building higher-order nanostructures 38,39. Additionally, there are a variety of features that make DNA self-assembly attractive: At first, DNA sequence can be easily synthesized in a lab-scale using automated phosphoramidite chemistry. Second, availability of specific sites of DNA to covalently attach various functional groups allows DNA to contain functional reporters or biosensors. Lastly, the structural characteristics of DNA nanostructures can be manipulated in various ways: choice of restriction enzymes or DNA forms (A-DNA, B-DNA, and Z-DNA). These features of DNA self-assembly provide DNA with potential as versatile building blocks which can be tailored to make different architectures.
Figure 2.
DNA Self-assembly by multi-tile complexation. (A) DNA tiles formed nanosheets, and further self-assembled into nanotubes. Complementary segments in yellow tiles stick to the counterpart in blue tiles. Adapted with permission from 38, Copyright 2004, American Chemical Society. (B) Self-assembled nanoarray by DNA multi-tile complexation. (top) Schemes of each DNA tile. (bottom) Length and orientation of the nanoarray can be tuned by the stepwise involvement of dsDNA bridges between DNA tiles. Schematic drawing and AFM image of nanoarray. Adapted with permission from 39, Copyright 2008, American Chemical Society.
4.2. Development of DNA self-assembly
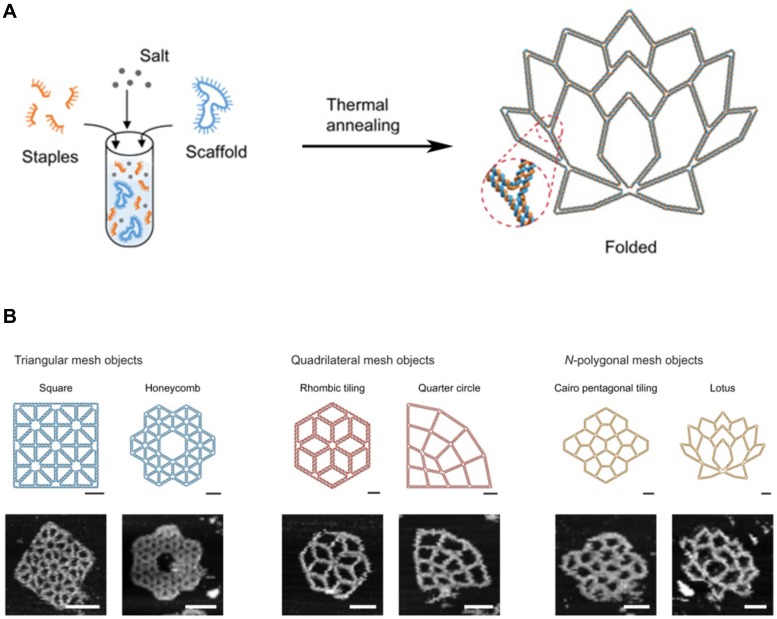
Generally, DNA self-assembly is based on the complementary base pairing between two single-stranded “sticky” ends. In early examples of DNA self-assembly, DNA tiles with unique stick ends self-assemble in particular patterns to form the entire structures 28,40. Each tile can form the secondary multi-tiles that further form higher-order structures 41,42. From the relatively simple polyhedrons to the sophisticated 3D structures, a lot of DNA nanostructures were constructed. With combining computational sciences with DNA nanotechnology, DNA tiles were programmed with built-in instructions regulating the next layer of tiles to self-assemble on the previous layer following algorithmic rules 43,44. This algorithmic DNA self-assembly allowed to generate complex nanoscale DNA patterns. Another approach to build complex DNA assemblies was the nucleated self-assembly where a longer strand acts as a nucleation site for the ligation of the corresponding strands to generate a high order structure. Shih et al. reported that a long piece of single-stranded DNA (1.7 kb) could be folded into an octahedron structure by a few short synthetic oligonucleotides 45. In 2006, Rothemund pioneered a method that causes long, single-stranded DNA molecules to self-assemble into the unique nucleated complex nanostructures, and thus begun the field of DNA origami 46. This folding process for DNA origami involves multiple small “staple” strands and a long single-stranded DNA (Figure 3A, B). The short staples complex with the long single-stranded DNA in multiple positions, forming an elegantly orchestrated 2D or 3D shapes (Figure 3B) 47,48. Recently, Benson et al. introduced a graph theory and a relaxation algorithm to fully automate the design process and precisely simulate the deployment of the long strands over the final structures 49. Because DNA origami is designed from the basic pieces and assembled in the complex structures, bottom-up self-assembly methods started to be considered economical and scalable. And with the ability to synthesize a plethora of architectures using parallel synthesis of nanostructures, DNA engineering has become relatively simple and thus been applied to multiple areas including subcellular delivery 50,51.
Figure 3.
DNA Origami. (A) DNA scaffold and staples were assembled into the programmed 2D DNA origami using the standard one-pot synthesis. (B) Programmed 2D frames and the corresponding AFM images are displayed. (left) Triangular blocks formed a square and a honeycomb lattice. (middle) Quadrilateral blocks formed a rhombic tiling and a quarter circle. (right) 5- and 7-polygonal objects formed a Cairo pentagonal tiling and a lotus, respectively. Scale bars, 20 nm (schematic frames) and 50 nm (AFM images). Adapted with permission from 48, Copyright 2019, American Association for the Advancement of Science (AAAS).
Another DNA self-assembly strategy is based on π-π stacking of the terminal base pairs in dsDNA, which is called as the blunt-end stacking. It was proposed by Woo and Rothemund in 2011 that complementary base pairing in blunt-ends offers a geometric framework that provides stacking 52. Dietz and his colleagues recently constructed a three-dimensional DNA origami via DNA blunt-end stacking interactions with sizes up to 1.2 giga-daltons 53,54.
4.3. Intracellular delivery applications of DNA self-assembly
DNA-based self-assemblies provides several advantages for delivery purposes. First of all, DNA nanostructures are biocompatible and stable with maintaining the designed architectures in physiological conditions 55. Accumulated understanding about DNA dynamics, such as canonical base pairing, allows for manipulation of physicochemical properties of self-assembled architectures 56. Furthermore, DNA can be programmed to interact with other molecules, such as targeting ligands or imaging agents, to guide the DNA nanostructures to specific targets 55. Among various DNA-based nanostructures, DNA origami is a particularly attractive self-assembling nanomaterial for intracellular delivery. The ability to correctly predict the structures based on known sequences and conformations of staple strands using a software allows developing a precisely designed nanomaterials with tightly controlled size, shape, and structure and having functional sites at the desired locations which can be tuned at sites exactly available for ligand attachments. Materials solely made of DNA molecules can be prone to degradation in the presence of abundant nucleases present in the biological milieu however self-assembly of DNA into an origami shields such sites of nuclease activity and provide resistance against degradation. Qian et al. demonstrated that DNA origami nanoarray was much stable than natural DNA configurations in cell lysate for 12 hours at room temperature 57. Whereas, Castro and his colleagues tested the stability of DNA origami against various nucleases and found that T7 endonucleases I and DNase I, which is the most abundant nuclease in bloodstream, degraded the test materials quickly 58. These conflicting degradation rates perhaps suggest that the suprastructures of DNA can be engineered to prevent degradation. Mikkilä et al. reported that coating the surface of DNA origami with viral capsid proteins significantly enhanced the intracellular delivery of the carrier into the HEK293 cells 59. Therefore, highly packed structure and surface coating can be used to further improve the intracellular delivery of DNA nanostructures while preserving its integrity 58,60. Doxorubicin (DOX), an anticancer agent that intercalates with nuclear DNA and perturbs cell division causing cell death, can be packaged inside a DNA vector due to its ability to intercalate with DNA. Ding and his colleagues reported that DNA nanoparticles containing DOX can be delivered in MCF-7R tumor-bearing mice and a decrease in tumor volume can be achieved with limited side effects 61. Högberg et al. reported the twisted DNA origami displayed a sustained release of DOX than the regular DNA origami while maintaining similar encapsulation of drugs, suggesting release profiles of drugs can be controlled through varying the structures 62. DNA nanostructures can form hybrid delivery system by loading other nanoparticles such as gold nanoparticles or carbon nanotubes 63,64. The incorporation of gold nanoparticles inside DNA architectures allows the hybrid system to be used for optoacoustic imaging and photothermal therapy 64. Also, DNA origami has been used to deliver aptamers 65, proteins 66, quantum dots 67, and immunostimulants 68, thus the modular platforms that can package various cargos can reach their targets.
Other than DNA origami, DNA-based “nanoswitches” have been devised. These are based on self-assembly responsive to exogenous stimuli 69. Krishnan and her coworkers have constructed DNA-based reporters that act as sensors to monitor pH, chloride, hypochlorous acid, and calcium ion inside organelles of living cells 70,71. Such nanostructures have shown to be powerful tools to study intracellular processes in vitro and might forge a new era to understand trafficking processes in vivo. They also developed cell-targetable DNA-based icosahedral nanocapsules encapsulating a neuroactive steroid by self-assembly and chemical conjugation of the DNA blocks 72. DNA strands can be assembled in the shape of a tetrahedron nanoparticles containing the site-specific hybridization with siRNA. These monodisperse nanoparticles are easily programmable to have desirable size, spatial orientation, and the number of targeting ligands. Lee et al. showed that, through the attachment of folate to these nanoparticles, the cancer-specific targeting and gene silencing can be achieved in in vitro and in vivo 73.
However, nucleotide-based nanocarriers are challenged from various factors. One challenge is the ceaseless assault of nucleases present in in vivo environment and lysosomal compartments. Although self-assembly of nucleotides enhances the nuclease resistance of final structures, many nucleotide-based nanocarriers are still much susceptible to degradation than other types of nanoparticles. Chemical modification of nucleotides can extend the serum stability of DNA nanostructures. Phosphorothioate modification, a replacement of a sulfur atom with an oxygen atom in the phosphate backbone, is widely used to enhance enzymatic resistance of nucleic acids 74. Boranophosphate substitution is also another option to improve serum stability of oligonucleotides 75. The stability of nucleotide-based nanocarriers can be increased by crosslinking as well. Cassinelli et al. used the click chemistry to cross-link between oligonucleotides to form DNA nanotubes with the better thermodynamic as well as enzymatic stabilities 76. Layer-by-layer approach can also be used to enhance the circulation time of DNA nanostructures. Perrault and Shih reported that packaging DNA nanoparticles with lipid bilayers not only extended the half-life of oligonucleotides, but also reduced the immunogenicity of the nanoparticles 77. Another challenge underlying the intracellular delivery using self-assembling DNA is their cellular trafficking. Considering the inherent negative charges in DNA molecule, it is impressive that a considerable amount of self-assembled DNA nanoparticles overcome charge repulsive forces and enter cells. As with other nanocarriers, self-assembling DNA nanostructures enter cells via endocytosis. Multiple routes of endocytosis, including caveolin-mediated 78 and clathrin-mediated endocytosis 79 and macropinocytosis 80, are reported as the mechanisms of cellular entry of DNA nanostructures. Although the rules governing the endocytosis of DNA structures are unclear, these variable trafficking suggests the possibility that the uptake kinetics of self-assembled DNA can be modulated by their physicochemical properties and surface compositions.
5. RNA self-assembly
Like DNA, ease of designing RNA has largely contributed to the advent of RNA nanotechnologies. RNA self-assembly also refers to the spontaneous folding of the nucleotide sequence into a complex hierarchy based on the combinations of various non-covalent interactions between distant nucleotides. Although RNA shares many similarities with DNA, RNA is regarded much versatile and thermodynamically stable than DNA due to the presence of non-canonical interactions. Furthermore, RNA molecules are single stranded and generally exist in a variety of complex structures whereas most DNA molecules predominantly form base-paired double helices. This diversity on RNA engineering enables RNA molecules to become self-assembled RNA architectures for a wide range of applications.
5.1. Structural features of RNA contributing self-assembly
As noted, the structure of RNA molecule is nearly identical to that of DNA molecule, except for several differences: RNA has a ribose instead of deoxyribose, RNA nucleotides include uracil but not thymine. These fundamental differences confer dynamic interactions upon RNA molecules, folding themselves into labyrinthine architectures. One additional hydroxyl group in the RNA structure increases the number of hydrogen bonds that RNA molecule forms, thereby contributing to both self-assembly and thermodynamic stability of RNA structures. Other than hydrogen bonds and canonical base pairing, RNA self-assembly is dependent on multiple non-canonical base pairing, including the wobble base pairing, imino- and sheared GA pairing, the reverse Hoogsteene AU pairing of polynucleotide strands, and base stacking. These inherent features of RNA allow themselves to assemble into numerous intricate architectures ranging from the secondary to quaternary structures. Over the past decades, extensive research about natural RNA biomolecules revealed that the recurrent structural motifs are related to localized design of nucleotides. These repeating structures are thought to act as modules that make specific architectures (or folds) to play the programmed operations including various biological and mechanical functions. One additional unique characteristic of RNA as a building block for nanostructures is that RNA self-assembly is affected by ionic compounds. Divalent metal cations, like Mg2+, neutralize the phosphate backbone of RNA, facilitating them to fold into functional 3D structures. This attribute can be used to develop RNA-based sensors for the detection of metallic ions. Therefore, with proper understanding of RNA structures, a variety of self-assembling RNA nanoarchitectures have been precisely designed with higher order structures for diverse applications.
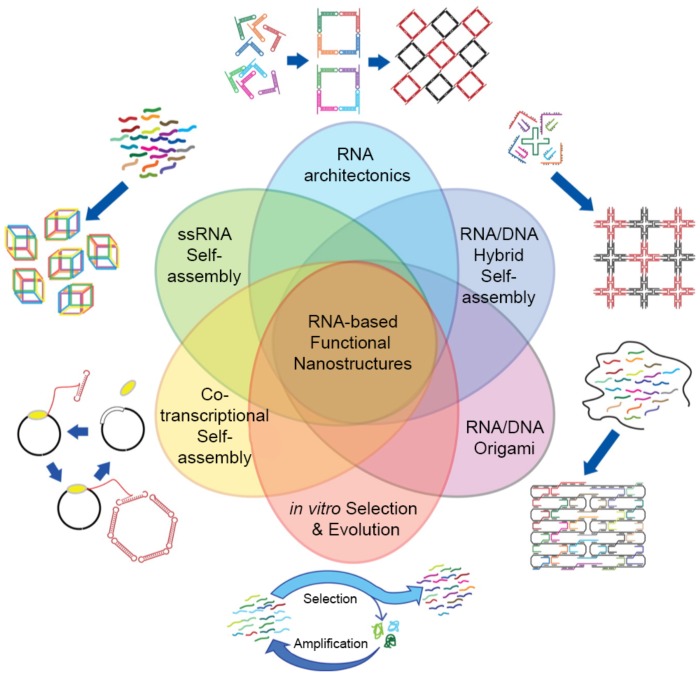
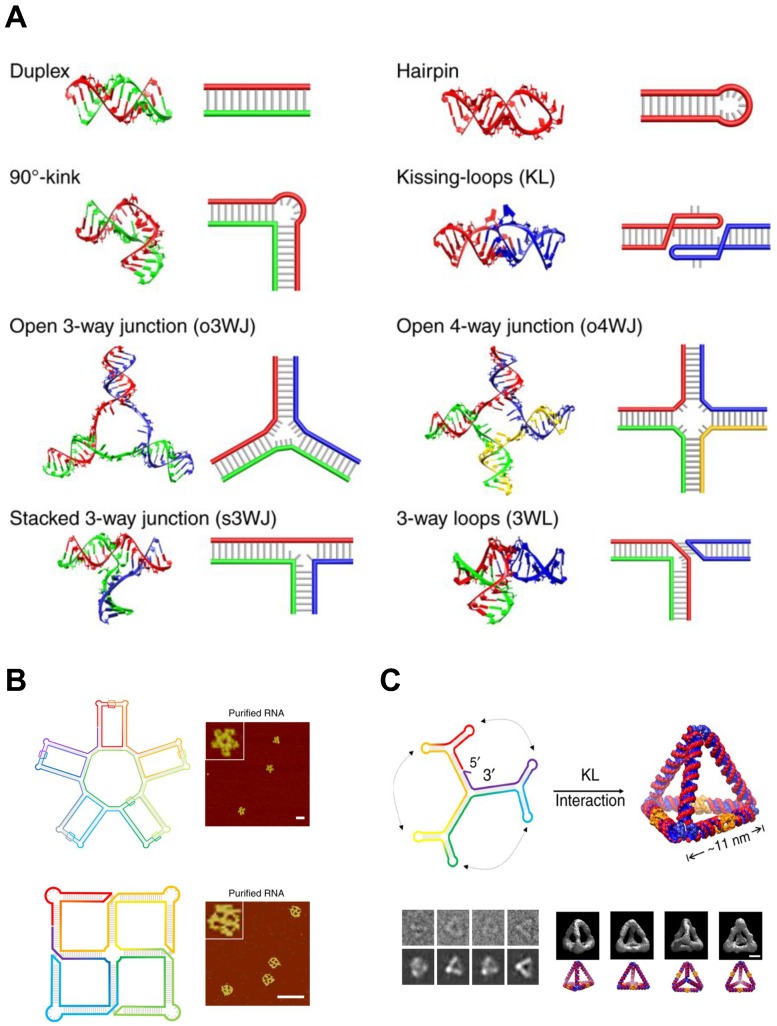
5.2. Classification of RNA self-assembly
RNA self-assembly can be categorized into four major types: (1) Tectonic RNA (2) Single-stranded RNA (3) Cotranscriptional RNA, and (4) Hybrid RNA self-assembly (Figure 4).
Figure 4.
Classifications and other design strategies for RNA self-assembly. RNA architectures can be prepared via various self-assembly strategies. Four major RNA self-assembly designs include Tectonic RNA, Single-stranded RNA, Cotranscriptional RNA, and Hybrid RNA self-assembly. RNA/DNA Origami and in vitro Selection and Evolution (or SELEX) are also used to design sophisticated RNA nanostructures. Even though each design strategy is distinct, they can be complementarily used to control self-assembly. Adapted with permission from 25. Copyright 2014 American Chemical Society.
RNA tectonics refer to the idea that RNA molecules can be used as building blocks to construct self-assembled RNA architectures with desirable structures and properties 81. To achieve the defined RNA structures, each small RNA block must be precisely designed and completely characterized. The knowledge obtained from naturally occurring RNA structures has strongly supported the establishment of principles and methodologies of RNA tectonics, enabling us to design a number of RNA modules. Especially, small RNA modules are useful bridging parts: In tectonic RNA assembly, RNA duplex structures act as backbones and small RNA modules connect and assemble the duplexes into 2D and 3D structures. In this regard, RNA tectonics are able to build a myriad of RNA nanoarchitectures, including cubes 82, polyhedrons 83, and squares 84.
In contrast to the RNA tectonics, single-stranded RNA self-assembly hires unstructured RNA strands to build architectures based on the classical Watson and Crick base pairing (Figure 5A). This approach is inspired from DNA nanotechnologies where the canonical base pairing mainly arranges the building blocks. The advantage of single-stranded RNA self-assembly lies in its relatively simple sequence design. Recent studies showed single-stranded RNAs assembled into various 2D and 3D structures (Figure 5B, C) 85,86.
Figure 5.
RNA Self-assembly: Design motifs and 2D/3D RNA structures. (A) Various motifs for RNA structures. A scheme and the corresponding 3D modeling are shown for each motif. (B) 2D RNA structures. (top) A structural scheme and the corresponding AFM images of an RNA 5-petal flower. Scale bar is 60 nm. (bottom) A structural scheme and the corresponding AFM image of an RNA tetra-square. Scale bar is 50 nm. (C) RNA nanostructure folded into an 3D RNA tetrahedron. (top) A structural scheme of the RNA tetrahedron. Color denotes the base index along the folding path; rainbow colored from 5' to 3'end. (bottom left) Comparisons between raw cryo-EM images of RNA tetrahedrons and the corresponding projection of the reconstructions. (bottom right) Four different viewpoints of the reconstruction of the RNA tetrahedron and the corresponding images of simulation. Scale bar, 5 nm. Adapted with permission from 86, Copyright 2019, Nature Publishing Group.
Cotranscriptional RNA self-assembly is much related to the time when RNA structures form. As the name suggests, self-assembly of RNA occurs while the RNA molecule is transcribed, suggesting that RNA folding can be genetically encoded and expressed in cells. No DNA cotranscriptional folding has been demonstrated to be self-assembled isothermally yet so this type of self-assembly is possibly limited to RNA self-assembly for now. Multiple RNA strands can be assembled into the nanostructures with a cotranscriptional manner 82, or RNA self-assembly can occur in living bacteria 87. This strategy has also been used to develop RNA origami 88 and RNA nanoparticles 89.
Hybrid RNA self-assembly indicates the RNA assembly together with DNA. One of the advantages of this hybrid strategy is the size of final structures. Even though RNA assemblies offer a variety of structures and functionalities, the size of architectures remains in nanoscale. Precisely controlled RNA scaffolds are generally a few hundred nanometers while DNA scaffolds can be built up to micron-sizes with tens of thousands of nucleotides. DNA design methods thereby have been introduced in RNA nanotechnologies to construct hybrid RNA-DNA assemblies with considering the structural features of A-form RNA duplexes (11 base pairs per turn and +19° inclination of base pair to axis). Moreover, by adopting DNA folding in origami, RNA-DNA hybrid origamis have been developed with a single stranded RNA template and multiple DNA staples 90,91. The hybrid RNA self-assembly has been used to build nanoscale scaffolds 82, multi-stranded structures 92, and micron size arrays 93, usually used for applications in drug delivery.
Recently, the advancement in the selection and evolution techniques (SELEX) enabled to efficiently discover RNA sequences with new structural or biological properties 25. It can expand the structural diversity of RNA architectures by providing artificial structural motifs to RNA toolbox, which may contribute to the discovery of novel RNA self-assembly.
5.3. Intracellular delivery applications of RNA self-assembly
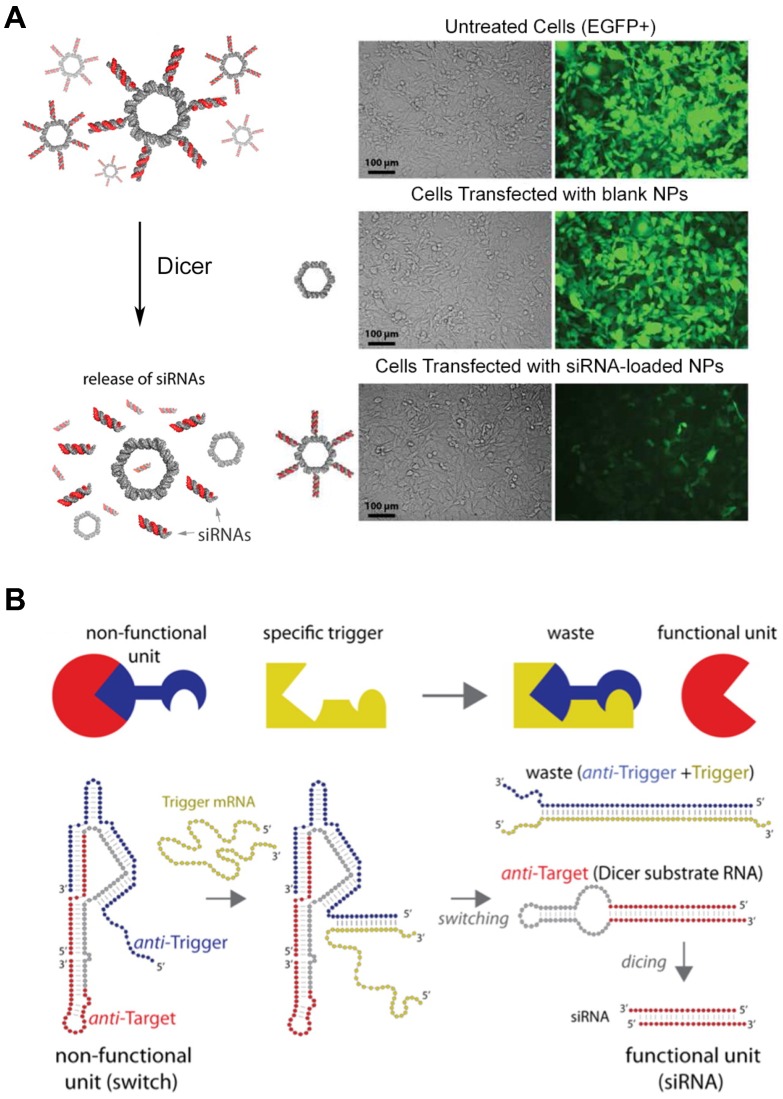
The versatility of RNA structures enables RNA molecules to become a fascinating material in the biomedical field. The feasibility of RNA structures as a delivery platform particularly draws a lot of attentions due to their biocompatibility, controllability, and ease of synthesis. Moreover, RNA nanoparticles are thought to be advantageous to deliver therapeutic RNA molecules, such as small interfering RNA (siRNA) and anti-micro RNA oligonucleotides (AMOs). Shapiro and his collaborators functionalized the surface of their RNA nanoparticles with siRNA (Figure 6A) 89, and they also developed the DNA-RNA hybrid system with splitting capability that triggered the release of the siRNA moiety causing RNA interference (Figure 6B) 94,95. Recently, the delivery of the AMO against oncogenic miRNA was assisted by RNA nanoparticles and displayed the inhibited tumor growth 96. Other than nucleic acid-based cargos, small molecules can be loaded in the RNA nanostructures. Doxorubicin-loaded RNA nanoparticles showed the enhanced cytotoxicity against ovarian cancer cells 97, and micellar RNA nanoparticles successfully loaded paclitaxel and induced cancer cell apoptosis 98. RNA nanoparticles also can be easily decorated with aptamers, nucleic acid-based ligands, for the targeted delivery. RNA nanoparticles functionalized with aptamers have been extensively studied for targeted cancer treatment 99,100. Other targeting ligands, such as folate or antibodies can be chemically conjugated to the terminal ends of RNA helices. RNA nanostructures are also used for immunotherapy. RNA nanoparticles incorporating immunostimulatory molecules (for example, CpG oligodeoxynucleotide) activated innate immune responses 101. Zhu et al. reported that self-assembled DNA-RNA hybrid nanoparticles delivered CpG and short hairpin RNA (shRNA) as well as neoantigens to antigen presenting cells 102. The structural features of RNA nanoparticles, such as size, shape, and sequence, can be tuned to stimulate different immunogenic pathways, suggesting the probability of immunomodulation. Guo et al. reported that structurally distinct RNA nanoparticles induced different levels of cytokines, including tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6), in macrophages 103. They showed that the immunogenicity of RNA nanoparticles increases from small linear to large 3D conformations. Also, several RNA sequences were reported to trigger immune responses by the interactions with toll-like receptors (TLRs) or cytosolic sensors (PKR, RIG-1, and MDA-5). However, the depth of understanding about how immune systems react with RNA structures still remain limited.
Figure 6.
siRNA-loaded RNA assemblies for gene silencing. (A) (left) RNA nanoparticles functionalized with siRNAs. Release of siRNA can be triggered by Dicer nuclease. (right) Human breast cancer cells that stably express EGFP (green) were treated with EGFP siRNA-loaded RNA nanoparticles. After 3 days of the treatment, EGFP expression was silenced, indicating siRNA-mediated knockdown as well as cellular entry of RNA nanoparticles. Adapted with permission from 89, Copyright 2012, American Chemical Society. (B) Hybrid RNA/DNA switch that changes its conformation by the presence of trigger mRNA and releases siRNA. Anti-target strand (red) and anti-trigger strand (blue) self-assemble into a non-functional unit. Once the trigger mRNA strand starts to interact with the anti-trigger strand, the interaction results to the release of shRNA that is a functional Dicer substrate RNA. Adapted with permission from 95, Copyright 2016, American Chemical Society.
In spite of the promising potential of RNA nanomaterials as a delivery platform, there are challenges related to their inherent characteristics. RNA-based nanocarriers are usually thought to possess better serum stability than DNA-based nanocarriers due to the presence of non-classical base interactions 104. However, RNA nanoparticles are still easily degraded in vivo than other materials due to the presence of RNases. Chemical modifications on RNA, such as 2'-O-methylation, improve the stability of RNA 105,106 but these chemical modifications also can alter its folding, compromising the final structures and biological functions. Another hurdle on self-assembling RNA nanostructures is their instability in dilution. RNA nanoparticles without covalent linkage go through extreme dilution when they are injected into the body, which generally accompanies the dissociation of the nanoparticles. Addition of cross-linking agents could rescue the stability of RNA nanoparticles; but further research is much needed to overcome the concerns about in vivo dissociation 107.
6. Supramolecular self-assembly of peptides
Supramolecular self-assembly is one of the most explored fields employed to fabricate a wide range of nanomaterials for drug delivery with precisely designed shapes, properties and efficacies. It is a bottom-up approach that the programmed organization of rationally designed individual molecules leads to the formation of distinct nanostructures. This assembly is based on the fine-tuned molecular interactions, such as hydrophobic, and electrostatic interactions, hydrogen bonding, van der Waals forces, and π-π stacking. The individual or combined interactions at specific conditions (pH, temperature, ionic strength, or polarity) influence the patterns of self-assembly, arranging into the unique nanostructures. Advantages of the self-assembling nanomaterials include structural versatility, atomic-scale resolution, simplicity of manufacturing, and controllability over morphology and biological functions.
Self-assembling peptides (SAPs) have been investigated over a few decades as a building block for nanostructures, especially for delivery purposes. Their chemical versatility (i.e., 20 types of natural amino acids) provided a wider design space on sequence, and the inherent biocompatibility became another advantage as a delivery system. Naturally occurring peptide motifs or in vitro designed peptides can self-assemble into nanostructures with diverse secondary structures including α helix, β sheet, and β turn. Amphiphilic peptides composed of hydrophilic and hydrophobic segments are also known to self-assemble. The hydrophilic bioactive domain can be modified to give the desired functionality, such as targeting, cell-penetrating ability, or stimuli-responsiveness. All proteins and peptides in nature consists of 20 natural amino acids. Except for glycine, all other natural amino acids are present as L-forms and the α carbon atom which has different R groups determines different characteristics of amino acids. Further, the R groups primarily affect the orientation of different secondary structures. By the careful design of peptide sequence, specific assembly patterns are achievable, which is the main idea of bottom-up fabrication of self-assembled nanostructures. Extensive research over the past few years have led to the formation of basic rules of peptide self-assembly that has allowed for the rational design of SAPs. Below are some examples of the characterized peptide-based nanostructures developed so far.
6.1. β sheet
In β sheets, multiple peptide strands form the secondary structures via interstrand hydrogen bonding between carbonyl group and amine group. β sheet structures can be subcategorized into two groups: Parallel β sheets and anti-parallel β sheets. Parallel β sheets are formed when two strands are arranged in the same direction. In contrast, anti-parallel β sheets are characterized by two strands meet each other in the opposite direction by hydrogen bonding. One way to differentiate two sheets is to count the number of atoms in a hydrogen bonded ring. The number of atoms in a hydrogen bonded ring in parallel β sheet is 12 but that of anti-parallel β sheet alternates 10 and 14. Since early 1990s, parallel and anti-parallel β sheets have been used to develop peptide-based nanomaterials. Both types of β sheets tend to be self-arranged into long filamentous structures. On self-assembling, hydrophilic residues of amino acids are located toward the exterior water phase while hydrophobic residues are buried in the core, contributing to the unique structures of β sheeted materials that are effective for small molecule or gene delivery applications (Figure 7A) 108. The first β sheet nanomaterial, RADA 16, has been utilized for drug delivery 109,110, regenerative medicine (neural stem cell differentiation) 111, in vivo brain damage repair and bone regeneration 112,113. The SAPs that form the nanostructures with a high aspect ratio, like ribbons, fibrils, and fibers, with varied numbers of β sheets in the final structures.
Figure 7.
Intracellular Delivery Using the Self-Assembling Peptides (SAPs). (A) The SAPs incorporating β sheets formed nanoparticles that were able to encapsulate small molecules or plasmid DNA by hydrophobic interactions. Reproduced with permission from 108, Copyright 2017, American Chemical Society. (B) Coiled coil-based SAPs encapsulating small molecule drugs assembled into nanofibers. These nanofibers would be implanted for the treatment of osteoarthritis. Reproduced with permission from 117, Copyright 2018, American Chemical Society.
6.2. α helix and coiled coil
α helix is another type of the secondary structures of proteins, which was first characterized by Pauling, Corey, and Branson 114. This type of SAP has a right hand-spiral conformation in which all amine groups in the backbone form hydrogen bonds with carbonyl groups located at earlier residues in the backbone. When two or more alpha helices gather, they form the unique structure called the coiled coil. This rope-like conformation is a basic folding pattern of natural proteins which are involved in many biological functions, such as cell signaling and gene expression. The coiled coil self-assembly generally contains a repeated motif composed of seven residues (abcdefg), which is known as a heptad repeat. Hydrophobic amino acids are almost always located at the first and fourth residues (the a and d positions) and oppositely charged amino acids are located at the fifth and seventh residues (the e and g positions), generating the hydrophobic and electrostatic interactions, respectively. The remaining residues (the b, c, and f) are not the main driving forces for assembly but can be used to decorate the SAPs 115. Coiled coil-based SAPs have been developed to deliver small molecules 116,117, biomacromolecules 118, imaging agents 119, or other nanoparticles 119,120. Yin et al. engineered a cartilage oligomeric matrix protein that forms the coiled-coil structure which enables encapsulation of a small molecule intended to prevent degeneration of joints (Figure 7B). Kopeček group developed a novel approach to induce apoptosis using coiled coil nanomaterials without small molecules, named as the drug-free macromolecular therapeutics. They used coiled coil peptides crosslinked with anti-CD20 antibody to target and induce apoptosis in CD20+ malignant B cells 121,122. Recently, it has been shown that a coiled coil CPP that were rigid and fibrous with a high aspect ratio showed higher propensity for entry into cells 123,124.
6.3. Peptide amphiphiles
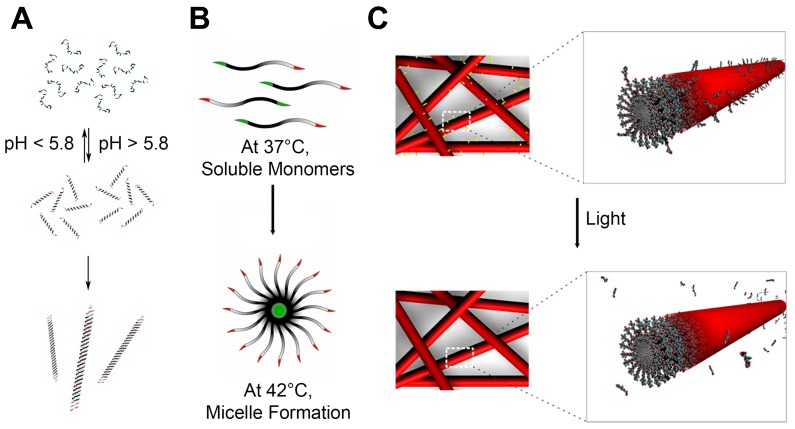
Peptide amphiphiles (PA) are typically composed of three regions: 1) hydrophobic chain mostly containing lipid tails, 2) β sheet sequence, and 3) bioactive head 125. On self-assembling, the hydrophobic chains are located at the core of nanostructures by intermolecular hydrophobic interactions while the peptide regions are at the periphery in aqueous solution. The surface peptides can be further modified and decorated with bioactive epitopes. The length of hydrophobic tails can be tuned to modulate the hydrophobicity. The peptide sequences can give structures and shapes to the nanomaterials. By a mere addition of replacement of 5 to about 20 amino acids, intermolecular hydrogen bonding forms β sheets which become 1D nanostructures, further followed by the entanglement into nanofibers with a cylindrical geometry 126,127. The head sequences can increase water solubility of whole structures, give biological functions to the surface of self-assembled PAs, or both. This last region is particularly useful to tailor the functions of PAs with negligible changes in the structures of self-assembly. For biological applications, the bioactive heads can be designed to facilitate cellular internalization. Inclusion of cell membrane adhesive sequence, such as RGD (Arginylglycylaspartic acid) peptides that are known to adhere to integrins, is amongst the most used sequences to improve the subcellular delivery of therapeutic molecules 10,128,129. The head of PAs can be designed to present antigenic epitopes as a vaccine platform. The PAs-based vaccines were investigated to induce immune responses against tumor 130,131 or bacteria 132. It was also tried to make the PAs responsive to exogenous stimuli, such as pH 133, enzyme 127, heat 134 and light 135, that can lead to dissociation of the assembled structures or specific epitopes which can be useful in controlled release of therapeutic molecules (Figure 8A-C).
Figure 8.
Peptide Self-assembly that are Stimuli-Responsive. (A) Coiled coil peptides undergo reversible pH-dependent self-assembly into nanofibers with a high aspect-ratio. Adapted with permission from 133, Copyright 2006, American Chemical Society. (B) Thermal-sensitive elastin-like polypeptide (ELPs): At 37°C, below the critical micelle temperature (CMT), it exists as soluble monomers. At 42°C, above the CMT, it self-assembles into micellar structures. Adapted with permission from 134, Copyright 2012, American Chemical Society. (C) Peptide amphiphiles including photocleavable linkers release epitopes upon exposure to light. Adapted with permission from 135, Copyright 2012, American Chemical Society.
6.4. Miscellaneous nanostructures
There are other types of SAPs including elastin like polypeptides, cyclic peptides, and short aromatic peptides. Elastin-like polypeptides (ELPs) are composed of repeats of short peptide sequences, and the most widely used repeat is VPGVG 134,136. Synthetic (VPGVG)n biopolymers have a unique feature that makes them an intriguing delivery platform. ELPs possess the low critical solution temperature which means that they are soluble below 25°C in water but they undergo self-complexation into a viscoelastic state at 37°C 137. This feature provides ELPs with potential as thermosensitive biomaterials. Shi et al. synthesized the ELP-conjugated polymers that formed nanomicelles with temperature-sensitive assembling behaviors, and they reported that hydrophobic molecules can be entrapped in the micelles that can undergo cell internalization 138. Devalliere et al. developed the ELPs-based biopolymers for co-delivery of a growth factor and a tissue-protective molecule that can lead to sustained delivery and facilitate wound healing 139. Cyclic peptides that contain a circular sequence also can self-assemble. Each cyclic peptide serves as a basic building block and self-assembles into the tubular structures where all amide groups and residues are facing outside 140,141. This outward arrangement is because D-type and L-type amino acids are alternately arranged. Hydrogen bonding between carbonyl and amine groups in adjacent rings drives cyclic peptides to the formation of tubular nanostructures. These nanotubes are known to form pores on the lipid bilayers, channeling the extracellular molecules to cytosol 142,143. In addition, the cyclic peptides-based nanotubes can be used to eradicate bacteria by forming pores on the bacterial membranes 144. Short aromatic peptides such as FF, FY, Fmoc-FF, and Nap-FF have shown that the incorporation of those peptide can cause self-assembly of peptides via strong aromatic π-π stacking 145. Recently, Ashwanikumar et al. reported that the structures of self-assembled peptide-based nanomaterials can be manipulated from a random coil, a distorted α-helix, a β-sheet, or to a pure α-helix by changing the number of phenylalanine in the sequence 109. The twisted drill-like nanostructures showed the highest internalization as compared to the other regular cylindrical counterparts in vitro and in vivo.
7. Cell penetrating peptides (CPPs)
Cell penetrating peptides (CPPs) are short sequenced peptides which facilitate cellular uptake of various molecules. It was discovered by two independent research groups in 1988 that the trans-activator of transcription (TAT) protein of human immunodeficiency virus type-1 (HIV-1) aids the cellular internalization of virus 146,147. The discovery of the first CPP, TAT protein, led to another natural CPP in Antennapedia 148. The structure-activity relationship of the peptide sequences to its ability to enter cells led to the identification of basic functional units. Arginine rich 16 amino acids of the third helix of Antennapedia homeodomain, named as Penetratin, are responsible for the cell penetration function 149. Since then, plenty of CPPs have been found and synthesized, and utilized to translocate a variety of cargos, such as small molecules 150, nucleic acids 151, nanoparticles 152, proteins 153, and imaging 154 and MRI contrast agents 155 in both preclinical studies and clinical trials 156.
The mechanism of action of CPPs was highly debated and it was postulated that these were capable to directly enter cells with an energy-independent manner, bypassing the endolysosomal barriers. However, it is now reported that 95% of TAT peptides and polyarginines enter cells via endocytosis 157,158. Another study proposed that direct penetration of CPPs into cells did exist only in specific conditions, i.e., it is dependent on hydrophobic moieties or cell types and is a rare event 159. Now, it is widely acknowledged that CPPs, especially CPP-conjugated nanoparticles, enter cells via endocytosis by interactions that trigger specific endocytic pathways 160,161. Among different endocytosis pathways, micropinocytosis is major route of the internalization of CPPs 162-164 with limited entry through clathrin-mediated endocytosis 165.
8. Applications of self-assembling CPP conjugates for intracellular delivery
Cell penetrating peptides remain as a valuable tool to deliver a variety of cargos, such as proteins, small molecules, nucleic acids, and imaging agents, and prove their capabilities in a lot of preclinical studies (Figure 9). Among a wide range of the applications of CPPs, the self-assembling CPPs have been considered as a promising material for intracellular delivery. CPPs can be self-assembled through conjugations with the self-assembling materials, including lipids, polymers, and peptides. Detailed reviews onto the self-assembly of various CPP conjugates can be found elsewhere 166. Through physicochemical variations in lipids, polymers, and peptides, we can manipulate the structures and functions of CPP-based nanostructures. Lipid is one of the frequently used partners for CPP conjugations (Figure 9A) 167,168. It was demonstrated that the chain length and density of lipid tails in the CPP conjugates influence self-assembly and cellular uptake of the CPP-lipid conjugates 169,170. Cui and his coworkers reported that the compositions of lipid and the order of amino acids in the CPP nanoparticles influenced structures and encapsulation efficiencies of the CPP-lipid conjugates 168,171. Other than linear fatty acids, cholesterol was also conjugated with TAT peptides. Li and Yang reported that the TAT-cholesterol conjugates self-assembled into the core-shell nanomicelles 172,173. The nanomicelles were distributed across blood-brain barrier (BBB) and displayed the strong antimicrobial activities against Staphylococcus aureus and Cryptococcus neoformans. The CPP-peptide conjugates have been extensively studied. It has been demonstrated that the addition of CPPs to variously structured peptides, such as α helix 174,175 and β sheet (Figure 9B) 176,177, helps the internalization of the nanostructures. Wada et al. proposed the location and the number of TAT peptides in the conjugates perhaps structurally related to the cellular uptake of the CPP-peptide/siRNA complexes 174,175. Hategan et al. reported that addition of TAT peptides to amyloid-β fibrils enhanced rigidity and mechanical resistances of the complexes by increasing β sheet conformations, accompanying with morphological changes and augmented neurotoxicity 178. Wu et al. pointed the roles of amphiphilicity and net charges of CPP-peptides in the self-assembly 179. CPPs have utilized to enhance the cellular uptake of polysaccharide-based nanoparticles, such as folic acid 180, chitosan 181, and hyaluronic acid derivatives 182. Bitton and his coworkers reported that the conjugation of CPPs to polysaccharides changed structures and rheological behaviors of the conjugates 182. Amphiphilic polymers have also been linked with CPPs to assemble of the polymer-CPP conjugates 12. Zhang et al. examined the effects of conjugation sites of the RGD peptide in a series of poly(ɛ-caprolactone-co-lactide)-PEG block copolymers, and reported the conjugation of RGD on PEG blocks showed the enhanced cell adhesion 183. Oligonucleotide-based nanoparticles can be conjugated with CPPs. Burns et al. conjugated their DNA origami with TAT peptide to enhance its subcellular delivery of a model protein (Figure 9C) 7. Qu et al. demonstrated that TAT-conjugated DNA dendrimers encoding CpG motifs displayed the better immunostimulation in macrophage cells, resulting in the higher secretion of cytokines 37. To overcome the lack of specificity of CPPs, a variety of targeting moieties have been conjugated. Sun et al. complexed CPPs with the acid-sensitive anionic oligopeptides to block the internalization at neutral pH 184. In acidic pH, the oligopeptide changed its conformation, exposing the CPP moiety to cells. Adipocyte targeting peptide was conjugated to polyarginines not only to increase cellular uptake, but also to complex with nucleic acids 185. In the following in vivo studies, the intravenously administered complexes reached mouse fat tissues, suggesting the potential as a targeting gene delivery carrier. MacEwan and Chilkoti developed the peptide-based thermosensitive nanomicelles 134,186. In the mild hyperthermic condition (42°C), it self-assembled into micelles with peripheral polyarginine whereas it remained as monomers in the physiological temperature (37°C) (Figure 8B). This controllability over the morphologies and the cellular internalization makes the CPP conjugates to become a promising material for local cancer therapy. Tu and Zhu synthesized the PEG-CPP conjugates which were composed of PEG, a matrix metalloproteinase 2 (MMP2)-sensitive peptide linker, TAT peptide, and DOX 187. They included the MMP2-sensitive link to deshield PEG corona once the nanomicelles encountered extracellular MMP2 which is known to be abundant in tumor microenvironment. It was shown the enzymatic cleavage of PEG corona exposed the TAT moiety, thereby enhancing in vitro cellular uptake of the DOX-loaded nanomicelles.
Figure 9.
Self-Assembling CPP Conjugates for Intracellular Delivery. (A) Lipid-CPP conjugates containing cyclic iRGD assembled into spherical nanovesicles. Hydrophobic small molecules can be loaded in the hydrophobic compartment of the nanovesicles. Adapted with permission from 167, Copyright 2018, American Chemical Society. (B) TAT-conjugated β sheet-forming peptides formed theranostic nanofibrils. Hydrophobic interface of the nanofibrils was able to load hydrophobic drugs. Reproduced with permission from 177, Copyright 2016, American Chemical Society. (C) TAT-decorated DNA origami for protein delivery. TAT peptides were conjugated to facilitate cellular uptake of DNA nanostructures. Reproduced with permission from 7, Copyright 2018, American Chemical Society.
As described above, CPPs with the self-assembling capability offer various advantages for intracellular delivery. It can enhance the cellular internalization of the nanoassemblies by increasing local density of CPPs, thereby assuring the higher possibility to interact with cell membrane 186. The CPP assemblies either physically or chemically constructed possess the larger capacity to package cargos inside. And the packaging of cargos inside protects their stability from degradation, which makes the CPP assemblies the better delivery systems. Furthermore, by making the self-assembling CPPs responsive to exogenous stimuli (e.g., temperature and ionic strength), we can modulate the functionality of the assemblies, posing potential as nanosensors 188.
Despite many advantages of SAP-mediated delivery, there is a concern that SAPs can be degraded by proteolytic enzymes before they reach their in vivo destinations. One way that has been tried to improve the stability of SAPs is to use D-form amino acids. Since natural proteinases degrade L-peptides promptly than D-peptides, using D-forms is effective to protect peptides from enzymatic damages 189. It was reported that D-form nanofibers had higher resistance against proteinases than L-form counterparts, resulting to the extended circulation time 190. However, because the introduction of D-form amino acids may ruin self-assembling behaviors or secondary structures of SAPs, careful consideration in peptide design is necessary to evade any conformational failure. Potential of SAP-assisted delivery is also limited due to lack of computational models for design. Even though a few studies have employed elegant computational methods to design SAPs 191-195, it is still challenging to predict secondary structures of SAP-based nanoarchitectures before synthesis. To come up with information-based peptide chemistry for manipulating self-assembly processes and physicochemical characteristics, it is essential to comprehend how SAP sequence encodes its structural and biological properties at the molecular level 196. Development of robust SAP computing will lead to accurate prediction of structures, and provide rational design for successful intracellular delivery.
9. Conclusion and future perspectives
Over the past several decades, supramolecular chemistry has been employed to enhance the precision of nanostructures, leading to the development of numerous self-assembling biomaterials. Understanding of supramolecular assembly in biological systems has ignited an interest in bioinspired materials. Advances in self-assembling nucleic acids (SANs) led to a new generation of materials with exquisite structural features. Beyond the nanoscale, micrometer-sized structures can be built with self-assembly of nucleic acids 6,197,198, and long chain nucleic acids can be folded into predestined architectures 45,54. Peptide-based self-assembly has been honed to equip more functions. Not only do several peptides act as delivery platforms, but by incorporating bioactive sequences they can have therapeutic effects. In addition to the traditional peptide domains such as CPP moieties, newly identified functional peptide domains have expanded the library that includes peptides with new biological functions 22,199. Several SAPs that form hydrogel scaffolds were reported to induce angiogenesis in wounds by mimicking the natural extracellular matrix, thereby facilitating the wound healing processes 200. Engineering supramolecular self-assembly of either DNA, RNA or peptide can lead to delivery of a wide range of cargos with favorable circulation, biodistribution, cellular uptake, and efficacy. Amphiphilicity introduced to initiate the self-assembling process has allowed for encapsulation of hydrophobic small molecules 109. To achieve the target-specific delivery, it was introduced in the self-assembled delivery systems either to have targeting ligands on the surface 99, or to change their conformations in response to external stimuli 127,135. Short genes for RNA interference can be loaded in either SANs or SAPs, and knockdown the target gene expression 73,201.
Recent applications of the supramolecular self-assembly have been extended to diverse fields. Stem cell therapy is one area where the self-assembling biomaterials can be of high benefit. Evidence from studies in stem cell biology indicates that both SAPs and SANs are able to regulate proliferation, differentiation, and migration of stem cells 202,203, implying their clinical potential in stem cell therapy while maintaining higher order of biocompatibility compared to synthetic counterparts. However, their rapid erosion, particularly in in vivo environment, limits their broader applications in the field, which raises the needs of methods to enhance the stability of self-assembling biomaterials.
Immunotherapy is an emerging field of cancer therapy, and self-assembling nanomaterials have been used to potentiate the immunogenicity. Most SANs were designed to contain immunogenic oligodeoxynucleotides, for example CpG, in their sequences so as to stimulate immune signaling pathways 68. On the other hand, SAPs were used to deliver neoantigen-encoded peptides 204. Combining these two systems into a single hybrid system could be used to co-deliver neoantigens and adjuvants for the personalized cancer treatment.
Gene therapy stands in the front line of modern therapeutics, and the inherent fragility of genetic cargos entails the involvement of various delivery vectors. Early examples of the gene delivery using SANs and SAPs are limited to the short nucleotides (siRNA, miRNA, and shRNA) delivery because of the difficulties to effectively package the massive nucleotides, such as mRNA and pDNA. Recently, it was reported that self-assembled peptide-poloxamine nanoparticles could deliver therapeutic pDNA and mRNA for the cystic fibrosis treatment in in vitro and in vivo, suggesting the possibility to deliver long genes using peptide- or nucleic acid-based materials that are self-assembling 205. The materials that assemble into hollow structures might be beneficial to package the large cargos. In addition, the fact that DNA and RNA building blocks can be designed to generate complementary pairing with the genetic cargos could be useful to ensure the effective protection against degradation by forming tight packaging. There are also lots of demands for the effective delivery systems in gene editing. Although some SANs have been used to deliver the CRISPR/Cas9 system for gene editing, low transfection efficiency and non-specific delivery (off-target effects) still remain as critical challenges 206,207.
There are multiple concerns regarding the intracellular delivery via SANs and SAPs. First of all, abundant enzymes that are present in in vivo milieu are huge threats to the nucleic acid- and peptide-based nanostructures. As mentioned in the review, efforts to increase resistance of nanostructures against digestion have been made by modifying their structures but it is still not enough to deliver a significant amount of cargos during the desirable length of time. Therefore, a novel approach is needed to improve the stability of the assemblies, for example by addition of mild crosslinking agents that are devoid of hampering the flexibility and dynamics of the nanoparticles. Another concern is related to their non-specific delivery. For ideal treatment for any disease, delivering cargos should be specific to the target sites. As already noted, the surface decoration of nanoparticles with targeting ligands has been attempted to guide nanoparticles to the specific loci. However, the relatively complicated preparation and low efficacy in in vivo conditions are limiting the therapeutic outcomes in the ligand-based targeting approaches. Recent efforts to provide molecular trigger systems within nanoassemblies shed light on the development of intelligent delivery systems. DNA nanorobots equipped with the dual-functional aptamers can target the specific cells and trigger the conformational changes that facilitate the release of cargos in specific conditions in in vitro 66 and in vivo 208. Further efforts to discover multifunctional aptamers are expected to improve the precision of delivery. Scaling-up SAPs and SANs is also an important consideration. The bottom-up approach entails mass production of building blocks without error. Even though several chemistries to accurately synthesize nucleotides and peptides have been developed so far, securing the effective methods to produce them in an industrial-scale is highly desirable to translate the supramolecular assembly into clinics 209,210.
Designing the elementary building blocks for supramolecular self-assembly is getting precise owing to the development of nanoinformatics. It is feasible to simulate the assembling patterns in the final architectures from monomers. Using computational and spatial simulations, the transformation and the oligomer formation can be predicted 49,211-213. Attempts to diversify the supramolecular structures are also in progress. Lately, small molecule-mediated self-assembly has drawn a lot of attentions. It was known that a few small molecules, for instance paclitaxel derivatives, tend to self-assemble and form nanoparticles 214,215. Cyanuric acid and sulfated indocyanine dyes were recently reported to help the self-assembly into the nanostructures 212,216. These findings are promising not only because it can increase the loading capacity of small molecules, but also because it can expand the structural design space and functionality of the final assemblies.
Despite exciting advances in understanding the fundamentals of intracellular delivery and designing new biomaterials that can overcome cellular barriers, the effective reach of drugs to exact targets remains farfetched. Low incidence of endosomal escape of nanoparticles remains one of the biggest challenges in intracellular delivery, especially for transfection, and our understanding of endosomal escape is meager. The ability to develop a diverse array of biomaterials through supramolecular assembly can open new avenues to reach intracellular targets. Nature has managed complex cellular structures and synchronous movement of endocytic vesicles within a cell using supramolecular proteins and peptides. And the cellular organelles are “organ-like” structures that contain unique environments. For example, the pH and the lipid and enzyme compositions of endosome and lysosome are distinctive from those of the Golgi apparatus and endoplasmic reticulum. One might surmise that these minute changes may be used to trigger the formation of a supramolecular architecture within the organelle to manipulate its function. Recent studies exploited intracellular enzymes to initiate in situ self-assembly of peptide monomers upon catalysis 217,218. Further, is it possible to let cells engulf small monomers that concentrate in an endosomal vesicle and, due to changes in pH or exposure to light, trigger assembly into a shape that causes endosomal rupture? Can we utilize the rates of assembly and disassembly to measure the environment inside cells-not just in vitro but in vivo? If supramolecular assembly occurs differently in diseased cells as compared to normal cells, could that be used to detect diseases at their earliest 219-221? Use of artificial intelligence and machine learning to design billions of new assemblies for effective delivery is already in the horizon. These questions can unleash infinite possibilities for drug delivery, nanotechnology and material sciences for new applications. This also implies that rational engineering of supramolecular assembly may overcome the challenges in intracellular delivery. With burgeoning collaboration with a variety of fields, including cell biology, biomedical engineering, and clinical medicine, we look forward to get through these scientific progresses to advance our ability to improve interactions of biomaterials at a cellular level, and rational engineering of supramolecular assembly may overcome the challenges in intracellular delivery.
Acknowledgments
G.S will like to thank, OSU College of Pharmacy startup funding (G.S), National Institute of Biomedical Imaging and Bioengineering (N.I.B.I.B) 1R15EB021581-01 (G.S) and Medical Research Foundation of Oregon for their generous support for this work.
Abbreviations
- RGD)
Arginylglycylaspartic acid
- CPP)
Cell-penetrating peptide
- CRISPR)
Clustered, regularly-interspaced short palindromic repeats
- dsDNA)
Double-stranded DNA
- DOX)
Doxorubicin
- EGFP)
Enhanced GFP
- ELP)
Elastin-like polypeptide
- GFP)
Green fluorescent protein
- MMP)
Matrix metalloproteinase
- mRNA)
Messenger RNA
- PA)
Peptide amphiphile
- pDNA)
Plasmid DNA
- PEG)
Polyethylene glycol
- SAN)
Self-assembling nucleic acid
- SAP)
Self-assembling peptide
- shRNA)
Short hairpin RNA
- siRNA)
Small interfering RNA
- TAT)
Trans-activator of transcription
References
- 1.Petkau-Milroy K, Brunsveld L. Supramolecular chemical biology; Bioactive synthetic self-assemblies. Org Biomol Chem. 2013;11:219–32. doi: 10.1039/c2ob26790j. [DOI] [PubMed] [Google Scholar]
- 2.Caulder DL, Raymond KN. Supermolecules by Design. Acc Chem Res. 1999;32:975–82. [Google Scholar]
- 3.Tu Y, Peng F, Adawy A. et al. Mimicking the Cell: Bio-inspired functions of supramolecular assemblies. Chem Rev. 2016;116:2023–78. doi: 10.1021/acs.chemrev.5b00344. [DOI] [PubMed] [Google Scholar]
- 4.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–51. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doolittle LK, Rosen MK, Padrick SB. Measurement and Analysis of In Vitro Actin Polymerization. In: Coutts AS, editor. Adhesion Protein Protocols. 3rd ed.Totowa, NJ: Humana Press; 2013. pp. 273–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tikhomirov G, Petersen P, Qian L. Triangular DNA Origami Tilings. J Am Chem Soc. 2018;140:17361–64. doi: 10.1021/jacs.8b10609. [DOI] [PubMed] [Google Scholar]
- 7.Burns JR, Lamarre B, Pyne ALB, Noble JE, Ryadnov MG. DNA Origami Inside-Out Viruses. ACS Synth Biol. 2018;7:767–73. doi: 10.1021/acssynbio.7b00278. [DOI] [PubMed] [Google Scholar]
- 8.Yin H, Wang H, Li Z, Shu D, Guo P. RNA Micelles for the Systemic Delivery of Anti-miRNA for Cancer Targeting and Inhibition without Ligand. ACS Nano. 2019;13:706–17. doi: 10.1021/acsnano.8b07948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bousmail D, Chidchob P, Sleiman HF. Cyanine-Mediated DNA Nanofiber Growth with Controlled Dimensionality. J Am Chem Soc. 2018;140:9518–30. doi: 10.1021/jacs.8b04157. [DOI] [PubMed] [Google Scholar]
- 10.Hartgerink JD, Beniash E, Stupp SI. Self-Assembly and Mineralization of Peptide-Amphiphile Nanofibers. Science. 2001;294:1684–88. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 11.Wang M, Wang J, Zhou P. et al. Nanoribbons self-assembled from short peptides demonstrate the formation of polar zippers between β-sheets. Nat Commun. 2018;9:5118. doi: 10.1038/s41467-018-07583-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brendel JC, Sanchis J, Catrouillet S. et al. Secondary Self-Assembly of Supramolecular Nanotubes into Tubisomes and Their Activity on Cells. Angew Chem Int Ed Engl. 2018;57:16678–82. doi: 10.1002/anie.201808543. [DOI] [PubMed] [Google Scholar]
- 13.Stewart MP, Sharei A, Ding X. et al. In vitro and ex vivo strategies for intracellular delivery. Nature. 2016;538:183–92. doi: 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- 14.Sahay G, Alakhova DY, Kabanov A V. Endocytosis of nanomedicines. J Control Release. 2010;145:182–95. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang W, Kim BYS, Rutka JT, Chan WCW. Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol. 2008;3:145–50. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 16.Stewart MP, Lorenz A, Dahlman J, Sahay G. Challenges in carrier-mediated intracellular delivery: moving beyond endosomal barriers. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2016;8:465–78. doi: 10.1002/wnan.1377. [DOI] [PubMed] [Google Scholar]
- 17.Gilleron J, Querbes W, Zeigerer A. et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638–46. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 18.Sahay G, Querbes W, Alabi C. et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol. 2013;31:653–58. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol. 2017;35:222–29. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 20.Patel S, Ashwanikumar N, Robinson E. et al. Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated mRNA. Nano Lett. 2017;17:5711–18. doi: 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–28. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Lönn P, Kacsinta AD, Cui X-S. et al. Enhancing Endosomal Escape for Intracellular Delivery of Macromolecular Biologic Therapeutics. Sci Rep. 2016;6:32301. doi: 10.1038/srep32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auffinger P, Ennifar E. Silver-wired DNA. Nat Chem. 2017;9:932–34. doi: 10.1038/nchem.2869. [DOI] [PubMed] [Google Scholar]
- 24.Evans CG, Winfree E. Physical principles for DNA tile self-assembly. Chem Soc Rev. 2017;46:3808–29. doi: 10.1039/c6cs00745g. [DOI] [PubMed] [Google Scholar]
- 25.Grabow WW, Jaeger L. RNA Self-Assembly and RNA Nanotechnology. Acc Chem Res. 2014;47:1871–80. doi: 10.1021/ar500076k. [DOI] [PubMed] [Google Scholar]
- 26.Han D, Pal S, Nangreave J. et al. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342–46. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 27.Seeman NC. Nucleic acid junctions and lattices. J Theor Biol. 1982;99:237–47. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–33. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 29.Kopperger E, List J, Madhira S. et al. A self-assembled nanoscale robotic arm controlled by electric fields. Science. 2018;359:296–301. doi: 10.1126/science.aao4284. [DOI] [PubMed] [Google Scholar]
- 30.Tan SJ, Campolongo MJ, Luo D, Cheng W. Building plasmonic nanostructures with DNA. Nat Nanotechnol. 2011;6:268–76. doi: 10.1038/nnano.2011.49. [DOI] [PubMed] [Google Scholar]
- 31.Kaminska I, Bohlen J, Mackowski S, Tinnefeld P, Acuna GP. Strong Plasmonic Enhancement of a Single Peridinin-Chlorophyll a -Protein Complex on DNA Origami-Based Optical Antennas. ACS Nano. 2018;12:1650–55. doi: 10.1021/acsnano.7b08233. [DOI] [PubMed] [Google Scholar]
- 32.Wilner OI, Weizmann Y, Gill R. et al. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat Nanotechnol. 2009;4:249–54. doi: 10.1038/nnano.2009.50. [DOI] [PubMed] [Google Scholar]
- 33.Dutta PK, Varghese R, Nangreave J. et al. DNA-directed artificial light-harvesting antenna. J Am Chem Soc. 2011;133:11985–93. doi: 10.1021/ja1115138. [DOI] [PubMed] [Google Scholar]
- 34.Funke JJ, Dietz H. Placing molecules with Bohr radius resolution using DNA origami. Nat Nanotechnol. 2016;11:47–52. doi: 10.1038/nnano.2015.240. [DOI] [PubMed] [Google Scholar]
- 35.Edwardson TGW, Lau KL, Bousmail D, Serpell CJ, Sleiman HF. Transfer of molecular recognition information from DNA nanostructures to gold nanoparticles. Nat Chem. 2016;8:162–70. doi: 10.1038/nchem.2420. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Pei H, Zhu B. et al. Self-Assembled Multivalent DNA Nanostructures for Noninvasive Intracellular Delivery of Immunostimulatory CpG Oligonucleotides. ACS Nano. 2011;5:8783–89. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 37.Qu Y, Yang J, Zhan P. et al. Self-Assembled DNA Dendrimer Nanoparticle for Efficient Delivery of Immunostimulatory CpG Motifs. ACS Appl Mater Interfaces. 2017;9:20324–29. doi: 10.1021/acsami.7b05890. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell JC, Harris JR, Malo J, Bath J, Turberfield AJ. Self-Assembly of Chiral DNA Nanotubes. J Am Chem Soc. 2004;126:16342–43. doi: 10.1021/ja043890h. [DOI] [PubMed] [Google Scholar]
- 39.Park SH, Finkelstein G, LaBean TH. Stepwise Self-Assembly of DNA Tile Lattices Using dsDNA Bridges. J Am Chem Soc. 2008;130:40–41. doi: 10.1021/ja078122f. [DOI] [PubMed] [Google Scholar]
- 40.Edwardson TGW, Carneiro KMM, McLaughlin CK, Serpell CJ, Sleiman HF. Site-specific positioning of dendritic alkyl chains on DNA cages enables their geometry-dependent self-assembly. Nat Chem. 2013;5:868–75. doi: 10.1038/nchem.1745. [DOI] [PubMed] [Google Scholar]
- 41.He Y, Ye T, Su M. et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Ke Y, Yan H. Self-Assembly of Symmetric Finite-Size DNA Nanoarrays. J Am Chem Soc. 2005;127:17140–41. doi: 10.1021/ja055614o. [DOI] [PubMed] [Google Scholar]
- 43.Mao C, LaBean TH, Reif JH, Seeman NC. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature. 2000;407:493–96. doi: 10.1038/35035038. [DOI] [PubMed] [Google Scholar]
- 44.Rothemund PWK, Papadakis N, Winfree E. Algorithmic Self-Assembly of DNA Sierpinski Triangles. PLoS Biol. 2004;2:e424. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–21. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 46.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 47.Douglas SM, Dietz H, Liedl T. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–18. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jun H, Zhang F, Shepherd T. et al. Autonomously designed free-form 2D DNA origami. Sci Adv. 2019;5:eaav0655. doi: 10.1126/sciadv.aav0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benson E, Mohammed A, Gardell J. et al. DNA rendering of polyhedral meshes at the nanoscale. Nature. 2015;523:441–44. doi: 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- 50.Carlson R. The changing economics of DNA synthesis. Nat Biotechnol. 2009;27:1091–94. doi: 10.1038/nbt1209-1091. [DOI] [PubMed] [Google Scholar]
- 51.Marchi AN, Saaem I, Vogen BN, Brown S, LaBean TH. Toward Larger DNA Origami. Nano Lett. 2014;14:5740–47. doi: 10.1021/nl502626s. [DOI] [PubMed] [Google Scholar]
- 52.Woo S, Rothemund PWK. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat Chem. 2011;3:620–27. doi: 10.1038/nchem.1070. [DOI] [PubMed] [Google Scholar]
- 53.Gerling T, Wagenbauer KF, Neuner AM, Dietz H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science. 2015;347:1446–52. doi: 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]
- 54.Wagenbauer KF, Sigl C, Dietz H. Gigadalton-scale shape-programmable DNA assemblies. Nature. 2017;552:78–83. doi: 10.1038/nature24651. [DOI] [PubMed] [Google Scholar]
- 55.Madhanagopal BR, Zhang S, Demirel E, Wady H, Chandrasekaran AR. DNA Nanocarriers: Programmed to Deliver. Trends Biochem Sci. 2018;43:997–1013. doi: 10.1016/j.tibs.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y-J, Groves B, Muscat RA, Seelig G. DNA nanotechnology from the test tube to the cell. Nat Nanotechnol. 2015;10:748–60. doi: 10.1038/nnano.2015.195. [DOI] [PubMed] [Google Scholar]
- 57.Mei Q, Wei X, Su F. et al. Stability of DNA Origami Nanoarrays in Cell Lysate. Nano Lett. 2011;11:1477–82. doi: 10.1021/nl1040836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castro CE, Kilchherr F, Kim D-N. et al. A primer to scaffolded DNA origami. Nat Methods. 2011;8:221–29. doi: 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- 59.Mikkilä J, Eskelinen A-P, Niemelä EH. et al. Virus-Encapsulated DNA Origami Nanostructures for Cellular Delivery. Nano Lett. 2014;14:2196–200. doi: 10.1021/nl500677j. [DOI] [PubMed] [Google Scholar]
- 60.Ahmadi Y, De Llano E, Barišić I. (Poly)cation-induced protection of conventional and wireframe DNA origami nanostructures. Nanoscale. 2018;10:7494–504. doi: 10.1039/c7nr09461b. [DOI] [PubMed] [Google Scholar]
- 61.Liu J, Song L, Liu S. et al. A DNA-Based Nanocarrier for Efficient Gene Delivery and Combined Cancer Therapy. Nano Lett. 2018;18:3328–34. doi: 10.1021/acs.nanolett.7b04812. [DOI] [PubMed] [Google Scholar]
- 62.Zhao Y-X, Shaw A, Zeng X. et al. DNA Origami Delivery System for Cancer Therapy with Tunable Release Properties. ACS Nano. 2012;6:8684–91. doi: 10.1021/nn3022662. [DOI] [PubMed] [Google Scholar]
- 63.Eskelinen A-P, Kuzyk A, Kaltiaisenaho TK. et al. Assembly of Single-Walled Carbon Nanotubes on DNA-Origami Templates through Streptavidin-Biotin Interaction. Small. 2011;7:746–50. doi: 10.1002/smll.201001750. [DOI] [PubMed] [Google Scholar]
- 64.Du Y, Jiang Q, Beziere N. et al. DNA-Nanostructure-Gold-Nanorod Hybrids for Enhanced In Vivo Optoacoustic Imaging and Photothermal Therapy. Adv Mater. 2016;28:10000–07. doi: 10.1002/adma.201601710. [DOI] [PubMed] [Google Scholar]
- 65.Chhabra R, Sharma J, Ke Y. et al. Spatially Addressable Multiprotein Nanoarrays Templated by Aptamer-Tagged DNA Nanoarchitectures. J Am Chem Soc. 2007;129:10304–05. doi: 10.1021/ja072410u. [DOI] [PubMed] [Google Scholar]
- 66.Douglas SM, Bachelet I, Church GM. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science. 2012;335:831–34. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 67.Schreiber R, Do J, Roller E-M. et al. Hierarchical assembly of metal nanoparticles, quantum dots and organic dyes using DNA origami scaffolds. Nat Nanotechnol. 2014;9:74–78. doi: 10.1038/nnano.2013.253. [DOI] [PubMed] [Google Scholar]
- 68.Schüller VJ, Heidegger S, Sandholzer N. et al. Cellular Immunostimulation by CpG-Sequence-Coated DNA Origami Structures. ACS Nano. 2011;5:9696–702. doi: 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 69.Chandrasekaran AR, Zavala J, Halvorsen K. Programmable DNA Nanoswitches for Detection of Nucleic Acid Sequences. ACS Sensors. 2016;1:120–23. [Google Scholar]
- 70.Leung K, Chakraborty K, Saminathan A, Krishnan Y. A DNA nanomachine chemically resolves lysosomes in live cells. Nat Nanotechnol. 2019;14:176–83. doi: 10.1038/s41565-018-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thekkan S, Jani MS, Cui C, A DNA-based fluorescent reporter maps HOCl production in the maturing phagosome. Nat Chem Biol; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Veetil AT, Chakraborty K, Xiao K. et al. Cell-targetable DNA nanocapsules for spatiotemporal release of caged bioactive small molecules. Nat Nanotechnol. 2017;12:1183–89. doi: 10.1038/nnano.2017.159. [DOI] [PubMed] [Google Scholar]
- 73.Lee H, Lytton-Jean AKR, Chen Y. et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7:389–93. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eckstein F. Phosphorothioates, Essential Components of Therapeutic Oligonucleotides. Nucleic Acid Ther. 2014;24:374–87. doi: 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- 75.Li P, Sergueeva ZA, Dobrikov M, Shaw BR. Nucleoside and Oligonucleoside Boranophosphates: Chemistry and Properties. Chem Rev. 2007;107:4746–96. doi: 10.1021/cr050009p. [DOI] [PubMed] [Google Scholar]
- 76.Cassinelli V, Oberleitner B, Sobotta J. et al. One-Step Formation of “Chain-Armor”-Stabilized DNA Nanostructures. Angew Chem Int Ed Engl. 2015;54:7795–98. doi: 10.1002/anie.201500561. [DOI] [PubMed] [Google Scholar]
- 77.Perrault SD, Shih WM. Virus-Inspired Membrane Encapsulation of DNA Nanostructures To Achieve In Vivo Stability. ACS Nano. 2014;8:5132–40. doi: 10.1021/nn5011914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liang L, Li J, Li Q. et al. Single-Particle Tracking and Modulation of Cell Entry Pathways of a Tetrahedral DNA Nanostructure in Live Cells. Angew Chem Int Ed Engl. 2014;53:7745–50. doi: 10.1002/anie.201403236. [DOI] [PubMed] [Google Scholar]
- 79.Bastings MMC, Anastassacos FM, Ponnuswamy N. et al. Modulation of the Cellular Uptake of DNA Origami through Control over Mass and Shape. Nano Lett. 2018;18:3557–64. doi: 10.1021/acs.nanolett.8b00660. [DOI] [PubMed] [Google Scholar]
- 80.Kim K-R, Kim D-R, Lee T. et al. Drug delivery by a self-assembled DNA tetrahedron for overcoming drug resistance in breast cancer cells. Chem Commun. 2013;49:2010. doi: 10.1039/c3cc38693g. [DOI] [PubMed] [Google Scholar]
- 81.Westhof E, Masquida B, Jaeger L. RNA tectonics: towards RNA design. Fold Des. 1996;1:R78–88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- 82.Afonin KA, Bindewald E, Yaghoubian AJ. et al. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol. 2010;5:676–82. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Severcan I, Geary C, Chworos A. et al. A polyhedron made of tRNAs. Nat Chem. 2010;2:772–79. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dibrov SM, McLean J, Parsons J, Hermann T. Self-assembling RNA square. Proc Natl Acad Sci. 2011;108:6405–08. doi: 10.1073/pnas.1017999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Han D, Qi X, Myhrvold C. et al. Single-stranded DNA and RNA origami. Science. 2017;358:eaao2648. doi: 10.1126/science.aao2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li M, Zheng M, Wu S. et al. In vivo production of RNA nanostructures via programmed folding of single-stranded RNAs. Nat Commun. 2018;9:2196. doi: 10.1038/s41467-018-04652-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of Intracellular Reactions with Rationally Designed RNA Assemblies. Science. 2011;333:470–74. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 88.Geary C, Rothemund PWK, Andersen ES. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science. 2014;345:799–804. doi: 10.1126/science.1253920. [DOI] [PubMed] [Google Scholar]
- 89.Afonin KA, Kireeva M, Grabow WW. et al. Co-transcriptional Assembly of Chemically Modified RNA Nanoparticles Functionalized with siRNAs. Nano Lett. 2012;12:5192–95. doi: 10.1021/nl302302e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Endo M, Yamamoto S, Tatsumi K. et al. RNA-templated DNA origami structures. Chem Commun. 2013;49:2879–81. doi: 10.1039/c3cc38804b. [DOI] [PubMed] [Google Scholar]
- 91.Wang P, Hyeon Ko S, Tian C, Hao C, Mao C. RNA-DNA hybrid origami: folding of a long RNA single strand into complex nanostructures using short DNA helper strands. Chem Commun. 2013;49:5462–64. doi: 10.1039/c3cc41707g. [DOI] [PubMed] [Google Scholar]
- 92.Ko SH, Su M, Zhang C. et al. Synergistic self-assembly of RNA and DNA molecules. Nat Chem. 2010;2:1050–55. doi: 10.1038/nchem.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stewart JM, Subramanian HKK, Franco E. Self-assembly of multi-stranded RNA motifs into lattices and tubular structures. Nucleic Acids Res. 2017;45:5449–57. doi: 10.1093/nar/gkx063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Afonin KA, Viard M, Martins AN. et al. Activation of different split functionalities on re-association of RNA-DNA hybrids. Nat Nanotechnol. 2013;8:296–304. doi: 10.1038/nnano.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bindewald E, Afonin KA, Viard M. et al. Multistrand Structure Prediction of Nucleic Acid Assemblies and Design of RNA Switches. Nano Lett. 2016;16:1726–35. doi: 10.1021/acs.nanolett.5b04651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee TJ, Yoo JY, Shu D. et al. RNA Nanoparticle-Based Targeted Therapy for Glioblastoma through Inhibition of Oncogenic miR-21. Mol Ther. 2017;25:1544–55. doi: 10.1016/j.ymthe.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pi F, Zhang H, Li H. et al. RNA nanoparticles harboring annexin A2 aptamer can target ovarian cancer for tumor-specific doxorubicin delivery. Nanomedicine. 2017;13:1183–93. doi: 10.1016/j.nano.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shu Y, Yin H, Rajabi M. et al. RNA-based micelles: A novel platform for paclitaxel loading and delivery. J Control Release. 2018;276:17–29. doi: 10.1016/j.jconrel.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Porciani D, Cardwell LN, Tawiah KD. et al. Modular cell-internalizing aptamer nanostructure enables targeted delivery of large functional RNAs in cancer cell lines. Nat Commun. 2018;9:2283. doi: 10.1038/s41467-018-04691-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Afonin KA, Viard M, Koyfman AY. et al. Multifunctional RNA Nanoparticles. Nano Lett. 2014;14:5662–71. doi: 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Khisamutdinov EF, Jasinski DL, Li H. et al. Fabrication of RNA 3D Nanoprisms for Loading and Protection of Small RNAs and Model Drugs. Adv Mater. 2016;28:10079–87. doi: 10.1002/adma.201603180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu G, Mei L, Vishwasrao HD. et al. Intertwining DNA-RNA nanocapsules loaded with tumor neoantigens as synergistic nanovaccines for cancer immunotherapy. Nat Commun. 2017;8:1482. doi: 10.1038/s41467-017-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo S, Li H, Ma M. et al. Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles. Mol Ther - Nucleic Acids. 2017;9:399–408. doi: 10.1016/j.omtn.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guo P. The emerging field of RNA nanotechnology. Nat Nanotechnol. 2010;5:833–42. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jasinski D, Haque F, Binzel DW, Guo P. Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano. 2017;11:1142–64. doi: 10.1021/acsnano.6b05737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ku SH, Jo SD, Lee YK, Kim K, Kim SH. Chemical and structural modifications of RNAi therapeutics. Adv Drug Deliv Rev. 2016;104:16–28. doi: 10.1016/j.addr.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 107.Shu Y, Pi F, Sharma A. et al. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv Drug Deliv Rev. 2014;66:74–89. doi: 10.1016/j.addr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ping Y, Ding D, Ramos RANS. et al. Supramolecular β-Sheets Stabilized Protein Nanocarriers for Drug Delivery and Gene Transfection. ACS Nano. 2017;11:4528–41. doi: 10.1021/acsnano.6b08393. [DOI] [PubMed] [Google Scholar]
- 109.Ashwanikumar N, Plaut JS, Mostofian B. et al. Supramolecular self assembly of nanodrill-like structures for intracellular delivery. J Control Release. 2018;282:76–89. doi: 10.1016/j.jconrel.2018.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ashwanikumar N, Kumar NA, Saneesh Babu PS. et al. Self-assembling peptide nanofibers containing phenylalanine for the controlled release of 5-fluorouracil. Int J Nanomedicine. 2016;11:5583–94. doi: 10.2147/IJN.S104707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Koss K, Tsui C, Unsworth LD. Induced Neural Differentiation of MMP-2 Cleaved (RADA)4 Drug Delivery Systems. J Control Release. 2016;243:204–13. doi: 10.1016/j.jconrel.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 112.Cheng T-Y, Chen M-H, Chang W-H, Huang M-Y, Wang T-W. Neural stem cells encapsulated in a functionalized self-assembling peptide hydrogel for brain tissue engineering. Biomaterials. 2013;34:2005–16. doi: 10.1016/j.biomaterials.2012.11.043. [DOI] [PubMed] [Google Scholar]
- 113.Sun X, Yin H, Wang Y. et al. In Situ Articular Cartilage Regeneration through Endogenous Reparative Cell Homing Using a Functional Bone Marrow-Specific Scaffolding System. ACS Appl Mater Interfaces. 2018;10:38715–28. doi: 10.1021/acsami.8b11687. [DOI] [PubMed] [Google Scholar]
- 114.Pauling L, Corey RB, Branson HR. The structure of proteins: Two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci. 1951;37:205–11. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fletcher JM, Horner KA, Bartlett GJ. et al. De novo coiled-coil peptides as scaffolds for disrupting protein-protein interactions. Chem Sci. 2018;9:7656–65. doi: 10.1039/c8sc02643b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eriksson M, Hassan S, Larsson R. et al. Utilization of a Right-handed Coiled-coil Protein from Archaebacterium Staphylothermus marinus as a Carrier for Cisplatin. Anticancer Res. 2009;29:11–18. [PubMed] [Google Scholar]
- 117.Yin L, Agustinus AS, Yuvienco C. et al. Engineered Coiled-Coil Protein for Delivery of Inverse Agonist for Osteoarthritis. Biomacromolecules. 2018;19:1614–24. doi: 10.1021/acs.biomac.8b00158. [DOI] [PubMed] [Google Scholar]
- 118.Assal Y, Mizuguchi Y, Mie M, Kobatake E. Growth Factor Tethering to Protein Nanoparticles via Coiled-Coil Formation for Targeted Drug Delivery. Bioconjug Chem. 2015;26:1672–77. doi: 10.1021/acs.bioconjchem.5b00266. [DOI] [PubMed] [Google Scholar]
- 119.Martelli G, Zope HR, Bròvia Capell M, Kros A. Coiled-coil peptide motifs as thermoresponsive valves for mesoporous silica nanoparticles. Chem Commun. 2013;49:9932–34. doi: 10.1039/c3cc45790g. [DOI] [PubMed] [Google Scholar]
- 120.Yang J, Shimada Y, Olsthoorn RCL. et al. Application of Coiled Coil Peptides in Liposomal Anticancer Drug Delivery Using a Zebrafish Xenograft Model. ACS Nano. 2016;10:7428–35. doi: 10.1021/acsnano.6b01410. [DOI] [PubMed] [Google Scholar]
- 121.Wu K, Liu J, Johnson RN, Yang J, Kopeček J. Drug-Free Macromolecular Therapeutics: Induction of Apoptosis by Coiled-Coil-Mediated Cross-Linking of Antigens on the Cell Surface. Angew Chem Int Ed Engl. 2010;49:1451–55. doi: 10.1002/anie.200906232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang R, Yang J, Chu T-W, Hartley JM, Kopeček J. Multimodality Imaging of Coiled-Coil Mediated Self-Assembly in a “Drug-Free” Therapeutic System. Adv Healthc Mater. 2015;4:1054–65. doi: 10.1002/adhm.201400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nakayama N, Hagiwara K, Ito Y. et al. Superior Cell Penetration by a Rigid and Anisotropic Synthetic Protein. Langmuir. 2015;31:2826–32. doi: 10.1021/la504494x. [DOI] [PubMed] [Google Scholar]
- 124.Nakayama N, Takaoka S, Ota M, Takagaki K, Sano K-I. Effect of the Aspect Ratio of Coiled-Coil Protein Carriers on Cellular Uptake. Langmuir. 2018;34:14286–93. doi: 10.1021/acs.langmuir.8b02616. [DOI] [PubMed] [Google Scholar]
- 125.Woolfson DN, Mahmoud ZN. More than just bare scaffolds: towards multi-component and decorated fibrous biomaterials. Chem Soc Rev. 2010;39:3464. doi: 10.1039/c0cs00032a. [DOI] [PubMed] [Google Scholar]
- 126.Paramonov SE, Jun H-W, Hartgerink JD. Self-Assembly of Peptide-Amphiphile Nanofibers: The Roles of Hydrogen Bonding and Amphiphilic Packing. J Am Chem Soc. 2006;128:7291–98. doi: 10.1021/ja060573x. [DOI] [PubMed] [Google Scholar]
- 127.Webber MJ, Newcomb CJ, Bitton R, Stupp SI. Switching of self-assembly in a peptide nanostructure with a specific enzyme. Soft Matter. 2011;7:9665. doi: 10.1039/c1sm05610g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hartgerink JD, Beniash E, Stupp SI. Peptide-amphiphile nanofibers: A versatile scaffold for the preparation of self-assembling materials. Proc Natl Acad Sci. 2002;99:5133–38. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xiao Y, Zhang Q, Wang Y. et al. Dual-functional protein for one-step production of a soluble and targeted fluorescent dye. Theranostics. 2018;8:3111–25. doi: 10.7150/thno.24613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Black M, Trent A, Kostenko Y. et al. Self-Assembled Peptide Amphiphile Micelles Containing a Cytotoxic T-Cell Epitope Promote a Protective Immune Response In Vivo. Adv Mater. 2012;24:3845–49. doi: 10.1002/adma.201200209. [DOI] [PubMed] [Google Scholar]
- 131.Chen J, Pompano RR, Santiago FW. et al. The use of self-adjuvanting nanofiber vaccines to elicit high-affinity B cell responses to peptide antigens without inflammation. Biomaterials. 2013;34:8776–85. doi: 10.1016/j.biomaterials.2013.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Trent A, Ulery BD, Black MJ. et al. Peptide Amphiphile Micelles Self-Adjuvant Group A Streptococcal Vaccination. AAPS J. 2015;17:380–88. doi: 10.1208/s12248-014-9707-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zimenkov Y, Dublin SN, Ni R. et al. Rational Design of a Reversible pH-Responsive Switch for Peptide Self-Assembly. J Am Chem Soc. 2006;128:6770–71. doi: 10.1021/ja0605974. [DOI] [PubMed] [Google Scholar]
- 134.MacEwan SR, Chilkoti A. Digital Switching of Local Arginine Density in a Genetically Encoded Self-Assembled Polypeptide Nanoparticle Controls Cellular Uptake. Nano Lett. 2012;12:3322–28. doi: 10.1021/nl301529p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sur S, Matson JB, Webber MJ, Newcomb CJ, Stupp SI. Photodynamic Control of Bioactivity in a Nanofiber Matrix. ACS Nano. 2012;6:10776–85. doi: 10.1021/nn304101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Suyama K, Tatsubo D, Iwasaki W. et al. Enhancement of Self-Aggregation Properties of Linear Elastin-Derived Short Peptides by Simple Cyclization: Strong Self-Aggregation Properties of Cyclo[FPGVG]n, Consisting Only of Natural Amino Acids. Biomacromolecules. 2018;19:3201–11. doi: 10.1021/acs.biomac.8b00353. [DOI] [PubMed] [Google Scholar]
- 137.Despanie J, Dhandhukia JP, Hamm-Alvarez SF, MacKay JA. Elastin-like polypeptides: Therapeutic applications for an emerging class of nanomedicines. J Control Release. 2016;240:93–108. doi: 10.1016/j.jconrel.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shi P, Aluri S, Lin Y-A. et al. Elastin-based protein polymer nanoparticles carrying drug at both corona and core suppress tumor growth in vivo. J Control Release. 2013;171:330–38. doi: 10.1016/j.jconrel.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Devalliere J, Dooley K, Hu Y. et al. Co-delivery of a growth factor and a tissue-protective molecule using elastin biopolymers accelerates wound healing in diabetic mice. Biomaterials. 2017;141:149–60. doi: 10.1016/j.biomaterials.2017.06.043. [DOI] [PubMed] [Google Scholar]
- 140.Ghadiri MR, Granja JR, Milligan RA, McRee DE, Khazanovich N. Self-assembling organic nanotubes based on a cyclic peptide architecture. Nature. 1993;366:324–27. doi: 10.1038/366324a0. [DOI] [PubMed] [Google Scholar]
- 141.Kim HS, Hartgerink JD, Ghadiri MR. Oriented Self-Assembly of Cyclic Peptide Nanotubes in Lipid Membranes. J Am Chem Soc. 1998;120:4417–24. [Google Scholar]
- 142.Chen J, Zhang B, Xia F. et al. Transmembrane delivery of anticancer drugs through self-assembly of cyclic peptide nanotubes. Nanoscale. 2016;8:7127–36. doi: 10.1039/c5nr06804e. [DOI] [PubMed] [Google Scholar]
- 143.Larnaudie SC, Brendel JC, Romero-Canelón I. et al. Cyclic Peptide-Polymer Nanotubes as Efficient and Highly Potent Drug Delivery Systems for Organometallic Anticancer Complexes. Biomacromolecules. 2017;19:239–47. doi: 10.1021/acs.biomac.7b01491. [DOI] [PubMed] [Google Scholar]
- 144.Barbosa SC, Nobre TM, Volpati D. et al. The importance of cyclic structure for Labaditin on its antimicrobial activity against Staphylococcus aureus. Colloids Surf B Biointerfaces. 2016;148:453–59. doi: 10.1016/j.colsurfb.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 145.Ashwanikumar N, Kumar NA, Nair SA, Kumar GSV. Phenylalanine-containing self-assembling peptide nanofibrous hydrogel for the controlled release of 5-fluorouracil and leucovorin. RSC Adv. 2014;4:29157–64. [Google Scholar]
- 146.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–93. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 147.Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55:1179–88. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 148.Joliot A, Pernelle C, Deagostini-Bazin H, Prochiantz A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc Natl Acad Sci. 1991;88:1864–68. doi: 10.1073/pnas.88.5.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–50. [PubMed] [Google Scholar]
- 150.Sawant RR, Torchilin VP. Enhanced cytotoxicity of TATp-bearing paclitaxel-loaded micelles in vitro and in vivo. Int J Pharm. 2009;374:114–18. doi: 10.1016/j.ijpharm.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ye J, Liu E, Gong J. et al. High-Yield Synthesis of Monomeric LMWP(CPP)-siRNA Covalent Conjugate for Effective Cytosolic Delivery of siRNA. Theranostics. 2017;7:2495–508. doi: 10.7150/thno.19863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhao Y, Ren W, Zhong T. et al. Tumor-specific pH-responsive peptide-modified pH-sensitive liposomes containing doxorubicin for enhancing glioma targeting and anti-tumor activity. J Control Release. 2016;222:56–66. doi: 10.1016/j.jconrel.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 153.Hu J-W, Liu BR, Wu C-Y, Lu S-W, Lee H-J. Protein transport in human cells mediated by covalently and noncovalently conjugated arginine-rich intracellular delivery peptides. Peptides. 2009;30:1669–78. doi: 10.1016/j.peptides.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 154.Liu BR, Huang Y, Winiarz JG, Chiang H-J, Lee H-J. Intracellular delivery of quantum dots mediated by a histidine-and arginine-rich HR9 cell-penetrating peptide through the direct membrane translocation mechanism. Biomaterials. 2011;32:3520–37. doi: 10.1016/j.biomaterials.2011.01.041. [DOI] [PubMed] [Google Scholar]
- 155.Mishra R, Su W, Pohmann R. et al. Cell-penetrating peptides and peptide nucleic acid-coupled MRI contrast agents: evaluation of cellular delivery and target binding. Bioconjug Chem. 2009;20:1860–68. doi: 10.1021/bc9000454. [DOI] [PubMed] [Google Scholar]
- 156.Guidotti G, Brambilla L, Rossi D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol Sci. 2017;38:406–24. doi: 10.1016/j.tips.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 157.Wadia JS, Stan R V, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10:310–15. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 158.Nakase I, Niwa M, Takeuchi T. et al. Cellular Uptake of Arginine-Rich Peptides: Roles for Macropinocytosis and Actin Rearrangement. Mol Ther. 2004;10:1011–22. doi: 10.1016/j.ymthe.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 159.Hirose H, Takeuchi T, Osakada H. et al. Transient focal membrane deformation induced by arginine-rich peptides leads to their direct penetration into cells. Mol Ther. 2012;20:984–93. doi: 10.1038/mt.2011.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Palm-Apergi C, Lönn P, Dowdy SF. Do cell-penetrating peptides actually “penetrate” cellular membranes? Mol Ther. 2012;20:695–97. doi: 10.1038/mt.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.LeCher JC, Nowak SJ, McMurry JL. Breaking in and busting out: cell-penetrating peptides and the endosomal escape problem. Biomol Concepts. 2017;8:1011–14. doi: 10.1515/bmc-2017-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kaplan IM, Wadia JS, Dowdy SF. Cationic TAT peptide transduction domain enters cells by macropinocytosis. J Control Release. 2005;102:247–53. doi: 10.1016/j.jconrel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 163.Nakase I, Tadokoro A, Kawabata N. et al. Interaction of Arginine-Rich Peptides with Membrane-Associated Proteoglycans Is Crucial for Induction of Actin Organization and Macropinocytosis. Biochemistry. 2007;46:492–501. doi: 10.1021/bi0612824. [DOI] [PubMed] [Google Scholar]
- 164.Wadia JS, Schaller M, Williamson RA, Dowdy SF. Pathologic Prion Protein Infects Cells by Lipid-Raft Dependent Macropinocytosis. PLoS One. 2008;3:e3314. doi: 10.1371/journal.pone.0003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Richard JP, Melikov K, Brooks H. et al. Cellular Uptake of Unconjugated TAT Peptide Involves Clathrin-dependent Endocytosis and Heparan Sulfate Receptors. J Biol Chem. 2005;280:15300–06. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 166.Shi Y, Conde J, Azevedo HS. Sunna A, Care A, Bergquist PL, eds. Peptides and Peptide-based Biomaterials and their Biomedical Applications, Springer, Cham; 2017. Empowering the Potential of Cell-Penetrating Peptides for Targeted Intracellular Delivery via Molecular Self-Assembly; pp. 265–78. [DOI] [PubMed] [Google Scholar]
- 167.Jiang Y, Pang X, Liu R. et al. Design of an Amphiphilic iRGD Peptide and Self-Assembling Nanovesicles for Improving Tumor Accumulation and Penetration and the Photodynamic Efficacy of the Photosensitizer. ACS Appl Mater Interfaces. 2018;10:31674–85. doi: 10.1021/acsami.8b11699. [DOI] [PubMed] [Google Scholar]
- 168.Cui H, Cheetham AG, Pashuck ET, Stupp SI. Amino Acid Sequence in Constitutionally Isomeric Tetrapeptide Amphiphiles Dictates Architecture of One-Dimensional Nanostructures. J Am Chem Soc. 2014;136:12461–68. doi: 10.1021/ja507051w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bode SA, Thévenin M, Bechara C. et al. Self-assembling mini cell-penetrating peptides enter by both direct translocation and glycosaminoglycan-dependent endocytosis. Chem Commun. 2012;48:7179–81. doi: 10.1039/c2cc33240j. [DOI] [PubMed] [Google Scholar]
- 170.Lim Y, Lee E, Lee M. Controlled Bioactive Nanostructures from Self-Assembly of Peptide Building Blocks. Angew Chem Int Ed Engl. 2007;46:9011–14. doi: 10.1002/anie.200702732. [DOI] [PubMed] [Google Scholar]
- 171.Zhang P, Cheetham AG, Lin YA, Cui H. Self-assembled tat nanofibers as effective drug carrier and transporter. ACS Nano. 2013;7:5965–77. doi: 10.1021/nn401667z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu L, Xu K, Wang H. et al. Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent. Nat Nanotechnol. 2009;4:457. doi: 10.1038/nnano.2009.153. [DOI] [PubMed] [Google Scholar]
- 173.Wang H, Xu K, Liu L. et al. The efficacy of self-assembled cationic antimicrobial peptide nanoparticles against Cryptococcus neoformans for the treatment of meningitis. Biomaterials. 2010;31:2874–81. doi: 10.1016/j.biomaterials.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 174.Wada S, Iwata M, Ozaki Y. et al. Design of cyclic RGD-conjugated Aib-containing amphipathic helical peptides for targeted delivery of small interfering RNA. Bioorg Med Chem. 2016;24:4478–85. doi: 10.1016/j.bmc.2016.07.040. [DOI] [PubMed] [Google Scholar]
- 175.Wada S, Takesada A, Nagamura Y. et al. Structure-activity relationship study of Aib-containing amphipathic helical peptide-cyclic RGD conjugates as carriers for siRNA delivery. Bioorg Med Chem Lett. 2017;27:5378–81. doi: 10.1016/j.bmcl.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 176.Lim Y, Lee E, Lee M. Cell-Penetrating-Peptide-Coated Nanoribbons for Intracellular Nanocarriers. Angew Chem Int Ed Engl. 2007;46:3475–78. doi: 10.1002/anie.200604576. [DOI] [PubMed] [Google Scholar]
- 177.Kim I, Han EH, Ryu J. et al. One-Dimensional Supramolecular Nanoplatforms for Theranostics Based on Co-Assembly of Peptide Amphiphiles. Biomacromolecules. 2016;17:3234–43. doi: 10.1021/acs.biomac.6b00966. [DOI] [PubMed] [Google Scholar]
- 178.Hategan A, Bianchet MA, Steiner J. et al. HIV Tat protein and amyloid-β peptide form multifibrillar structures that cause neurotoxicity. Nat Struct Mol Biol. 2017;24:379–86. doi: 10.1038/nsmb.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wu D, Zhang S, Zhao Y. et al. The effects of motif net charge and amphiphilicity on the self-assembly of functionally designer RADA16-I peptides. Biomed Mater. 2018;13:035011. doi: 10.1088/1748-605X/aab2fd. [DOI] [PubMed] [Google Scholar]
- 180.Yan C, Gu J, Hou D. et al. Synthesis of Tat tagged and folate modified N-succinyl-chitosan self-assembly nanoparticles as a novel gene vector. Int J Biol Macromol. 2015;72:751–56. doi: 10.1016/j.ijbiomac.2014.09.031. [DOI] [PubMed] [Google Scholar]
- 181.Yan C, Gu J, Jing H, Taishi J, Lee RJ. Tat-Tagged and Folate-Modified N-Succinyl-chitosan (Tat-Suc-FA) Self-assembly Nanoparticle for Therapeutic Delivery OGX-011 to A549 Cells. Mol Pharm. 2017;14:1898–905. doi: 10.1021/acs.molpharmaceut.6b01167. [DOI] [PubMed] [Google Scholar]
- 182.Bernstein-Levi O, Ochbaum G, Bitton R. The effect of covalently linked RGD peptide on the conformation of polysaccharides in aqueous solutions. Colloids Surf B Biointerfaces. 2016;137:214–20. doi: 10.1016/j.colsurfb.2015.06.042. [DOI] [PubMed] [Google Scholar]
- 183.Zhang Z, Lai Y, Yu L, Ding J. Effects of immobilizing sites of RGD peptides in amphiphilic block copolymers on efficacy of cell adhesion. Biomaterials. 2010;31:7873–82. doi: 10.1016/j.biomaterials.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 184.Sun C, Shen W-C, Tu J, Zaro JL. Interaction between Cell-Penetrating Peptides and Acid-Sensitive Anionic Oligopeptides as a Model for the Design of Targeted Drug Carriers. Mol Pharm. 2014;11:1583–90. doi: 10.1021/mp400747k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Won Y-W, Adhikary PP, Lim KS. et al. Oligopeptide complex for targeted non-viral gene delivery to adipocytes. Nat Mater. 2014;13:1157–64. doi: 10.1038/nmat4092. [DOI] [PubMed] [Google Scholar]
- 186.Macewan SR, Chilkoti A. Controlled Apoptosis by a Thermally Toggled Nanoscale Amplifier of Cellular Uptake. Nano Lett. 2014;14:2058–64. doi: 10.1021/nl5002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tu Y, Zhu L. Enhancing cancer targeting and anticancer activity by a stimulus-sensitive multifunctional polymer-drug conjugate. J Control Release. 2015;212:94–102. doi: 10.1016/j.jconrel.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 188.Lock LL, Cheetham AG, Zhang P, Cui H. Design and Construction of Supramolecular Nanobeacons for Enzyme Detection. ACS Nano. 2013;7:4924–32. doi: 10.1021/nn400218a. [DOI] [PubMed] [Google Scholar]
- 189.Tugyi R, Uray K, Ivan D. et al. Partial D-amino acid substitution: Improved enzymatic stability and preserved Ab recognition of a MUC2 epitope peptide. Proc Natl Acad Sci. 2005;102:413–18. doi: 10.1073/pnas.0407677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Liu J, Liu J, Chu L. et al. Self-Assembling Peptide of d-Amino Acids Boosts Selectivity and Antitumor Efficacy of 10-Hydroxycamptothecin. ACS Appl Mater Interfaces. 2014;6:5558–65. doi: 10.1021/am406007g. [DOI] [PubMed] [Google Scholar]
- 191.Colombo G, Soto P, Gazit E. Peptide self-assembly at the nanoscale: a challenging target for computational and experimental biotechnology. Trends Biotechnol. 2007;25:211–18. doi: 10.1016/j.tibtech.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 192.Slyngborg M, Fojan P. A computational study of the self-assembly of the RFFFR peptide. Phys Chem Chem Phys. 2015;17:30023–36. doi: 10.1039/c5cp01324k. [DOI] [PubMed] [Google Scholar]
- 193.Zhang HV, Polzer F, Haider MJ. et al. Computationally designed peptides for self-assembly of nanostructured lattices. Sci Adv. 2016;2:e1600307. doi: 10.1126/sciadv.1600307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shen H, Fallas JA, Lynch E. et al. De novo design of self-assembling helical protein filaments. Science. 2018;362:705–09. doi: 10.1126/science.aau3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Marcos E, Chidyausiku TM, McShan AC. et al. De novo design of a non-local β-sheet protein with high stability and accuracy. Nat Struct Mol Biol. 2018;25:1028–34. doi: 10.1038/s41594-018-0141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Woods D, Doty D, Myhrvold C. et al. Diverse and robust molecular algorithms using reprogrammable DNA self-assembly. Nature. 2019;567:366–72. doi: 10.1038/s41586-019-1014-9. [DOI] [PubMed] [Google Scholar]
- 197.Tikhomirov G, Petersen P, Qian L. Programmable disorder in random DNA tilings. Nat Nanotechnol. 2017;12:251–59. doi: 10.1038/nnano.2016.256. [DOI] [PubMed] [Google Scholar]
- 198.Tikhomirov G, Petersen P, Qian L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature. 2017;552:67–71. doi: 10.1038/nature24655. [DOI] [PubMed] [Google Scholar]
- 199.Wang Z, Zhang RX, Zhang T. et al. In Situ Proapoptotic Peptide-Generating Rapeseed Protein-Based Nanocomplexes Synergize Chemotherapy for Cathepsin-B Overexpressing Breast Cancer. ACS Appl Mater Interfaces. 2018;10:41056–69. doi: 10.1021/acsami.8b14001. [DOI] [PubMed] [Google Scholar]
- 200.Albadr A, Coulter S, Porter S, Thakur R, Laverty G. Ultrashort Self-Assembling Peptide Hydrogel for the Treatment of Fungal Infections. Gels. 2018;4:48. doi: 10.3390/gels4020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Chen G, Liu D, He C. et al. Enzymatic Synthesis of Periodic DNA Nanoribbons for Intracellular pH Sensing and Gene Silencing. J Am Chem Soc. 2015;137:3844–51. doi: 10.1021/ja512665z. [DOI] [PubMed] [Google Scholar]
- 202.Park HS, Choi GH, Kim D. et al. Use of Self-Assembling Peptides to Enhance Stem Cell Function for Therapeutic Angiogenesis. Stem Cells Int. 2018;2018:1–12. doi: 10.1155/2018/4162075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Li S, Tian T, Zhang T, Cai X, Lin Y. Advances in biological applications of self-assembled DNA tetrahedral nanostructures. Mater Today. 2019;24:57–68. [Google Scholar]
- 204.Rad-Malekshahi M, Fransen MF, Krawczyk M. et al. Self-Assembling Peptide Epitopes as Novel Platform for Anticancer Vaccination. Mol Pharm. 2017;14:1482–93. doi: 10.1021/acs.molpharmaceut.6b01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Guan S, Munder A, Hedtfeld S. et al. Self-assembled peptide-poloxamine nanoparticles enable in vitro and in vivo genome restoration for cystic fibrosis. Nat Nanotechnol. 2019;14:287–97. doi: 10.1038/s41565-018-0358-x. [DOI] [PubMed] [Google Scholar]
- 206.Sun W, Ji W, Hall JM. et al. Self-Assembled DNA Nanoclews for the Efficient Delivery of CRISPR-Cas9 for Genome Editing. Angew Chem Int Ed Engl. 2015;54:12029–33. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kim SM, Shin SC, Kim EE. et al. Simple in Vivo Gene Editing via Direct Self-Assembly of Cas9 Ribonucleoprotein Complexes for Cancer Treatment. ACS Nano. 2018;12:7750–60. doi: 10.1021/acsnano.8b01670. [DOI] [PubMed] [Google Scholar]
- 208.Li S, Jiang Q, Liu S. et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat Biotechnol. 2018;36:258–64. doi: 10.1038/nbt.4071. [DOI] [PubMed] [Google Scholar]
- 209.Praetorius F, Kick B, Behler KL. et al. Biotechnological mass production of DNA origami. Nature. 2017;552:84–87. doi: 10.1038/nature24650. [DOI] [PubMed] [Google Scholar]
- 210.Lönnberg H. Synthesis of oligonucleotides on a soluble support. Beilstein J Org Chem. 2017;13:1368–87. doi: 10.3762/bjoc.13.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Veneziano R, Ratanalert S, Zhang K. et al. Designer nanoscale DNA assemblies programmed from the top down. Science. 2016;352:1534–1534. doi: 10.1126/science.aaf4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Shamay Y, Shah J, Işık M. et al. Quantitative self-assembly prediction yields targeted nanomedicines. Nat Mater. 2018;17:361–68. doi: 10.1038/s41563-017-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhao X, Liao C, Ma Y-T. et al. Top-down Multiscale Approach To Simulate Peptide Self-Assembly from Monomers. J Chem Theory Comput. 2019;15:1514–22. doi: 10.1021/acs.jctc.8b01025. [DOI] [PubMed] [Google Scholar]
- 214.Tian R, Wang H, Niu R, Ding D. Drug delivery with nanospherical supramolecular cell penetrating peptide-taxol conjugates containing a high drug loading. J Colloid Interface Sci. 2015;453:15–20. doi: 10.1016/j.jcis.2015.04.028. [DOI] [PubMed] [Google Scholar]
- 215.Song Q, Zhang R, Lei L, Li X. Self-Assembly of Succinated Paclitaxel into Supramolecular Hydrogel for Local Cancer Chemotherapy. J Biomed Nanotechnol. 2018;14:1471–76. doi: 10.1166/jbn.2018.2595. [DOI] [PubMed] [Google Scholar]
- 216.Avakyan N, Greschner AA, Aldaye F. et al. Reprogramming the assembly of unmodified DNA with a small molecule. Nat Chem. 2016;8:368–76. doi: 10.1038/nchem.2451. [DOI] [PubMed] [Google Scholar]
- 217.Li J, Kuang Y, Shi J. et al. Enzyme-Instructed Intracellular Molecular Self-Assembly to Boost Activity of Cisplatin against Drug-Resistant Ovarian Cancer Cells. Angew Chem Int Ed Engl. 2015;54:13307–11. doi: 10.1002/anie.201507157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Li L-L, Qiao S-L, Liu W-J. et al. Intracellular construction of topology-controlled polypeptide nanostructures with diverse biological functions. Nat Commun. 2017;8:1276. doi: 10.1038/s41467-017-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ye D, Shuhendler AJ, Cui L. et al. Bioorthogonal cyclization-mediated in situ self-assembly of small-molecule probes for imaging caspase activity in vivo. Nat Chem. 2014;6:519–26. doi: 10.1038/nchem.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Nejadnik H, Ye D, Lenkov OD. et al. Magnetic Resonance Imaging of Stem Cell Apoptosis in Arthritic Joints with a Caspase Activatable Contrast Agent. ACS Nano. 2015;9:1150–60. doi: 10.1021/nn504494c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wei Y, Zhou W, Li X. et al. Coupling hybridization chain reaction with catalytic hairpin assembly enables non-enzymatic and sensitive fluorescent detection of microRNA cancer biomarkers. Biosens Bioelectron. 2016;77:416–20. doi: 10.1016/j.bios.2015.09.053. [DOI] [PubMed] [Google Scholar]