Abstract
Healthcare provider organizations (HPOs) increasingly participate in large-scale research efforts sponsored by external organizations that require use of consent management systems that may not integrate seamlessly with local workflows. The resulting inefficiency can hinder the ability of HPOs to participate in studies. To overcome this challenge, we developed a method using REDCap, a widely adopted electronic data capture system, and novel middleware that can potentially generalize to other settings. In this paper, we describe the method, illustrate its use to support the NIHAll of Us Research Program and PCORI ADAPTABLE studies at our HPO, and encourage other HPOs to test replicability of the method to facilitate similar research efforts. Code is available on GitHub at https://github.com/wcmc-research-informatics/.
Introduction
Healthcare provider organizations (HPOs) increasingly participate in large-scale research studies, including biobanks and pragmatic clinical trials, that require management of patient consent and extraction of electronic health record (EHR) data. For biobanks, efforts include those led by individual HPOs, such as Geisinger MyCode and Vanderbilt BioVU (1–4), as well as by external sponsors, such as the China Kadoorie Biobank and the United States National Institutes of Health (NIH) All of Us Research Program (AoU) (5–8). For pragmatic clinical trials, a major investigation led by an external sponsor, the Patient Centered Outcomes Research Institute’s (PCORI) ADAPTABLE study (9), involves the participation of 36 HPOs in the United States. Large-scale studies led by HPOs have succeeded in part due to individual institutions designing new research processes to integrate with existing clinical and research workflows and systems (2,10,11). However, in our experience, external sponsors have designed large-scale studies to primarily support research goals and nominally integrate with HPOs’ existing institutional systems and workflows, which has hindered the ability of HPOs to participate in studies.
For AoU and ADAPTABLE, which aim to enroll one million and 20,000 participants, respectively, participants manage consent for study activities in systems overseen by external sponsors that do not integrate in an automated fashion with the local internal informatics infrastructure of HPOs. Using external consent systems, study team members from HPOs can view demographics, consent status, and other study data of individual participants as well as export participant records in bulk as a comma separated value (CSV) file.
Based on our experience, external consent management systems pose three barriers to large-scale study participation at HPOs. First, although the external consent systems assign a unique research identifier (ID) for each study participant, they do not capture the corresponding unique clinical ID of the participant as a patient in the healthcare provider organization, which is frequently a medical record number (MRN). As a result, study teams must manually associate each participant’s unique research ID with a local MRN, a process with potential for data entry error and downstream consequences. If a study team member associates a participant’s research ID with the MRN of a different patient, the study team may subsequently extract and share EHR data with an external research sponsor for a patient who did not consent to study participation in violation of patient privacy and confidentiality laws. Second, study team members may opt to use shadow applications (e.g., Microsoft Excel) that lack security and automated integration with enterprise clinical and research systems to support manual association of research IDs and MRNs as well as collection of other participant details not captured in the external consent system (e.g., scheduled appointments). With study consent data segregated in non-enterprise applications, EHR systems cannot display patients’ research participation status to inform clinician awareness and facilitate billing compliance for large-scale studies. Finally, to combine local shadow records with bulk records from external consent system CSV files, study team members must manually manipulate data, which is time-consuming and error-prone.
To address these barriers, we developed a method using a widely adopted electronic data capture system and novel middleware that can potentially generalize to other settings. The goal of this paper is to describe the method, illustrate its use to support AoU and ADAPTABLE at our HPO, and encourage other HPOs to test replicability of the method to support similar research efforts.
Methods
As described below, we developed a method for integrating our HPO’s existing clinical and research information systems with external consent management systems. The method consisted of semi-automated participant identity management, automated individual participant record transmission, and automated bulk participant record ingestion. We aimed to minimize study team effort and disruption of existing clinical and research systems and workflows.
Setting
At Weill Cornell Medicine (WCM), a multispecialty outpatient practice organization in New York City affiliated with NewYork-Presbyterian Hospital, clinicians used the EpicCare Ambulatory EHR system for care and billing for more than 2 million patients. A clinical trials management system (CTMS) integrated with the EHR and electronic institutional review board (eIRB) system served as the institutional system of record for all patients who consented to enroll as participants in research studies (12). Notably, for quality assurance purposes, institutional policy required manual review of each participant consent status update prior to data entry into the CTMS. Upon entry of data, the CTMS updated the EHR in real time with patients’ consent statuses for research studies. To support researchers with electronic patient data, the institution aggregated and transformed data from numerous clinical and research information systems into a data warehouse to support specific purposes, such as i2b2 and custom data marts (13). To support collection of novel measures absent from the EHR, Weill Cornell Medicine made REDCap, an electronic data capture system used widely among academic medical centers (14), available to all institutional faculty and staff participating in research. Separate teams oversaw clinical informatics, including the EHR and CTMS, and research informatics, including research data aggregation and REDCap. The research informatics team led the current effort.
For consent management for AoU and ADAPTABLE, study team members accessed two external systems hosted in secure cloud environments, the NIH-developed HealthPro for AoU and the commercial Mytrus for ADAPTABLE. Although the external consent systems exported participant data for our institution in bulk as a CSV file, they lacked application programming interfaces (APIs) for the study team to automatically obtain data.
For AoU and ADAPTABLE, eligibility criteria and enrollment workflows differed. For AoU, eligible patients included anyone 18 years of age and older, and study team members associated research IDs from HealthPro with local MRNs after patients consented to participate in the study. For ADAPTABLE, eligible patients included only patients meeting a cohort definition based on a computable phenotype algorithm executed against EHR data, and study team members associated research IDs from Mytrus with local MRNs before patients consented to participate in the study. Of note, ADAPTABLE required association of research IDs prior to consent to enable a paper mailing that described the study and instructed interested individuals to use the “Golden Ticket” research ID included in the mailing as a code to enroll in the study via the Mytrus portal. Both studies involved in-person visits by participants to our HPO to complete enrollment.
Semi-automated participant identity management
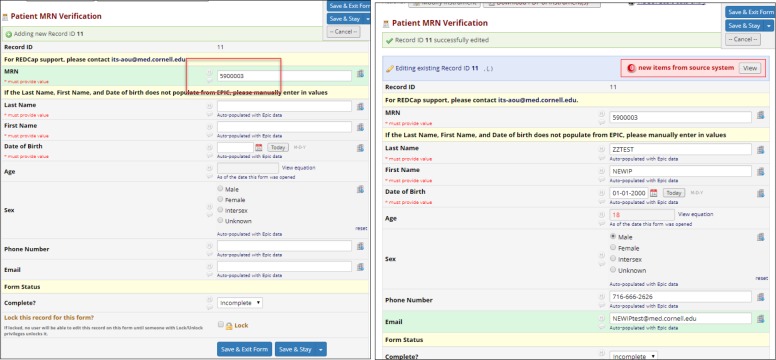
To manage participant identity, we created a semi-automated workflow using REDCap that consisted of case report forms configured for use with the dynamic data pull (DDP) plugin. As described elsewhere, DDP allows users to populate REDCap fields with data automatically extracted from an external database, such as an EHR system or data warehouse (15), based on entry of a unique identifier. To associate a local MRN with a unique research ID for a study, a study team member copied-and-pasted a patient’s MRN from the EHR into a REDCap case report form field. Based on entry of the MRN, the DDP plugin retrieved demographics—including first name, last name, date of birth, and sex—from the institutional data warehouse for adjudication by a study team member prior to saving into designated fields in REDCap (Figure 1).
Figure 1.
Entry of MRN (left) followed by retrieval and adjudication of values from EHR (right).
In addition to obtaining local demographics from the EHR, a study team member manually transcribed research ID and demographic values from the external consent system to REDCap. The semi-automated workflow ensured that a study coordinator reviewed and confirmed each association of a research ID with an MRN, while the DDP plugin reduced double data entry and risk of error for obtaining data from the EHR. For each study, we created a separate REDCap project.
Automated individual participant record transmission
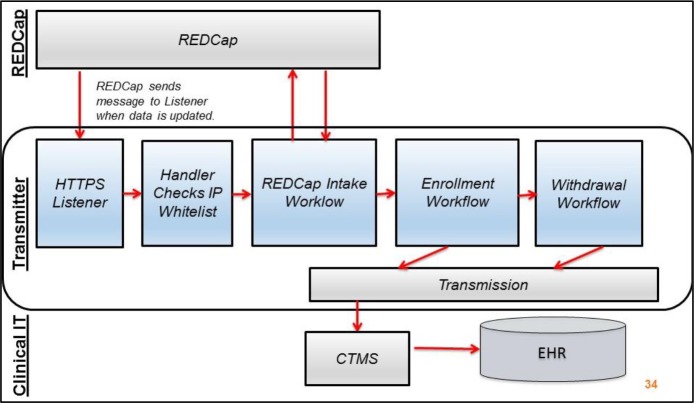
To make participant identity management data stored in REDCap available to other clinical and research information systems, we automated participant record transmission using novel middleware called Transmitter. Based on participant data saved in a REDCap project, Transmitter sent individual participant records from REDCap to the CTMS (Figure 2). Specifically, we configured the REDCap data entry trigger (DET) to send messages to Transmitter each time a study team member saved a record in a REDCap project representing a study. Upon receipt of a DET message, which identified the REDCap project and record ID responsible for the message, Transmitter called the REDCap API to retrieve all data for the record ID.
Figure 2.
Components of Transmitter and relationship to REDCap and clinical IT systems.
Transmitter then evaluated the returned values against study-specific rules, which required configuration by informatics staff, to determine a course of action, such as enrollment or withdrawal. For example, for a newly enrolled research participant in a particular study, a study team member saved values in REDCap for research ID, MRN, enrollment start date, date of birth from the EHR, and date of birth from the consent management system. Using the values obtained from REDCap, Transmitter determined whether the five fields contained valid input, including if the date of birth values from the EHR and external consent system matched. Upon satisfaction of study-specific rules, Transmitter sent a new enrollment record to the CTMS.
Automated bulk participant record ingestion
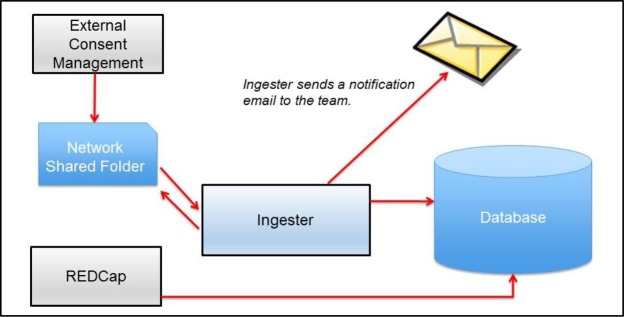
To augment participant identity management data stored in REDCap with external consent management system data, we automated ingestion of CSV files exported from external systems using novel middleware called Ingester (Figure 3). Rather than requiring study team members to copy-and-paste all participant details from an external system to an internal participant management system, Ingester combined a bulk export file from the external consent management system with data captured in a local participant management system on the basis of a shared a key across data sets, such as a participant research ID.
Figure 3.
Relationship of Ingester to external consent management and internal systems.
We configured Ingester to monitor a file folder on a network volume for files with particular filenames generated by external consent systems. Upon deposit of a file with a filename meeting criteria configured by informatics staff, Ingester performed a quality check on contents, such as confirming whether the number and names of fields were expected. If the contents met specifications, Ingester processed the deposited file for storage in a relational database management system. For files that did not meet screening criteria, Ingester discarded them and did not store contents in a relational database as a security precaution. After processing a file, Ingester notified informatics staff whether ingestion succeeded or failed. With ingested data from external consent systems available in the same database as participant identity management records from REDCap, informatics staff had the ability to combine data in an automated fashion to generate reports to support the study team.
Results
At our healthcare provider organization, the method has successfully supported the NIH All of Us Research Program and PCORI ADAPTABLE studies. Although workflows differed across the two studies, application of the method’s components—semi-automated participant identity management, automated participant record transmission, and automated ingestion of CSV files—required minimal customization by informatics staff. Study team members and institutional administrators have expressed positive feedback about automation produced by the method. On Github we have shared source code and documentation for Transmitter (https://github.com/wcmc-research-informatics/transmitter) and Ingester (https://github.com/wcmc-research-informatics/ingester) as well as the REDCap data dictionaries for AoU (https://github.com/wcmc-research-informatics/nih-pmi-data-dictionary) and ADAPTABLE (https://github.com/wcmc-research-informatics/adaptable-data-dictionary).
NIH All of Us Research Program
We initially developed the method to support our HPO’s participation in AoU. We implemented participant identity management for study go-live in July 2017 followed by automated ingestion of CSV files in October 2017 and automated transmission of individual participant records in December 2017.
Semi-automated participant identity management for the NIH All of Us Research Program has supported enrollment of more than 2,000 WCM patients as participants in the study’s first year. Because eligibility criteria for AoU are broad (i.e., 18 years of age and older), we configured REDCap DDP to retrieve demographics for MRNs of any adult patients at the institution stored in the research data warehouse. To obtain MRNs, study team members accessed the EHR and searched using patient name. Study team members copied-and-pasted research ID and other demographics from HealthPro to REDCap to ensure alignment with local MRN-based demographics. As described elsewhere in these proceedings, REDCap also enabled study team members to maintain appointment scheduling, compensation, and customer relationship management records linked to the external research ID and internal MRN of each participant (16). Although the method automated retrieval of demographics from the EHR, occasionally DDP failed to return demographics due to exogenous interface issues between the EHR and data warehouse, which led study team members to manually copy-and-paste demographics from the EHR.
Automated ingestion of CSV files exported from the NIH HealthPro system has reduced double data entry for study team members and enabled reports combining participant identity and relationship management data from REDCap with consent management system data from HealthPro. For example, rather than copy-and-paste dates of biospecimen collection and survey completion from HealthPro to REDCap for each participant, study team members obtained the data through ingestion of the CSV file, which joined with REDCap data using each participant’s unique PMI ID. To date, Ingester has processed 78 files successfully and 31 files unsuccessfully for AoU. Failures to process have been due to unexpected contents or filenames. In response to NIH announcements of changes to the HealthPro CSV format, informatics staff updated Ingester to meet new criteria, such as additional fields. Additionally, for AoU, we configured Ingester to automatically refresh data from REDCap when a user deposited a HealthPro CSV file. The deposit-CSV-file-to-refresh-REDCap-data mechanism served as a “button” for the study team to ensure records in reports were up to date as needed. As of this writing, study team members have accessed five different types of reports that combined data from REDCap and HealthPro to support AoU activities, such as compensating full participants.
Automated transmission of individual participant records from REDCap to the CTMS has enabled more than 1,400 AoU enrollments to display in the EHR and support billing compliance activities. Of note, we migrated about 630 enrollments from REDCap to the CTMS that predated Transmitter go-live. Additionally, Transmitter has processed 10 withdrawal messages to the CTMS. Informatics staff have regularly monitored Transmitter logs and communicated with CTMS staff to identify instances where MRNs from REDCap do not match patient identities in the EHR, allowing for manual reconciliation of edge cases.
PCORI ADAPTABLE
After deploying the method to support AoU, we extended it to ADAPTABLE, a study which launched at our HPO with manual workflows in January 2017. To replace manual processes, we implemented semi-automated participant identity management and automated ingestion of CSV files in July 2018.
Semi-automated participant identity management using REDCap replaced a manual Microsoft Excel-based approach and supported recruitment of 49 new potential participants within the first two weeks of system availability. Of note, we migrated more than 2,000 recruitment records from the legacy Microsoft Excel-based approach to REDCap prior to go-live. Rather than configure REDCap DDP to retrieve demographics for MRNs of any patient at the institution as in AoU, we configured it to retrieve demographics only for patients meeting the ADAPTABLE computable phenotype, which we stored in a separate data mart in the data warehouse. To obtain MRNs, study team members accessed the data mart via the web-based Microsoft SQL Server Reporting Services. After importing demographics from the data mart, study team members used REDCap to document contact with potential participants (e.g., telephone calls) to inform follow-up contacts.
Automated ingestion of CSV files from Mytrus has eliminated double data entry and enabled study team members to understand which patients from Weill Cornell Medicine enrolled in ADAPTABLE. In contrast to AoU, the ADAPTABLE study team associated research IDs created by Mytrus with each MRN of eligible Weill Cornell Medicine patients prior to contacting patients about participating in the study. When patients enrolled in the study by accessing an online participant portal hosted by Mytrus, they entered their pre-assigned research ID. As a result, the Mytrus CSV file exported by the study team indicated which research IDs had enrolled and not enrolled in the study. Through Ingester, the CSV file combined with the REDCap data to enable reporting to support study team recruitment activities. As of the time of this writing, automated individual participant record transmission from REDCap to the CTMS awaits institutional approval.
Discussion
We developed a method to bridge the gap between external research sponsor and internal clinical and research systems that has supported two large-scale studies, the NIH All of Us Research Program and PCORI ADAPTABLE, at our healthcare provider organization. The method, which combined a widely used electronic data capture system (14) with novel middleware we have made publicly available on Github, may generalize to other studies and settings. Adoption of the method in other healthcare provider organizations may enable informatics personnel to more quickly support study teams and streamline activities for study teams to meet scientific objectives.
After developing the method for AoU, we successfully extended it to ADAPTABLE. Notably, we designed the method to support AoU beginning with study launch while we retrofitted the method to ADAPTABLE after eighteen months of manual study processes with little informatics effort. Additionally, the modular nature of the method enabled us to disable automated participant transmission for ADAPTABLE and support a different enrollment workflow. Future work can evaluate whether the method can generalize to other studies, including industry-initiated clinical trials requiring use of external systems for consent and investigator-initiated studies from local investigators that may use REDCap for patient consent (17,18).
Other institutions have begun to adopt elements of the method, and the method can extend to support integration with additional systems and workflows. For AoU, Columbia University Medical Center has implemented Ingester, and at least one other AoU HPO has implemented the semi-automated identity management technique using REDCap DDP. Because of the modular nature of the method, it may also extend to systems beyond those used at our institution. We have demonstrated use of Transmitter with REDCap and our CTMS, but other institutions can generate new Transmitter libraries to integrate with other EDC (e.g., OpenClinica), CTMS (e.g., Forte OnCore), and EHR systems. Similarly, other institutions can extend Ingester to store records in Postgres, Oracle, and database management systems other than Microsoft SQL Server. Furthermore, institutions can extend Transmitter to support workflows beyond subject enrollment that require participant data, such as biospecimen laboratory processing. Future work includes containerizing (19) Ingester and Transmitter to ease installation and maintenance.
A limitation of the method is the time required of study team members for semi-automated participant identity management. Although automated approaches for patient identity management exist, they require customization in for implementation in local settings and require human effort for identity verification because they are not free from error (20,21). The semi-automated approach using REDCap has enabled study teams to verify identity at initial recruitment and again at final extraction of EHR data for submission to external research sponsors. In addition to the importance of adhering to privacy and confidentiality laws, correct data lays the best foundation for good science; misalignment of data (e.g., one participant’s biobanking data being incorrectly associated with another’s EHR data) threatens validity of analysis. Although automated identity management may require less time than the semi-automated approach described here, the need for human verification for EHR data extraction influenced our decision. Compared to a Microsoft Excel-based approach, the REDCap-based semi-automated approach is more scalable.
Following AoU launch, the NIH released an API for automated secure retrieval of data from the external consent system HealthPro. As of the time of this writing, we do not have API access and will explore its potential use as part of future research. Making use of the API could mean expending significant programmatic effort, which some healthcare provider organizations may lack. In other cases, HPOs might prefer to maintain a buffer between automated intake of data from NIH and enterprise systems. In light of such considerations, our method offers a viable approach for sites wanting to maintain a buffer, and the Ingester may obviate the need for custom API programming for sites that are content to use a CSV-based approach to external data integration. Regardless, external research sponsors need to consider integration with local systems and workflows at HPOs to ensure successful study participation.
Although institutions could seek to manage participant identity in a CTMS rather than REDCap, the local workflow at our institution—with only central research administrators and no study team members using the CTMS—dictated our design. However, based on our experience with AoU and ADAPTABLE teams at sites other than our own, use of REDCap or Excel is common for participant identity management. Although sites could choose to use a CTMS for identity management, REDCap (or OpenClinica) provides flexibility for additional workflows, such as relationship management (i.e., call log) and compensation management. REDCap offers form customization capabilities that can be implemented by a study team (or research informatics staff) with little or no programming in order to facilitate study-specific data capture and workflows; such functionality might not be available in a CTMS (or would require significant programming effort).
Storing both identity management records from REDCap and external consent system records automatically processed through Ingester in the same relational database has enabled reporting and data analysis that would otherwise be difficult to implement. Additionally, reporting functionality in external consent systems might not be sufficient for study team needs as external systems may lack local details (e.g., clinic site of consent). In the case of AoU, reporting that combines HealthPro and REDCap data has informed AoU executive decision-making with regard to targeted sites for accrual as enrollment efforts expanded over time, and we anticipate this to be the case for ADAPTABLE as well.
Conclusion
We developed a method consisting of three components—semi-automated identity management, automated participant record transmission, and automated bulk participant record ingestion—that integrated external consent management systems with local clinical and research systems and workflows to support two large-scale studies at our healthcare provider organization. Extending the method from one to two studies required little effort by informatics staff, and study teams have responded favorably to the automation enabled by the method. We encourage other settings to test generalizability of the method for supporting other healthcare provider organizations and studies.
Acknowledgments
Research reported in this publication was supported by the National Institutes of Health (NIH) Office of the Director (UG3 OD023163-01), NIH National Center for Advancing Translational Sciences (UL1TR000457), and the Patient-Centered Outcomes Research Institute (PCORI) (CDRN-1306-03961). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or PCORI.
References
- 1.Carey DJ, Fetterolf SN, Davis FD, Faucett WA, Kirchner HL, Mirshahi U. The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research. Genet Med. 2016 Feb 11;18(9):906–913. doi: 10.1038/gim.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008 Sep;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boutin N, Holzbach A, Mahanta L, Aldama J, Cerretani X, Embree K. The information technology infrastructure for the translational genomics core and the partners biobank at partners personalized medicine. J Pers Med. 2016 Jan 21;6(1) doi: 10.3390/jpm6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016 Feb;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Chen J, Collins R, Guo Y, Peto R, Wu F. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol. 2011 Dec;40(6):1652–1666. doi: 10.1093/ije/dyr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leitsalu L, Alavere H, Tammesoo M-L, Leego E, Metspalu A. Linking a population biobank with national health registries-the estonian experience. J Pers Med. 2015 Apr 16;5(2):96–106. doi: 10.3390/jpm5020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015 Mar 31;12(3):e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015 Feb 26;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernandez AF, Fleurence RL, Rothman RL. The ADAPTABLE trial and pcornet: shining light on a new research paradigm. Ann Intern Med. 2015 Oct 20;163(8):635–636. doi: 10.7326/M15-1460. [DOI] [PubMed] [Google Scholar]
- 10.Boutin NT, Mathieu K, Hoffnagle AG, Allen NL, Castro VM, Morash M. Implementation of electronic consent at a biobank: an opportunity for precision medicine research. J Pers Med. 2016 Jun 9;6(2) doi: 10.3390/jpm6020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Avolio L, Ferguson R, Goryachev S, Woods P, Sabin T, O’Neil J. Implementation of the Department of Veterans Affairs’ first point-of-care clinical trial. J Am Med Inform Assoc. 2012 Jun;19(e1):e170–6. doi: 10.1136/amiajnl-2011-000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campion TR, Blau VL, Brown SW, Izcovich D, Cole CL. Implementing a clinical research management system: one institution’s successful approach following previous failures. AMIA Jt Summits Transl Sci Proc. 2014 Apr 7;2014:12–17. [PMC free article] [PubMed] [Google Scholar]
- 13.Sholle E, Kabariti J, Johnson S, Leonard J, Pathak J, Varughese V. Secondary Use of Patients’ Electronic Records (SUPER): An Approach for Meeting Specific Data Needs of Clinical and Translational Researchers. AMIA Annu Symp Proc. 2017. pp. 1581–1588. [PMC free article] [PubMed]
- 14.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)-- a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009 Apr;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campion TR, Sholle ET, Davila MA. Generalizable middleware to support use of redcap dynamic data pull for integrating clinical and research data. AMIA Jt Summits Transl Sci Proc. 2017 Jul 26;2017:76–81. [PMC free article] [PubMed] [Google Scholar]
- 16.Turner SP, Pompea ST, Williams KL, Sholle ET, Cole CL, Kaushal R. AMIA Jt Summits Transl Sci Proc; 2019. Implementation of Informatics to Support the NIH All of Us Research Program in a Healthcare Provider Organization. [PMC free article] [PubMed] [Google Scholar]
- 17.Frelich MJ, Bosler ME, Gould JC. Research Electronic Data Capture (REDCap) electronic Informed Consent Form (eICF) is compliant and feasible in a clinical research setting. Int J Clin Trials. 2015 Aug 13;2(3):51. [Google Scholar]
- 18.Haussen DC, Doppelheuer S, Schindler K, Grossberg JA, Bouslama M, Schultz M. Utilization of a smartphone platform for electronic informed consent in acute stroke trials. Stroke. 2017 Oct 6;48(11):3156–3160. doi: 10.1161/STROKEAHA.117.018380. [DOI] [PubMed] [Google Scholar]
- 19.Wagholikar KB, Dessai P, Sanz J, Mendis ME, Bell DS, Murphy SN. Implementation of informatics for integrating biology and the bedside (i2b2) platform as Docker containers. BMC Med Inform Decis Mak. 2018 Jul 16;18(1):66. doi: 10.1186/s12911-018-0646-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kho AN, Cashy JP, Jackson KL, Pah AR, Goel S, Boehnke J. Design and implementation of a privacy preserving electronic health record linkage tool in Chicago. J Am Med Inform Assoc. 2015 Sep;22(5):1072–1080. doi: 10.1093/jamia/ocv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DuVall SL, Fraser AM, Rowe K, Thomas A, Mineau GP. Evaluation of record linkage between a large healthcare provider and the Utah Population Database. J Am Med Inform Assoc. 2012 Jun;19(e1):e54–9. doi: 10.1136/amiajnl-2011-000335. [DOI] [PMC free article] [PubMed] [Google Scholar]