Abstract
Genome-wide association studies (GWAS) and fine-mapping efforts to date have identified more than 100 prostate cancer (PrCa)-susceptibility loci. We meta-analyzed genotype data from a custom high-density array of 46,939 PrCa cases and 27,910 controls of European ancestry with previously genotyped data of 32,255 PrCa cases and 33,202 controls of European ancestry. Our analysis identified 62 novel loci associated (P<5.0×10−8) with PrCa and one locus significantly associated with early-onset PrCa (≤55 years). Our findings include missense variants rs1800057 (odds ratio (OR) = 1.16; P = 8.2×10−9; G>C, p.Pro1054Arg) in ATM and rs2066827 (OR = 1.06; P = 2.3 × 10−9; T>G, p.Val109Gly) in CDKN1B. The combination of all loci captured 28.4% of the PrCa familial relative risk, and a polygenic risk score conferred an elevated PrCa risk for men in the ninetieth to ninety-ninth percentiles (relative risk = 2.69; 95% confidence interval (CI): 2.55–2.82) and first percentile (relative risk = 5.71; 95% CI: 5.04–6.48) risk stratum compared with the population average. These findings improve risk prediction, enhance fine-mapping, and provide insight into the underlying biology of PrCa1.
Although PrCa is the most common noncutaneous cancer among men in the Western world, and one in seven men will be diagnosed during their lifetime2, very few modifiable risk factors have been established3. Epidemiological studies have identified age, positive family history, and ancestry as the most prominent risk factors for PrCa4–7. PrCa incidence is highest among men of African ancestry, followed by men of European and Asian ancestries. These observations of ancestral differences in PrCa risk, in conjunction with studies demonstrating the influence of family history8,9, highlight the contribution of genetics to PrCa etiology10. Our previous work, using a multiplicative model, has estimated that more than 1,800 common SNPs independently contribute to PrCa risk among populations of European ancestry11. GWAS have reported more than 100 of these PrCa variants across multiancestral populations, most of which were identified in populations of European ancestry12–29.
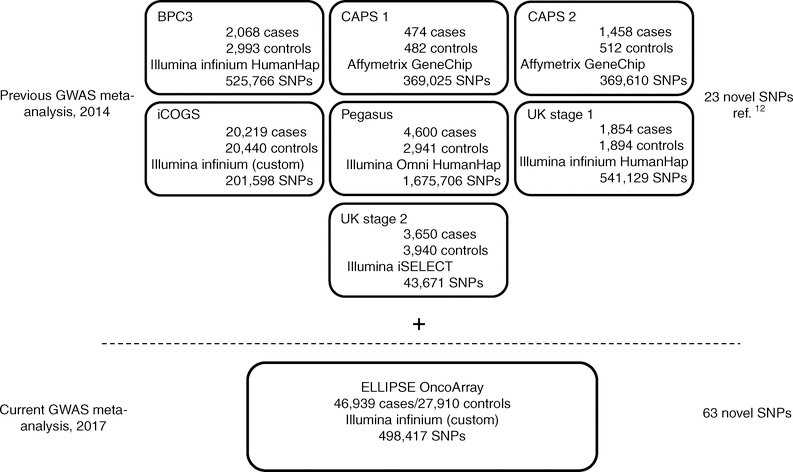
To facilitate additional discovery of PrCa genetic risk factors, we developed a custom high-density genotyping array, the OncoArray, including a 260,000-SNP backbone designed to adequately tag most common genetic variants (minor allele frequency (MAF) >5% in Europeans), and 310,000 SNPs from meta-analyses of five cancers (breast, colorectal, lung, ovarian, and prostate)30. Approximately 80,000 PrCa-specific markers derived from our previous multiancestral meta-analysis12 (including populations of European, African American, Japanese, and Latino ancestry), fine-mapping of known PrCa loci, and candidate SNPs nominated by study collaborators were included on the OncoArray. We assembled a new PrCa sample series from 52 studies to genotype with the OncoArray (Supplementary Tables 1 and 2). After application of rigorous quality control (QC) criteria and removal of overlapping samples from previous studies, our OncoArray sample yielded 46,939 PrCa cases and 27,910 controls without a known diagnosis of PrCa and of European ancestry for analysis (Methods and Supplementary Table 3). Genotypes were phased and imputed to the cosmopolitan panel of the 1000 Genomes Project (1KGP; June 2014 release) in SHAPEIT31 and IMPUTEv2 (ref.32) software (Methods and Supplementary Table 3). We performed a fixed-effects meta-analysis combining the summary statistics from our OncoArray analysis and seven previous PrCa GWAS or high-density SNP panels of European ancestry imputed to the 1KGP The final meta-analysis included 79,194 PrCa cases and 61,112 controls without a known diagnosis of PrCa (Fig. 1).
Fig. 1 |. ELLIPSE/PRACTICAL study overview of PrCa GWAS meta-analysis.
The top section describes the PrCa GWAS meta-analysis published in 2014, in which 23 novel variants were identified12. The current PrCa GWAS meta-analysis incorporated an additional 46,939 PrCa cases and 27,910 controls independent of the meta-analyses. The current meta-analysis discovered 62 novel variants associated with overall PrCa and 1 novel variant associated with early-onset PrCa.
We performed study- and consortia-specific meta-analyses to identify novel PrCa risk loci. We established a P-value threshold of 5.0 × 10−8 to determine genome-wide significance. Our large sample size enabled several stratified meta-analyses focusing on key clinical and biological parameters (Methods and Supplementary Tables 4 and 5). All analyses used a likelihood-ratio test to minimize bias from rare variants, and a logistic-regression framework was used for all analyses, except for the Gleason score, for which linear regression was used. The genotype doses were incorporated in an allelic genetic model. The average λ1000, an inflation statistic calibrated to a sample size of 1,000 cases and 1,000 controls33, across the eight GWAS studies was 1.02 (range 0.98–1.09) and 1.00 for the overall meta-analysis (Supplementary Table 6). Our novel findings excluded variants within defined fine-mapped regions of previously reported PrCa risk loci (Supplementary Table 7).
After the exclusion of all known susceptibility regions (fine-mapping coordinates provided in Supplementary Table 7 and Supplementary Note), we identified 64 loci associated with overall PrCa susceptibility and 1 locus associated with early-onset PrCa (P < 5.0 × 10−8) in the meta-analysis (Supplementary Fig. 1), of which 53 were imputed, and 12 were genotyped with the OncoArray. The cluster plots for the genotyped makers are presented in Supplementary Fig. 2. Although most of the imputed markers were of high quality, with an average imputed r2 >0.80 for 61 of the 65 loci across all contributing GWAS (Supplementary Table 8), we closely examined four variants with a poor imputation quality score (r2 <0.80) in the OncoArray samples by inspecting linkage disequilibrium (LD) plots including only genotyped SNPs from the OncoArray and performing an imputation QC assessment (Methods). After reviewing the LD plots and the imputation QC, we determined that loci rs6602880 and rs144166867 were probably false positives due to imputation artifacts (Supplementary Fig. 3 and Supplementary Table 9). Overall, we identified 62 novel loci associated with overall PrCa risk and one novel locus associated with early-onset PrCa (Table 1). The consortia-specific associations were consistent across the eight contributing GWAS studies (Supplementary Table 10).
Table 1 |.
Prostate cancer OncoArray and GWAS meta-analysis for 63 novel regions
| SNP | Reference RAFa | Band | Position | Nearest gene | Allelesb | RAF | OR | 95% CI | Pc |
|---|---|---|---|---|---|---|---|---|---|
| Novel loci associated with overall prostate cancer | |||||||||
| rs56391074 | 0.329 | 1p22.3 | 88210715 | RP11–60A14.1 | AT/A | 0.38 | 1.05 | 1.03–1.06 | 1.7 × 10−8 |
| rs34579442 | 0.316 | 1q21.3 | 153899900 | DENND4B | C/CT | 0.34 | 1.07 | 1.05–1.09 | 4.5 × 10−14 |
| rs62106670 | 0.400 | 2p25.1 | 8597123 | AC011747.3 | T/C | 0.38 | 1.05 | 1.04–1.07 | 7.1 × 10−9 |
| rs74702681 | 0.024 | 2p14 | 66652885 | MEIS1-AS3 | T/C | 0.02 | 1.17 | 1.11–1.23 | 2.0 × 10−9 |
| rs11691517 | 0.750 | 2q13 | 111893096 | BCL2L11 | T/G | 0.74 | 1.07 | 1.05–1.08 | 3.5 × 10−12 |
| rs34925593 | 0.481 | 2q31.1 | 174234547 | CDCA7 | C/T | 0.48 | 1.05 | 1.03–1.07 | 2.8 × 10−8 |
| rs59308963 | 0.726 | 2q33.1 | 202123479 | CASP8 | T/TATTCTGTC | 0.73 | 1.05 | 1.03–1.07 | 2.4 × 10−8 |
| rs1283104 | 0.407 | 3q13.12 | 106962521 | DUBR | G/C | 0.38 | 1.05 | 1.03–1.07 | 8.8 × 10−9 |
| rs182314334 | 0.888 | 3q25.1 | 152004202 | MBNL1 | T/C | 0.90 | 1.09 | 1.06–1.12 | 4.1 × 10−11 |
| rs142436749 | 0.012 | 3q26.2 | 169093100 | MECOM | G/A | 0.01 | 1.25 | 1.16–1.34 | 4.7 × 10−9 |
| rs10793821 | 0.580 | 5q31.1 | 133836209 | RNU6–456P | T/C | 0.57 | 1.05 | 1.04–1.07 | 5.4 × 10−11 |
| rs76551843 | 0.991 | 5q35.1 | 169172133 | DOCK2 | A/G | 0.99 | 1.31 | 1.19–1.44 | 1.7 × 10−8 |
| rs4976790 | 0.096 | 5q35.3 | 177968915 | COL23A1 | T/G | 0.11 | 1.08 | 1.05–1.10 | 6.7 × 10−9 |
| rs12665339 | 0.148 | 6p21.33 | 30601232 | ATAT1 | G/A | 0.17 | 1.06 | 1.04–1.08 | 5.6 × 10−9 |
| rs9296068 | 0.645 | 6p21.32 | 32988695 | HLA-DOA | T/G | 0.65 | 1.05 | 1.03–1.07 | 1.3 × 10−8 |
| rs9469899 | 0.356 | 6p21.31 | 34793124 | UHRF1BP1 | A/G | 0.36 | 1.05 | 1.03–1.07 | 5.3 × 10−9 |
| rs4711748 | 0.232 | 6p21.1 | 43694598 | RP1–261G23.5 | T/C | 0.23 | 1.05 | 1.03–1.07 | 3.4 × 10−8 |
| rs527510716 | 0.251 | 7p22.3 | 1944537 | MAD1L1 | C/G | 0.24 | 1.06 | 1.04–1.08 | 4.9 × 10−8 |
| rs11452686 | 0.567 | 7p21.1 | 20414110 | ITGB8 | T/TA | 0.56 | 1.05 | 1.03–1.07 | 7.8 × 10−9 |
| rs17621345 | 0.758 | 7p14.1 | 40875192 | SUGCT | A/C | 0.74 | 1.07 | 1.05–1.09 | 6.7 × 10−14 |
| rs1048169 | 0.367 | 9p22.1 | 19055965 | HAUS6 | C/T | 0.38 | 1.06 | 1.05–1.08 | 6.5 × 10−14 |
| rs10122495 | 0.296 | 9p13.3 | 34049779 | RN7SKP114 | T/A | 0.31 | 1.05 | 1.03–1.07 | 1.3 × 10−8 |
| rs1182 | 0.258 | 9q34.11 | 132576060 | TOR1A | A/C | 0.22 | 1.06 | 1.04–1.08 | 1.1 × 10−9 |
| rs141536087 | 0.166 | 10p15.3 | 854691 | LARP4B | GCGCA/G | 0.15 | 1.08 | 1.06–1.11 | 9.0 × 10−13 |
| rs1935581 | 0.605 | 10q23.31 | 90195149 | RNLS | C/T | 0.63 | 1.05 | 1.03–1.07 | 6.5 × 10−9 |
| rs7094871 | 0.540 | 10q25.2 | 114712154 | TCF7L2 | G/C | 0.54 | 1.04 | 1.03–1.06 | 4.8 × 10−8 |
| rs1881502 | 0.193 | 11p15.5 | 1507512 | MOB2 | T/C | 0.19 | 1.06 | 1.04–1.08 | 7.4 × 10−9 |
| rs61890184d | 0.088 | 11p15.4 | 7547587 | PPFIBP2 | A/G | 0.12 | 1.07 | 1.05–1.10 | 6.6 × 10−9 |
| rs547171081 | 0.468 | 11p11.2 | 47421962 | RP11–750H9.5 | CGG/C | 0.47 | 1.05 | 1.03–1.07 | 3.4 × 10−8 |
| rs2277283 | 0.300 | 11q12.3 | 61908440 | INCENP | C/T | 0.31 | 1.06 | 1.04–1.08 | 3.0 × 10−10 |
| rs12785905 | 0.051 | 11q13.2 | 66951965 | KDM2A | C/G | 0.05 | 1.12 | 1.08–1.17 | 7.8 × 10−9 |
| rs11290954 | 0.688 | 11q13.5 | 76260543 | C11orf30 | AC/A | 0.68 | 1.06 | 1.05–1.08 | 7.4 × 10−13 |
| rs1800057 | 0.031 | 11q22.3 | 108143456 | ATM | G/C | 0.02 | 1.16 | 1.10–1.22 | 8.1 × 10−9 |
| rs138466039 | 0.009 | 11q24.2 | 125054793 | PKNOX2 | T/C | 0.01 | 1.32 | 1.22–1.44 | 2.0 × 10−11 |
| rs878987 | 0.143 | 11q25 | 134266372 | B3GAT1 | G/A | 0.15 | 1.07 | 1.04–1.09 | 4.8 × 10−8 |
| rs2066827 | 0.757 | 12p13.1 | 12871099 | CDKN1B | T/G | 0.76 | 1.06 | 1.04–1.08 | 2.3 × 10−9 |
| rs10845938 | 0.554 | 12p13.1 | 14416918 | RNU6–491P | G/A | 0.55 | 1.06 | 1.04–1.08 | 9.8 × 10−13 |
| rs7968403 | 0.655 | 12q14.2 | 65012824 | RASSF3 | T/C | 0.64 | 1.06 | 1.04–1.08 | 3.4 × 10−12 |
| rs5799921 | 0.697 | 12q21.33 | 90160530 | RNU6–148P | GA/G | 0.68 | 1.06 | 1.04–1.08 | 7.0 × 10−12 |
| rs7295014 | 0.342 | 12q24.33 | 133067989 | FBRSL1 | G/A | 0.35 | 1.05 | 1.04–1.07 | 9.5 × 10−10 |
| rs1004030 | 0.581 | 14q11.2 | 23305649 | MMP14 | T/C | 0.58 | 1.05 | 1.03–1.06 | 1.5 × 10−8 |
| rs11629412 | 0.582 | 14q13.3 | 37138294 | PAX9 | C/G | 0.58 | 1.06 | 1.04–1.08 | 2.3 × 10−12 |
| rs4924487 | 0.836 | 15q15.1 | 40922915 | CASC5 | C/G | 0.81 | 1.06 | 1.04–1.09 | 1.3 × 10−8 |
| rs33984059 | 0.982 | 15q21.3 | 56385868 | RFX7 | A/G | 0.98 | 1.19 | 1.12–1.27 | 1.1 × 10−8 |
| rs112293876 | 0.280 | 15q22.31 | 66764641 | MAP2K1 | C/CA | 0.29 | 1.06 | 1.04–1.08 | 3.5 × 10−10 |
| rs11863709 | 0.945 | 16q21 | 57654576 | GPR56 | C/T | 0.96 | 1.16 | 1.11–1.21 | 1.8 × 10−11 |
| rs201158093 | 0.435 | 16q23.3 | 82178893 | RP11–510J16.5 | TAA/TA | 0.44 | 1.05 | 1.03–1.07 | 9.1 × 10−9 |
| rs28441558 | 0.050 | 17p13.1 | 7803118 | CHD3 | C/T | 0.05 | 1.16 | 1.12–1.20 | 1.0 × 10−16 |
| rs142444269 | 0.798 | 17q11.2 | 30098749 | RP11–805L22.3 | C/T | 0.78 | 1.07 | 1.05–1.09 | 3.2 × 10−10 |
| rs2680708 | 0.623 | 17q22 | 56456120 | RNF43 | G/A | 0.61 | 1.05 | 1.03–1.06 | 1.6 × 10−8 |
| rs8093601 | 0.459 | 18q21.2 | 51772473 | MBD2 | C/G | 0.44 | 1.05 | 1.03–1.06 | 2.3 × 10−8 |
| rs28607662 | 0.085 | 18q21.2 | 53230859 | TCF4 | C/T | 0.10 | 1.08 | 1.05–1.11 | 2.8 × 10−8 |
| rs12956892 | 0.300 | 18q21.32 | 56746315 | OACYLP | T/G | 0.30 | 1.05 | 1.03–1.07 | 7.7 × 10−9 |
| rs533722308 | 0.390 | 18q21.33 | 60961193 | BCL2 | CT/C | 0.42 | 1.05 | 1.03–1.07 | 1.2 × 10−8 |
| rs10460109 | 0.414 | 18q22.3 | 73036165 | TSHZ1 | T/C | 0.42 | 1.05 | 1.03–1.06 | 3.5 × 10−8 |
| rs11666569 | 0.728 | 19p13.11 | 17214073 | MYO9B | C/T | 0.71 | 1.05 | 1.03–1.07 | 8.2 × 10−9 |
| rs118005503 | 0.912 | 19q12 | 32167803 | THEG5 | G/C | 0.91 | 1.09 | 1.06–1.13 | 7.3 × 10−9 |
| rs61088131 | 0.848 | 19q13.2 | 42700947 | POU2F2 | T/C | 0.82 | 1.06 | 1.04–1.09 | 8.8 × 10−9 |
| rs11480453 | 0.641 | 20q11.21 | 31347512 | DNMT3B | C/CA | 0.60 | 1.05 | 1.03–1.06 | 3.2 × 10−8 |
| rs6091758 | 0.465 | 20q13.2 | 52455205 | BCAS1 | G/A | 0.47 | 1.07 | 1.06–1.09 | 6.4 × 10−18 |
| rs9625483 | 0.026 | 22q12.1 | 28888939 | TTC28 | A/G | 0.03 | 1.14 | 1.09–1.20 | 2.4 × 10−8 |
| rs17321482 | 0.873 | 23p22.2 | 11482634 | ARHGAP6 | C/T | 0.87 | 1.07 | 1.05–1.09 | 2.1 × 10−13 |
| Novel locus associated with early-onset prostate cancer | |||||||||
| rs138004030 | 0.920 | 6q27 | 170475879 | LOC154449 | G/A | 0.91 | 1.27 | 1.17–1.38 | 2.9 × 10−8 |
Risk-allele frequency (RAF) in 1KGP Europeans.
Risk allele/reference allele.
P values generated from likelihood-ratio tests.
Region previously reported by Wang et al.49, rs12791447; rs61890184-rs12791447 r2 (EUR) = 0.41.
We performed several stratified analyses defined by clinical and population parameters. We detected a novel variant, rs138004030, which was significantly associated with early-onset disease (Table 1) but was only nominally significant for overall PrCa risk (P = 0.02). In addition, we detected four markers significantly associated (P < 5 × 10−8) with advanced PrCa and two markers associated with early-onset PrCa (Supplementary Table 11). However, the case-only analyses of these markers indicated marginal statistical significance (P < 1.0 × 10−3). Additionally, these markers were in LD with nearby index markers associated with overall PrCa and were not significantly associated with overall aggressive disease after adjustment for the index marker (Supplementary Table 11). A similar association pattern was observed for rs111599055, which was in LD with rs7295014 (r2 = 0.54), a marker associated with overall disease. The early-onset marker rs77777548 was independent of novel and known PrCa-risk loci. However, the marker was relatively rare (effect-allele frequency <0.02), was indicated as monomorphic in the 1KGP, and had a moderate imputation quality score (average r2 = 0.57); hence, we did not include it in further analyses.
Among the 63 novel associations, 38 variants were found to be located within gene-rich regions (Supplementary Table 12): intronic (32 SNPs), missense (4 SNPs), and 3′ untranslated region (UTR) (2 SNPs). Analyses of expression quantitative trait loci (eQTL) in The Cancer Genome Atlas (TCGA) database identified statistically significant associations (P < 0.05; Supplementary Table 12) in normal PrCa tissue for 17 of the novel associations, including both 3′-UTR SNPs and 11 of the 32 intronic SNPs. Cis-eQTL associations were identified for 3′-UTR variant rs1048169 with HAUS6 (3′-UTR) and intronic variants rs182314334 with MBNL1, rs4976790 with COL23A1, rs9469899 with UHRF1BP1, rs878987 with B3GAT1, rs11629412 with PAX9, and rs11666569 with MYO9B. The eQTL associations were consistent with the observed PrCa-SNP associations, given that we assessed colocalization between the GWAS and eQTL SNPs. The TCGA data analysis did not identify an eQTL association with any of the four missense SNPs.
We assessed the association of our newly discovered loci with prostate-specific antigen (PSA) levels by using a series of disease-free controls (n = 9,090; Methods). Among the 48 available loci, we observed a significant association for rs8093601 (P = 5.0 × 10−4; Supplementary Table 13) after correction for multiple testing (P = 0.05/48 = 1.0 × 10−3). This marker lies near MBD2 (encoding methyl-CpG binding domain protein 2) and has not previously been associated with either PrCa risk or PSA levels. The effect estimates of PrCa clinical features and overall PrCa did not differ (Supplementary Table 14). LD plots incorporating several functional annotation features for each of the 63 novel markers are presented in Supplementary Fig. 4.
Several strong candidate genes were identified among the PrCa-susceptibility loci, including ATM, a key gene within the DNA-damage response pathway, in which truncating variants contribute to PrCa susceptibility and progression, particularly aggressive PrCa34,35. The index variant within this region is the missense variant rs1800057, exerting a modestly increased risk of PrCa (OR = 1.16; P = 8.15 × 10−9; G>C, p.Pro1054Arg; Fig. 2a). Although rs1800057 is designated ‘benign’ by ClinVar (see URLs), it has been suggested to be associated with a twofold-increased risk of early-onset PrCa in a small clinical series and has been found to be unassociated with morbidity after treatment36. In addition to the ATM region, we identified missense variants at three separate loci: rs2066827 within CDKN1B, encoding a cyclin-dependent-kinase inhibitor that controls cell-cycle progression; rs33984059 within RFX7, encoding a transcription factor; and rs2277283 within INCENP, encoding a centromere-interacting protein.
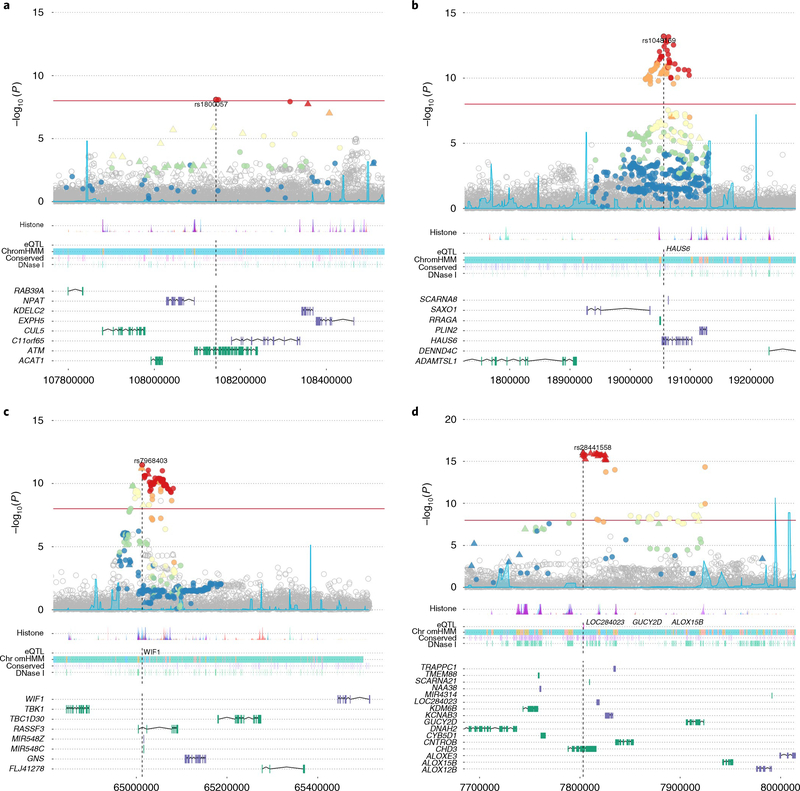
Fig. 2 |. Locus Explorer plots depicting the statistical association with PrCa and biological context of variants from four of the newly identified PrCa-risk loci (n = 74,849 biologically independent samples).
a-d, Top, Manhattan plots of variant −log10 P values (y axis), with the Index SNP labeled. Variants that were directly genotyped with the OncoArray are represented as triangles, and imputed variants are represented as circles. Variants in LD with the index SNP are denoted by color (red, r2 >0.8; orange, 0.6 < r2 < 0.8; yellow, 0.4 < r2 < 0.6; green, 0.2 < r2 < 0.4, blue, r2 ≤0.2). Middle, relative locations of selected biological annotations: histone marks within seven cell lines from the ENCODE project; genes for which the index SNP is an eQTL in TCGA prostate adenocarcinoma dataset; chromatin state annotation by ChromHMM in PrEC cells; conserved elements within the genome; and DNAse I-hypersensitivity sites in ENCODE prostate cell lines. Bottom, positions of genes within the region, with genes on the positive and negative strands marked in green and purple, respectively. The horizontal axis represents genomic coordinates in the hg19 reference genome. a, rs1800057 (chromosome (chr) 11: 107643000–108644000). The index variant is a nonsynonymous SNP in ATM. b, rs1048160 (chr 9: 18556000–19557000). The index variant is located within the 3′ UTR of HAUS6 and is an eQTL for HAUS6. c, rs7968403 (chr 12: 64513000–65514000). The signal is centered on RASSF3, and the index variant is located within the first intron. This SNP is also situated within a region annotated for multiple regulatory markers and is an eQTL for the more distant WIF1 gene. d, rs28441558 (chr 17: 7303000–8304000). The signal implicates a cluster of highly correlated variants centered on CHD3. The index SNP is also an eQTL for three other more distantly located genes.
rs1048169 at 9p22 is located in the 3′ UTR of HAUS6 (Fig. 2b), which encodes a subunit of augmin, a protein complex required for proper microtubule formation and chromosome segregation during cell division37. rs1048169 is also an eQTL for HAUS6 expression. Interestingly, an additional lead SNP identified in this study, rs11666569 at 19p13, was found to be an eQTL for two genes, including HAUS8, which encodes another member of the augmin complex. These discoveries may implicate a potential role of augmin in PrCa susceptibility.
rs7968403 (OR = 1.06; P = 3.38 × 10−12; Fig. 2c) is situated within the first intron of RASSF3. Members of the Ras-association-domain family (RASSF) are putative tumor suppressors implicated in a range of biological processes38. RASSF3 is ubiquitously expressed across tissue types and has been observed to arrest the cell cycle in the G1 phase and to induce apoptosis through the p53 pathway39. A PrCa-risk locus, ~100 kb away, within RASSF6 has been identified in a previous study11. However, rs7968403 was also an eQTL for the distant WIF1 (encoding WNT-inhibitory factor 1; Fig. 2c). WIF1 inhibits Wnt signaling and is frequently downregulated in PrCa40, whereas aberrant activation of Wnt signaling is common in many solid tumor types. Restoration of WIF1 expression has also been demonstrated to decrease cell motility and invasiveness in a metastatic PrCa cell line and to reduce tumor growth in a mouse xenograft model41. Both RASSF3 and WIF1 therefore are plausible mechanisms for the modulation of PrCa risk at this locus.
rs28441558 at 17p13 was the lead variant for a cluster of highly correlated SNPs centered on the CHD3 gene (Fig. 2d). CHD3 encodes an ATPase that forms a component of the nucleosome-remodeling and deacetylase (NuRD) histone deacetylase complex, which is involved in chromatin remodeling. NuRD plays an important role in regulating gene expression, as both a silencer and an activator of transcription, in addition to its roles in maintaining genomic integrity and in the DNA-damage response42. Alterations in NuRD function have been implicated in several cancer types and found to act in a highly complex manner43,44. However, rs28441558 was also observed to be an eQTL for three genes: LOC284023, encoding a currently uncharacterized noncoding RNA transcript; GUCY2D, encoding a guanylate cyclase enzyme expressed predominantly in the retina; and ALOX15B, encoding a member of the lipoxygenase family of enzymes that produce fatty acid hyperoxides. Although CHD3 appears to be the most biologically plausible candidate gene for this locus, we cannot exclude roles of any of these genes.
Our pathway analysis based on mapping each SNP to the nearest gene (Methods) by using the meta-analysis summary association statistic identified several pathways implicated in PrCa susceptibility. The top 53 pathways detected (enrichment score (ES) >0.50) are provided in Supplementary Table 15. The most significant pathway detected was PD-1 signaling (ID: 389948), ES = 0.74, as defined by the REACTOME database (Supplementary Fig. 5). This pathway is intriguing, given the therapeutic potential of several checkpoint inhibitors focusing on the PD-1 signaling pathway to enhance immune responses45.
In summary, we identified 63 novel PrCa-susceptibility variants, including strong candidate loci highlighting the DNA-repair and cell-cycle pathways. Previous studies have probably overestimated the effect estimates of PrCa loci as a result of the ‘winner’s curse’, thus yielding a biased familial relative risk (FRR) and polygenic risk score (PRS). Here, we applied a weighted Bayesian correction approach and demonstrated that our large sample size minimized the winner’s curse bias46 (Methods and Supplementary Fig. 6). We applied the beta estimates calculated in our overall meta-analysis to the OncoArray sample set to calculate the FRR and PRS risk models (Supplementary Table 16). Our prediction models included 85 previously reported PrCa loci replicating in our overall meta-analysis and our 62 novel loci associated with overall PrCa risk. Assuming a familial risk estimate of 2.5 for PrCa47,48, we demonstrated that our 147 loci captured 28.4% of the FRR (Supplementary Table 17). The 62 newly identified PrCa loci increased the FRR by 4.4%. On the basis of the assumption of a log-additive model, the estimated RR for PrCa relative to men in the twenty-fifth to seventy-fifth PRS percentiles (baseline group) was 5.71 (95% CI: 5.04–6.48) for men in the top first percentile of the PRS distribution and 2.69 (95% CI: 2.55–2.82) for individuals in the ninetieth to ninety-ninth percentiles of the PRS distribution (Table 2). The PRS score was positively associated with overall PrCa risk (OR = 1.86; 95% CI: 1.83–1.89; Supplementary Table 18). Our novel associations highlight several biological pathways that warrant further investigation. The increased PRS can be used to improve the identification of men at high risk for PrCa and therefore inform PSA guidelines for screening and management to reduce the burden of over-testing.
Table 2 |.
Estimation of polygenic risk scores by using 147 prostate cancer-susceptibility variants
| Risk category percentilea | Relative risk | 95% CI |
|---|---|---|
| <1 | 0.15 | 0.11–0.20 |
| 1–10 | 0.35 | 0.32–0.37 |
| 10–25 | 0.54 | 0.51–0.57 |
| 25–75 | 1.00 (baseline) | |
| 75–90 | 1.74 | 1.67–1.82 |
| 90–99 | 2.69 | 2.55–2.82 |
| ≥99 | 5.71 | 5.04–6.48 |
PRS percentiles based on the cumulative score distributed among controls. The beta coefficients computed from the European overall meta-analysis were applied to determine the PRS risk among individuals in the OncoArray study.
Methods
Methods, including statements of data availability and any associated accession codes and references, are available at https://doi.org/10.1038/s41588-018-0142-8.
Methods
Study subjects
A brief overview and study details for participating PrCa studies in the newly genotyped OncoArray project are provided in Supplementary Table 1 for men of European ancestry. All studies were approved by the appropriate ethics committees (as described in the references for each study listed in Supplementary Table 1), and informed consent was obtained from all participants. Supplementary Table 2 summarizes the PrCa sample series of the Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE) consortium contributing both newly obtained genotyping data for the OncoArray and previous GWAS. Most of the studies contributing to the OncoArray were case-control studies primarily based in either the United States or Europe. In total, 52 new studies provided core data on disease status, age at diagnosis (age at observation or questionnaire for controls), family history of PrCa, and clinical factors for cases (for example, PSA at diagnosis and Gleason score) for 48,455 PrCa cases and 28,321 disease-free controls. Previous GWAS contributed an additional 32,255 PrCa cases and 33,202 disease-free controls of European ancestry to the overall meta-analysis12. Supplementary Table 3 provides QC information by consortia (e.g., OncoArray project, UK GWAS, and so forth) for both samples and SNPs. After removal of all overlapping samples, the OncoArray contribution for newly genotyped samples was 46,939 PrCa cases and 27,910 disease-free controls.
Several strata-specific analyses were implemented to evaluate the effects of genetic variation on PrCa disease aggressiveness. Supplementary Table 4 describes the analysis title, outcome and reference groups, and the statistical model used. Several classification schemes (low aggressiveness, intermediate aggressiveness, and so forth) were implemented to better assess the spectrum of genetic involvement. All classification schemes incorporated the diagnostic clinical features PSA, tumor stage, and Gleason score. To compare the results with those from previous PrCa aggressive analyses12 by our research group, we included the ‘advanced (plus death due to PrCa)’ classification. Contributing study groups missing clinical features were excluded (Supplementary Table 2). Individuals with missing or granular clinical information were excluded. The strata-specific sample sizes from the PrCa GWAS consortium are provided in Supplementary Table 5. Furthermore, we analyzed Gleason score as a continuous variable.
OncoArray SNP selection
The NCI GAME-ON consortium (http://epi.grants.cancer.gov/gameon/) provided SNPs to be included in the Illumina OncoArray. Approximately 50% of the OncoArray was a compilation of SNP lists by the GAME-ON disease consortium of cancer (breast, colorectal, lung, ovarian, and prostate), a common set of variants for common risk regions, other related traits (BMI, age at menarche, and so forth), pharmacogenetics, and candidates30. The remaining content of the OncoArray was selected as a ‘GWAS backbone’ (Illumina HumanCore), which aimed to provide high coverage for most common variants through imputation. Approximately 79,000 SNPs were selected specifically for their relevance to PrCa, on the basis of prior evidence of association with overall or subtype-specific disease, fine-mapping of known PrCa regions, and candidate submissions (survival, exome sequencing, and so forth). To maximize the efficiency of the array, cancer-specific candidate lists were merged to remove redundant genetic variation30.
Genotype calling and quality control
Details of the genotype calling and QC for the iCOGS and GWAS have been described elsewhere11–28.
Of the 568,712 variants selected for genotyping on the OncoArray, 533,631 were successfully manufactured on the array (including 778 duplicate probes). OncoArray genotyping of ELLIPSE studies was conducted at five sites (Cambridge, CIDR, Copenhagen, USC, and NCI). Details of the genotype calling for the OncoArray have been described in more detail elsewhere30. Briefly, we developed a single calling pipeline that was applied to more than 500,000 samples across the GAME-ON consortium. An initial cluster file was generated by using 56,284 samples selected from all major genotyping centers and ancestries, with the Gentrain2 algorithm. Variants likely to have problematic clusters were selected for manual inspection on the basis of the following criteria: call rate <99%, MAF <0.001, poor Illumina intensity and clustering metrics, deviation from the MAF observed in the 1KGP, by using the criterion , where p0 and p1 are the minor frequencies in the 1KGP and OncoArray datasets, respectively, and C = 0.008. This procedure resulted in manual adjustment of the cluster file for 3,964 variants and the exclusion of 16,526 variants. The final cluster file was then applied to the full dataset.
Our QC pipeline for ELLIPSE excluded SNPs with a call rate <95% by study, not in Hardy-Weinberg equilibrium (P < 10−7 in controls or P < 10−12 in cases) or with concordance <98% among 11,260 duplicate pairs. To minimize imputation errors, we additionally excluded SNPs with a MAF <1% and a call rate <98% in any study, SNPs that could not be linked to the 1KGP reference, those with MAF for Europeans that differed from that for the 1KGP, and a further 16,526 SNPs for which the cluster plot was judged to be not ideal. Of the 533,631 manufactured SNPs on the OncoArray, we retained 498,417 SNPs among our samples of European ancestry after QC.
We excluded duplicate samples and first-degree relatives within each study, duplicates across studies, samples with a call rate <95%, and samples with extreme heterozygosity (>4.9 s.d. from the mean for the reported ancestry). We excluded duplicated samples as well as first-degree relatives across the GWAS studies CAPS1, CAPS2, UK Stage 1, UK Stage 2, and iCOGS. Duplicate and first-degree-related samples were assessed across the BPC3 and Pegasus GWAS studies as well. Ancestry was computed through principal component analysis using 2,318 informative markers on a subset of ~47,000 samples and projected onto the complete OncoArray dataset. The current analysis was restricted to men of European ancestry, defined as individuals with an estimated proportion of European ancestry >0.8, with reference to the HapMap populations, on the basis of the first two principal components. Of the 78,182 samples genotyped (regardless of ancestry), the final dataset consisted of 74,849 samples, of which 46,939 PrCa cases and 27,910 disease-free controls (Supplementary Table 3), after exclusion of overlap samples, were meta-analyzed with previous studies.
Imputation
We imputed genotypes for ~70 million SNPs for all samples by using the October 2014 (Phase 3) release of the 1KGP data as the reference panel. We imputed the OncoArray and GWAS datasets through a two-stage imputation approach, using SHAPEIT31 for phasing and IMPUTEv2 (ref. 32) for imputation. The imputation was performed in 5-Mb nonoverlapping intervals. All subjects were split into subsets of ~10,000 samples, with subjects from the same group in the subset. We imputed genotypes for all SNPs that were polymorphic (MAF > 0.1%) in European samples. We excluded data for all monomorphic SNPs and those with an imputation r2 <0.3, thus leaving a total of 20,370,935 SNPs across chromosomes 1–22 and chromosome X. Of the SNPs imputed, 49.3% had a MAF <1%, 15.2% had a MAF ranging between 1% and 5%, and 35.5% had a MAF ≥5%.
Statistical analyses
Per-allele odds ratios and standard errors were generated for the OncoArray and each GWAS, with adjustment for principal components and study-relevant covariates through logistic regression. The OncoArray and iCOGS analyses were additionally stratified by country and study, respectively. We used the first seven principal components in our analysis of individuals of European ancestry, because additional components did not further decrease inflation in the test statistics.
OR estimates were derived with either SNPTEST (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html) or an in-house C++ program (Supplementary Table 3). OR estimates and standard errors were combined by a fixed-effects inverse variance meta-analysis in METAL50. All statistical tests conducted were two sided.
Our analyses included overall PrCa and several clinically relevant strata. These strata comprised: (i) high versus low aggressive PrCa; (ii) high versus low/intermediate aggressive PrCa; (iii) advanced versus nonadvanced PrCa; (iv) advanced PrCa versus controls; (v) early-onset PrCa (≤55 years) versus controls; and (iv) Gleason score (Supplementary Tables 4 and 5). We defined low aggressive as tumor stage ≤T1 and Gleason score ≤6 and PSA <10 ng/mL; intermediate aggressive as tumor stage T2 or Gleason score = 7 or PSA 10–20 ng/mL; high aggressive as tumor stage T3/T4 or N1 or M1, or Gleason score ≥8 or PSA >20 ng/mL; and advanced as either metastatic disease, Gleason score ≥8, PSA > 100 or PrCa-related death (Supplementary Table 4).
Definition of newly associated loci
To search for novel loci, we assessed all SNPs excluding those within a known PrCa locus, defined by current fine-mapping assessments (Supplementary Table 7). SNPs that were associated with disease risk at P < 5 × 10−8 in the meta-analysis (GWAS and OncoArray) were considered novel. The SNP with the lowest P value in a region was considered the lead SNP. Imputation quality was assessed on the basis of IMPUTE2 imputation r2 in the OncoArray dataset (Supplementary Table 8).
For ten regions where the newly identified locus was near a previously known region, we reported a novel association if the pairwise r2 between the new and the previously known SNP was <0.2. For novel PrCa associations for which the variant was imputed in the OncoArray study sample series and had an imputed quality score <0.70, we assessed the quality of the imputation by masking the variant in a subset of the 1KGP European sample and calculating the concordance after reimputation in the remaining 1KGP samples.
Reliability of imputation
Novel SNPs with an IMPUTE2 r2 <0.80 among the OncoArray sample series (Supplementary Table 8) were flagged for further investigation to minimize the probability of false positives. First, we examined LD plots (http://locuszoom.org/) for poorly imputed SNPs (±500 kb), including only genotyped SNPs within the region. The imputed index SNP was included in the plot to determine the strength of LD with nearby signals and to assess a pattern of association. Furthermore, we performed an imputation experiment using the 2,504 1KGP Phase 3 samples. We split this sample into two parts: a random sample of 259 individuals of European ancestry (excluding Finnish individuals) and a mixed-population reference panel of 2,245 individuals. The random sample of 259 individuals of European ancestry was filtered to include only the genetic variants available from the OncoArray after QC. This procedure ensured that the same imputation input was used in the overall imputation. The 259 individuals were imputed by using 2,245 individuals as the reference panel. A 5-Mb segment of the genome was selected on the basis of the target SNP (±250 Mb). SHAPEIT2 was used for prephasing, and IMPUTE2 was used for imputation. Customized imputation settings included an effect size of 20,000, allowance of large-region imputation and a random seed of 12345. A weighted linear kappa statistic was calculated to determine the correlation of the imputation with the true genotypes.
We evaluated four SNPs whose IMPUTE2 r2 was <0.80 in the OncoArray sample series: rs527510716 (chr 7), rs6602880 (chr 10), rs533722308 (chr 18), and rs144166867 (chr X). Supplementary Fig. 3 includes the LD plots for three of the poorly imputed SNPs. The variant rs144166867 (chr X) could not be plotted, because no genotype SNPs were available within ±500 kb on the OncoArray. Both LD plots for markers rs527510716 (chr 7) and rs533722308 (chr 18) showed significant associations (P < 1 × 10−3) for several genotype markers with moderate LD of the index SNP The kappa coefficients for markers rs527510716 (chr 7) and rs533722308 (chr 18) were 0.911 and 0.931, respectively (Supplementary Table 9). The marker rs6602880 (chr 10) had a kappa coefficient of 0.812 and was the only significant variant in the LD plot. The kappa coefficient for marker rs144166867 (chr X) was 0.665 (Supplementary Table 9). The markers rs6602880 (chr 10) and rs144166867 (chr X) were probably false positives due to poor imputation for these regions.
Proportion of familial risk explained
The contribution of the known SNPs to the familial risk of PrCa, under a multiplicative model, was computed with the formula
where λ0 is the observed familial risk to first-degree relatives of PrCa cases47,48, assumed to be 2.5, and λk is the familial relative risk due to locus k, given by:
where pk is the frequency of the risk allele for locus k, qk = 1−pk, and rk is the estimated per-allele odds ratio.
On the basis of the assumption of a log-additive model, we constructed a PRS from the summed risk-allelic doses weighted by the per-allele log ORs. Thus, for each individual j, we derived:
where N is the number of SNPs, gij is the allele dose at SNPi for individual j, and βi is the per-allele log-odds ratio of SNPi.
The risk of PrCa was estimated for the percentiles of the distribution of the PRS (<1, 1–10, 10–25, 25–75, 75–90, 90–99, >99 and <10, 10–25, 25–75, 75–90, >90) for which cumulative score thresholds were determined according to the observed distribution among controls. We applied effect sizes and allele frequencies obtained from the overall meta-analysis of Europeans to estimate risk scores for individuals of European ancestry in the OncoArray study51. A standardized PRS score was calculated by dividing the observed PRS score by the s.d. of the PRS score among controls. A logistic-regression framework was used to evaluate the percentile comparisons and to determine the risk estimate. The models were adjusted for the first seven principal components to account for population stratification and stratified by country.
The FRR and PRS risk estimation was limited to the variants for which our overall meta-analysis indicated a statistically significant association. In total, we included 147 PrCa index SNPs in our risk-score modeling, including 85 previously published associations and the 62 novel findings reported here. To correct for potential bias in effect estimation of newly discovered variants, we implemented a fully Bayesian version of a weighted correction given in equation (3).4 in ref. 46. Specifically, we placed a normal prior distribution on MLE effect estimates of the form βm ~ N (βCor, τ2). Here, βm is the log OR from the overall meta-analysis; βCor is the bias-corrected estimate calculated with the expectation-adjusted estimator from equation (3).1 in in ref. 46; and τ is a prespecified variance of the effect distribution reflecting the bias and is defined as .
eQTL analyses
Genotype and gene expression data were downloaded from TCGA for 494 samples with PrCa (https://gdc-portal.nci.nih.gov/). QC was performed on both these datasets as follows: on the genotype, we filtered out samples with high heterozygosity (mean heterozygosity ±2 s.d.) and missing genotypes and duplicated or related samples. We then performed principal component analysis on the 494 samples plus 2,506 samples from 1KGP to infer the ancestry of the TCGA samples; samples of non-European ancestry were removed. We also filtered out variants with missing call rate >5%. For the expression data, samples from two plates had, on average, much higher expression values than did the remaining samples and therefore were excluded. We also filtered genes with mean expression across samples ≤6 counts. Finally, expression values were quantile-normalized by samples and rank-transformed by genes. After QC, we used the data from 359 samples. For the eQTL analysis, 35 PEER factors from the top 10,000 expressed genes were used as covariates, plus three genotyping PCs (which explained 18% of total variation). eQTL analysis was performed in FastQTL with 1,000 permutations over the 85 regions. We used a window of 1 Mb (upstream/downstream) from the transcription start site of each gene.
Gene set enrichment analyses
The file Human_GOBP_AllPathways_no_GO_iea_September_01_2016_symbol.gmt (http://baderlab.org/GeneSets/) from the GeneSets database52, was used for all analyses. This database contains pathways from Reactome53, NCI Pathway Interaction Database54, Gene Ontology (GO) biological process55, HumanCyc56, MSigdb57, NetPath58, and Panther59. We manually corrected several pathways in which the c gene was entered as PDK1. GO pathways inferred from electronic annotation terms were excluded. The same pathway (for example, apoptosis) may be defined in two or more databases with potentially different sets of genes, and all versions of these duplicate/overlapping pathways were included. Pathway size was determined by the total number of genes in the pathway to which SNPs in the imputed GWAS dataset could be mapped. To provide more biologically meaningful results, and to reduce false positives, only pathways that contained between 10 and 200 genes were considered.
Gene information (hg19) was downloaded from the ANNOVAR60 website (http://annovar.openbioinformatics.org/). SNPs were mapped to the nearest gene within 500-kb windows; those that were further away from any gene were excluded. Gene significance was calculated by assigning the lowest P value observed across all SNPs assigned to a gene61,62, on the basis of the combined European meta-analysis (previous GWAS and OncoArray).
The gene-set enrichment analysis (GSEA)52 algorithm, as implemented in the GenGen package (http://gengen.openbioinformatics.org/en/latest/)62,63, was used to perform pathway analysis. Briefly, the algorithm calculates an ES for each pathway on the basis of a weighted Kolmogorov-Smirnov statistic63. To calculate the ES, we performed 100 permutations and averaged the final score. Pathways with most of their genes at the top of the ranked list of genes obtain higher ES values. Only pathways with positive ES and at least one gene with P < 5 × 10−8 were retained for subsequent analysis. An enrichment map was created in the Enrichment Map (EM) v 2.1.0 application52 in Cytoscape v3.40 (ref. 64), with application of force-directed layout, in weighted mode. We restricted our pathway analysis to those with an ES ≥ 0.50 to ensure a true-positive rate >0.20 and a false-positive rate <0.15.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The OncoArray genotype data and relevant covariate information (ancestry, country, principal components, and so forth) generated during this study have been deposited in dbGaP under accession code phs001391.v1.p1. In total, 47 of the 52 OncoArray studies encompassing nearly 90% of the individual samples will be available (Supplementary Table 19). The previous meta-analysis summary results and genotype data12 are available in dbGaP under accession code phs001081.v1.p1. The complete meta-analysis summary associations statistics are publicly available at the PRACTICAL website (http://practical.icr.ac.uk/blog/).
Supplementary Material
Acknowledgements
We pay tribute to Brian Henderson for his vision and leadership; he was a driving force behind the OncoArray project and he unfortunately passed away before seeing its fruition. We also thank the individuals who participated in these studies enabling this work.
Genotyping of the OncoArray was funded by the US National Institutes of Health (NIH) (U19 CA 148537 for the ELLIPSE project and X01HG007492 to the Center for Inherited Disease Research (CIDR) under contract no. HHSN268201200008I. Additional analytical support was provided by NIH NCI U01 CA188392 (to F.R.S.).
Funding for the iCOGS infrastructure came from the European Community’s Seventh Framework Programme under grant agreement no. 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, and C8197/A16565), the NIH (CA128978) and Post-Cancer GWAS Initiative (1U19 CA148537, 1U19 CA148065, and 1U19 CA148112; the GAME-ON initiative), the Department of Defense (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund.
This work was supported by the Canadian Institutes of Health Research; the European Commission’s Seventh Framework Programme grant agreement no. 223175 (HEALTH-F2-2009-223175); Cancer Research UK grants C5047/A7357, C1287/A10118, C1287/A16563, C5047/A3354, C5047/A10692, and C16913/A6135; and NIH Cancer Post-Cancer GWAS initiative grant no. 1 U19 CA 148537-01 (the GAME-ON initiative).
We also thank the following for funding support: the Institute of Cancer Research and the Everyman Campaign, the Prostate Cancer Research Foundation, Prostate Research Campaign UK (now Prostate Action), the Orchid Cancer Appeal, the National Cancer Research Network UK, and the National Cancer Research Institute (NCRI) UK. We are grateful for the support of NIHR funding to the NIHR Biomedical Research Centre at the Institute of Cancer Research and the Royal Marsden NHS Foundation Trust.
The Prostate Cancer Program of Cancer Council Victoria also acknowledges grant support from the National Health and Medical Research Council, Australia (126402, 209057, 251533, 396414, 450104, 504700, 504702, 504715, 623204, 940394, and 614296), VicHealth, Cancer Council Victoria, the Prostate Cancer Foundation of Australia, the Whitten Foundation, PricewaterhouseCoopers, and Tattersall’s. E.A.O., D.M.K., and E.M.K. acknowledge the Intramural Program of the National Human Genome Research Institute for support.
The BPC3 was supported by the NIH, National Cancer Institute (cooperative agreements U01-CA98233 to D.J.H., U01-CA98710 to S.M.G., U01-CA98216 to E.R., and U01-CA98758 to B.E.H., and the Intramural Research Program of the NIH/National Cancer Institute, Division of Cancer Epidemiology and Genetics).
The CAPS GWAS study was supported by the Swedish Cancer Foundation (grant nos. 09-0677, 11-484, and 12-823), the Cancer Risk Prediction Center (CRisP; http://ki.se/en/meb/crisp/), a Linneus Centre grant (contract ID 70867902) financed by the Swedish Research Council, and the Swedish Research Council (grant nos. K2010- 70X-20430-04-3, and 2014-2269).
PEGASUS was supported by the Intramural Research Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH.
A full description of funding and acknowledgements can be found in the Supplementary Note.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41588-018-0142-8.
URLs. ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/.
References
- 1.Goh CL et al. Genetic variants associated with predisposition to prostate cancer and potential clinical implications. J. Intern. Med 271, 353–365 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD & Jemal A Cancer statistics, 2016. CA Cancer J. Clin 66, 7–30 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Cuzick J et al. Prevention and early detection of prostate cancer. Lancet Oncol. 15, e484–e492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altekruse SF et al. Spatial patterns of localized-stage prostate cancer incidence among white and black men in the southeastern United States, 1999–2001. Cancer Epidemiol. Biomarkers Prev 19, 1460–1467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanford JL & Ostrander EA Familial prostate cancer. Epidemiol. Rev 23, 19–23 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Bunker CH et al. High prevalence of screening-detected prostate cancer among Afro-Caribbeans: the Tobago Prostate Cancer Survey. Cancer Epidemiol. Biomarkers Prev 11, 726–729 (2002). [PubMed] [Google Scholar]
- 7.Ghadirian P, Howe GR, Hislop TG & Maisonneuve P Family history of prostate cancer: a multi-center case-control study in Canada. Int. J. Cancer 70, 679–681 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Gronberg H, Damber L & Damber JE Familial prostate cancer in Sweden: a nationwide register cohort study. Cancer 77, 138–143 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Matikaine MP et al. Relatives of prostate cancer patients have an increased risk of prostate and stomach cancers: a population-based, cancer registry study in Finland. Cancer Causes Control 12, 223–230 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Eeles R et al. The genetic epidemiology of prostate cancer and its clinical implications. Nat. Rev. Urol 11, 18–31 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Eeles RA et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet 45, 385–391 (2013). e1-e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al Olama AA et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat. Genet 46, 1103–1109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al Olama AA et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet 41, 1058–1060 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Amundadottir LT et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet 38, 652–658 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Eeles RA et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet 41, 1116–1121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eeles RA et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 40, 316–321 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Gudmundsson J et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet 41, 1122–1126 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gudmundsson J et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet 39, 631–637 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Gudmundsson J et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat. Genet. 40, 281–283 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudmundsson J et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet 39, 977–983 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Haiman CA et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat. Genet. 43, 570–573 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kote-Jarai Z et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat. Genet 43, 785–791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumacher FR et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum. Mol. Genet 20, 3867–3875 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J et al. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat. Genet 40, 1153–1155 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takata R et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat. Genet 42, 751–754 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Thomas G et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet 40, 310–315 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Yeager M et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet 39, 645–649 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Duggan D et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J. Natl. Cancer Inst 99, 1836–1844 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Al Olama Amin A. et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum. Mol. Genet 22, 408–415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amos CI et al. The OncoArray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol. Biomarkers Prev. 26, 126–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaneau O, Marchini J & Zagury JF A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Howie BN, Donnelly P & Marchini J A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Bakker PI et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 17, R122–R128 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leongamornlert D et al. Frequent germline deleterious mutations in DNA repair genes in familial prostate cancer cases are associated with advanced disease. Br. J. Cancer 110, 1663–1672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mateo J et al. DNA-repair defects and Olaparib in metastatic prostate cancer. N. Engl. J. Med 373, 1697–1708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer A et al. ATM missense variant P1054R predisposes to prostate cancer. Radiother. Oncol 83, 283–288 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Sanchez-Huertas C & Luders J The augmin connection in the geometry of microtubule networks. Curr. Biol 25, R294–R299 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Volodko N, Gordon M, Salla M, Ghazaleh HA & Baksh S RASSF tumor suppressor gene family: biological functions and regulation. FEBS Lett. 588, 2671–2684 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Kudo T et al. The RASSF3 candidate tumor suppressor induces apoptosis and G1-S cell-cycle arrest via p53. Cancer Res. 72, 2901–2911 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Wissmann C et al. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. J. Pathol. 201, 204–212 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Yee DS et al. The Wnt inhibitory factor 1 restoration in prostate cancer cells was associated with reduced tumor growth, decreased capacity of cell migration and invasion and a reversal of epithelial to mesenchymal transition. Mol. Cancer 9, 162 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allen HF, Wade PA & Kutateladze TG The NuRD architecture. Cell. Mol. Life Sci 70, 3513–3524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai AY & Wade PA Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer 11, 588–596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basta J & Rauchman M The nucleosome remodeling and deacetylase complex in development and disease. Transl. Res 165, 36–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDermott DF & Atkins MB PD-1 as a potential target in cancer therapy. Cancer Med. 2, 662–673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong H & Prentice RL Bias-reduced estimators and confidence intervals for odds ratios in genome-wide association studies. Biostatistics 9, 621–634 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiciński M, Vangronsveld J & Nawrot TS An epidemiological reappraisal of the familial aggregation of prostate cancer: a meta-analysis. PLoS One 6, e27130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albright F et al. Prostate cancer risk prediction based on complete prostate cancer family history. Prostate 75, 390–398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang M et al. Large-scale association analysis in Asians identifies new susceptibility loci for prostate cancer. Nat. Commun 6, 8469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willer CJ, Li Y & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al Olama Amin A. et al. Risk analysis of prostate cancer in PRACTICAL, a multinational consortium, using 25 known prostate cancer susceptibility loci. Cancer Epidemiol. Biomarkers Prev 24, 1121–1129 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merico D, Isserlin R, Stueker O, Emili A & Bader GD Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joshi-Tope G et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 33, D428–D432 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaefer CF et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 37, D674–D679 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashburner M et al. Gene ontology: tool for the unification of biology. Nat. Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romero P et al. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 6, R2 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kandasamy K et al. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 11, R3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas PD et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang K, Li M & Hakonarson H ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Jia P, Wolfinger RD, Chen X & Zhao Z Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics 98, 1–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang K, Li M & Hakonarson H Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet 11, 843–854 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Wang K, Li M & Bucan M Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet 81, 1278–1283 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The OncoArray genotype data and relevant covariate information (ancestry, country, principal components, and so forth) generated during this study have been deposited in dbGaP under accession code phs001391.v1.p1. In total, 47 of the 52 OncoArray studies encompassing nearly 90% of the individual samples will be available (Supplementary Table 19). The previous meta-analysis summary results and genotype data12 are available in dbGaP under accession code phs001081.v1.p1. The complete meta-analysis summary associations statistics are publicly available at the PRACTICAL website (http://practical.icr.ac.uk/blog/).