Abstract
The highly contagious disease tuberculosis (TB) is caused by the bacterium Mycobacterium tuberculosis (Mtb), which has been evolving drug resistance at an alarming rate. Like all human pathogens, Mtb requires iron for growth and virulence. Consequently, Mtb iron transport is an emerging drug target. However, the development of anti-TB drugs aimed at these metabolic pathways have been restricted by the dearth of information on Mtb iron acquisition. In this review, we describe the multiple strategies utilized by Mtb to acquire ferric iron and heme-iron. Mtb iron uptake is a complex process, requiring biosynthesis and subsequent export of Mtb siderophores, followed by ferric iron scavenging and ferric-siderophore import into Mtb. Additionally, Mtb possesses two possible heme uptake pathways, and an Mtb-specific mechanism of heme degradation that yields iron and novel heme-degradation products. We conclude with perspectives for potential therapeutics that could directly target Mtb heme and iron uptake machineries. We also highlight how hijacking Mtb heme and iron acquisition pathways for drug import may facilitate drug transport through the notoriously impregnable Mtb cell-wall.
Graphical Abstract
1. Introduction
1.1. Tuberculosis
Tuberculosis (TB) is one of the deadliest infectious diseases known to man. TB has plagued the world since ancient times, and has been discovered in ancient Egyptian mummies.1 Prior to the advent of anti-TB drugs in the 1940s, a TB diagnosis was considered a death sentence.2 Mycobacterium tuberculosis (Mtb), the etiological agent of TB, is a slow-growing, acid-fast bacterium.2 Mtb has evolved multiple mechanisms to evade host immune defenses, and upon host assault and antibiotic treatment Mtb often moves into a latent state within the human host.3 Notably, latent TB is asymptomatic, but can be activated by malnutrition or in immunocompromised individuals.4 Before the discovery of anti-TB drugs, TB patients were sent to isolated resorts to be quarantined and treated, which at the time consisted of little more than bedrest.2 Following the discovery of penicillin, the first anti-TB drug streptomycin was discovered in 1943 from Streptomyces griseus.2 Other anti-TB drugs soon followed including isoniazid, rifampicin, pyrazinamide and ethambutol, and by the end of the 1970s, it was thought that TB could be eradicated globally.2 Unfortunately, this was not to be the case. Today, TB remains one of the most deadly infectious diseases worldwide.
1.2. The need for novel anti-TB drugs
With the onset of AIDS in the 1980s, TB cases started to re-emerge.5 HIV and TB are synergistic. AIDS weakens the immune system, allowing for latent TB to be reactivated into a full-blown active TB infection. An estimated one-third of the world’s population is infected with latent TB, and those living with HIV are 20 to 30 times more likely to develop active TB than those without HIV.6 In 2016, 10.4 million people were diagnosed and treated for TB, and there were 1.7 million TB-related deaths. Notably 95% of these deaths were in low- or medium-income countries.6
Following the discovery of streptomycin in 1943, an array of anti-TB drugs was developed over the next several decades. First line drugs such as isoniazid, rifampicin, pyrazinamide, ethambutol, and streptomycin are used in the standard cocktail to treat TB patients. These drugs are administered together as a drug regimen cocktail in accordance with guidelines set forth by the World Health Organization (WHO).6 The usual TB treatment regimen lasts more than 6 months, and up to 18 months in the case of drug resistant isolates.6 Unfortunately, many anti-TB drugs cause harsh side effects or result in medical complications such as liver damage. Moreover, the prolonged period of treatment can result in therapeutic noncompliance by patients that leads to the development of drug resistant strains.6
The TB epidemic is compounded by the rise of multidrug resistant TB (MDR-TB).7 Over the last two decades, the number of cases of MDR-TB resistant to anti-TB drugs isoniazid and rifampicin has been steadily increasing, with WHO currently estimating ~600,000 new MDR-TB cases per year. Even more concerning, however, is the emergence of extensively drug resistant TB (XDR-TB) which, like MDR-TB, is resistant to isoniazid and rifampin, but has additional resistance to fluoroquinolone and second-generation anti-TB drugs (such as amikacin, kanamycin, or capreomycin). To date, only 30% of XDR-TB cases are successfully treated. The MDR-TB and XDR-TB burden largely falls in India, China and the Russian Federation.6 With the emergence and proliferation of MDR-TB and XDR-TB, the development of new classes of anti-TB therapeutics is critical. Drug discovery efforts are on-going; however, only one ‘new’ anti-TB drug, the highly toxic bedaquiline, has been approved in the last several decades. Due to its particularly harsh side effects, bedaquiline is only used to treat the most severe cases of MDR-TB and XDR-TB.8
1.3. Mtb iron assimilation
Because of its role in many essential biological processes, iron is essential for the survival of all living organisms including bacterial pathogens.9–11 To acquire iron, bacteria have evolved pathways to take up both host heme and non-heme iron (simply referred to as iron henceforth). As iron and/or heme acquisition are essential, the mechanisms of pathogen iron and heme uptake are thought to represent potential drug targets.
In this review, we will focus on the Mtb siderophore-dependent iron and heme uptake pathways. We will start by describing sources of iron that Mtb is likely to encounter during active infection within humans. As Mtb is neither a Gram-negative or Gram-positive bacterium, we will briefly discuss general bacterial siderophore-dependent iron and heme uptake mechanisms. Then, we will review the Mtb iron and heme uptake pathways in detail. We will include structural and functional analyses, and discuss the relationship between Mtb iron and heme uptake pathways with those of Gram-negative and Gram-positive organisms. The topics of siderophore biosynthesis and regulation of siderophore-dependent iron and heme uptake pathways will be reviewed albeit only briefly. While it has been suggested that Mtb may internalize holo-iron-binding proteins, transferrin (Tf) and lactoferrin (Lf),12,13 the molecular mechanism of internalization of holo-Tf and holo-Lf proteins is unknown and will not be discussed further. Finally, we will present ideas on how to exploit Mtb heme and iron uptake systems as a target for novel anti-Mtb drugs.
2. Overview of Iron in the Host
2.1. Role of iron in life
Metals are an integral part of life. Within the human body, the most abundant metal ions are Na+, K+, Mg2+ and Ca2+, while the most abundant transition metal is iron. Iron performs a multitude of extremely diverse roles in biology, with well-known roles in biological catalysis14,15 electron transport,16 and oxygen transport and storage.17
During pathogenesis, the limited availability of free iron and heme-iron in the host represents a serious challenge to the pathogen. Although iron is a useful catalyst when coordinated to a protein, ‘free’ iron has toxic effects. In an aqueous environment, uncomplexed iron, predominately ferrous (Fe2+), is a potent activator of free-radical chemistry through the Fenton reaction, catalyzing the formation of highly reactive hydroperoxy and hydroxy radicals.18 Further, Fe2+ readily oxidizes to its ferric form (Fe3+), which has extremely low solubility (10−18M) and results in ferric iron precipitate.19 As a consequence of iron’s reactivity and potential insolubility, mammals maintain very low concentrations of free, unbound iron. Indeed, the majority of iron is tightly coordinated to high affinity iron transport, storage and scavenging proteins.20
Furthermore, iron-sequestration is a key component of the mammalian innate immune system, and humans have dedicated proteins whose sole role is to scavenge extracellular iron to prevent its uptake by pathogens.21,22 The battle for iron between the human host and bacteria has evolved into a sophisticated cat and mouse game, where bacteria have developed complex pathways to steal iron from host proteins, and to circumvent this, humans have evolved proteins to neutralize bacterial iron-scavenging pathways.23,24
2.2. Sources of host iron
The human body contains approximately 5 g of iron, which is divided into ferric (Fe3+) and ferrous (Fe2+) iron ions, and heme-iron, where iron is coordinated to protoporphyrin IX (PPIX).
The majority of human non-heme iron is circulating in the bloodstream, where most of circulating Fe3+ ions are complexed by transferrin (Tf) and lactoferrin (Lf). Tf coordinates two Fe3+ ions tightly (Ka = 1023 M−1) and shuttles iron between tissues.25 Lf is a glycosylated Fe3+ ion transporter that is part of the innate immune system and is predominantly found in secretory fluids.21 Importantly, Tf serves a key antimicrobial role by establishing iron-replete conditions.21 Non-circulating iron is found within tissues, where iron is coordinated to proteins such as, iron-containing enzymes, regulatory proteins, and ferrous iron is stored in large ferritin nanocages.26
2.3. Sources of host heme
Heme-iron represents the most abundant form of circulating iron in the human body. Similar to free iron, heme is highly reactive. Therefore, the concentration of free heme is kept extremely low and heme is tightly bound to proteins (Ka = 108 - 1013 M−1).28,31 Approximately 80% of all human iron is incorporated into the heme-containing oxygen carrier hemoglobin (Hb) in erythrocytes,32,33 making Hb the greatest human reservoir of any type of iron.34 Hb exists in two oxidation states with very different biophysical properties. Reduced Hb is an α2/β2 tetramer that can transport O2 and binds ferrous heme very tightly. In contrast, oxidized Hb (metHb) breaks down into α/β heterodimers and no longer binds O2.27,28 The binding affinity of metHb for ferric heme is considerably diminished, where the half-life of heme dissociation is approximately 2–3 minutes for the β subunit of metHb, Table 1.27 Thus, for heme bound to metHb, heme dissociates passively on a timescale that is rapid enough to create a pool of labile heme that can be utilized by bacterial heme uptake systems. In a healthy human, a mere 1–2% of Hb is in the metHb form. However, the availability of metHb may rise during bacterial infection as many pathogens secrete hemolysins that lyse erythrocyte cells, resulting in Hb release into the blood serum.35 Hb released under hemolytic conditions is subject to rapid oxidation and thus heme is more susceptible to dissociation and acquisition by a pathogen.27
Table 1:
Heme binding properties of host heme sources. The rate kon refers to the bimoeclular rate constant of heme binding to apo-protein in dilute conditions. The rate koff represents the unimolecular rate of heme dissociation. The relationship between kon, koff and the association constants Ka is Ka = kon/koff. This is the same for Tables 2 & 3.
| Protein | Coordination | kon (μM−1s−1) | koff (s−1) | Ka (M−1) | Ref |
|---|---|---|---|---|---|
| metHb α- chain (pH 7) | 6c His/H2O | 100 | 1.6 × 10−4 | 6.0 × 1011 | 27,28 |
| metHb β- chain (pH 7) | 6c His/H2O | 100 | 4.2 × 10−3 | 2.4 × 1010 | 27,28 |
| Serum albumin | 5c Tyr | ~ 50 | 1.1 × 10−2 | 4 × 106 | 28 |
| Hemopexin | 6c His/His | ND | ND | 109 − 1.9 × 1014 | 29,30 |
ND = not determined, 5 c = 5-coordinate, 6c = 6-coordinate
To prevent heme loss to pathogens, mammals have evolved mechanisms to recover heme dissociated from metHb. There are three primary proteins involved in metHb and free heme recovery: haptoglobin (Hp),22,36 hemopexin (Hx),37,38 and serum albumin.39 Hp is a tetrameric serum glycoprotein that forms a tight interaction with metHb to prevent heme dissociation.22,37 Hx is a monomeric glycoprotein that binds free heme from the bloodstream with an extremely high affinity, Table 1. While serum albumin binds heme with relatively low affinity (Ka ~ 10 × 106 M−1),39,28 due to its overall high abundance, it also plays an important role in reducing the free heme concentration in serum by binding free heme and transferring it to Hx.
2.4. Potential sources of host iron for Mtb
TB is contracted via inhalation of Mtb into the lungs. Mtb is primarily an intracellular organism, residing in the phagosome of lung macrophages during initial infection, Figure 1. Additionally, TB has an asymptomatic, latent stage, where Mtb can remain dormant in granulomas for decades.4 During active infection, Mtb will extracellularly disseminate via the bloodstream from its primary infection site to secondary ones, such as the central nervous and lymphatic systems.
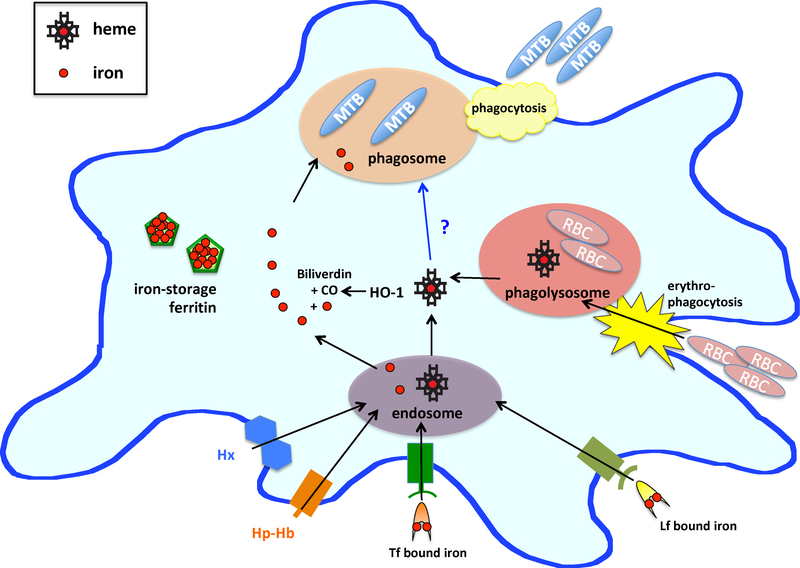
Figure 1.
Iron availability in macrophages. Macrophages uptake iron by sequestering Tf/Lf bound iron and storing released iron in ferritin nanocages. Heme is either uptaken by Hx and Hp-Hb complex receptors where the proteins are degraded in the endosome, or by erythophagocytosis of senescent red blood cells (RBC). In both cases, heme is released into the macrophage cytosol. There, heme is degraded by heme oxygenase (HO-1) to release iron, carbon monoxide (CO) and biliverdin – discussed in 5.1. Mtb residing in the macrophage phagosome will encounter iron and likely also heme and Hb.
In the macrophage phagosome, Mtb will encounter extremely low free iron levels,40 even though the macrophage cytosol contains relatively high iron levels (Figure 1).33 To overcome this, it has been suggested that Mtb siderophores promote iron diffusion across the phagosomal membrane.41 This hypothesis is supported by 59Fe labeling experiments that demonstrate iron accumulation in Mtb residing in phagosomes.42,43 Moreover, macrophages degrade senescent red blood cells in the phagolysosome,44 making Hb a promising source of heme-iron for macrophage-residing Mtb (Figure 1). Indeed, a recent study has established that the cytosolic concentration of the bioavialable heme pool within human lung cells is 400–600 nM.45
When Mtb is extracellular, such as in the bloodstream, Mtb may sequester iron from Tf, Lf and ferritin. Extracellular Mtb will also encounter heme. Mtb possesses hemolysins that lyse erythrocyte cells35, thereby releaseing Hb into the bloodstream as metHb and creating a pool of labile heme as well as heme tha is weakly bound to serum albumin. Notably, in severe Mtb infection blood accumulates in the lung cavities, possibly resulting in elevated heme levels in the lungs.
3. Bacterial Siderophore-dependent Iron Uptake Pathways
3.1. Siderophore-dependent iron uptake
Siderophore-mediated iron uptake is part of the iron-uptake repertoire in both Gram-negative and Gram-positive bacteria. Siderophores are iron-chelating small molecules (Mw ranging from 100 to 2000 Da) secreted by microorganisms to facilitate iron scavenging, transport and uptake.46–48 The defining feature of siderophores is their extremely high affinity for Fe3+ ions with Ka values up to 1052 M−1.49 Siderophores are opportunistic iron scavengers that are not known to display a preference regarding their iron source, as they remove iron from Tf, Lf, ferritin, and other human iron binding proteins. Once bound to ferric iron, ferric-siderophores are imported into bacteria by a variety of mechanisms.
3.2. Siderophore structure
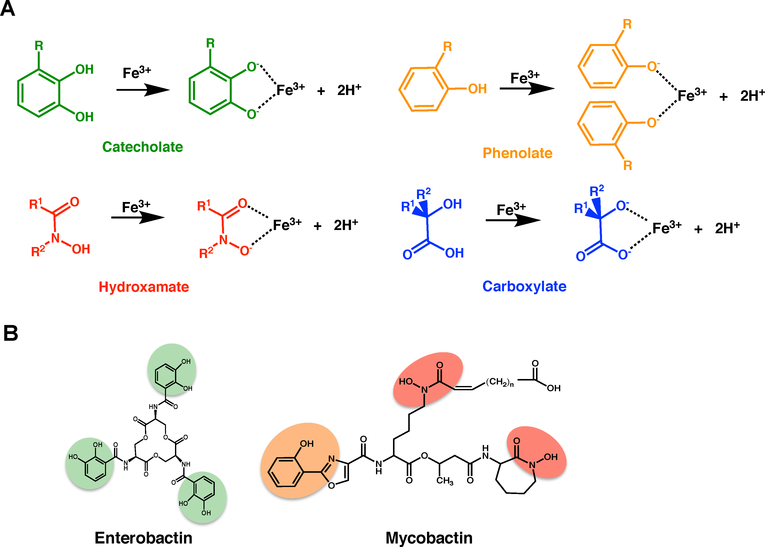
The majority of siderophores form hexadentate, octahedral complexes with ferric iron. Siderophores can be classified into four major categories based on the ferric iron-binding moiety: carboxylates, catecholates, hydroxamates and phenolates (Figure 2A), all of which are hard Lewis bases. Moreover, some siderophores contain multiple binding moieties.48
Figure 2.
Overview of the diversity in siderophore structures A. The following four moieties confer siderophore iron-binding capacity: catecholate (green), hydroxamate (red), phenoloate (orange) and carboxylate (blue). B. E. coli and Mtb siderophores, enterobactin and mycobactin (respectively), with the iron-binding moieties highlighted.
Pathogenic Mtb produce two siderophores termed ‘mycobactins’, mcyobactin and carboxymycobactin Figures 2B & 3, which are mixed-type siderophores with both phenolate and hydroxamate moieties (Figure 2).40 While the two siderophores share an identical core, consisting of five amino acids with a characteristic oxazolidine ring derived from salicylate (Figure 3),40,50 they differ in hydrophobicity and cellular localization. Owing to its long aliphatic tail, mycobactin is hydrophobic and cell-wall and membrane associated. In contrast, the secreted carboxymycobactin features a shorter tail that terminates with carboxylate group, rendering it hydrophilic (Figure 3).51,52
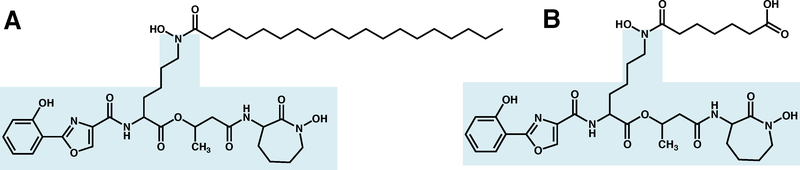
Figure 3.
Structure of A. hydrophobic Mtb mycobactin T (where the acyl chain on the central modified lysine can vary in length and modifications) and B. hydrophilic Mtb carboxymycobactin. The mycobactin core is highlighted in light blue.
Other mycobacterial species produce a variety of mycobactins, which all retain the same structural core (Figure 3) but are derivatized differently. While Mtb has been shown to produce mycobactin T, Figure 3A, Mycobacterium smegmatis and Mycobacterium paratuberculosis produce their own specific siderophores, mycobactin S and mycobactin J, respectively.40 Non-pathogenic, saprophytic mycobacteria also produce a different extracellular siderophore that does not have the mycobactin structural core; a formylated pentapeptide called exochelin;53 however in this review, we shall only discuss Mtb mycobactins (see Figure 3).
3.3. Siderophore biosynthesis
3.3.1. Overview
Biosynthetic pathways of siderophore production can be classified into two types: nonribosomal peptide synthetase (NRPS)-dependent,54 and NRPS-independent55 siderophore biosynthetic pathways.46,48,56 NRPSs are large multienzyme complexes that synthesize small cyclic or linear polypeptide products without requiring an mRNA template, and usually utilize a thiotemplate mechanism for biosynthesis. NRPSs have been proposed to be responsible for the synthesis of many aryl-capped (or aromatic ring capped) siderophores such as Mtb mycobactin (Figure 3).48
3.3.2. Mtb mycobactin biosynthesis
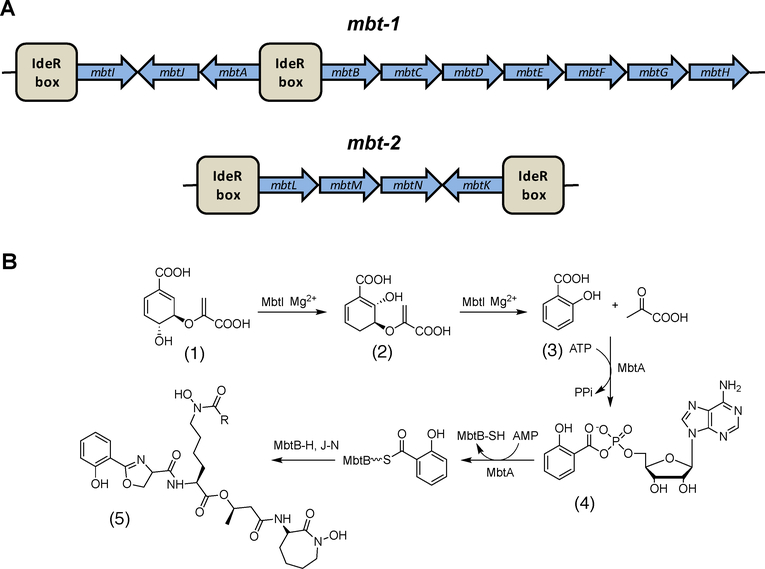
Biosynthesis of mycobactin occurs in an NRPS-dependent manner. As the biochemistry and structures within this pathway have been recently reviewed,57,58 we will only present a brief summary of the pathway. In Mtb, mycobactin biosynthetic proteins are encoded by two gene clusters, mbt-1 and mbt-2, where each locus includes genes mbtA to mbtJ and mbtK to mbtN, respectively (Figure 4A).59–60 The proteins encoded by mbt-1 assemble the core of the mycobactin molecule, whereas those encoded by mbt-2 assemble the aliphatic hydrophobic side chain of mycobactin T.
Figure 4.
Biosynthesis of mycobactin. A. Organization of the mbt genes in the mbt-1 and mbt-2 loci and the location of the IdeR boxes. B. Biosynthetic pathway of mycobactin (5) with the first two steps catalyzed by MbtI and MbtA followed by the reaction carried out by the megasynthase complex and then derivatization of the lysine molecule. Chorismate (1), isochorismate (2), salicyclic acid (3) and salicyl adenylate intermediate (4).
In the mycobactin biosynthetic pathway, Figure 4B, several enzymes produce and activate precursor components. MbtI catalyzes the conversion of chorismate (1) to salicylic acid (3). Then MbtA activates salicylic acid and catalyzes its transfer to the thiolate domain of MbtB through an acyl adenylate intermediate, Figure 4B (4). MbtB is part of the megasynthase comprised of three NRPSs (MbtB, MbtE and MbtF) and two polyketide synthases (MbtC and MbtD), which then assembles the core mycobactin structure (Figure 3, light blue).61 Not part of the megasynthase are MbtG which catalyzes the hydroxylation of L-lysine to N6-hydroxy-L-lysine,60 and MbtH that chaperones the correct folding of the NRPSs (MbtB, MbtE, and MbtF).62 MbtJ is the only protein with an unknown function encoded by the mbt-1 locus. The proteins encoded by mbt-2 produce the functional mycobactin T, Figure 4B. First, the aliphatic, hydrophobic, acyl chain is assembled by the enzymes MtbL, MbtM, and MbtN. Second, MbtK catalyzes the linkage of the acyl chain to the mycobactin core.59
3.4.3. Regulation of mycobactin biosynthesis
Mycobactin synthesis is regulated by the iron-dependent regulator (IdeR), Figure 4A, which has sequence similarity to the iron-dependent Diphtheria Toxin Repressor from Corynebacterium diphtheria.63,64 IdeR represses genes associated with mycobactin synthesis in the presence of iron, while acting as a positive regulator for the Mtb iron-storage ferritin proteins BfrA and BfrB63,64 IdeR also regulates many genes that encode proteins involved in Mtb mycobactin export and import (see sections 3.5 & 3.6).63,65 Importantly, an Mtb ideR deletion mutant does not survive in macrophages or a mouse model, suggesting that ideR is an essential gene for Mtb virulence,66 possibly because of its central role in regulating iron homeostasis.
3.4. Bacterial siderophore export and import
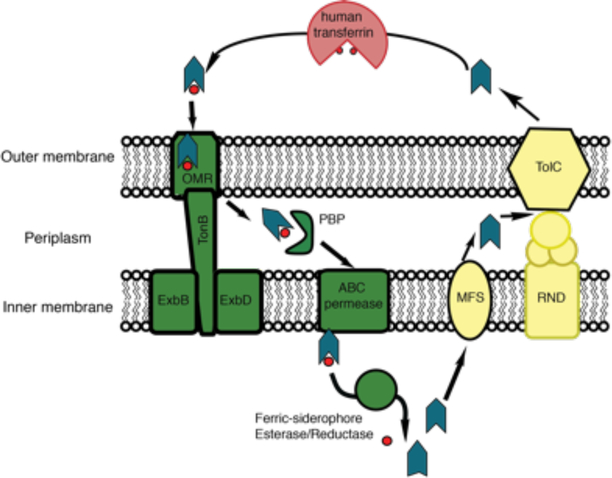
Siderophore-mediated iron uptake has been extensively studied in E. coli, and therefore warrants discussion to illustrate how bacteria may acquire iron and shuttle it across the outer and inner membranes to the cytoplasm.67–69 E. coli enterobactin (Figure 2B) is synthesized in the cytoplasm, and is then translocated into the periplasm by a transmembrane protein, which is a member of the ATPase major facilitator superfamily (MFS).70 The multifunctional efflux channel TolC then exports enterobactin from the periplasm into the extracellular space.70,71 TolC-mediated export of enterobactin also requires the membrane fusion protein and resistance nodulation cell division (RND) transporters (Figure 5).71
Figure 5.
E. coli siderophore export and import mechanism. The siderophore is the green-blue arrowhead and the red spheres are iron. Proteins involved in siderophore export are colored in yellow, while proteins involved in import of ferric-siderophores are colored in green. In the cytoplasm, the ferric-siderophore esterase induces the release of iron from E. coli enterobactin by modification of the siderophore, whereas the ferric-siderophore reductase reduces the iron in non-E. coli siderophores to induce its release. In E. coli, siderophores are recycled.
Upon scavenging extracellular ferric iron, E. coli ferric-enterobactin (FeEnt) is imported into the periplasm utilizing a specific β-barrel outer membrane receptor (OMR) through a TonB-dependent transport mechanism, Figure 5.72–74 In the periplasm, a periplasmic binding protein (PBP) binds and delivers FeEnt to the inner membrane ATP-binding cassette (ABC) transporter complex, so it can be translocated into the cytoplasm where the siderophore is modified by FeEnt esterase to promote the release of iron,75–78 Figure 5. Moreover, E. coli has also evolved to import ferric siderophores from other bacteria using specific siderophore-binding TonB-dependent receptors, such as ferrichrome from Aspergillus.79 Once in the cytoplasm, an E. coli ferric-siderophore reductase induces iron release by reducing ferric iron to ferrous iron, resulting in iron dissociation.80
In Gram-positive bacteria, siderophore export and import is simplified since only one membrane needs to be traversed. However, these export and import pathways have been relatively understudied. While it is known that ferric-siderophores are recognized by siderophore binding proteins (homologus to PBPs) located in the cell membrane, and that a permase assists in the translocation of the ferric-siderophore across the membrane,81,82 the molecular mechanisms of siderophore transport through the Gram-positive cell-wall are poorly understood.
3.5. Mtb siderophore export
3.5.1. Overview
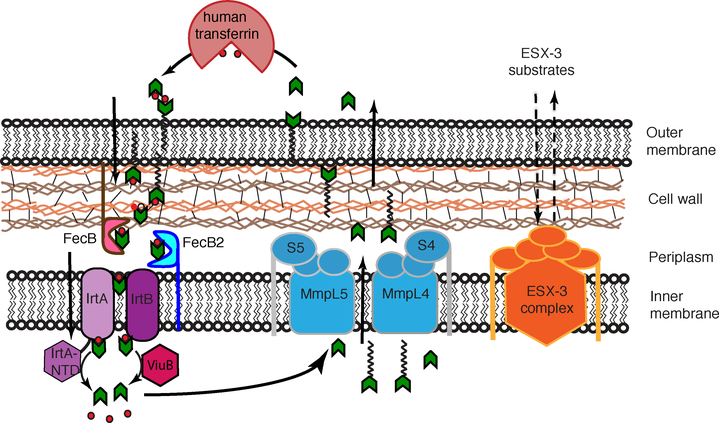
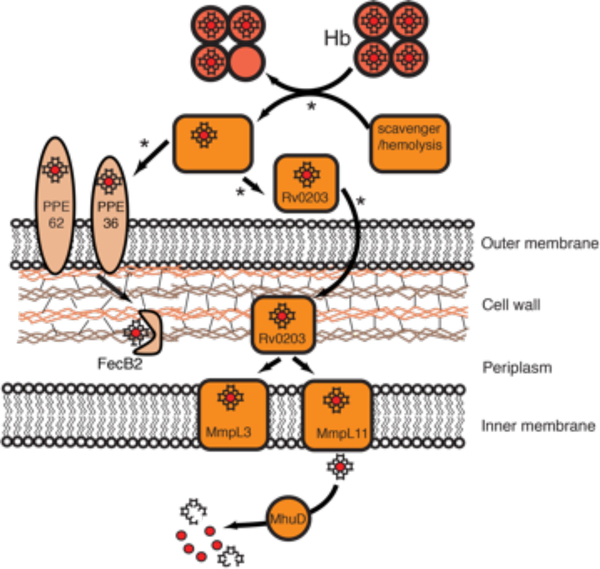
The two Mtb siderophores, carboxymycobactin and mycobactin (Figure 3) are utilized differently. Carboxymycobactins are relatively soluble and are exported into the extracellular environment, where they scavenge iron from human iron-containing proteins such as Tf and Lf.58 Mycobactins, on the other hand, due to their insolubility, are anchored to the Mtb outer membrane and cell-wall environments. Export of mycobactin and carboxymycobactin across the inner membrane is dependent on MmpL (Mycobacterial membrane protein Large) proteins, MmpL4 and MmpL5, along with their small membrane-associated accessory proteins, MmpS (Mycobacterial membrane protein Small) proteins, MmpS4 and MmpS5 (see Figure 8).83 More recently, it was proposed that ESX-3, one of the five Type VII secretion systems in Mtb, also contributes to iron acquisition as ESX-3 mutants display defects in ferric-carboxymycobactin uptake and accumulation of cell-wall associated mycobactins (see Figure 8).84,85 However, the precise role ESX-3 plays in iron import/export is unknown and thus will not be further discussed in this review.
Figure 8.
Mtb mycobactin-dependent iron uptake. Green arrows are carboxymycobactins and the ones with acyl tails are mycobactins. MmpL4/S4 and MmpL5/S5 systems export mycobactin. Carboxymycobactins scavenge iron from host iron-binding proteins, and then putatively transfer iron to cell-wall associated mycobactins. PBP proteins, FecB and FecB2, are proposed to shuttle iron to the periplasmic side of the inner membrane, where the IrtA/IrtB complex shuttle ferric-carboxymycobactin across the inner membrane. Iron is released into the cytoplasm by ferric reductases, IrtA-NTD and ViuB, upon which carboxymycobactin is recycled. ESX-3 is involved in mycobactin import-export by an unknown mechanism.
3.5.2. Mtb MmpL proteins
MmpL proteins appear to act both in siderophore export and heme import pathways. We will therefore present a broad description of this important class of Mtb proteins. MmpL proteins belong to the RND permease superfamily of transmembrane transporters,86 which associate with outer membrane factors. This assembly is stabilized by periplasmic membrane fusion proteins to form a three-component efflux pump (extensively reviewed in 87), which is driven by proton motive force. Mtb MmpL proteins are functionally important and have been shown to mediate the transport of a variety of substrates across the mycobacterial inner membrane, including Mtb lipids and siderophores.88,89 Mutational analyses revealed that one of the MmpL proteins, MmpL3, is essential for Mtb viability,86,90 and several other MmpL proteins are necessary for Mtb virulence in mice infections including MmpL4, MmpL5, MmpL7, MmpL8, MmpL10 and MmpL11.86,91
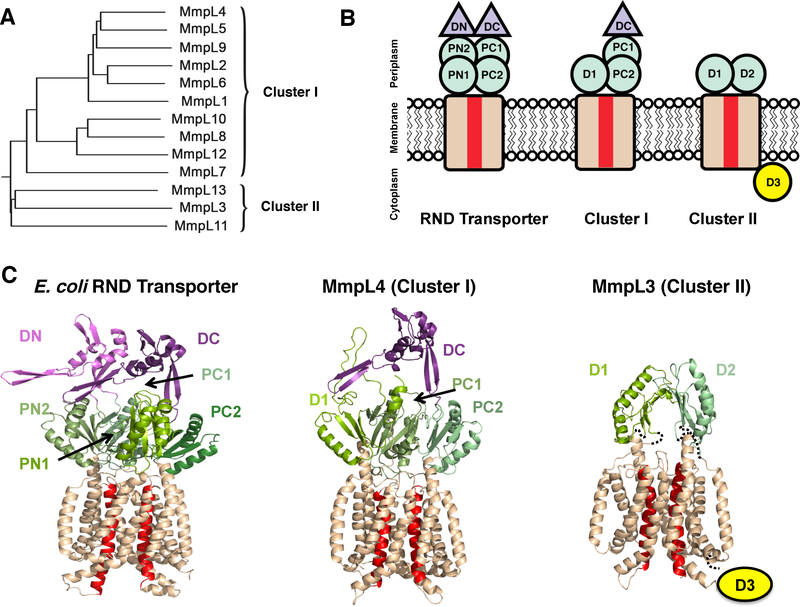
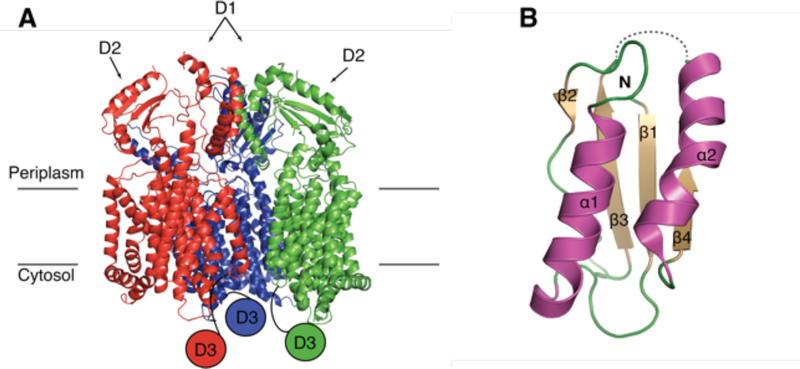
The structures of canonical Gram-negative RND transporters are homotrimeric, where each monomer harbors twelve transmembrane helices (TM) with N-terminal and C-terminal periplasmic domains inserted between TM1 and TM2 and between TM7 and TM8, respectively. Each periplasmic domain comprises two structurally similar porter subdomains (N-terminal porter subdomains, PN1 and PN2, and C-terminal porter subdomains, PC1 and PC2, containing a βαββαβ motif) and a ‘middle’ docking subdomain (DN or DC), Figures 6B&C.87
Figure 6.
Mtb MmpL family. A. The 13 Mtb MmpL proteins fall into two distinct clusters. B. Mtb MmpL proteins are members of the RND superfamily; PN, PC, D1 and D2 are porter subdomains (green) and DN and DC are docking domains (purple). The 12 TM domain is colored wheat, where red TMs are those involved in proton motive force. C. Cartoon representation of monomeric CusA (PDB: 4DNT) and Phyre2 predictions of MmpL4 (Cluster I) and MmpL3 (Cluster II) structures. The MmpL Cluster II cytoplasmic D3 domain has no structural prediction.
Mtb has 13 MmpL members of the RND family, all of which are actinobacteria-specific inner membrane proteins of approximately 1,000 residues. Mtb MmpL proteins structurally cluster into two distinct subfamilies of RND transporters,92 Figure 6A. MmpL Cluster I proteins have an N-terminal domain similar to an RND porter subdomain, and a C-terminal domain that resembles that of RND proteins; comprising two porter subdomains and a docking subdomain Figure 6B&C.92 In contrast, MmpL Cluster II proteins have minimal periplasmic architecture, whereby the N- and C-terminal domains each contain a single porter subdomain, Figure 6B&C. Unlike other RND or MmpL Cluster I proteins, each MmpL Cluster II member has a C-terminal cytoplasmic domain,92 Figure 6B&C. In addition, some of the Cluster I MmpL members are encoded along with a small membrane-associated accessory protein, MmpS.88,89
3.5.3. Structures of MmpL4/5 and MmpS4/5
Both proteins involved in siderophore export, MmpL4 and MmpL5, are members of MmpL Cluster I (see above section 3.5.2, Figure 6A). Thus, they are predicted to contain two periplasmic domains, a D1 porter subdomain and a larger D2 domain comprised of two porter and one docking subdomains (Figures 6B&C).
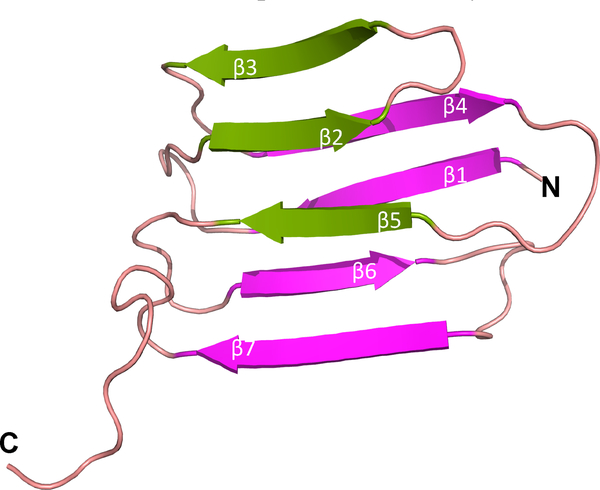
MmpL4 and MmpL5 associate with MmpS4 and MmpS5, respectively. The structure of the mature form of MmpS4 (without its N-terminal signal peptide or single transmembrane tether) was solved by NMR and revealed an immunoglobulin-like fold comprising a 3- and 4-stranded β-sheet along with a disordered C-terminal, Figure 7.83 The sequence identity between MmpS4 and MmpS5 is greater than 50%, suggesting similar structural folds. It was demonstrated that MmpS4 interacts only with the D1 domain of MmpL4.83
Figure 7.
Structure of MmpS4 (PDB: 2LW3).
MmpS4 is shown to interact with MmpL4-D1.
3.5.4. Mtb siderophore export mechanism
MmpL 4/5 are needed for mycobactin and carboxymycobactin export (see Figure 8). In addition, several experiments suggest the accessory proteins MmpS4 and MmpS5 are also required for mycobactin and carboxymycobactin secretion.83,93 First, it was demonstrated that an MtbΔmmpS4/5 double mutant is not viable under low iron conditions, suggesting both proteins take part in iron acquisition. Interestingly, the growth of the single MtbΔmmpS4 and MtbΔmmpS5 mutants are not attenuated in low iron conditions, suggesting MmpS4 and MmpS5 have redundant roles. While MmpS4 and MmpS5 are likely to be involved in ferric iron acquisition, they do not participate in heme uptake since the MtbΔmmpS4/5 double mutant grows in media supplemented with heme.83 Next, it was shown that MtbΔmmpS4/5 can take up exogenous iron-loaded mycobactin, indicating that the MmpL4/S4 and MmpL5/S5 systems are involved specifically in mycobactin export, but are not required for mycobactin import.83 Biosynthesis of mycobactin and its secretion may occur in a coordinated manner since MmpS4/5 localize to the inner membrane along with the mycobactin biosynthetic enzyme, MtbG.83 This led to speculation that the MmpL4/S4 and MmpL5/S5 systems form a multienzyme complex with MtbG to couple mycobactin biosynthesis and its export. Similar coordinated export has been previously observed for M. smegmatis MmpS4 and MbtH in the biosynthesis of glycopeptidolipids.94
An assessment of Mtb MmpL4 and MmpL5 deletion mutants within mice revealed that MmpL4 is required for full virulence,86 while MmpL5 is essential for Mtb survival in the lung.91 With respect to MmpS4 and MmpS5, the growth of the single MtbΔmmpS4 and MtbΔmmpS5 mutants were attenuated in mice lungs. More strikingly, for the MtbΔmmpS4/5 double mutant there was 100% survival over a 180-day period compared to those mice infected with wild-type Mtb that had a 0% survival rate in the same time period.83 This is the strongest attenuation observed in Mtb mutants lacking iron utilization genes, suggesting that MmpL4/S4 and MmpL5/S5 mycobactin export systems are required for Mtb virulence, and represent promising drug targets.
3.5.5. Mtb siderophore recycling
In some bacteria, including in P. aeruginosa and E. coli, siderophores are recycled.95,96 A recent study demonstrated that both Mtb siderophores, mycobactin and carboxymycobactin, are also recycled, Figure 8.99 Moreover, it was observed that carboxymycobactins only transiently accumulate within the cell,99 suggesting an equilibrium between the secreted carboxymycobactin influx and efflux, a phenomenon also observed for aerobactin in E. coli95. In addition, it was demonstrated that when mycobactin is presented to the MtbΔmmpS4/5 mutant, mycobactins accumulate within the cytoplasm to the point where they become toxic to Mtb. 99 However, it remains to be seen if this toxic build-up of mycobactins arises from their iron-bound or apo form. Taken together, MmpL4/S4 and MmpL5/S5 systems are involved in both siderophore export and mycobactin and carboxymycobactin recycling.99 Inhibiting these pathways would produce a two-fold effect: (1) Restriction of iron uptake and (2) toxic build-up of mycobactins and carboxymycobactins in the cytoplasm.
3.6. Mtb ferric-siderophore import
3.6.1. Overview
Iron assimilation is essential for Mtb growth and virulence.50,97 However, import mechanisms for ferric mycobactins are poorly defined, as several necessary steps required for the import of mycobactins are unknown. One aspect of importing mycobactin that has been relatively well characterized is the role of IrtA/IrtB. IrtA/IrtB is a heterdimeric protein involved in both the import of ferric-carboxymycobactin into the Mtb cytoplasm and the reduction of iron in the ferric-carboxymycobactin complex to release iron into the cytoplasm.97,98
3.6.2. Potential protein players in the periplasm
The current consensus is that hydrophobic mycobactins are cell-surface receptors or cell-wall associated and are involved in transporting ferric iron from the extracellular environment through the cell-wall environment. In support of this hypothesis, experiments show that ferric-carboxymycobactin can transfer its ferric iron cargo to mycobactin.40,51 Next, two predicted ferric-siderophore PBPs, FecB and FecB2, may shuttle ferric-mycobactin or ferric-carboxymycobactin from the Mtb cell-wall environment to the periplasmic side of the ItrA/ItrB transporter complex in the inner membrane, Figure 8. FecB is predicted to be part of the Mtb iron-regulated IdeR regulon65 and a study in Mycobacterium avium shows that M. avium FecB is upregulated in the presence of low-iron conditions,99 suggesting this protein plays a role in iron acqusition.
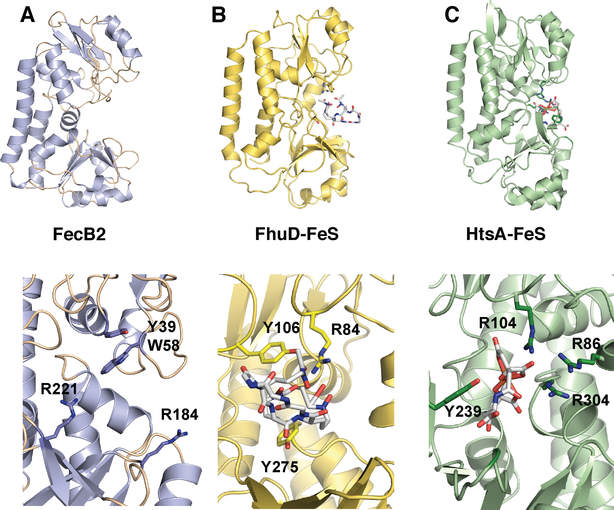
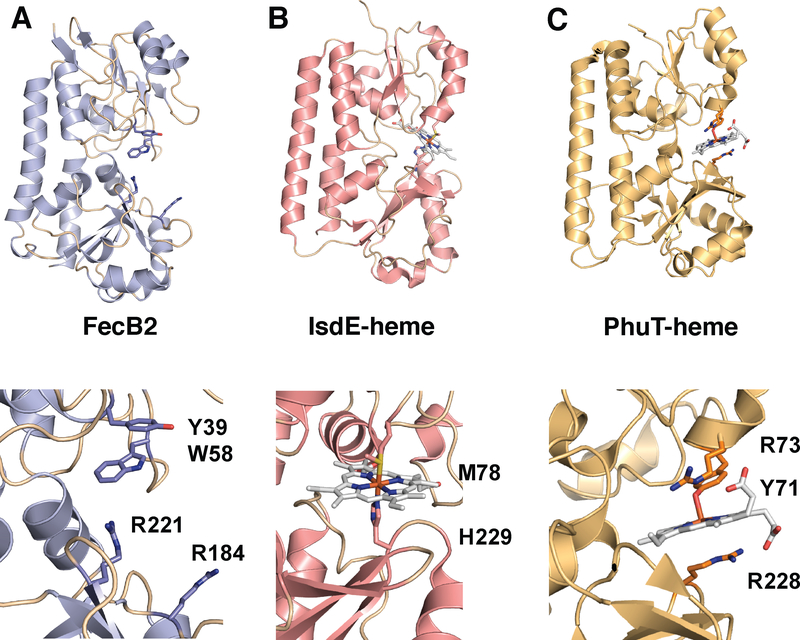
Mtb FecB and FecB2 are predicted to belong to the Type III PBP superfamily.100 Mtb FecB has moderate sequence identity to other ferric-siderophore PBPs such as E. coli FhuD (Fe3+-ferrichrome),101 whereas the sequence of Mtb FecB2 is most similar to E. coli FepB (Fe3+-enterobactin - catechol).102 The structure of Mtb FecB2 has been solved (PDB: 4PM4, Figure 9A), and is composed of N- and C-terminal lobes consisting of mixed α/β structures, where the two lobes are linked by a rigid “backbone” helix, characteristic of PBPs.100 Indeed, Mtb FecB2 has high structural homology to other ferric-siderophore and heme PBPs (such as IsdE and PhuT).100 Mtb FecB2 has highest structural similarity with Staphylococcus aureus HtsA,103 which binds an hydroxamate-type siderophore, staphyloferrin A. Within the HtsA-ferric-staphyloferrin A complex, iron is not directly coordinated by HtsA residues, Figure 9C, and like the E coli FhuD-gallichrome complex, Figure 9B, HtsA utilizes a combination of tyrosine and arginine residues to bind its siderophore noncovalently.101,103 The binding cleft of FecB2 contains two arginines (Arg184, Arg221) and a tyrosine residue (Tyr39) that could be involved in stabilizing the mycobactin backbone and/or coordinating ferric iron. However, the structure of Mtb FecB2 featured an empty binding site that does not shed light on the protein’s potential cargo or mode of binding.
Figure 9.
Top panels are ribbon diagrams of the PBPs and the bottom panels are rotated 90° and zoomed in. Ferric-siderophores (FeS) are shown in white stick and active site Tyr and Arg residues are marked. A. Mtb FecB2 (PDB: 4PM4; blue), B. E. coli gallichrome:FhuD complex (PDB:1EFD; yellow), and C. S. aureus ferric-staphyloferrinA:HtsA complex (PDB:3EIW; green).
Mtb FecB and FecB2 have 24% sequence identity and may be functionally redundant. FecB2 has a predicted 35-residue signal peptide whereas FecB is predicted to have a 68-residue N-terminal extension containing both a signal peptide and a membrane lipid attachment site, suggesting that FecB and FecB2 localize to different cellular locations and may work in tandem to shuttle ferric-carboxymycobactin and/or ferric-mycobactin through the lipid-rich cell-wall environment to the periplasmic side of the inner membrane. There are two potential mechanisms of PBP-dependent siderophore transport through the periplasm and/or the cell-wall. They may use an affinity gradient manner to ensure directional transport similar to how S. aureus imports heme.104 Alternatively, a more complicated mechanism is possible where iron dissociates from ferric-mycobactin or ferric-carboxymycobactin within the periplasm as observed in pyoverdine-mediated iron transport, where pyoverdine is one of the P. aeruginosa siderophores.105–108
3.6.3. Ferric-carboxymycobactin inner membrane permase - IrtA/IrtB
It has been shown that IrtA/IrtB is an inner membrane ABC heterodimeric transporter, which is critical for the import of Fe3+-carboxymycobactin from the Mtb periplasm to the cytoplasm,97 Figure 8. We propose that ferric-carboxymycobactin bound to Mtb FecB or Mtb FecB2 directly transfers its ferric-carboxymycobactin cargo to the IrtA/IrtB complex for translocation to the cytoplasm. The genes that encode for IrtA/IrtB are co-operonic and are downstream of a predicted IdeR binding site.109,110 IrtA and IrtB are predicted to have six and five TMs, respectively, and both have C-terminal ATPase domains.97 IrtA and IrtB likely form a heterodimeric ABC transporter based on their similarity to the Y. pestis iron transporter, YbtPQ.111 IrtA has an additional domain at its N-terminus, which is predicted to be a cytoplasmic ferric-siderophore reductase domain (IrtA-NTD) (discussed further in section 3.6.4).97,110
Mutational analysis demonstrates that both IrtA and IrtB are required for fully functional Mtb iron import, however, the exact role of IrtA and IrtB are still not fully resolved.97 Under iron deplete conditions, inactivation of IrtB alone or both IrtA/IrtB led to significant growth defects. The effects were more severe with inactivated IrtA/IrtB than inactivated IrtB alone, suggesting that IrtA can form a homodimer that retains partial iron import activity. As the carboxymycobactin concentration in the culture filtrate was measured to be equivalent in the MtΔirtA/irtB mutant and wild-type strains, IrtA/IrtB does not act as a carboxymycobactin exporter. Further, as the addition of ferric-carboxymycobactin as the sole iron source did not restore growth in the MtbΔirtA/irtB double mutant, it is clear that the IrtA/IrtB complex is involved in carboxybactin-dependent iron uptake. Notably, the MtbΔirtA/irtB mutant resulted in impaired replication in both macrophages and mouse lungs, suggesting that IrtA/IrtB is required for virulence.97
In contrast, a study by Farhana et al, also suggests that M. smegmatis IrtA is involved in mycobactin export; however, compared to the study in Mtb, M. smegmatis IrtB alone is involved in ferric-mycobactin import.110 To resolve this apparent contradiction between studies, the precise function of IrtA and IrtB requires further investigation to determine whether the proteins form functional homodimers or a heterodimer.
3.6.4. Cytoplasmic ferric-carboxymycobactin reduction
Once ferric-carboxymycobactins reach the Mtb cytoplasm, iron is released for utilization or storage. In some Gram-negative bacteria, the release of iron from ferric-siderophores is achieved through the flavin-dependent reduction of siderophore bound iron by an iron-siderophore reductase.112,113 As discussed earlier, the affinity of ferrous iron to the siderophore is substantially lower compared to that of ferric iron. Thus iron reduction in the ferric-siderophore complex results in the release of ferrous iron.112 IrtA-NTD is predicted to be a ferric-siderophore reductase.98 Additionally, IrtA-NTD possesses a RXYS(T) motif, which has been associated with flavin adenine dinucleotide (FAD) binding in ferric oxidoreductases, Figure 8.98 Notably, unlike IrtA, IrtB lacks an analogous domain.97
Results from in vitro binding experiments show that IrtA-NTD binds FAD in a 1:1 ratio.98 To show that the FAD-dependent IrtA-NTD domain is required for fully functional IrtA/IrtB carboxymycobactin-dependent iron uptake, a mutational analysis of the conversed FAD-binding motif (Arg70, Tyr72 and Thr73) was carried out. The results demonstrated that mutating IrtA-NTD Tyr72 and Thr73 to Ala resulted in diminished growth under low iron conditions, while the IrtA-NTD Arg70Ala mutation did not affect growth, suggesting that Tyr72 and Thr73 are critical for IrtA-NTD FAD binding and activity. Taken together the results suggest that FAD binding to IrtA-NTD is required for fully functional IrtA/IrtB, and, furthermore, since IrtA-NTD is located on the cytoplasmic side of the inner membrane, IrtA-NTD is a cytoplasmic ferric-siderophore reductase.98
In addition to IrtA-NTD, the Mtb proteome is predicted to encode another ferric-siderophore reductase, ViuB, (Figure 8), which is homologous to IrtA-NTD98 and a ferric-siderophore reductase ViuB from Vibrio cholerea.113 Mtb ViuB is induced under low iron conditions110 and in bacillus Calmette-Guérin (BCG, an attenuated strain of Mycobacterium bovis) replicating in mouse macrophages.114 However, the Mtb ViuB knockout mutant showed no phenotype when grown in low iron conditions or macrophages.115 Thus, further studies of Mtb ViuB are still required to determine if it plays a functional role in siderophore reduction.
4. Bacterial Heme Uptake Pathways
4.1. Heme uptake in Gram-negative bacteria
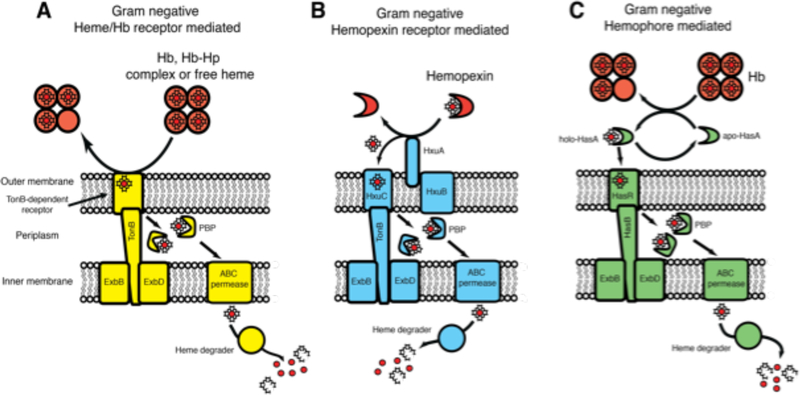
Heme uptake pathways have been discovered in a large number of Gram-negative bacteria, and have been extensively studied in P. aeruginosa, Serratia marscescens, Yersinia pestis, and Haemophilus influenza.116–122 Two general mechanisms of heme uptake are found in Gram-negative bacteria. Heme is either acquired through outer membrane receptors that bind host heme and/or heme proteins such as Hb or Hx (Figures 10A and 10B),118,120–121,122 or through the action of secreted heme scavenging proteins called hemophores (Figure 10C).116,117,119 After heme has been acquired from the host, it is transported across the bacterial outer membrane using a TonB-dependent outer membrane receptor (Figure 10).118 Once in the periplasm, heme is transferred to PBPs, which shuttle heme to an inner membrane ABC transporter.118,123 In some species, two separate heme uptake pathways are present. In such cases, the individual heme uptake pathways are proposed to have complementary roles to enable heme scavenging from alternate heme sources or are differentially utilized depending on host heme availability.124
Figure 10.
Heme uptake mechanisms in Gram-negative bacteria. Heme is acquired using an outer membrane receptor either specific for A. heme or a heme protein such as Hb, B. hemopexin or C. a secreted hemophore. Heme is transferred across the outer membrane in a TonB-dependent manner, before being shuttled to the inner membrane by a PBP, where an ABC permease translocates heme into the cytoplasm. Heme is then degraded by a heme degrading protein to release iron.
4.2. Heme uptake in Gram-positive bacteria
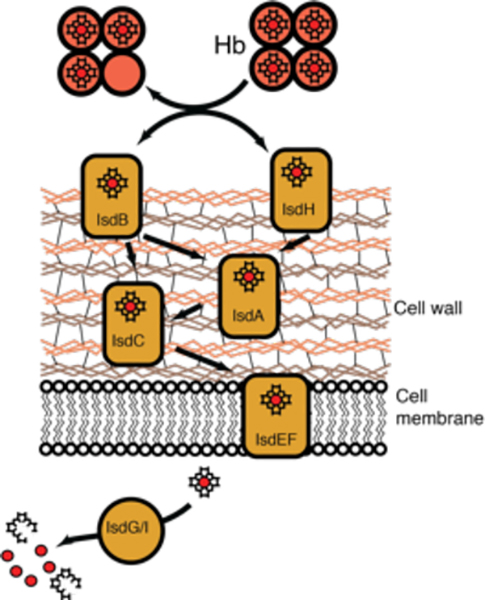
Heme uptake pathways have also been discovered in several Gram-positive species, where those of S. aureus,125,126 Streptococcus pyogenes,127 and Bacillus anthracis have been studied extensively.128,129 These organisms use a sophisticated cascade of heme transport proteins to chaperone heme through the bacterium’s thick peptidoglycan cell-wall and an ABC transporter to translocate heme across the inner membrane, Figure 11.
Figure 11.
Heme uptake in Gram-positive S. aureus. Heme is acquired using cell surface receptors that directly interact with and scavenge heme from Hb. Heme is then shuttled across the cell-wall through a series of heme transporters via protein-protein interactions. Heme is finally translocated across the membrane using an ABC transporter and degraded via the IsdG-type heme degraders, IsdG and IsdI, see section 5.3.
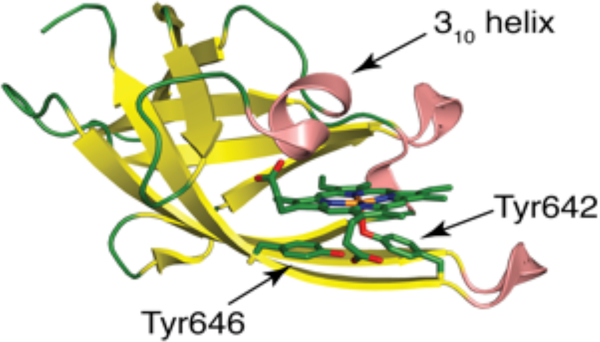
A characteristic feature in all Gram-positive heme uptake pathways studied to date is the presence of heme uptake proteins containing structurally conserved NEAT domains (Figure 12).130,131 While NEAT domains are not conserved at the amino acid level, they all share a common, superimposable immunoglobulin-like fold.34,132 NEAT domains serve many functional roles and can act as hemophores (eg. B. anthracis IsdX1 and IsdX2),128,133,134 cell surface receptors (eg. S. aureus IsdB and IsdH),126,135,136 and cell-wall anchored heme-transporters (eg. S. aureus IsdC and IsdA).104,137 Some NEAT domain proteins contain a single domain (eg. IsdX1),128 while others are modular, with some featuring multiple NEAT domains in a single polypeptide chain, such as IsdX2.133 The functional properties of a NEAT domain are determined by the immediate heme coordination environment, secondary coordination sphere, and a 310-helix (Figure 12) that is located above the NEAT heme binding site and mediates protein-protein interactions.128 S. aureus heme extraction from metHb is achieved via direct protein-protein interactions between NEAT domains and metHb.151 For a comprehensive review on NEAT domains we direct the reader to 130.
Figure 12.
Structure of the third NEAT domain of the S. aureus heme scavenger IsdH in its heme-bound form (PDB: 2Z6F).
4.3. Mtb heme uptake
4.3.1. Overview
Mtb heme uptake is distinct from other bacteria, lacking close homology to proteins associated with either Gram-negative or Gram-positive bacterial heme uptake systems, apart from homologs of PBPs that might play roles in both siderophore-dependent iron and heme uptake pathways in Mtb. Here we describe two putative heme uptake pathways in Mtb, see Figure 13. The first comprises mycobacterial proteins found in the Mtb non-operonic genomic region between genes Rv0202c and Rv0207c.90 The second pathway involves two surface exposed proteins and one predicted small-molecule PBP.147
Figure 13.
Mtb heme uptake. In the first pathway, the proposed proteins are colored in orange, in the second they are light orange. In the first pathway, heme uptake involves a secreted protein, Rv0203. The heme source for Rv0203 is still not known. Rv0203 may bind free heme, or work in combination with a hitherto unidentified partner that facilitates heme release from Hb. Heme transport across the mycobacterial outer membrane occurs via an unknown mechanism. Heme uptake across the inner membrane involves heme transfer between Rv0203 and two RND transmembrane transporters, MmpL3 and MmpL11 that accept heme from Rv0203 through protein-protein interactions. Finally, heme in the cytosol is degraded by MhuD to release iron. In the second pathway, PPE36 and PPE62 are cell-surface associated and predicted to bind free extracellular heme. FecB2 is a PBP protein, also predicted to bind heme, which shuttles heme through the Mtb periplasm. Asterisks indicate proposed steps.
4.3.2. Evidence of mycobacterial heme uptake
(i). Initial evidence
Indirect evidence of an Mtb heme uptake pathway was first found in research unrelated to heme uptake. It was observed that a recombinant BCG strain harboring a defective mycobactin biosynthetic pathway was able slowly replicate in SCID mice suggesting it uses an alternative iron source, likely heme-iron.148 It was also been shown that the heme-analogue gallium-protoporphyrin IX (GaPPIX) is lethal to M. smegmatis. This implies that mycobacteria can acquire GaPPIX and process it in the cytoplasm to release toxic Ga metal, or that it is directly incorporated into heme proteins whereby it disrupts essential pathways.149 This result further demonstrated that mycobacteria can take up metalloporphyrins (MPPs), such as heme.
(ii). Demonstration of a heme uptake pathway
Heme uptake was directly examined using an Mtb strain (MtbΔmbtB) that has a disrupted mycobactin biosynthetic pathway. This mutant strain replicates poorly in media containing ferric iron as the sole iron source, indicating that mycobactin-mediated iron uptake is non-functional.90 In contrast, addition of heme or Hb to the media, results in a restoration of growth, comparable to growth of MtbΔmbtB supplemented with exogenous mycobactin. These results indicate that Mtb has a heme uptake system.
4.4. The first heme uptake pathway
4.4.1. Identification of protein players
A single 10.6 kDa protein, Rv0203 was identified from pulldown experiments on Mtb culture filtrate using heme-agarose beads followed by mass spectrometry analysis, suggesting Rv0203 may represent an extracellular heme binding protein.90 The search for additional Mtb heme uptake proteins focused on the genomic vicinity surrounding Rv0203. Two other proteins, MmpL3 (Rv0206c) and MmpL11 (Rv0202c), were hypothesized to have a role in heme transport across the inner membrane as they are predicted to be large RND-like transmembrane transporters, Figure 13.
To directly probe the roles of Rv0203, MmpL3 and MmpL11 in heme uptake, knockout mutants were constructed in the mycobactin-deficient strain, MtbΔmbtB. The MtbΔmbtBΔmmpL3 mutant could not be made, as mmpL3 is an Mtb essential gene.150 Both MtbΔmbtBΔrv0203 and MtbΔmbtBΔmmpL11 displayed normal growth kinetics when grown in media containing ferric iron supplemented with exogenous mycobactin. However, both mutants displayed a significant growth defect compared to MtbΔmbtB when grown in media with both iron and heme present, but without exogenous mycobactin. This strongly suggests that both Rv0203 and MmpL11 are required for Mtb to utilize heme as an iron source. MmpL3 shares approximately 30% sequence homology with MmpL11; and therefore may also play a role in Mtb heme uptake.90
4.4.2. Structural biology of Mtb heme uptake proteins
(i). Rv0203 structure
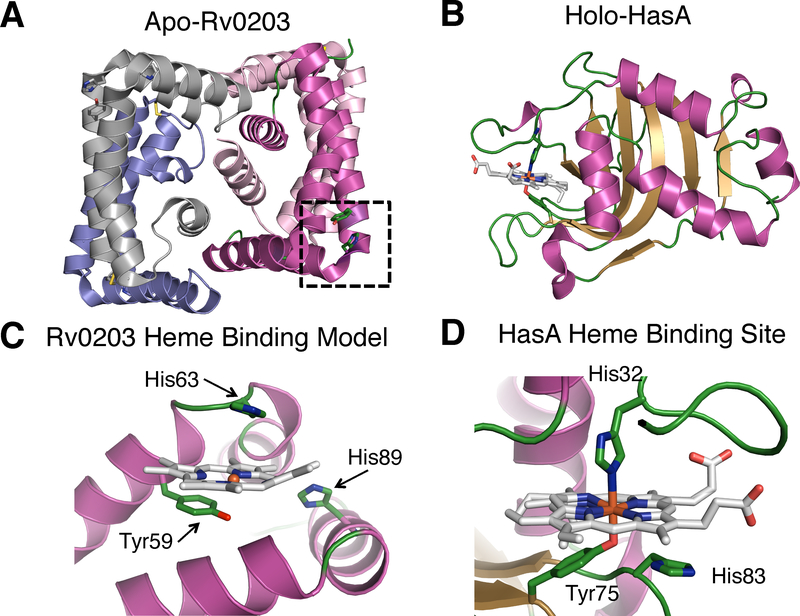
The crystal structure of apo-Rv0203 reveals an entirely α-helical protein that crystallizes as a dimer of dimers, where each individual Rv0203 protomer consists of five α-helices, Figure 14A.90 Each promoter in the tetramer structure is L-shaped and arranged around a central core composed of 4 α-helices. The central helices (α5) represent the Rv0203 C-termini and are critical in stabilizing dimer-dimer interactions. Tetrameric Rv0203 is also observed in solution, where Rv0203 exists in a concentration dependent equilibrium with the dimeric form.
Figure 14 –
Structure of Rv0203 (PDB: 3MAY) in comparison with HasA (PDB:1DKH). A. Tetrameric structure of Rv0203. The proposed heme binding site is boxed for one monomer. B. Structure of HasA, C. a model for holo-Rv0203, and D. the heme binding site of HasA.
A structural homology search revealed that Rv0203 features a unique fold since there were no other proteins with an overall similar fold. Interestingly, structural comparison with the S. marcescens hemophore, HasA, which has no structural or sequence homology to Rv0203 (Figures 14A & B), reveals that Rv0203 surface residues Tyr59, His63 and His89 are arranged in a similar motif as the HasA heme binding residues His32, Tyr75 and His83, Figure 14.116 The three Rv0203 residues are located in α-helices 1 and 3 within a hinge region on the protein surface that may be able to accommodate a heme molecule, leading to the hypothesis that Rv0203 residues Tyr59, His63 and His89 represent the Rv0203 heme binding site (Figure 14C).
(ii). Role of Rv0203
Mutagenesis studies demonstrated that the proposed heme binding residue Tyr59 is required for heme binding. Rv0203 variants Tyr59Ala and Tyr59Phe both bind heme poorly, suggesting this residue is essential in heme binding. Further evidence for Tyr-heme-iron ligation is found in the spectroscopic signatures of Rv0203, which have greatest similarity with a high spin phenolate ligated heme molecule.151 While the heme-binding properties of Rv0203 are known, the holo-Rv0203 crystal structure has not yet been determined and several aspects of the Rv0203 heme binding mechanism remain unclear. The cleft between α-helices 1 and 3 in apo-Rv0203 is too small to fully accommodate a heme molecule. It remains to be seen if heme binding induces a conformation change to fully accommodate heme within the cleft. Kinetic studies of heme binding to Rv0203 suggest that the interaction is dominated by the Tyr heme-iron ligation, with little added contribution from non-covalent interactions. Compared to heme transport proteins in S. aureus and S. marcescens, Rv0203 binds heme relatively weakly as the rate of heme dissociation, koff, is 0.08 s−1 (Tables 2 and 3),151 precluding affinity driven heme acquisition from metHb. Further experiments showed that Rv0203 does not steal heme from metHb through protein-protein interactions, suggesting Rv0203 may represent a hemophore that binds free heme, akin to the hemophore-like protein HmuY from P. gingivalis.152 Alternatively, Rv0203 may function as a heme transport protein that binds free heme and shuttles its cargo to the outer membrane and/or through the cell-wall and periplasm.151,153
Table 2:
Heme binding properties of bacterial heme uptake.
| Protein | Coordination | kon (μM−1s−1) | koff (s−1) | Ka (M−1) | Ref. |
|---|---|---|---|---|---|
| NEAT domain proteins | |||||
| IsdA | 5c Tyr | 100 | 2.6 × 10−4 | 3.8 × 1011 | 137 |
| IsdC | 5c Tyr | ND | ≥ 1 × 1012 | 137 | |
| Shp | 6c Met/Met | 1.6 | 3 × 10−4 | 5.9 × 109 | 138 |
| Gram negative heme scavengers | |||||
| HasA | 6c Tyr/His | 16 | 3 × 10−4 | 5.3 × 109 | 139,140 |
| HusA | Unknown | ND | ND | 1.4 × 109 | 141 |
| TonB dependent receptors | |||||
| HasR | 6c His/His | ND | ND | 5 × 106 | 142 |
| HmuR | 6c His/HS | ND | ND | 4.2 × 104 | 143,144 |
| Periplasmic heme transport proteins | |||||
| HmuT | 5c Tyr | ND | ND | 3.5 × 109 | 145,146 |
ND = not determined, 5c = 5-coordinate, 6c = 6-coordinate
Table 3:
Heme binding properties of proposed Mtb heme uptake proteins.
| Protein | Coordination | kon (μM−1s−1) | koff (s−1) | Ka (M−1) | Ref |
|---|---|---|---|---|---|
| Rv0203 | 5 c O−/Tyr | 133 | 0.0082 | 1.6 × 109 | 151 |
| Mmp3-D1 | 5c/6c unknown ligands | 29a | bkf: 0.0036 (44%) ks: 0.0004 (56%) | 8.1 × 109 | 153 |
| MmpL11-D1 | 5c/6c unknown ligands | 53 | 0.34 | 1.6 × 109 | 153 |
| PE22-PPE36 | unknown | ND | ND | 250 | 147 |
| PPE62 | unknown | ND | ND | 2500 | 147 |
| Rv0265c (FecB2) | unknown | ND | ND | 3333 | 147 |
on rate refers to ferrous CO-heme binding.
kf and ks represent the fast and slow rates of heme dissociation, respectively.
ND = not determined, 5c = 5-coordinate, 6c = 6-coordinate
(iii). Structures of MmpL3 and MmpL11
MmpL3 and MmpL11 are both members of the RND superfamily of transmembrane transporters,90,92 as described in section 3.5.2. MmpL3 and MmpL11 have at least two roles in Mtb. Both are proposed to be involved in heme uptake and are also required for the export of important Mtb cell-wall lipids, including mycolic acids.90,154–158 There are no X-ray structures of full-length MmpL3 or MmpL11, however, a structure exists for the D2 soluble domain from MmpL11, Figure 15B.92 The MmpL11 D2 structure consists of two anti-parallel α-helices situated above the antiparallel β-sheet forming a βαββαβ fold similar to RND porter subdomains, Figure 15B.92
Figure 15.
A. A predicted structural model of MmpL3 and B. the structure of MmpL11-D2 porter domain (PDB:4Y0L).
Further information on the structure and mechanism of MmpL3 and MmpL11 can be gleaned from low-resolution cryo-EM data. The 18 Å electron density map of a close MmpL3 homolog, Corynebacterium glutamicum CmpL1 forms a trimer with C3 symmetry, analogous to previously characterized Gram-negative RND transporters.87 Based on the low resolution CmpL1 structure, a model of MmpL3 was constructed, revealing the protein’s main structural elements, Figure 15A.159 The MmpL3 model consists of 12 TMs, and three soluble domains, consistent with the predicted topology of the MmpL Cluster II subfamily (see Figure 6). The D1 and D2 porter subdomains interact on the periplasmic side, while the large D3 domain is located on the cytoplasmic side.159 Based on the CmpL1 structure, the MmpL3 trimer model is arranged around the central channel, presumed to be involved in substrate transport, and also features a periplasmic “head” domain located above the channel, Figure 15A. Notably, no density was observed for the cytoplasmic D3 domain. Since MmpL11 has a similar topological prediction as MmpL3, one expects MmpL11 to feature a similar overall architecture as MmpL3.92 MmpL3 and MmpL11 have minimal periplasmic domain architecture compared to other RND family proteins (as described in section 3.5.2) that has been proposed to facilitate export of large lipidic substrates.92
4.4.3. Heme uptake by Rv0203, MmpL3 and MmpL11
(i). Heme binding properties of MmpL3 and MmpL11
Both MmpL3 and MmpL11 D1 domains bind heme. Their heme binding properties are similar, and both are characteristic of a mixed spin system with a combination of His, Tyr and His/Tyr heme-iron ligation. Heme coordination to both D1 domains induces domain oligimerization, possibly along the heme interface. Given that MmpL3 and MmpL11 assemble into trimers, it is feasible that heme-induced D1 domain oligomerization represents a functionally relevant behavior that occurs during inner membrane heme transfer.153
The heme binding kinetics of both MmpL3 and MmpL11 D1 domains are complex, and are dependent on both the heme redox and D1 oligomeric states. Ferrous-CO heme binding to both MmpL3 and MmpL11 D1 domains displays the typical linear relationship between heme concentration and binding rate, yielding association rates similar to those of other heme transport proteins. In contrast, ferric heme binding to D1 domains is multiphasic. Ferric heme binding reaction likely represents a composite of multiple events, including heme binding to D1 domains in various oligomeric states and heme-D1 oligomerization.153 The Ka of MmpL3 and MmpL11 D1 domains are higher than that of Rv0203, suggesting directional transfer from Rv0203 to D1 domains is possible (Table 3) based on a heme affinity gradient.
(ii). Heme transfer between Rv0203 and MmpL3 and MmpL11 D1 domains
Rv0203 transfers heme to both MmpL3 and MmpL11 D1 domains. The transfer is both rapid and unidirectional. Stopped flow experiments demonstrate that the rate of heme transfer between Rv0203 and both D1 domains is faster than passive heme dissociation from Rv0203, indicative that heme transfer is accelerated by protein-protein interactions.153 In contrast, the reverse transfer proceeds at the same slow rate of passive release by holo-D1 indicating D1 domains do not transfer heme to Rv0203. To determine the efficiency of heme transfer, biotinylated heme-Rv0203 and MmpL3 and MmpL11 D1 domains were incubated together, and after separation of Rv0203 and D1 domains, approximately 80% of heme bound to Rv0203 was transferred to the respective D1 domain,153 reinforcing the hypothesis that Rv0203 transfers heme directionally to MmpL3 and MmpL11 D1.
4.5. The second heme uptake pathway
4.5.1. Identification of protein players
More recently the Niederweis group utilized an innovative methodology to identify potential cell-surface and periplasmic proteins thought to be involved in heme uptake. An Mtb transposon library screen was used to identify mutants with greater resistance to toxic heme analog GaPPIX than wild-type Mtb.147 These experiments identified two mycobacteria-specific proteins (PPE36 and PPE62) and one previously described (see 3.6.2) predicted periplasmic protein (FecB2) whose attenuated expression resulted in Mtb mutants with resistance to GaPPIX.147 This suggests that these three proteins may play a role in GaPPIX transport to its protein target, or could be direct targets of the resulting GaPPIX toxicity. Moreover, the Mtb transposon mutants exhibited reduced growth in the presence of heme, as compared to wild-type Mtb, further suggesting that these proteins play a role in heme uptake. In support of these results, the MtbΔppe62 and MtbΔfecB2 mutants also displayed attenuated growth in heme alone, while the MtbΔppe36 mutant was non-viable with heme as the sole iron source. Interestingly, the MtbΔfecB2 mutant also had reduced growth in the presence of ferric-carboxymycobactin, suggesting that FecB2 might play a role in both ferric-siderophore and heme acquisition pathways.
4.5.2. Predicted protein functions
Further biochemical analysis of PPE36, PPE62, and FecB2, demonstrated that both PPE proteins are likely cell surface-exposed, while FecB2 is a membrane-tethered PBP. Additionally, it was shown by surface plasmon resonance that these proteins bind heme; albeit only in the low millimolar range (Table 3).
Interestingly, the PPE proteins are unique to mycobacteria and usually form a heterodimer with their operonic mycobacteria-specific PE partner (they are named for conserved PE and PPE N-terminal motifs). Members of the Mtb PE/PPE protein family have been shown to be important for secreted or cell-wall associated virulence and immunogenicity factors.160,161 PPE proteins usually have a conserved N-terminal 180-residue domain that forms a 4-helical bundle with its operonic PE protein partner. The PPE protein C-terminal regions are polymorphic and have a variety of different functions, such as enzymatic activities or contain large repeat domains or attachment domains that interact with other Mtb proteins.162 PPE36, which is required for growth in heme supplemented media, is a 243-residue protein and has a predicted 180-residue N-terminal PPE helical domain and a short disordered C-terminal domain. However, the functional role of PPE36 in heme uptake remains elusive.
PPE62 is a much larger protein than PPE36, comprising 582 residues. The N-terminal domain is predicted to adopt the expected PPE helical structure. The initial half of the C-terminal domain shows a right-handed β-helix comprised of three parallel β-sheets repeats, which is structurally reminiscent of HxuA,163 a cell-surface Hx receptor in H. influenza involved in removing heme from Hx (Figure 10C). Thus, it is possible that PPE62 has a similar Hx-binding function as HxuA. However, it should be noted that growth of the PPE62 deletion mutant was only tested in media with free heme as the only iron source and thus it seems unlikely that the sole function of PPE62 is to steal heme from Hx, like HxuA.
FecB2 is a putative heme binding PBP chaperone. The structure of Mtb FecB2 has been solved in its apo-form (PDB code: 4PM4, Figure 16A), as described in section 3.6.2. FecB2 has similar fold to PBP IsdE from S. aureus126 and PhuT from P. aeruginosa,164 both of which chaperone heme through the periplasm. Heme-iron in IsdE is six-coordinate with methionine and histidine ligands (Met78 and His229), Figure 16B,126 whereas, PhuT donates a tyrosine ligand (Tyr71) to heme-iron which is further stabilized by two arginines, Arg73 and Arg228, Figure 16C.164 Within the FecB2 small-molecule binding site there are two arginines (Arg184 and Arg221) and a tyrosine Tyr39, which may be involved in heme-iron coordination, Figure 16A. FecB2 contains a predicted N-terminal signal peptide followed by a cysteine residue still present in its mature form,165 leading to the speculation that it is a lipoprotein localized to the periplasmic side of the inner membrane. Thus, it has been proposed that FecB2 may be an inner membrane localized PBP that chaperones heme through the periplasm to an inner membrane heme transporter.147
Figure 16.
Top panels are ribbon diagrams of the PBPs. The bottom panels are close-ups of the substrate binding sites. Siderophores are shown in white stick and active site Tyr and Arg residues are indicated. A. Mtb FecB2 (PDB: 4PM4; blue), B. S. aureus heme:IsdE complex (PDB:2Q8Q; pink), and C. P. aeruginosa heme:PhuT complex (PDB:2R79; orange).
4.6. Problems with the potential components of Mtb heme uptake
4.6.1. MmpL proteins are small molecule exporters
As mentioned above, Rv0203 binds heme with a lower affinity than the secreted hemophore HasA and does not acquire heme from Hb. Nevertheless, Rv0203 was shown to be critical in efficient Mtb heme uptake.90 We proposed that Rv0203 is a free heme scavenger, however, it is also possible that an unidentified protein assists Rv0203 in scavenging heme from Hb, similar to the HmuY system described in P. gingivalis, where heme release from metHb is catalyzed by Hb proteases that partially degrade metHb.152
MmpL proteins have been shown to export small molecules including lipids and siderophores.166 MmpL3 has been shown to export trahalose monomycolate155 and also acts as a flippase for mycolic acids.158 Additionally, MmpL11 has been shown to export lipids in Mtb and M. smegmatis.156 These data raise the question of whether MmpL3 and MmpL11 are really bidirectional transporters. An alternative role for MmpL3 and MmpL11 in heme metabolism has been presented, whereby the proteins’ function is to prevent heme toxicity within the Mtb cytoplasm by enabling heme secretion.147 However, experimental investigation of this hypothesis is required.
4.6.2. Proteins identified by GaPPIX screen have a low affinity to heme
All three proteins, PPE36, PEE62 and FecB2, which were identified through the aforementioned GaPPIX Mtb transposon library experiment, bind heme with millimolar affinity.147 The heme-binding affinity of these proteins is at least 1000-fold lower as compared to other known proteins involved in bacterial heme uptake systems (Tables 2&3),137 raising uncertainty about their proposed roles in GaPPIX or heme transport. Based on the low heme binding affinities, it is possible that these proteins are not the direct target of GaPPIX but play an indirect role in GaPPIX import and Ga toxicity.
FecB2 is proposed to be a PBP heme-transporter protein similar to S. aureus IsdE,137 however, heme binding to IsdE is at least 6 orders of magnitude tighter than binding to FecB2.167 FecB2 has a lone Tyr39 in its small-molecule binding pocket that could potentially coordinate heme-iron, analogous to P. aeruginosa PhuT.164 However, in the FecB2 apo-structure, this tyrosine appears to be shielded from coordinating ferric iron as it π-stacks with Trp58, Figure 16A. Since FecB2 has an extremely low affinity for heme compared to S. aureus IsdE, one could speculate that FecB2 does not function as a periplasmic heme chaperone.
As mentioned in section 4.5.2, the C-terminal region of PPE62 is predicted to have a β-propeller like fold similar to HxuA, a cell-surface Hx receptor involved in Gram-negative heme uptake.168 The low heme binding affinity of PPE62 is consistent with its proposed structure, as HxuA binds Hx tightly, but does not bind heme itself and HxuA is not required in free heme uptake.163 It therefore remains to be seen if PPE62, which was proposed to be part of a free heme acquisition pathway,147 is a bifunctional protein with dual Hx and heme binding properties, or if PPE62 is in fact an Hx-binding protein.
5. Cytoplasmic Heme-Degradation to Release Iron
5.1. Overview
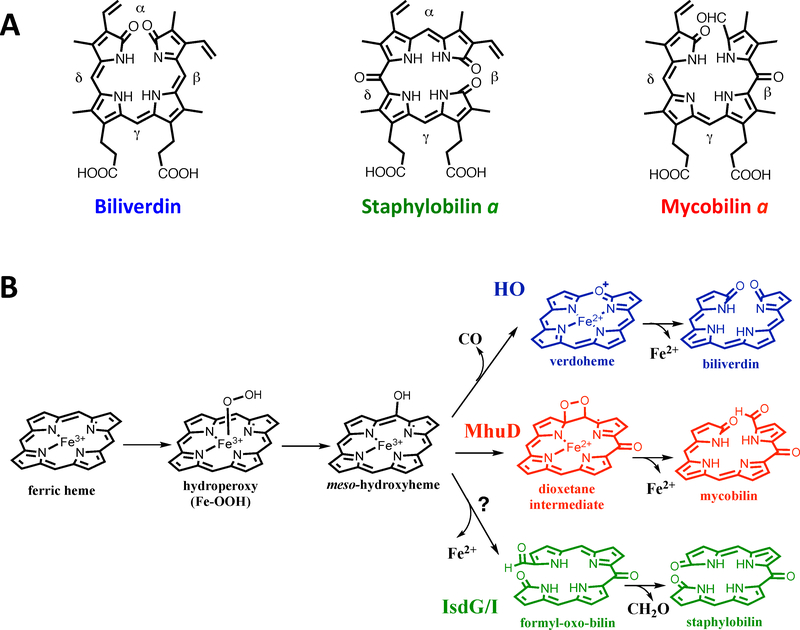
The first discovered and most extensively studied heme degrading enzyme is human heme oxygenase 1 (hHO-1), which oxidatively degrades heme to release ferrous iron, Figure 17. 169 Heme oxygenase (HO) is the canonical heme degrading protein in eukaryotes and has also been found in several bacteria including P. aeruginosa, Corynebacterium diphtheriae, and Neisseria meningitidis.170–172 In the last decade, a novel class of non-HO heme degraders has been discovered, the IsdG-type branch.173 Notably, Mtb also harbors a non-canonical IsdG-type heme degrading protein, MhuD. However, the MhuD and IsdG heme degradation mechanisms differ substantially and efforts have been made to understand the unique steps in MhuD heme degradation.
Figure 17.
A: Structures of chromophore products of bacterial heme degradation. B: Proposed mechanisms of oxidative heme degradation. In the first monooxygenase step production of the intermediate meso-hydroxyheme is common to HO, MhuD and IsdG/I. The HO (blue), MhuD (red) and IsdG/I (green) products branch off after this step to produce different chromophores and side-products and iron.
5.2. Heme oxygenase
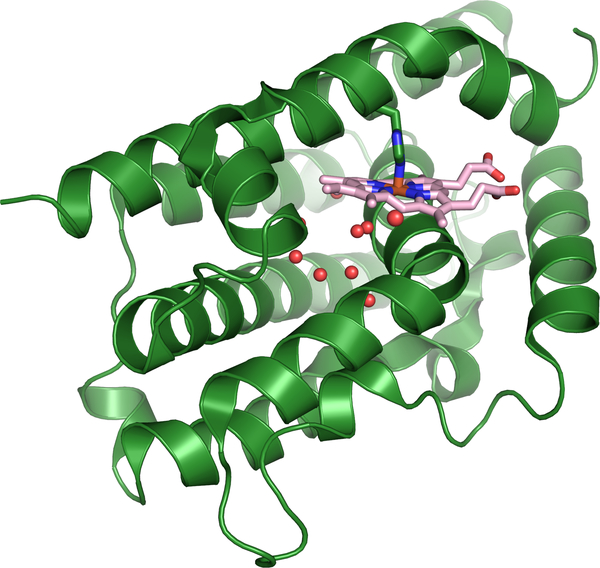
HOs degrade heme into the chromophore biliverdin IXα, carbon monoxide (CO) and ferrous iron, Figure 17.169 Multiple HO structures have been solved across many different species.173–175 The structure of canonical HO is comprised of an entirely α-helical fold, Figure 18.174 The heme binding pocket is composed of proximal and distal helices, where the heme iron is coordinated by a histidine residue on the proximal side with a network of ordered waters on the distal side.174 Heme is anchored in the HO active site by Arg and Lys residues that interact with the heme propionates. Moreover, the heme molecule is slightly solvent exposed and nearly planar.174
Figure 18.
Structure of hHO-1 (PDB:1N45) showing the water network surrounding the distal heme pocket.
The mechanism for HO-mediated heme degradation occurs via three successive monooxygenation reactions, reviewed extensively in176. The first monooxygenation step is activation of molecular oxygen by the heme iron center. The heme ferric iron center is reduced to its ferrous form while coordinating dioxygen and is converted into the activated ferric hydroperoxy [Fe3+-O2H] intermediate that is stabilized by the network of water molecules surrounding the distal heme pocket.174,177–179 The Fe3+-O2H species is cleaved resulting in the production of the α-meso-hydroxyheme intermediate.180,181 In the second monooxygenase step, the α-meso-hydroxyheme reacts with molecular oxygen to yield the second intermediate, α-verdoheme, and also results in the release of CO.182 Finally, a second round of molecular oxygen activation occurs during the third and final monooxygenation step, followed by hydroxylation of α-verdoheme to produce the final product, biliverdin and to release ferrous iron.176
5.3. IsdG-type heme oxygenases
5.3.1. Overview
The non-canonical IsdG-type heme degrading protein family is the other major class of heme degrading proteins, and the primary sequence and structure of IsdG-type enzymes are quite distinct from canonical HOs.173 IsdG-type heme degrading proteins are primarily found in bacteria, although recently a branch has been discovered in lower eukaryotes.183 The family is named after IsdG, which was first characterized in S. aureus. IsdG and its homolog IsdI are part of the S. aureus isd heme uptake operon.184
5.3.2. Structure and mechanism of IsdG-type proteins
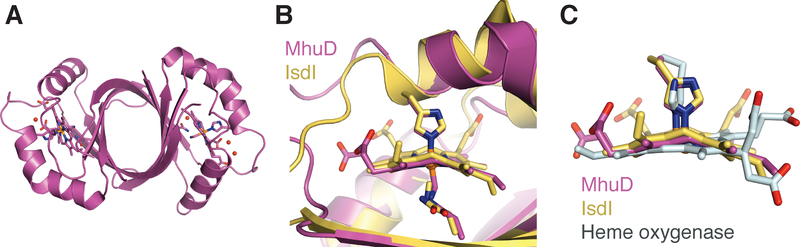
IsdG-type heme degrading enzymes are structurally distinct from HOs,185,186 and consist of a homodimer exhibiting a ferrodoxin-like α + β-barrel fold, a similar fold to Figure 19A.185,186 There are two predominately hydrophobic heme binding active sites per homodimer. Like HO, the heme-iron is coordinated through a histdine residue (His75 in S. aureus IsdG numbering) on the proximal side; however, on the distal side the water network is absent, Figure 19B. Instead, the heme-iron on the distal side is coordinated to a chloride ion that is in turn is coordinated by Asn6.186
Figure 19.
Structure of MhuD-monoheme A. MhuD bound to a single heme molecule (PDB:4NL5). Comparison of B. MhuD and IsdI active sites (PDB:3LGN), and C. heme ruffling in MhuD, IsdI and hHO-1 (PDB:1N45).
IsdG and IsdI convert heme to novel chromophores, staphylobilin isomers187 and formaldehyde, Figure 17.188 The staphylobilin isomers result from cleavage of the tetrapyrrole ring at the β/δ-meso carbon and an oxygenation reaction at the δ/β-meso carbon, respectively, followed by the release of the β/δ-meso carbon as formaldehyde.
5.3.3. Heme ruffling is required for heme activation in IsdG-type heme degraders
A defining feature in S. aureus IsdG-type enzymes is the presence of a highly ruffled heme in their active sites.186 The S. aureus IsdG-N7A and IsdI structures feature an extraordinary out-ofplane heme distortion of 1.9 and 2.3 Å, respectively (Figure 19C).186 NMR experiments show that the observed heme ruffling appears to alter the electronic structure of heme, possibly sensitizing the heme meso-carbons to nucleophilic attack.189 The requirement for heme ruffling in IsdG-type heme degradation was demonstrated by mutational analysis. Within the IsdG/I heme binding pocket, Trp66 forms a hydrophobic interaction with the heme protoporphyrin ring to promote ruffling. An IsdG-Trp66Ala variant resulted in a reduction of heme ruffling, which correlated with a severe attenuation in heme degradation. Moveover, the heme electronic structure in IsdG-Trp66Ala is different from that of wild-type MhuD, such that it has been proposed that the meso-carbons are not activated.190 Together, these results suggest that heme ruffling in IsdG-type heme degrading proteins is required to increase the heme molecule’s reactivity in the hydrophobic active site.190
Despite, knowing the heme-bound IsdG and IsdI structures, our understanding of the intermediate steps in IsdG/I catalyzed heme degradation is poor. However, several elegant studies partially elucidate the IsdG mechanism and suggest that the first oxidation step forms the β/δ-meso-hydroxyheme intermediates.191,192
5.3.4. IsdG-type proteins require an electron donor
IsdG and IsdI require reducing equivalents to degrade heme. It has been shown that S. aureus proteins, NtrA and IruO, donate electrons to IsdG and IsdI heme degrading proteins.193–195 IruO is a FAD-containing NADPH-dependent reductase, while NtrA is thought to be a novel nitroreductase capable of S-nitrosoglutathione activity.
5.4. Mtb MhuD heme degradation
5.4.1. Overview
An IsdG-type protein is also found in Mtb, MhuD (Mycobacterial heme utilizing Degrader). 196,197 MhuD shares high sequence and structural homology with IsdG-type heme degrading proteins,196,197 however MhuD degrades heme into unique chromophores, mycobilin isomers, (Figure 17) and iron.198
5.4.2. The structure of MhuD
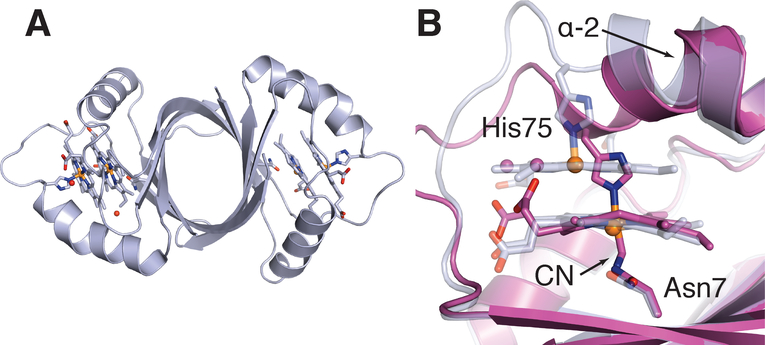
Similar to IsdG and IsdI, MhuD forms a homodimeric ferrodoxin-type α + β-barrel fold, Figures 19A & 20A.196,197 The structure of MhuD has been solved in both a diheme form (discussed below) and a cyanide-inhibited monoheme form. In the cyanide-inhibited MhuD-monoheme structure, heme-iron is coordinated by proximal His75 and a distal cyanide.197 Most notably, the heme molecule in the MhuD active site is rotated approximately 90° about the axis normal to the tetrapyrrole ring compared to the heme orientation observed in IsdG and IsdI.197 Examination of the MhuD-monoheme structure revealed that the heme was distorted by 1.4 Å,197 a decrease from the 1.9 Å and 2.3 Å heme distortion observed in the IsdG-Asn7Ala and IsdI structures, Figure 19C.186 Heme distortion, like in IsdG/I, appears to be induced by the neighboring Trp66 residue, and mutation of Trp into a less bulky hydrophobic residue results in reduced heme degrading activity.190,199 Variable temperature magnetic circular dichroism, NMR and electronic spectroscopy experiments in MhuD indicate that ruffling of the heme delocalizes spin density from the central iron and pyrrole rings, as also seen in IsdI, potentially making the meso carbons more susceptible to nucleophilic attack.189,197,199
Figure 20.
Structure of MhuD-diheme A. MhuD dimer bound to two heme molecules (PDB:3HX9). B. Comparison of MhuD monoheme (purple) and diheme (light grey) active sites.
5.4.3. Mechanism of MhuD heme degradation
MhuD heme degradation can be divided into two steps. It was suggested that the first step in MhuD heme degradation is similar to the monooxygenation step of IsdI and HO (Figure 17). In this mechanism, MhuD is proposed to oxidize heme into β/δ-meso-hydroxyheme intermediates via formation of a [Fe3+-O2−] species.198 It was also proposed that there is no release of CO in MhuD-dependent heme degradation, and so it follows that there is no formation of the verdoheme intermediate.198 Once the β/δ-meso-hydroxyheme intermediate is formed, Figure 17, it has been demonstrated that meso-hydroxyheme is converted into mycobilin by a dioxygenase step.200 For the second step to occur, meso-hydroxyheme is thought to undergo radical localization to the α-meso carbon before dioxygenation at the α-meso position into a dioxetane intermediate, which is proposed to spontaneously decompose into Fe-mycobilin, Figure 17.200 Thus, MhuD degradation of heme is highly unusual as its active site performs two different reactions, a monooxygenase reaction followed by a dioxygenase reaction. The first monooxygenation reaction, which requires a proton-donating environment, is possibly the rate-limiting step in the hydrophobic MhuD heme binding pocket. However it is thought that distal Asn7, one of the only polar residues in the distal heme pocket, forms a hydrogen bond to the terminal hydroxyl group of Fe3+-O2H to stabilize the intermediate. The second dioxygenase reaction step, where the ruffled heme may allow radical localization on the α-meso carbon, is likely enabled by the MhuD hydrophobic active site environment. Furthermore, heme ruffling may also promote the initial monooxygenase reaction by changing the electronic structure of heme-iron.189,197 Thus, it appears that heme ruffling is required for the coupled mono- and di-oxygenase reactions within the MhuD active site.200
5.4.4. The unique diheme form of MhuD
MhuD is capable of binding up to two hemes per active site, Figure 20A. The feature has not been observed in any other IsdG-type protein studied to-date. In the MhuD-diheme structure, the protoporphyrin rings are stacked 3.45 Å apart and oriented such that the propionate groups from one heme are rotated approximately 80° relative to those from the second heme, Figure 20B.196 The more solvent exposed heme-iron is coordinated by His75 while the solvent protected heme-iron is coordinated by a chloride ion and further stabilized by Asn7.196 The MhuD active site can accommodate two heme molecules through the straightening of α-helix 2 that is kinked in the MhuD-monoheme structure, together with a conformational change in the flexible loop region following α-helix 2, Figure 20B. Furthermore, both heme molecules in each MhuD-diheme active site are essentially planar.196 As one might anticipate, the MhuD-diheme form had no heme degrading activity. Loss of activity could be due to three possible scenarios (1) the planarity of the two heme molecules resulting in a difference in heme’s electronic structure, (2) different protein-heme non-covalent interactions and (3) limited room to accommodate a hydroperoxo species. The biological significance of MhuD binding two molecules of hemes per monomer is unknown. However, when Mtb resides in an environment where the heme-iron supply is not limited, MhuD’s ability to accommodate two hemes per monomer could provide an avenue for heme storage analogous the iron-storage ferritin proteins, Mtb BfrA and BfrB.201 Furthermore, MhuD-diheme inactivation may also provide a regulatory mechanism to prevent heme degradation under iron rich conditions.
5.4.5. Potential Mtb MhuD electron donors
An electron donor is essential for MhuD-dependent heme cleavage. There are numerous Mtb candidates that could be potential electron donors based on homology to S. aureus IsdG electron donor proteins, IruO and NtrA.193,195 All candidates, however, remain to be tested to elucidate the electron-donor for MhuD.
6. Iron Acquisition as Anti-TB Drug Targets
6.1. Targeting the mycobactin biosynthetic pathway
Over the last several decades, there have been tremendous efforts to design inhibitors to interrupt mycobactin-mediated iron acquisition, and in particular, to inhibit mycobactin biosynthesis (Figure 4).57,202 Although fourteen proteins are responsible for the biosynthesis of mycobactin, only two have studied extensively as potential drug targets, MbtA and MbtI. Both MbtA and MbtI catalyze initial steps in the biosynthetic pathway, Figure 4B. MbtI catalyzes the conversion of chorismate to salicylic acid, the substrate of the Mbt megasynthase complex. MbtA activates salicylic acid and catalyzes its transfer to the thiolate domain of MbtB through an acyl adenylate intermediate, Figure 4B (4). Many of the designed inhibitors are based on chorismate (Figure 4B (1)) and iso-chorismate analogues (Figure 4B (2)) for MbtI and variations of the acyl-adenylate intermediate (Figure 4B (4)) for MbtA. Additionally, some progress has been made on developing inhibitors for MbtM, which is involved in fatty acid chain activation for linkage to the mycobactin core structure.59 For more in depth information on inhibitors for MbtA, MbtI and MbtM, we direct the reader to a detailed review by Meneghetti & Villa et al.57
A potential new anti-TB drug target within the mycobactin biosynthetic pathway is MbtH. It is required for the correct folding of the three NRPSs, MbtB, MbtE and MbtF, which are constituents of the megasynthase involved in mycobactin core synthesis.62 Thus, interruption of MbtH chaperone activity for the Mtb NRPSs would disrupt mycobactin synthesis and consequently inhibit iron acquisition and Mtb growth.
6.2. Targeting iron uptake pathways
The proteins involved in Mtb mycobactin export and import represent both a new set of anti-TB drug targets and a potential gateway to introduce antibacterials via a ‘Trojan horse-like’ mechanism.
As siderophore receptors and import/export machinery are highly selective, compounds with a similar structure to the mycobactin core (Figure 3) could have an inhibitory effect on the mycobactin-dependent iron uptake pathway. In fact, M. smegmatis mycobactin S (a stereoisomer of Mtb mycobactin T) is toxic to Mtb.203 A recent example further highlights the potential of applying a “Trojan Horse Strategy” to inhibit Mtb growth. In one study, mycobactin was conjugated with artemisinin, Figure 21A, a natural product isolated from the plant Artemisia annua and a known treatment for Plasmodium falciparum malaria.204 In P. falciparum, artemisinin is activated by heme and results in its cleaveage and generation of free radicals that in turn damage many susceptible proteins.205 The mycobactin-artemisinin conjugate (Figure 21B) has strong and selective inhibitory effects against Mtb (MIC = 0.39 μg/mL), as well as MDR and XDR strains 206, thus demonstrating that mycobactin can be utilized in a ‘Trojan-horse-like’ manner. Another mycobactin analogue of particular interest is a recently synthesized mycobactin-maleimide analogue that already has inhibitory effects against Mtb (MIC90 = 0.88 μg/mL), and will allow the eventual synthesis of a series of novel mycobactin-drug conjugates.207 Finally, siderophores coordinated to toxic metals have been shown to inhibit bacterial growth. For example, when E. coli uptakes the scandium-enterobactin complex bacterial growth is attenuated,208 suggesting this might be an alternate avenue for exploiting Mtb mycobactins as therapeutics.
Figure 21.
The structure of A. artemisinin and B. mycobactin-artemisinin conjugate.
Another approach to attenuate Mtb growth is to design iron chelators that target the Mtb cytoplasm. A novel compound, pyrazolopyrimidinone (PZP) Figure 22, was found to attenuate Mtb growth209; however mutations in the ESX-3 complex (see Figure 8) resulted in resistance to PZP 210. Since ESX-3 is thought to play a role in mycobactin transport, there was speculation that the PZP acts by interfering with mycobactin-dependent iron acquisition.85,211,212 However, further investigation revealed that PZP acts as an Mtb cytoplasmic iron chelator, and does not interrupt mycobactin-dependent iron acquisition pathways.210
Figure 22.
The structure of pyrazolopyrimidinone (PZP)
Notably, in a comprehensive study identifying genes involved in Mtb antibiotic resistance, FecB was central to all antibiotics tested.213 Mtb FecB is a proposed PBP involved in siderophore-mediated iron uptake - see section 3.6.2. A MtbΔfecB mutant was hypersensitive to the uptake of variety of antibiotics, including vancomycin and verapamil.213 Thus, FecB may be a good anti-TB drug target as its inhibition may prevent the development of new MDR and XDR Mtb strains.
6.3. Targeting heme uptake pathways
As Mtb heme uptake was only discovered in 2011, these pathways have not been actively investigated as anti-TB drug targets; however heme-dependent iron uptake pathways may be suited for Trojan horse strategies or be targeted directly.48,214,215 A successful drug candidate would specifically target Mtb heme transport proteins, while having low affinity to host heme binding proteins.215
Heme analogues including metal-substituted protoporphyrins (MPPs) have antibacterial activity. Nearly two decades ago, it was demonstrated that GaPPIX was toxic against various bacteria including M. smegmatis.149 Further, MPP analogues were shown to inhibit heme import ABC transporters in Trypanosoma cruzi as well as disrupt the ability of S. aureus NEAT domain proteins to shuttle heme to the inner membrane-associated transport protein complex.216,217 Within the last five years, efforts have refocused on MPPs as potential anti-bacterials. Notably, in P. aeruginosa it was shown that GaPPIX inhibits growth by targeting cytochromes and likely interfers with cellular respiration.218 It was also demonstrated that both Ga- and Sn-PPIXs inhibit growth of Mycobacterium abscessus.154,219 Additionally, in M. abscessus-infected macrophages, treatment with MPPs inhibited mycobacterial growth but did not exhibit toxicity towards host macrophages. Moreover, 1 μM GaPPIX inhibited intracellular M. abscessus growth to a level significantly greater than 25 μM Ga(NO3)3, demonstrating that GaPPIX has greater antimycobacterial power than the toxic metal, Ga3+, alone.
In the Sher laboratory, MPPs have been tested as antibacterials against Mtb infection. Mtb-infected mice treated with SnPPIX in combination with conventional anti-TB drugs had a significantly reduced bacterial load compared to Mtb-infected mice treated with anti-TB drugs alone.220 The overall effect of this combination SnPPIX therapy was to increase the rate of Mtb clearance from 21 weeks for the conventional anti-TB therapy alone to 17 weeks. However, SnPPIX was shown to be a potent inhibitor of host hHO-1 in vitro. In contrast, the Mtb heme degrading protein MhuD was not inhibited by SnPPIX.220 Furthermore, SnPPIX does not inhibit Mtb growth in vitro with an MIC50 of <200 μM. Taken together these results suggest that the in vivo SnPPIX effect against Mtb is due to SnPPIX acting as a host-directed therapy upon mammalian HO rather than an Mtb-directed therapy.220 Unlike SnPPIX, GaPPIX does inhibit Mtb growth in vitro with an MIC50 of 1.5 μM.147 So it remains to be seen if GaPPIX may act as both a host-directed and an Mtb-directed therapy.
In addition to their potential as anti-TB agents, heme analogues may provide insights into the mechanism of heme uptake and be utilized to identify new players in the Mtb heme acquisition pathways. This has, in part, already been demonstrated with the identification of transposon mutants that are resistant to GaPPIX - as discussed in section 4.5.1. Moreover, heme analogues could be used to assess the importance of heme acquisition in infectivity and virulence.
Significantly, over the last seven years, MmpL3 has become the most successful anti-TB drug target. High-throughput whole cell screens identified several potent anti-mycobacterial agents that target MmpL3, including BM212 and SQ109.155,215,221,222 However, these agents attenuate Mtb growth by acting upon MmpL3 mycolic acid flippase function or by dissipating the energy of proton motive force across the inner membrane.158 Thus, it is unlikely that the effect of the MmpL3 inhibitors is due to the interruption of heme uptake. Compounds that target MmpL3 have been extensively reviewed in 155,215,221,222.
7. Conclusions
Over the last two decades, bacterial heme and iron acquisition pathways have been discovered and mapped out in a number of organisms.117,118,124,132,223 Nevertheless, the relationship between heme uptake and virulence is not always straightforward. The contribution of heme acquisition to virulence has been firmly established in only a select number of bacteria. Most notably, elegant work by Skaar and others demonstrated that in S. aureus heme-iron is the preferred iron source over transferrin iron in the early stage of S. aureus infection.224 Additionally, in uropathogenic E. coli it has been shown that heme uptake is necessary for urinary tract colonization, 225 and in P. gingivalis, which lacks an endogenous heme biosynthesis pathway, heme uptake is a critical virulence factor.226 These observations suggest that heme acquisition pathways have excellent potential as antibacterial targets. However, except for the cases referenced above, the interplay between heme and iron uptake during early, active and secondary pathogen infections or dormancy remains largely unknown.
Two mechanisms of iron acquisition have been discovered in Mtb, iron and heme uptake. Iron acquisition is dependent on mycobactin siderophores. Mycobactins are required for initial stages of Mtb infection 40,60. Thus, targeting Mtb mycobactin metabolism represents a promising strategy in designing novel anti-TB drugs, and, to date, three mycobactin biosynthesis enzymes, MbtA, MbtI and MbtM are being investigated as drug targets. However, further research is needed to create a broader understanding of Mtb iron metabolism since there are still large knowledge gaps surrounding the proteins involved in siderophore export and import. Unlike siderophore mediated iron uptake, a clear link between heme uptake and infection has not yet been established in Mtb. To effectively understand how Mtb heme uptake can be used as a drug target, it is imperative that future research focuses on understanding if heme uptake is predominately utilized in the initial stage of Mtb infection, in secondary infection or even in the dormant state.
Acknowledgements
This study was funded by NIH grant AI095208 (C.W.G.), National Science Foundation grant NSF-GRFP DGE-1321846 (A.C.), and USDA-NIFA 2015-67012-22895 (C.P.O). We’d also like to thank Nick Chim, Kalistyn Burley, Bonnie Cuthbert and Sumer Abdul-Hafiz for critical reading of this manuscript.
Abbreviations
In order of appearance
- TB
Tuberculosis
- Mtb
Mycobacterium tuberculosis
- AIDS
Acquired immune deficiency syndrom
- HIV
Human immunodeficiency virus
- WHO
World Health Organization
- MDR-TB
Multidrug Resistant TB
- XDR-TB
Extensively-drug Resistant TB
- Hb
hemoglobin
- PBP
periplasmic binding protein
- Hp
haptoglobin
- Hx
hemopexin
- SPR
Surface Plasmon Resonance
- ITC
Isothermal titration calorimetry
- ABC
ATP-binding cassette
- Tf
transferrin
- PPIX
protoporphyrin IX
- Lf
lactoferrin
- NRPS
nonribosomal peptide synthetase
- IdeR
iron-dependent regulator
- MFS
major facilitator superfamily
- RND
resistance nodulation cell division
- FeEnt
ferric-enterobactin
- MmpL
Mycobacterial membrane protein Large
- MmpS
Mycobacterial membrane protein Small
- TM
transmembrane helix
- NTD
N-terminal domain
- FAD
flavin adenine dinucleotide
- NEAT
Near Iron Transport
- CO
carbon monoxide
- Omp
outer membrane porin
- GaPPIX
gallium-protoporphyrin IX
- hHO-1
human heme oxygenase 1
- HO
heme oxygenase
- SAM
S-adenosylmethionine
- MhuD
Mycobacterial heme utilizing degrader
- PZP
pyrazolopyrimidinone
- SnPPIX
tin-protoporphyrin IX
- MMP
metalloprotoporphyrin IX
Biography
Alex Chao
Alex received his Bachelors degree in Chemistry from Arizona State University, where he performed undergraduate research examining the insulin signaling pathway as well as studying a bacterial surface adhesion protein. Alex is now a Ph.D. candidate at the UC Irvine in the Department of Molecular Biology and Biochemistry and is a recipient of the prestigious National Science Foundation Graduate Research Fellowship. Currently, he is working to biochemically and structurally characterize the Mtb heme degradation enzyme. Additionally, he is working to determine the structures of proteins involved in Mtb iron uptake.
Paul Sieminski
Paul studied glycerol-water mixtures at various compositions, using physical and thermodynamic methods to elucidate phase transitions in the system during his Bachelors degree in Biology at Pennsylvania State University. Interested in applying these principles to biological systems, using X-ray crystallography and biochemical methods, he initiated characterization of an important protein in Mtb siderophore trafficking during his Master’s degree at UC Irvine. Currently, Paul is a doctoral student studying structural biology and biophysics in the Department of Chemistry & Biochemistry at UC Los Angeles (UCLA).
Cedric Owens
Cedric is an Assistant Professor of Biochemistry at Chapman University in Orange, CA. After growing up and completing his Abitur in Bayreuth, Germany, he moved to Waterville, ME to obtain B.A. degrees in Chemistry and Mathematics Sciences from Colby College. For his Ph.D., he joined the lab of Dr. Celia Goulding at UC Irvine, where he characterized mycobacterial heme uptake. From there, he moved to UC San Diego to investigate electron transfer in nitrogenase in the lab of Dr. Akif Tezcan as a USDA NIFA postdoctoral fellow. Currently, Cedric’s research interests focus on characterizing and engineering regulation of nitrogen fixation in agriculturally relevant bacteria.
Celia Goulding
Celia is a Professor of Molecular Biology and Biochemistry and Pharmaceutical Sciences at the University of California, Irvine. She received a B.Sc(Hons) in chemistry and mathematics and a Ph.D. in physical organic chemistry from King’s College, London. After leaving the UK for the US, she was a postdoctoral fellow with Professor Rowena Matthews at the University of Michigan studying metalloprotein enzymology. Moving further west to UCLA, as research faculty under the supervision of Professor David Eisenberg, Celia became interested in the mechanisms by which Mtb is able to survive in human hostile environments, employing X-ray crystallography and other biophysical and biochemistry techniques. During her time at UCLA, she also spearheaded the Tuberculosis Structural Genomics Consortium, which has resulted in approximately 350 Mtb protein structures deposited in the Protein Databank since its inception in 2001. Celia eventually accepted an independent position at UC Irvine where she and her group investigate the function and structure of metalloproteins involved in heme and iron uptake pathways in Mtb.
References
- (1).Zink AR; Grabner W; Nerlich AG Molecular identification of human tuberculosis in recent and historic bone tissue samples: The role of molecular techniques for the study of historic tuberculosis. Am. J. Phys. Anthropol 2005, 126, 32–47. [DOI] [PubMed] [Google Scholar]
- (2).Gazit C In American Experience Boston, Ma, 2015.
- (3).Kiran D; Podell BK; Chambers M; Basaraba RJ Host-directed therapy targeting the Mycobacterium tuberculosis granuloma: a review. Semin. Immunopathol 2016, 38, 167–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ernst JD The immunological life cycle of tuberculosis. Nat. Rev. Immunol 2012, 12, 581–591. [DOI] [PubMed] [Google Scholar]
- (5).Phillips L Infectious disease: TB’s revenge. Nature 2013, 493, 14–16. [DOI] [PubMed] [Google Scholar]
- (6).The sWorld Health Organization, Global tuberculosis report 2017, World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- (7).Dheda K; Cox H; Esmail A; Wasserman S; Chang KC; Lange C Recent controversies about MDR and XDR-TB: Global implementation of the WHO shorter MDR-TB regimen and bedaquiline for all with MDR-TB? Respirology 2018, 23, 36–45. [DOI] [PubMed] [Google Scholar]
- (8).Tiberi S; du Plessis N; Walzl G; Vjecha MJ; Rao M; Ntoumi F; Mfinanga S; Kapata N; Mwaba P; McHugh TD et al. Tuberculosis: progress and advances in development of new drugs, treatment regimens, and host-directed therapies. Lancet Infect. Dis 2018, DOI: 10.1016/S1473-3099(18)30110-510.1016/S1473-3099(18)30110-5. [DOI] [PubMed] [Google Scholar]
- (9).Cairo G; Bernuzzi F; Recalcati S A precious metal: Iron, an essential nutrient for all cells. Genes Nutr. 2006, 1, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Skaar EP The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010, 6, e1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Doherty CP Host-pathogen interactions: the role of iron. J. Nutr 2007, 137, 1341–1344. [DOI] [PubMed] [Google Scholar]
- (12).Boradia VM; Malhotra H; Thakkar JS; Tillu VA; Vuppala B; Patil P; Sheokand N; Sharma P; Chauhan AS; Raje M et al. Mycobacterium tuberculosis acquires iron by cell-surface sequestration and internalization of human holo-transferrin. Nat. Commun 2014, 5, 4730. [DOI] [PubMed] [Google Scholar]
- (13).Malhotra H; Patidar A; Boradia VM; Kumar R; Nimbalkar RD; Kumar A; Gani Z; Kaur R; Garg P; Raje M et al. Mycobacterium tuberculosis Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) Functions as a Receptor for Human Lactoferrin. Front. Cell. Infect. Microbiol 2017, 7, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Beinert H; Holm RH; Munck E Iron-sulfur clusters: nature’s modular, multipurpose structures. Science 1997, 277, 653–659. [DOI] [PubMed] [Google Scholar]
- (15).Poulos TL Heme enzyme structure and function. Chem. Rev 2014, 114, 3919–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Gray HB; Winkler JR Electron transfer in proteins. Annu. Rev. Biochem 1996, 65, 537–561. [DOI] [PubMed] [Google Scholar]
- (17).Perutz MF Mechanisms regulating the reactions of human hemoglobin with oxygen and carbon monoxide. Annu. Rev. Physiol 1990, 52, 1–25. [DOI] [PubMed] [Google Scholar]
- (18).Prousek J Fenton chemistry in biology and medicine. Pure Appl. Chem 2007, 79, 2325–2338. [Google Scholar]
- (19).Lyons T; Eide D In Bioinorganic Chemistry; Bertini I;Gray H;Lippard S;Valentine J, Eds.; University Science Books: Mill Valley, CA, USA, 1994. [Google Scholar]
- (20).Conrad ME; Umbreit JN; Moore EG Iron absorption and transport. Am. J. Med. Sci 1999, 318, 213–229. [DOI] [PubMed] [Google Scholar]
- (21).Gonzalez-Chavez SA; Arevalo-Gallegos S; Rascon-Cruz Q Lactoferrin: structure, function and applications. Int. J. Antimicrob. Agents 2009, 33, 301 e301–308. [DOI] [PubMed] [Google Scholar]
- (22).Schaer DJ; Vinchi F; Ingoglia G; Tolosano E; Buehler PW Haptoglobin, hemopexin, and related defense pathways-basic science, clinical perspectives, and drug development. Front. Physiol 2014, 5, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Hoette TM; Clifton MC; Zawadzka AM; Holmes MA; Strong RK; Raymond KN Immune Interference in Mycobacterium tuberculosisIntracellular Iron Acquisition through Siderocalin Recognition of Carboxymycobactins. ACS Chem. Biol 2011, 6, 1327–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Johnson EE; Srikanth CV; Sandgren A; Harrington L; Trebicka E; Wang L; Borregaard N; Murray M; Cherayil BJ Siderocalin inhibits the intracellular replication of Mycobacterium tuberculosis in macrophages. FEMS Immunol. Med. Microbiol 2010, 58, 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Waldvogel-Abramowski S; Waeber G; Gassner C; Buser A; Frey BM; Favrat B; Tissot JD Physiology of iron metabolism. Transfus. Med. Hemother 2014, 41, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Arosio P; Elia L; Poli M Ferritin, cellular iron storage and regulation. IUBMB Life 2017, 69, 414–422. [DOI] [PubMed] [Google Scholar]
- (27).Hargrove MS Quaternary Structure Regulates Hemin Dissociation from Human Hemoglobin. J. Biol. Chem 1997, 272, 17385–17389. [DOI] [PubMed] [Google Scholar]
- (28).Hargrove MS; Barrick D; Olson JS The association rate constant for heme binding to globin is independent of protein structure. Biochemistry 1996, 35, 11293–11299. [DOI] [PubMed] [Google Scholar]
- (29).Hrkal Z; Vodrazka Z; Kalousek I Transfer of heme from ferrihemoglobin and ferrihemoglobin isolated chains to hemopexin. Eur. J. Biochem 1974, 43, 73–78. [DOI] [PubMed] [Google Scholar]
- (30).Seery VL; Muller-Eberhard U Binding of porphyrins to rabbit hemopexin and albumin. J. Biol. Chem 1973, 248, 3796–3800. [PubMed] [Google Scholar]
- (31).Hargrove MS; Singleton EW; Quillin ML; Ortiz LA; Phillips GN; Olson JS; Mathews AJ His64 (E7)--> Tyr apomyoglobin as a reagent for measuring rates of hemin dissociation. 1994, 269, 4207–4214. [DOI] [PubMed] [Google Scholar]
- (32).Crichton R Inorganic Biochemistry of Iron Metabolism: From Molecular Mechansims to Clinical Consequences; Wiley: West Sussex, UK, 2001. [Google Scholar]
- (33).Lane DJ; Merlot AM; Huang ML; Bae DH; Jansson PJ; Sahni S; Kalinowski DS; Richardson DR Cellular iron uptake, trafficking and metabolism: Key molecules and mechanisms and their roles in disease. Biochim. Biophys. Acta 2015, 1853, 1130–1144. [DOI] [PubMed] [Google Scholar]
- (34).Tong Y; Guo M Bacterial heme-transport proteins and their heme-coordination modes. Arch. Biochem. Biophys 2009, 481, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wren BW; Stabler RA; Das SS; Butcher PD; Mangan JA; Clarke JD; Casali N; Parish T; Stoker NG Characterization of a haemolysin from Mycobacterium tuberculosis with homology to a virulence factor of Serpulina hyodysenteriae. Microbiology 1998, 144 (Pt 5), 1205–1211. [DOI] [PubMed] [Google Scholar]
- (36).Andersen CB; Torvund-Jensen M; Nielsen MJ; de Oliveira CL; Hersleth HP; Andersen NH; Pedersen JS; Andersen GR; Moestrup SK Structure of the haptoglobin-haemoglobin complex. Nature 2012, 489, 456–459. [DOI] [PubMed] [Google Scholar]
- (37).Smith A; McCulloh RJ Hemopexin and haptoglobin: allies against heme toxicity from hemoglobin not contenders. Front. Physiol 2015, 6, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Paoli M; Anderson BF; Baker HM; Morgan WT; Smith A; Baker EN Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two betapropeller domains. Nat. Struct. Biol 1999, 6, 926–931. [DOI] [PubMed] [Google Scholar]
- (39).Bunn HF; Jandl JH Exchange of heme among hemoglobins and between hemoglobin and albumin. J. Biol. Chem 1968, 243, 465–475. [PubMed] [Google Scholar]
- (40).Ratledge C Iron, mycobacteria and tuberculosis. Tuberculosis 2004, 84, 110–130. [DOI] [PubMed] [Google Scholar]
- (41).Luo M; Fadeev EA; Groves JT Mycobactin-mediated iron acquisition within macrophages. Nat. Chem. Biol. 2005, 1, 149–153. [DOI] [PubMed] [Google Scholar]
- (42).Olakanmi O; Schlesinger LS; Ahmed A; Britigan BE Intraphagosomal Mycobacterium tuberculosis acquires iron from both extracellular transferrin and intracellular iron pools. Impact of interferon-gamma and hemochromatosis. J. Biol. Chem 2002, 277, 49727–49734. [DOI] [PubMed] [Google Scholar]
- (43).Wagner D; Maser J; Lai B; Cai Z; Barry CE 3rd; Honer Zu Bentrup K; Russell DG; Bermudez LE Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J. Immunol 2005, 174, 1491–1500. [DOI] [PubMed] [Google Scholar]
- (44).Soe-Lin S; Apte SS; Andriopoulos B Jr.; Andrews MC; Schranzhofer M; Kahawita T; Garcia-Santos D; Ponka P Nramp1 promotes efficient macrophage recycling of iron following erythrophagocytosis in vivo. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 5960–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Yuan X; Rietzschel N; Kwon H; Walter Nuno AB; Hanna DA; Phillips JD; Raven EL; Reddi AR; Hamza I Regulation of intracellular heme trafficking revealed by subcellular reporters. Proc. Natl. Acad. Sci. U. S. A 2016, 113, E5144–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Saha R; Saha N; Donofrio RS; Bestervelt LL Microbial siderophores: a mini review. J. Basic Microbiol 2013, 53, 303–317. [DOI] [PubMed] [Google Scholar]
- (47).Miethke M; Marahiel MA Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev 2007, 71, 413–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wilson BR; Bogdan AR; Miyazawa M; Hashimoto K; Tsuji Y Siderophores in Iron Metabolism: From Mechanism to Therapy Potential. Trends Mol. Med 2016, 22, 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Zheng T; Nolan EM Siderophore-based detection of Fe(III) and microbial pathogens. Metallomics 2012, 4, 866–880. [DOI] [PubMed] [Google Scholar]
- (50).De Voss JJ; Rutter K; Schroeder BG; Barry CE 3rd. Iron acquisition and metabolism by mycobacteria. J. Bacteriol 1999, 181, 4443–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Gobin J; Moore CH; Reeve JR Jr.; Wong DK; Gibson BW; Horwitz MA Iron acquisition by Mycobacterium tuberculosis: isolation and characterization of a family of iron-binding exochelins. Proc. Natl. Acad. Sci. U. S. A 1995, 92, 5189–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Wong DK; Gobin J; Horwitz MA; Gibson BW Characterization of exochelins of Mycobacterium avium: evidence for saturated and unsaturated and for acid and ester forms. J. Bacteriol 1996, 178, 6394–6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Sharman GJ; Williams DH; Ewing DF; Ratledge C Isolation, purification and structure of exochelin MS, the extracellular siderophore from Mycobacterium smegmatis. Biochem. J 1995, 305 (Pt 1), 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Crosa JH; Walsh CT Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev 2002, 66, 223–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Carroll CS; Moore MM Ironing out siderophore biosynthesis: a review of non- ribosomal peptide synthetase (NRPS)-independent siderophore synthetases. Crit. Rev. Biochem. Mol. Biol 2018, 53, 356–381. [DOI] [PubMed] [Google Scholar]
- (56).Challis GL A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. ChemBioChem 2005, 6, 601–611. [DOI] [PubMed] [Google Scholar]
- (57).Meneghetti F; Villa S; Gelain A; Barlocco D; Chiarelli LR; Pasca MR; Costantino L Iron Acquisition Pathways as Targets for Antitubercular Drugs. Curr. Med. Chem 2016, 23, 4009–4026. [DOI] [PubMed] [Google Scholar]
- (58).Sritharan M Iron Homeostasis in Mycobacterium tuberculosis: Mechanistic Insights into Siderophore-Mediated Iron Uptake. J. Bacteriol 2016, 198, 2399–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Krithika R; Marathe U; Saxena P; Ansari MZ; Mohanty D; Gokhale RS A genetic locus required for iron acquisition in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 2069–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Quadri LE; Sello J; Keating TA; Weinreb PH; Walsh CT Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol 1998, 5, 631–645. [DOI] [PubMed] [Google Scholar]
- (61).Chavadi SS; Stirrett KL; Edupuganti UR; Vergnolle O; Sadhanandan G; Marchiano E; Martin C; Qiu WG; Soll CE; Quadri LE Mutational and phylogenetic analyses of the mycobacterial mbt gene cluster. J. Bacteriol 2011, 193, 5905–5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).McMahon MD; Rush JS; Thomas MG Analyses of MbtB, MbtE, and MbtF suggest revisions to the mycobactin biosynthesis pathway in Mycobacterium tuberculosis. J. Bacteriol 2012, 194, 2809–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Gold B; Rodriguez GM; Marras SA; Pentecost M; Smith I The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol. Microbiol 2001, 42, 851–865. [DOI] [PubMed] [Google Scholar]
- (64).Schmitt MP; Predich M; Doukhan L; Smith I; Holmes RK Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect. Immun 1995, 63, 4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Prakash P; Yellaboina S; Ranjan A; Hasnain SE Computational prediction and experimental verification of novel IdeR binding sites in the upstream sequences of Mycobacterium tuberculosis open reading frames. Bioinformatics 2005, 21, 2161–2166. [DOI] [PubMed] [Google Scholar]
- (66).Pandey R; Rodriguez GM IdeR is required for iron homeostasis and virulence in Mycobacterium tuberculosis. Mol. Microbiol 2014, 91, 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Raymond KN; Dertz EA; Kim SS Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 3584–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Cao J; Woodhall MR; Alvarez J; Cartron ML; Andrews SC EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol. Microbiol. 2007, 65, 857–875. [DOI] [PubMed] [Google Scholar]
- (69).Cartron ML; Maddocks S; Gillingham P; Craven CJ; Andrews SC Feo-- transport of ferrous iron into bacteria. Biometals 2006, 19, 143–157. [DOI] [PubMed] [Google Scholar]
- (70).Bleuel C; Grosse C; Taudte N; Scherer J; Wesenberg D; Krauss GJ; Nies DH; Grass G TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J. Bacteriol 2005, 187, 6701–6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Horiyama T; Nishino K AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS One 2014, 9, e108642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Liu J; Rutz JM; Feix JB; Klebba PE Permeability properties of a large gated channel within the ferric enterobactin receptor, FepA. Proc. Natl. Acad. Sci. U. S. A 1993, 90, 10653–10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Zhou XH; van der Helm D; Adjimani J Purification of outer membrane iron transport receptors from Escherichia coli by fast protein liquid chromatography: FepA and FecA. Biometals 1993, 6, 25–35. [DOI] [PubMed] [Google Scholar]
- (74).Skare JT; Ahmer BM; Seachord CL; Darveau RP; Postle K Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem 1993, 268, 16302–16308. [PubMed] [Google Scholar]
- (75).Greenwood KT; Luke RK Enzymatic hydrolysis of enterochelin and its iron complex in Escherichia Coli K-12. Properties of enterochelin esterase. Biochim. Biophys. Acta 1978, 525, 209–218. [DOI] [PubMed] [Google Scholar]
- (76).Stephens DL; Choe MD; Earhart CF Escherichia coli periplasmic protein FepB binds ferrienterobactin. Microbiology 1995, 141 (Pt 7), 1647–1654. [DOI] [PubMed] [Google Scholar]
- (77).Pierce JR; Earhart CF Escherichia coli K-12 envelope proteins specifically required for ferrienterobactin uptake. J. Bacteriol 1986, 166, 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Shea CM; McIntosh MA Nucleotide sequence and genetic organization of the ferric enterobactin transport system: homology to other periplasmic binding protein-dependent systems in Escherichia coli. Mol. Microbiol 1991, 5, 1415–1428. [DOI] [PubMed] [Google Scholar]
- (79).Moeck GS; Coulton JW; Postle K Cell envelope signaling in Escherichia coli. Ligand binding to the ferrichrome-iron receptor fhua promotes interaction with the energy-transducing protein TonB. J. Biol. Chem 1997, 272, 28391–28397. [DOI] [PubMed] [Google Scholar]
- (80).Miethke M; Hou J; Marahiel MA The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli. Biochemistry 2011, 50, 10951–10964. [DOI] [PubMed] [Google Scholar]
- (81).Dale SE; Sebulsky MT; Heinrichs DE Involvement of SirABC in iron- siderophore import in Staphylococcus aureus. J. Bacteriol 2004, 186, 8356–8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Grigg JC; Cooper JD; Cheung J; Heinrichs DE; Murphy ME The Staphylococcus aureus siderophore receptor HtsA undergoes localized conformational changes to enclose staphyloferrin A in an arginine-rich binding pocket. J. Biol. Chem 2010, 285, 11162–11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Wells RM; Jones CM; Xi Z; Speer A; Danilchanka O; Doornbos KS; Sun P; Wu F; Tian C; Niederweis M Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoSPathog. 2013, 9, e1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Siegrist MS; Bertozzi CR Mycobacterial lipid logic. Cell Host Microbe 2014, 15, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Tufariello JM; Chapman JR; Kerantzas CA; Wong KW; Vilcheze C; Jones CM; Cole LE; Tinaztepe E; Thompson V; Fenyo D et al. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc. Natl. Acad. Sci. U. S. A 2016, 113, E348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Domenech P; Reed MB; Barry CE 3rd. Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun 2005, 73, 3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Ruggerone P; Murakami S; Pos KM; Vargiu AV RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr. Top. Med. Chem 2013, 13, 3079–3100. [DOI] [PubMed] [Google Scholar]
- (88).Chalut C MmpL transporter-mediated export of cell-wall associated lipids and siderophores in mycobacteria. Tuberculosis 2016, 100, 32–45. [DOI] [PubMed] [Google Scholar]
- (89).Viljoen A; Dubois V; Girard-Misguich F; Blaise M; Herrmann JL; Kremer L The diverse family of MmpL transporters in mycobacteria: from regulation to antimicrobial developments. Mol. Microbiol 2017, 104, 889–904. [DOI] [PubMed] [Google Scholar]
- (90).Tullius MV; Harmston CA; Owens CP; Chim N; Morse RP; McMath LM; Iniguez A; Kimmey JM; Sawaya MR; Whitelegge JP et al. Discovery and characterization of a unique mycobacterial heme acquisition system. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 5051–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Lamichhane G; Tyagi S; Bishai WR Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect. Immun 2005, 73, 2533–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Chim N; Torres R; Liu Y; Capri J; Batot G; Whitelegge JP; Goulding CW The Structure and Interactions of Periplasmic Domains of Crucial MmpL Membrane Proteins from Mycobacterium tuberculosis. Chem. Biol 2015, 22, 1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Jones CM; Wells RM; Madduri AV; Renfrow MB; Ratledge C; Moody DB; Niederweis M Self-poisoning of Mycobacterium tuberculosis by interrupting siderophore recycling. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 1945–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Deshayes C; Bach H; Euphrasie D; Attarian R; Coureuil M; Sougakoff W; Laval F; Av-Gay Y; Daffe M; Etienne G et al. MmpS4 promotes glycopeptidolipids biosynthesis and export in Mycobacterium smegmatis. Mol. Microbiol 2010, 78, 989–1003. [DOI] [PubMed] [Google Scholar]
- (95).Gross R; Engelbrecht F; Braun V Genetic and biochemical characterization of the aerobactin synthesis operon on pColV. Mol. Gen. Genet 1984, 196, 74–80. [DOI] [PubMed] [Google Scholar]
- (96).Imperi F; Tiburzi F; Visca P Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 20440–20445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Rodriguez GM; Smith I Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J. Bacteriol 2006, 188, 424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Ryndak MB; Wang S; Smith I; Rodriguez GM The Mycobacterium tuberculosis high-affinity iron importer, IrtA, contains an FAD-binding domain. J. Bacteriol 2010, 192, 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Wagner D; Sangari FJ; Parker A; Bermudez LE fecB, a gene potentially involved in iron transport in Mycobacterium avium, is not induced within macrophages. FEMS Microbiol. Lett 2005, 247, 185–191. [DOI] [PubMed] [Google Scholar]
- (100).Chu BC; Vogel HJ A structural and functional analysis of type III periplasmic and substrate binding proteins: their role in bacterial siderophore and heme transport. Biol. Chem 2011, 392, 39–52. [DOI] [PubMed] [Google Scholar]
- (101).Clarke TE; Ku SY; Dougan DR; Vogel HJ; Tari LW The structure of the ferric siderophore binding protein FhuD complexed with gallichrome. Nat. Struct. Biol 2000, 7, 287–291. [DOI] [PubMed] [Google Scholar]
- (102).Chu BC; Otten R; Krewulak KD; Mulder FA; Vogel HJ The solution structure, binding properties, and dynamics of the bacterial siderophore-binding protein FepB. J. Biol. Chem 2014, 289, 29219–29234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Beasley FC; Vines ED; Grigg JC; Zheng Q; Liu S; Lajoie GA; Murphy ME; Heinrichs DE Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol. Microbiol 2009, 72, 947–963. [DOI] [PubMed] [Google Scholar]
- (104).Zhu H; Xie G; Liu M; Olson JS; Fabian M; Dooley DM; Lei B Pathway for Heme Uptake from Human Methemoglobin by the Iron-regulated Surface Determinants System of Staphylococcus aureus. J. Biol. Chem 2008, 283, 18450–18460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Brillet K; Ruffenach F; Adams H; Journet L; Gasser V; Hoegy F; Guillon L; Hannauer M; Page A; Schalk IJ An ABC transporter with two periplasmic binding proteins involved in iron acquisition in Pseudomonas aeruginosa. ACS Chem. Biol 2012, 7, 2036–2045. [DOI] [PubMed] [Google Scholar]
- (106).Ganne G; Brillet K; Basta B; Roche B; Hoegy F; Gasser V; Schalk IJ Iron Release from the Siderophore Pyoverdine in Pseudomonas aeruginosa Involves Three New Actors: FpvC, FpvG, and FpvH. ACS Chem. Biol 2017, 12, 1056–1065. [DOI] [PubMed] [Google Scholar]
- (107).Schalk IJ; Abdallah MA; Pattus F Recycling of pyoverdin on the FpvA receptor after ferric pyoverdin uptake and dissociation in Pseudomonas aeruginosa. Biochemistry 2002, 41, 1663–1671. [DOI] [PubMed] [Google Scholar]
- (108).Yeterian E; Martin LW; Lamont IL; Schalk IJ An efflux pump is required for siderophore recycling by Pseudomonas aeruginosa. Environ. Microbiol. Rep 2010, 2, 412–418. [DOI] [PubMed] [Google Scholar]
- (109).Rodriguez GM; Voskuil MI; Gold B; Schoolnik GK; Smith I ideR, An essential gene in mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun 2002, 70, 3371–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Farhana A; Kumar S; Rathore SS; Ghosh PC; Ehtesham NZ; Tyagi AK; Hasnain SE Mechanistic insights into a novel exporter-importer system of Mycobacterium tuberculosis unravel its role in trafficking of iron. PLoS One 2008, 3, e2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Fetherston JD; Bertolino VJ; Perry RD YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol 1999, 32, 289–299. [DOI] [PubMed] [Google Scholar]
- (112).Schroder I; Johnson E; de Vries S Microbial ferric iron reductases. FEMS. Microbiol. Rev 2003, 27, 427–447. [DOI] [PubMed] [Google Scholar]
- (113).Wyckoff EE; Allred BE; Raymond KN; Payne SM Catechol Siderophore Transport by Vibrio cholerae. J. Bacteriol 2015, 197, 2840–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Srivastava V; Rouanet C; Srivastava R; Ramalingam B; Locht C; Srivastava BS Macrophage-specific Mycobacterium tuberculosis genes: identification by green fluorescent protein and kanamycin resistance selection. Microbiology 2007, 153, 659–666. [DOI] [PubMed] [Google Scholar]
- (115).Santhanagopalan SM; Rodriguez GM Examining the role of Rv2895c (ViuB) in iron acquisition in Mycobacterium tuberculosis. Tuberculosis 2012, 92, 60–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Arnoux P; Haser R; Izadi N; Lecroisey A; Delepierre M; Wandersman C; Czjzek M The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat. Struct. Biol 1999, 6, 516–520. [DOI] [PubMed] [Google Scholar]
- (117).Cescau S; Cwerman H; Letoffe S; Delepelaire P; Wandersman C; Biville F Heme acquisition by hemophores. Biometals 2007, 20, 603–613. [DOI] [PubMed] [Google Scholar]
- (118).Clarke TE; Tari LW; Vogel HJ Structural biology of bacterial iron uptake systems. Curr. Top. Med. Chem 2001, 1, 7–30. [DOI] [PubMed] [Google Scholar]
- (119).Izadi N; Henry Y; Haladjian J; Goldberg M, E.; Wandersman, C.; Delepierre, M.; Lecroisey, A. Purification and characterization of an extracellular heme-binding protein, HasA, involved in heme iron acquisition. Biochemistry 1997, 36, 7050–7057. [DOI] [PubMed] [Google Scholar]
- (120).Simpson W; Olczak T; Genco CA Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J. Bacteriol 2000, 182, 5737–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Whitby PW; Seale TW; VanWagoner TM; Morton DJ; Stull TL The iron/heme regulated genes of Haemophilus influenzae: comparative transcriptional profiling as a tool to define the species core modulon. BMC Genomics 2009, 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Wong JC; Patel R; Kendall D; Whitby PW; Smith A; Holland J; Williams P Affinity, conservation, and surface exposure of hemopexin-binding proteins in Haemophilus influenzae. Infect. Immun 1995, 63, 2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Contreras H; Chim N; Credali A; Goulding CW Heme uptake in bacterial pathogens. Curr. Opin. Chem. Biol. 2014, 19, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Smith AD; Modi AR; Sun S; Dawson JH; Wilks A Spectroscopic Determination of Distinct Heme Ligands in Outer-Membrane Receptors PhuR and HasR of Pseudomonas aeruginosa. Biochemistry 2015, 54, 2601–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Skaar EP; Humayun M; Bae T; DeBord KL; Schneewind O Iron-source preference of Staphylococcus aureus infections. Science (New York, N.Y.) 2004, 305, 1626–1628. [DOI] [PubMed] [Google Scholar]
- (126).Grigg JC; Vermeiren CL; Heinrichs DE; Murphy MEP Haem recognition by a Staphylococcus aureus NEAT domain. Mol. Microbiol. 2007, 63, 139–149. [DOI] [PubMed] [Google Scholar]
- (127).Lei B; Smoot LM; Menning HM; Voyich JM; Kala SV; Deleo FR; Reid SD; Musser JM Identification and Characterization of a Novel Heme-Associated Cell Surface Protein Made by Streptococcus pyogenes. Infect. Immun. 2002, 70, 4494–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Ekworomadu MT; Poor CB; Owens CP; Balderas MA; Fabian M; Olson JS; Murphy F; Balkabasi E; Honsa ES; He C et al. Differential function of lip residues in the mechanism and biology of an anthrax hemophore. PLoS Pathog. 2012, 8, e1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Gat O; Zaide G; Inbar I; Grosfeld H; Chitlaru T; Levy H; Shafferman A Characterization of Bacillus anthracis iron-regulated surface determinant (Isd) proteins containing NEAT domains. Mol. Microbiol 2008, 70, 983–999. [DOI] [PubMed] [Google Scholar]
- (130).Sheldon JR; Heinrichs DE Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS. Microbiol. Rev 2015, 39, 592–630. [DOI] [PubMed] [Google Scholar]
- (131).Nobles CL; Maresso AW The theft of host heme by Gram-positive pathogenic bacteria. Metallomics 2011, 3, 788–796. [DOI] [PubMed] [Google Scholar]
- (132).Grigg JC; Ukpabi G; Gaudin CFM; Murphy MEP Structural biology of heme binding in the Staphylococcus aureus Isd system. J. Biol. Inorg. Chem 2010, 104, 341–348. [DOI] [PubMed] [Google Scholar]
- (133).Honsa ES; Fabian M; Cardenas AM; Olson JS; Maresso AW The five neariron transporter (NEAT) domain anthrax hemophore, IsdX2, scavenges heme from hemoglobin and transfers heme to the surface protein IsdC. J. Biol. Chem 2011, 286, 33652–33660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Honsa ES; Maresso AW; Highlander SK Molecular and evolutionary analysis of NEAr-iron Transporter (NEAT) domains. PLoS One 2014, 9, e104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Krishna Kumar K; Jacques DA; Pishchany G; Caradoc-Davies T; Spirig T; Malmirchegini GR; Langley DB; Dickson CF; Mackay JP; Clubb RT et al. Structural Basis for Hemoglobin Capture by Staphylococcus aureus Cell-surface Protein, IsdH. J. Biol. Chem 2011, 286, 38439–38447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Torres VJ; Pishchany G; Humayun M; Schneewind O; Skaar EP Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol 2006, 188, 8421–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Liu M; Tanaka WN; Zhu H; Xie G; Dooley DM; Lei B Direct Hemin Transfer from IsdA to IsdC in the Iron-regulated Surface Determinant (Isd) Heme Acquisition System of Staphylococcus aureus. J. Biol. Chem 2008, 283, 6668–6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).Nygaard TK; Blouin GC; Liu M; Fukumura M; Olson JS; Fabian M; Dooley DM; Lei B The mechanism of direct heme transfer from the streptococcal cell surface protein Shp to HtsA of the HtsABC transporter. J. Biol. Chem 2006, 281, 20761–20771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Yukl ET; Jepkorir G; Alontaga AY; Pautsch L; Rodriguez JC; Rivera M; Moenne-Loccoz P Kinetic and spectroscopic studies of hemin acquisition in the hemophore HasAp from Pseudomonas aeruginosa. Biochemistry 2010, 49, 6646–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Deniau C; Gilli R; Izadi-Pruneyre N; Letoffe S; Delepierre M; Wandersman C; Briand C; Lecroisey A Thermodynamics of heme binding to the HasA(SM) hemophore: effect of mutations at three key residues for heme uptake. Biochemistry 2003, 42, 10627–10633. [DOI] [PubMed] [Google Scholar]
- (141).Gao JL; Nguyen KA; Hunter N Characterization of a hemophore-like protein from Porphyromonas gingivalis. J. Biol. Chem 2010, 285, 40028–40038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Izadi-Pruneyre N; Huche F; Lukat-Rodgers GS; Lecroisey A; Gilli R; Rodgers KR; Wandersman C; Delepelaire P The heme transfer from the soluble HasA hemophore to its membrane-bound receptor HasR is driven by protein-protein interaction from a high to a lower affinity binding site. J. Biol. Chem 2006, 281, 25541–25550. [DOI] [PubMed] [Google Scholar]
- (143).Liu X; Olczak T; Guo HC; Dixon DW; Genco CA Identification of amino acid residues involved in heme binding and hemoprotein utilization in the Porphyromonas gingivalis heme receptor HmuR. Infect. Immun 2006, 74, 1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Olczak T; Dixon DW; Genco CA Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J. Bacteriol 2001, 183, 5599–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Mattle D; Zeltina A; Woo J-S; Goetz BA; Locher KP Two Stacked Heme Molecules in the Binding Pocket of the Periplasmic Heme-Binding Protein HmuT from Yersinia pestis. J. Mol. Biol 2010, 404, 220–231. [DOI] [PubMed] [Google Scholar]
- (146).Mattle D; Zeltina A; Woo JS; Goetz BA; Locher KP Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis. J. Mol. Biol 2010, 404, 220–231. [DOI] [PubMed] [Google Scholar]
- (147).Mitra A; Speer A; Lin K; Ehrt S; Niederweis M PPE Surface Proteins Are Required for Heme Utilization by Mycobacterium tuberculosis. MBio 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Tullius MV; Harth G; Maslesa-Galic S; Dillon BJ; Horwitz MA A Replication- Limited Recombinant Mycobacterium bovis BCG vaccine against tuberculosis designed for human immunodeficiency virus-positive persons is safer and more efficacious than BCG. Infect. Immun 2008, 76, 5200–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Stojiljkovic I; Kumar V; Srinivasan N Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol. Microbiol 1999, 31, 429–442. [DOI] [PubMed] [Google Scholar]
- (150).Sassetti CM; Boyd DH; Rubin EJ Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol 2003, 48, 77–84. [DOI] [PubMed] [Google Scholar]
- (151).Owens CP; Du J; Dawson JH; Goulding CW Characterization of heme ligation properties of Rv0203, a secreted heme binding protein involved in Mycobacterium tuberculosis heme uptake. Biochemistry 2012, 51, 1518–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Wojtowicz H; Wojaczynski J; Olczak M; Kroliczewski J; Latos-Grazynski L; Olczak T Heme environment in HmuY, the heme-binding protein of Porphyromonas gingivalis. Biochem. Biophys. Res. Commun 2009, 383, 178–182. [DOI] [PubMed] [Google Scholar]
- (153).Owens CP; Chim N; Graves AB; Harmston CA; Iniguez A; Contreras H; Liptak MD; Goulding CW The Mycobacterium tuberculosis secreted protein Rv0203 transfers heme to membrane proteins MmpL3 and MmpL11. J. Biol. Chem 2013, 288, 21714–21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Abdalla MY; Ahmad IM; Switzer B; Britigan BE Induction of heme oxygenase- 1 contributes to survival of Mycobacterium abscessus in human macrophages-like THP-1 cells. Redox Biol. 2015, 4, 328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Grzegorzewicz AE; Pham H; Gundi VA; Scherman MS; North EJ; Hess T; Jones V; Gruppo V; Born SE; Kordulakova J et al. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol 2012, 8, 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Pacheco SA; Hsu FF; Powers KM; Purdy GE MmpL11 protein transports mycolic acid-containing lipids to the mycobacterial cell wall and contributes to biofilm formation in Mycobacterium smegmatis. J. Biol. Chem 2013, 288, 24213–24222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (157).Varela C; Rittmann D; Singh A; Krumbach K; Bhatt K; Eggeling L; Besra GS; Bhatt A MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chem. Biol 2012, 19, 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).Xu Z; Meshcheryakov VA; Poce G; Chng SS MmpL3 is the flippase for mycolic acids in mycobacteria. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 7993–7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (159).Belardinelli JM; Yazidi A; Yang L; Fabre L; Li W; Jacques B; Angala SK; Rouiller I; Zgurskaya HI; Sygusch J et al. Structure-Function Profile of MmpL3, the Essential Mycolic Acid Transporter from Mycobacterium tuberculosis. ACS Infect. Dis 2016, 2, 702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (160).Brennan MJ The Enigmatic PE/PPE Multigene Family of Mycobacteria and Tuberculosis Vaccination. Infect. Immun 2017, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Fishbein S; van Wyk N; Warren RM; Sampson SL Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol. Microbiol 2015, 96, 901–916. [DOI] [PubMed] [Google Scholar]
- (162).Ekiert DC; Cox JS Structure of a PE-PPE-EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 14758–14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (163).Fournier C; Smith A; Delepelaire P Haem release from haemopexin by HxuA allows Haemophilus influenzae to escape host nutritional immunity. Mol. Microbiol 2011, 80, 133–148. [DOI] [PubMed] [Google Scholar]
- (164).Ho WW; Li H; Eakanunkul S; Tong Y; Wilks A; Guo M; Poulos TL Holo- and Apo-bound Structures of Bacterial Periplasmic Heme-binding Proteins. J. Biol. Chem 2007, 282, 35796–35802. [DOI] [PubMed] [Google Scholar]
- (165).Okuda S; Tokuda H Lipoprotein sorting in bacteria. Annu. Rev. Microbiol 2011, 65, 239–259. [DOI] [PubMed] [Google Scholar]
- (166).Sandhu P; Akhter Y Evolution of structural fitness and multifunctional aspects of mycobacterial RND family transporters. Arch. Microbiol. 2018, 200, 19–31. [DOI] [PubMed] [Google Scholar]
- (167).Tiedemann MT; Stillman MJ Heme binding to the IsdE(M78A; H229A) double mutant: challenging unidirectional heme transfer in the iron-regulated surface determinant protein heme transfer pathway of Staphylococcus aureus. J. Biol. Inorg. Chem 2012, 17, 995–1007. [DOI] [PubMed] [Google Scholar]
- (168).Zambolin S; Clantin B; Chami M; Hoos S; Haouz A; Villeret V; Delepelaire P Structural basis for haem piracy from host haemopexin by Haemophilus influenzae. Nat. Commun 2016, 7, 11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Tenhunen R; Marver HS; Schmid R Microsomal heme oxygenase. Characterization of the enzyme. J. Biol. Chem 1969, 244, 6388–6394. [PubMed] [Google Scholar]
- (170).Ratliff M; Zhu W; Deshmukh R; Wilks A; Stojiljkovic I Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol 2001, 183, 6394–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (171).Wilks A; Schmitt MP Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J. Biol. Chem 1998, 273, 837–841. [DOI] [PubMed] [Google Scholar]
- (172).Zhu W; Wilks A; Stojiljkovic I Degradation of heme in gram-negative bacteria: the product of the hemO gene of Neisseriae is a heme oxygenase. J. Bacteriol 2000, 182, 6783–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Wilks A; Ikeda-Saito M Heme utilization by pathogenic bacteria: not all pathways lead to biliverdin. Acc. Chem. Res. 2014, 47, 2291–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (174).Schuller DJ; Wilks a.; Ortiz de Montellano PR; Poulos TL. Crystal structure of human heme oxygenase-1. Nat. Struct. Biol 1999, 6, 860–867. [DOI] [PubMed] [Google Scholar]
- (175).Wilks A Heme oxygenase: evolution, structure, and mechanism. Antioxid. Redox Signal 2002, 4, 603–614. [DOI] [PubMed] [Google Scholar]
- (176).Matsui T; Unno M; Ikeda-Saito M Heme oxygenase reveals its strategy for catalyzing three successive oxygenation reactions. Acc. Chem. Res 2010, 43, 240–247. [DOI] [PubMed] [Google Scholar]
- (177).Liu Y; Koenigs Lightning L; Huang H; Moenne-Loccoz P; Schuller DJ; Poulos TL; Loehr TM; Ortiz de Montellano PR Replacement of the distal glycine 139 transforms human heme oxygenase-1 into a peroxidase. J. Biol. Chem 2000, 275, 34501–34507. [DOI] [PubMed] [Google Scholar]
- (178).Lightning LK; Huang H; Moenne-Loccoz P; Loehr TM; Schuller DJ; Poulos TL; de Montellano PR Disruption of an active site hydrogen bond converts human heme oxygenase-1 into a peroxidase. J. Biol. Chem 2001, 276, 10612–10619. [DOI] [PubMed] [Google Scholar]
- (179).Davydov R; Kofman V; Fujii H; Yoshida T; Ikeda-Saito M; Hoffman BM Catalytic mechanism of heme oxygenase through EPR and ENDOR of cryoreduced oxy- heme oxygenase and its Asp 140 mutants. J. Am. Chem. Soc 2002, 124, 1798–1808. [DOI] [PubMed] [Google Scholar]
- (180).Wilks A; Torpey J; Ortiz de Montellano PR Heme oxygenase (HO-1). Evidence for electrophilic oxygen addition to the porphyrin ring in the formation of alpha-meso-hydroxyheme. J. Biol. Chem. 1994, 269, 29553–29556. [PubMed] [Google Scholar]
- (181).Davydov RM; Tadashi Y; Ikeda-Saito M; Hoffman BM Hydroperoxy-Heme Oxygenase Generated by Cryoreduction Catalyzes the Formation of a-meso-Hydroxyheme as Detected by EPR and ENDOR. J. Am. Chem. Soc 1999, 121, 10656–10657. [Google Scholar]
- (182).Yoshida T; Noguchi M; Kikuchi G The step of carbon monoxide liberation in the sequence of heme degradation catalyzed by the reconstituted microsomal heme oxygenase system. J. Biol. Chem 1982, 257, 9345–9348. [PubMed] [Google Scholar]
- (183).Lojek LJ; Farrand AJ; Wisecaver JH; Blaby-Haas CE; Michel BW; Merchant SS; Rokas A; Skaar EP Chlamydomonas reinhardtii LFO1 Is an IsdG Family Heme Oxygenase. mSphere 2017, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (184).Mazmanian SK; Skaar EP; Gaspar AH; Humayun M; Gornicki P; Jelenska J; Joachmiak A; Missiakas DM; Schneewind O Passage of heme-iron across the envelope of Staphylococcus aureus. Science 2003, 299, 906–909. [DOI] [PubMed] [Google Scholar]
- (185).Wu R; Skaar EP; Zhang R; Joachimiak G; Gornicki P; Schneewind O; Joachimiak A Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J. Biol. Chem 2005, 280, 2840–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (186).Lee WC; Reniere ML; Skaar EP; Murphy ME Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J. Biol. Chem 2008, 283, 30957–30963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (187).Reniere ML; Ukpabi GN; Harry SR; Stec DF; Krull R; Wright DW; Bachmann BO; Murphy ME; Skaar EP The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol. Microbiol 2010, 75, 1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Matsui T; Nambu S; Ono Y; Goulding CW; Tsumoto K; Ikeda-Saito M Heme degradation by Staphylococcus aureus IsdG and IsdI liberates formaldehyde rather than carbon monoxide. Biochemistry 2013, 52, 3025–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (189).Takayama SJ; Ukpabi G; Murphy ME; Mauk AG Electronic properties of the highly ruffled heme bound to the heme degrading enzyme IsdI. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 13071–13076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (190).Ukpabi G; Takayama SJ; Mauk AG; Murphy ME Inactivation of the heme degrading enzyme IsdI by an active site substitution that diminishes heme ruffling. J. Biol. Chem 2012, 287, 34179–34188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Takayama SJ; Loutet SA; Mauk AG; Murphy ME A Ferric-Peroxo Intermediate in the Oxidation of Heme by IsdI. Biochemistry 2015, 54, 2613–2621. [DOI] [PubMed] [Google Scholar]
- (192).Streit BR; Kant R; Tokmina-Lukaszewska M; Celis AI; Machovina MM; Skaar EP; Bothner B; DuBois JL Time-resolved Studies of IsdG Protein Identify Molecular Signposts along the Non-canonical Heme Oxygenase Pathway. J. Biol. Chem 2016, 291, 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Hannauer M; Arifin AJ; Heinrichs DE Involvement of reductases IruO and NtrA in iron acquisition by Staphylococcus aureus. Mol. Microbiol 2015, 96, 1192–1210. [DOI] [PubMed] [Google Scholar]
- (194).Kobylarz MJ; Heieis GA; Loutet SA; Murphy MEP Iron Uptake Oxidoreductase (IruO) Uses a Flavin Adenine Dinucleotide Semiquinone Intermediate for Iron-Siderophore Reduction. ACS Chem. Biol 2017, 12, 1778–1786. [DOI] [PubMed] [Google Scholar]
- (195).Loutet SA; Kobylarz MJ; Chau CH; Murphy ME IruO is a reductase for heme degradation by IsdI and IsdG proteins in Staphylococcus aureus. J. Biol. Chem 2013, 288, 25749–25759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Chim N; Iniguez A; Nguyen TQ; Goulding CW Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. J. Mol. Biol 2010, 395, 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (197).Graves AB; Morse RP; Chao A; Iniguez A; Goulding CW; Liptak MD Crystallographic and spectroscopic insights into heme degradation by Mycobacterium tuberculosis MhuD. Inorg. Chem 2014, 53, 5931–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Nambu S; Matsui T; Goulding CW; Takahashi S; Ikeda-Saito M A new way to degrade heme: the Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO. J. Biol. Chem 2013, 288, 10101–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (199).Graves AB; Graves MT; Liptak MD Measurement of Heme Ruffling Changes in MhuD Using UV-vis Spectroscopy. J. Phys. Chem. B 2016, 120, 3844–3853. [DOI] [PubMed] [Google Scholar]
- (200).Matsui T; Nambu S; Goulding CW; Takahashi S; Fujii H; Ikeda-Saito M Unique coupling of mono- and dioxygenase chemistries in a single active site promotes heme degradation. Proc. Natl. Acad. Sci. U. S. A 2016, DOI: 10.1073/pnas.152333311310.1073/pnas.1523333113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (201).Reddy PV; Puri RV; Khera A; Tyagi AK Iron storage proteins are essential for the survival and pathogenesis of Mycobacterium tuberculosis in THP-1 macrophages and the guinea pig model of infection. J. Bacteriol 2012, 194, 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (202).Fang Z; Sampson SL; Warren RM; Gey van Pittius NC; Newton-Foot M Iron acquisition strategies in mycobacteria. Tuberculosis 2015, 95, 123–130. [DOI] [PubMed] [Google Scholar]
- (203).Hu J; Miller MJ Total Synthesis of a Mycobactin S, a Siderophore and Growth Promoter of Mycobacterium Smegmatis, and Determination of its Growth Inhibitory Activity against Mycobacterium tuberculosis. J. Am. Chem. Soc 1997, 119, 3462–3468. [Google Scholar]
- (204).Nosten F; White NJ Artemisinin-based combination treatment of falciparum malaria. Am. J. Trop. Med Hyg 2007, 77, 181–192. [PubMed] [Google Scholar]
- (205).Wang J; Zhang CJ; Chia WN; Loh CC; Li Z; Lee YM; He Y; Yuan LX; Lim TK; Liu M et al. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat. Commun 2015, 6, 10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Miller MJ; Walz AJ; Zhu H; Wu C; Moraski G; Mollmann U; Tristani EM; Crumbliss AL; Ferdig MT; Checkley L et al. Design, synthesis, and study of a mycobactin-artemisinin conjugate that has selective and potent activity against tuberculosis and malaria. J. Am. Chem. Soc 2011, 133, 2076–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (207).Juarez-Hernandez RE; Franzblau SG; Miller MJ Syntheses of mycobactin analogs as potent and selective inhibitors of Mycobacterium tuberculosis. Org. Biomol. Chem 2012, 10, 7584–7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (208).Plaha DS; Rogers HJ Antibacterial effect of the scandium complex of enterochelin. Studies of the mechanism of action. Biochim. Biophys. Acta 1983, 760, 246–255. [DOI] [PubMed] [Google Scholar]
- (209).Ioerger TR; O’Malley T; Liao R; Guinn KM; Hickey MJ; Mohaideen N; Murphy KC; Boshoff HI; Mizrahi V; Rubin EJ et al. Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS One 2013, 8, e75245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (210).Dragset MS; Poce G; Alfonso S; Padilla-Benavides T; Ioerger TR; Kaneko T; Sacchettini JC; Biava M; Parish T; Arguello JM et al. A novel antimycobacterial compound acts as an intracellular iron chelator. Antimicrob Agents Chemother 2015, 59, 2256–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (211).Siegrist MS; Steigedal M; Ahmad R; Mehra A; Dragset MS; Schuster BM; Philips JA; Carr SA; Rubin EJ Mycobacterial Esx-3 requires multiple components for iron acquisition. MBio 2014, 5, e01073–01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (212).Siegrist MS; Unnikrishnan M; McConnell MJ; Borowsky M; Cheng T-Y; Siddiqi N; Fortune SM; Moody DB; Rubin EJ Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 18792–18797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (213).Xu W; DeJesus MA; Rucker N; Engelhart CA; Wright MG; Healy C; Lin K; Wang R; Park SW; Ioerger TR et al. Chemical Genetic Interaction Profiling Reveals Determinants of Intrinsic Antibiotic Resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2017, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (214).Mislin GL; Schalk IJ Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Metallomics 2014, 6, 408–420. [DOI] [PubMed] [Google Scholar]
- (215).Owens CP; Chim N; Goulding CW Insights on how the Mycobacterium tuberculosis heme uptake pathway can be used as a drug target. Future Med. Chem 2013, 5, 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (216).Cupello MP; Souza CF; Buchensky C; Soares JB; Laranja GA; Coelho MG; Cricco JA; Paes MC The heme uptake process in Trypanosoma cruzi epimastigotes is inhibited by heme analogues and by inhibitors of ABC transporters. Acta Trop. 2011, 120, 211–218. [DOI] [PubMed] [Google Scholar]
- (217).Tiedemann MT; Pinter TB; Stillman MJ Insight into blocking heme transfer by exploiting molecular interactions in the core Isd heme transporters IsdA-NEAT, IsdC-NEAT, and IsdE of Staphylococcus aureus. Metallomics 2012, 4, 751–760. [DOI] [PubMed] [Google Scholar]
- (218).Hijazi S; Visca P; Frangipani E Gallium-Protoporphyrin IX Inhibits Pseudomonas aeruginosa Growth by Targeting Cytochromes. Front. Cell. Infect. Microbiol 2017, 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (219).Abdalla MY; Switzer BL; Goss CH; Aitken ML; Singh PK; Britigan BE Gallium Compounds Exhibit Potential as New Therapeutic Agents against Mycobacterium abscessus. Antimicrob Agents Chemother 2015, 59, 4826–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (220).Costa DL; Namasivayam S; Amaral EP; Arora K; Chao A; Mittereder LR; Maiga M; Boshoff HI; Barry CE 3rd; Goulding CW et al. Pharmacological Inhibition of Host Heme Oxygenase-1 Suppresses Mycobacterium tuberculosis Infection In Vivo by a Mechanism Dependent on T Lymphocytes. MBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (221).La Rosa V; Poce G; Canseco JO; Buroni S; Pasca MR; Biava M; Raju RM; Porretta GC; Alfonso S; Battilocchio C et al. MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob Agents Chemother 2012, 56, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (222).Tahlan K; Wilson R; Kastrinsky DB; Arora K; Nair V; Fischer E; Barnes SW; Walker JR; Alland D; Barry CE, 3rdet al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2012, 56, 1797–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (223).Anzaldi LL; Skaar EP Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect. Immun 2010, 78, 4977–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (224).Skaar EP; Humayun M; Bae T; DeBord KL; Schneewind O Iron-source preference of Staphylococcus aureus infections. Science 2004, 305, 1626–1628. [DOI] [PubMed] [Google Scholar]
- (225).Hagan EC; Mobley HL Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol. Microbiol 2009, 77, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Olczak T; Sosicka P; Olczak M HmuY is an important virulence factor for Porphyromonas gingivalis growth in the heme-limited host environment and infection of macrophages. Biochem. Biophys. Res. Commun 2015, 467, 748–753. [DOI] [PubMed] [Google Scholar]