Abstract
Acute Kidney Injury (AKI) in critical care is often a quickly-evolving clinical event with high morbidity and mortality. Early prediction of AKI risk in critical care setting can facilitate early interventions that are likely to provide ben- efit. Recently there have been some research on AKI prediction with patient Electronic Health Records (EHR). The class imbalance problem is encountered in such prediction setting where the number of AKI cases is usually much smaller than the controls. This study systematically investigates the impact of class imbalance on the performance of AKI prediction. We systematically investigate several class-balancing strategies to address class imbalance, includ- ing traditional statistical approaches and the proposed methods (case-control matching approach and individualized prediction approach). Our results show that the proposed class-balancing strategies can effectively improve the AKI prediction performance. Additionally, some important predictors (e.g., creatinine, chloride, and urine) for AKI can be found based on the proposed methods.
Introduction
Acute Kidney Injury (AKI) is a clinical event characterized by a sudden decrease in kidney function, which affected about 15% of all hospitalizations and more than 50% of patients in intensive care unit (ICU)1. Despite some precaution measures employed in hospitals, the incidence rate of AKI keeps increasing in recent years, which affects 13.3 million patients per year and resulting in 1.7 million deaths per year2. The mortality rate of AKI can reach 50% in the ICU and cause a considerable increase in healthcare expenditures that range from $5.4 to $24.0 billion in the United States3. In addition, AKI is associated with with end-stage renal disease, and chronic kidney disease, which might require ongoing dialysis and kidney replacement4.
According to the International Society of Nephrology5, identifying patients at risk of developing AKI may produce better outcomes than merely treating the established AKI. A direct method for AKI risk stratification is by analyzing any rise in serum creatinine level or decrease in urine output6. A number of biomarkers-based (e.g., NGAL, Cystatin C, and KIM-1, OPN, IL-18) methods were proposed for early detection of AKI by considering serum, plasma, or urine7. However, quantifying biomarkers is very expensive and time-consuming. Some score-based methods were used for the identification of AKI. Both the Acute Physiology and Chronic Health Evaluation (APACHE) II and Sequential Organ Failure Assessment (SOFA) severity scores were used to examine outcomes in patients with AKI8. Several studies assessed clinical decision support (CDS) tools for the identification of AKI9, 10. However, these methods mainly focused on the retrospectively identifying AKI patients, rather than on prospectively predicting AKI risk.
With the rapid development of computer hardware and software technologies, patient EHRs are becoming increas- ingly available. These data become a great resource for healthcare analytics. Recently, numerous data-driven models were proposed to predict AKI risk in different clinical settings11–16. Some of these methods used machine learning algorithms to analyze patient EHRs and develop models for the prediction of AKI in critical care15, 16.
Generally speaking, the prevalence of AKI is usually lower than 20%, which results in an imbalanced class ratio in collected patients dataset. This class imbalance could severely impact the prediction performance. This is because most machine learning classifiers are constructed to maximize the entire number of correct predictions, which are more sensitive to the majority class and less sensitive to the minority class17. Thus, if the imbalance issue is not properly addressed, the classification output can be biased towards majority class and lead to poor performance on AKI prediction. The misclassification of AKI prediction including false negative cases and false positive cases can affect the choice of therapeutic and the prognosis, which may increase the risk of condition deterioration and the overuse of medical resource, respectively. Most of previous studies on AKI prediction did not explicitly address the class imbalance problem. In addition, class imbalance is pervasive in many medical predictive modeling tasks. Thus, it is important to address the class imbalance problem and improve the performance of minority class prediction.
This study systematically addresses the class imbalance problem in AKI prediction. Specifically, the traditional statis- tical approaches, case-control matching approach, and individualized prediction approach are investigated. Our 5-fold cross-validation experimental results demonstrate that with class balancing techniques the prediction performance can be effectively improved. Compared with traditional statistical methods, case-control matching shows better perfor- mance, and individualized prediction approach performs the best.
Methods
Data Set
The patient data used in our study are from the Medical Information Mart for Intensive Care III (MIMIC-III) database18. The collection of MIMIC-III dataset is passive and de-identified, which is in compliance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule and does not produce significant impacts on patient safety15. This database was open sourced and freely accessible, and contained approximately sixty thousand admissions of pa- tients who stayed in critical care units of the Beth Israel Deaconess Medical Center between 2001 and 2012. In particular, this database included information such as patient demographics, vital signs, laboratory test results.
AKI Case Definition: In this study, we focus on stage-I AKI, which is defined by three criteria based on Kidney Disease Improving Global Outcomes (KDIGO) Clinical Practice Guideline19: (1) Increase in serum creatinine by ≥ 0.3 mg/dl (≥26.5 mol/l) within 48h; or (2) Increase in serum creatinine to ≥1.5 times baseline, which is known or presumed to have occurred within the prior 7 days; or (3) Urine volume < 0.5 ml/kg/h for 6h.
Patient Features: We extracted groups of features from MIMIC-III as follows. Additional details about features can be found at https://github.com/xuzhenxing2018/amia/blob/master/features_name.xlsx.
Demographics: Gender, age and ethnicity.
Medications: Medications20 that the patient take from the patients ICU admission until prediction time. We mainly considered the following categories: diuretics, Non-steroidal anti-inflammatory drugs (NSAID), radiocontrast agents, and angiotensin.
Comorbidities: Comorbidities that patients already have. We mainly considered the following categories: conges- tive heart failure, peripheral vascular, hypertension, diabetes, liver disease, myocardial infarction (MI), coronary artery disease (CAD), cirrhosis, and jaundice.
Chart-events: Vital signs measured at the bedside. We mainly considered diastolic blood pressure (DiasBP), glucose, heart rate, mean arterial blood pressure (MeanBP), respiration rate, SpO2, systolic blood pressure (SysBP), and temperature.
Lab-events: Laboratory test results. We considered bicarbonate, blood urea nitrogen (BUN), calcium, chloride, cre- atinine, hemoglobin, international normalized ratio (INR), platelet, potassium, prothrombin time (PT), partial throm- boplastin time (PTT), and white blood count (WBC).
We also consider the average of urine output and the minimum value of estimated glomerular filtration rate (eGFR).
Data Pre-Processing: If an ICU stay contained missing demographics and discrete variables, we deleted this ICU stay. For time-dependent continuous variables(e.g. lab, chart-events), we calculated statistics including the first, last, average, minimum, maximum, slope and the count based on observations during the observation window. Then we used mean imputation to fill missing continuous values and min-max scaler to normalize all these observations. And contained discrete variables (e.g. medication and comorbidities) were encoded as zero-one multi-hot vectors. In this way, each ICU stay is indexed by icustay id, and represented as a 147-dimension feature vector.
Experimental Setting
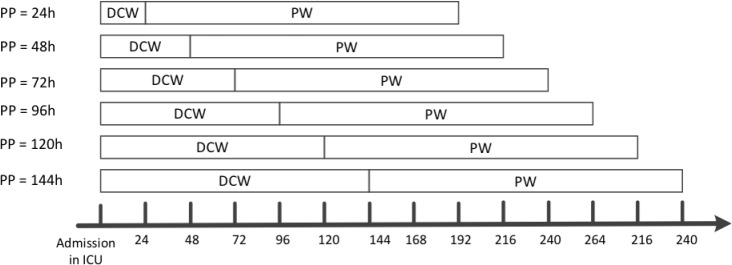
We adopted a predictive modeling setting with a rolling observational window design. Specifically, suppose t is the elapsed time (in hours) after the patient was admitted to ICU, we utilized the patient records in t to predict the AKI risk in the next 7 days, where t takes value in 24, 48, 72, 96, 120, 144 hours. An illustration of such rolling window design is shown in Figure 1. For each ICU stay, we obtained labels (AKI, or non-AKI) based on whether one of the three criteria in KDIGO definition was satisfied in the prediction window. Note that, if a patient met AKI criteria on admission to the ICU, we excluded the patient for prediction in order to avoid predicting AKI on top of AKI. We also excluded patients whose creatinine and urine data were missing for the whole time period that is being predicted.
Figure 1.
An illustration of AKI prediction. PP: prediction point; DCW: data collection window; PW: prediction window.
Handling the Class Imbalance Problem
The Class Imbalance Problem. The AKI class ratio is highly imbalanced in our data, which is detailed in Table 1. This may affect the prediction performance severely. We investigated the following strategies to handle such class imbalance problem. And all the class-balancing technique was applied to the training data after cross-validation.
Table 1.
The number of AKI and non-AKI samples in our dataset.
| 24h | 48h | 72h | 96h | 120h | 144h | |
|---|---|---|---|---|---|---|
| AKI | 8537 | 4778 | 3212 | 2239 | 1592 | 1195 |
| non-AKI | 30729 | 30703 | 30600 | 30486 | 30410 | 30337 |
| Total | 39266 | 35481 | 33812 | 32725 | 32002 | 31532 |
Traditional Class-Balancing Techniques. In this section, the classical under-sampling and over-sampling methods for addressing class imbalance problem are introduced.
The random under-sampling strategy (RU): One of the most common and simplest strategies to handle imbalanced data is uniformly random under-sampling of the controls to achieve equal number of cases and controls. This process will be repeated multiple times and a predictor is constructed for each created balanced dataset. Then these predictors will be combined through majority voting.
Cluster Centroids (CC): This technique uses K-means method to cluster the controls, and the cluster centroids will be extracted as the negative samples. The K was set as the number of cases to construct a balanced dataset.
Instance hardness thresholding (IH)21: This technique trains a classifier on the controls and removes the controls with lower probabilities. Due to the probability outputs, it is not always easy to acquire a specific number of samples. Classifier we selected was random forest classifier.
One-sided selection method (OS)22: This technique performs under-sampling method based on Tomeks links23 and 1-nearest neighbor rule to remove the noise samples from majority class samples.
Edited data set using nearest neighbors (ENN)24: This technique uses a nearest-neighbors algorithm and selection criteria to remove samples in the class to be under-sampled.
Synthetic Minority Oversampling Technique (SMOTE)25: This technique uses the existing minority class samples to synthesize elements for the minority class, which randomly chooses a point from the minority class and computes the k-nearest neighbors for this point, and then the synthetic points are added between the chosen point and its neighbors.
The combination of over- and under-sampling using SMOTE and Tomek links (ST)26: Since SMOTE does not consider any knowledge regarding the underlying distribution of samples, some noisy samples can be generated, e.g., when the different classes cannot be well separated. It is useful to employ an under-sampling algorithm to clean the noisy samples. Thus, the combination of SMOTE and Tomek links is performed, which uses SMOTE for over- sampling and Tomek links for cleaning.
The Case-Control Matching Strategy (MS). This method matches each case with a control based on the APACHE II score27 and Charlson comorbidity index28, and demographic information. Specifically, a matched control needs to (1) have the same gender as the case, and the age difference is within 5 years; (2) have the highest similarity score with the case measured by Manhattan distance based on APACHE II and Charlson comorbidity index. By this means, a resampled balanced training set was constructed with matched cases and controls.
Individualized Predictive Modeling (IS). Individualized predictive models are customized for a specific sample using the information acquired from similar samples. The similarity is measured with gower similarity which is one of the most popular measures of proximity for mixed data types29, 30. Compared to global models built on all samples, this strategy has the potential to find more individualized risk factors and obtain more accurate results. Specifically, the process for the individualized predictive modeling of a sample is shown as follows.
(1) Receive a sample S for testing and obtain feature vector which were summarized in the previous section of patient features. (2) Use gower similarity to find a cohort of K similar case samples and K similar control sample from all training samples, which constructs a cohort of 2K samples. (3) Build a model based on the similar sample cohort and predict label for sample S individually.
Predictive Models
For each predictive model, we used 5-fold cross validation to assess the performance. During each fold of iteration, we applied different balancing techniques to sample a balanced training dataset, on which classifier was trained. In general, we used global testing technique which means that all samples in testing dataset are predicted using the same classifier trained on the same training dataset. In particular, individualized predictive model used local testing technique, which means a customized classifier is trained on a customized training set and used to predict label for each sample in testing dataset.
We used several predictive models including £2 norm regularized Logistic Regression (Ridge)31, Elastic Net regu- larized Logistic Regression (EN)32, Random Forest (RF)33 and Gradient Boosting Decision Tree (GBDT)34. For the implementations of Ridge, EN, and RF, we used Scikit-learn software library35. For the GBDT, we chose XGBoost software library36. And the traditional balancing strategies were implemented with imbalanced-learn tool37. For class balancing techniques involving random (under or over) sampling, the sampling process were repeated 50 times. The gower similarity was calculated on the basis of the gower soft package38. We quantified our models with several robust measurements including AUC (the area under the receiver operating characteristic curve), recall, precision and F-score. For individualized predictive modeling, the size of the similar sample cohort were tuned from 200 to 600.
Results
Comparison of Different Methodologies with All Patient Features
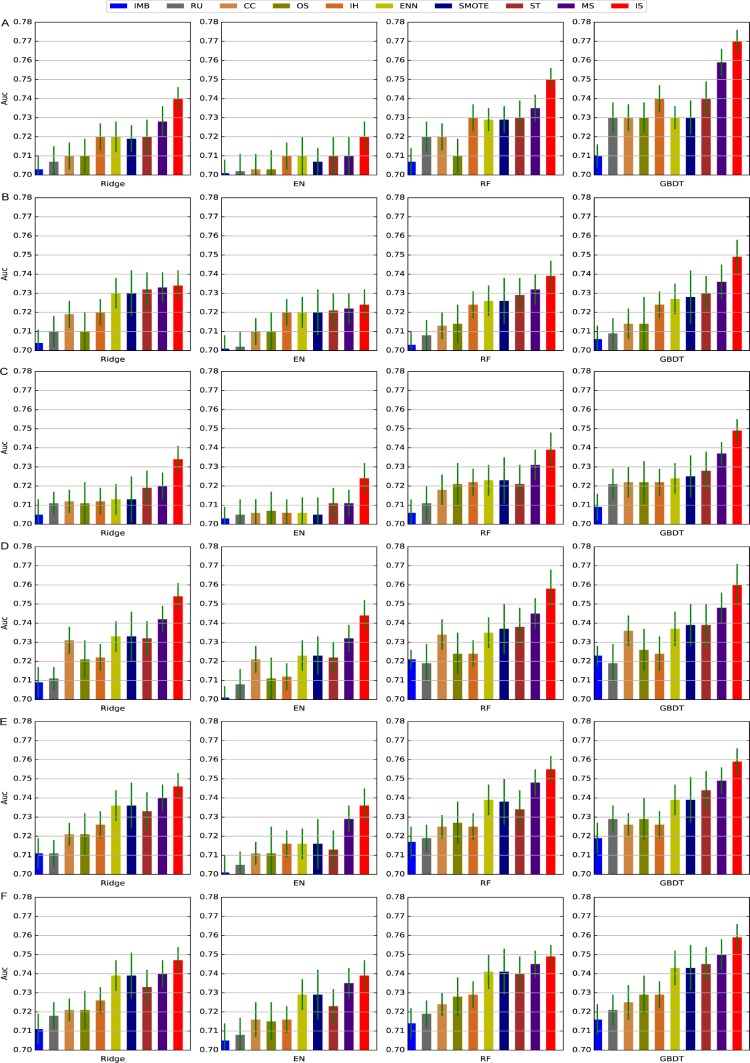
We tested the performance of different predictive models with different class-balancing techniques using all patient features with varying data collection windows. Figure 2 showed the performance of these approaches in terms of AUC. For performance in terms of precision, recall and F-score one can refer to https://github.com/xuzhenxing2018/amia.
Figure 2.
The AUC of different methods with 24-144 hours of data collection window. A-F denotes 24-144, respec- tively. IMB: imbalance data.
From Figure 2, we can observe that
Class balancing techniques can improve the prediction performance in general. This validates our assumption at the beginning that class imbalance will affect the performance of predictive models and class balancing techniques are necessary.
Case-control matching performs better than traditional class-balancing techniques. This suggests that determin- istic sampling is more effective than random sampling in this scenario.
Individualized predictive modeling performs the best. This implies the complexity of the AKI prediction prob- lem. Because of the complicated distribution of cases and controls, it is difficult to build a single model over the entire sample set with good prediction performance. Our individualized predictive modeling strategy can be viewed as an extreme case for local learning39, which is a learning strategy first divides the data space into different local regions and then build a predictor for each region. In our design, same number of case and control nearest neighbors are retrieved for each sample and thus the class imbalance problem no longer exists.
Comparing the performances of different predictors, we observe that GBDT obtains better performance.
Comparison of Different Feature Groups Using the GBDT
Five different groups of features were used to conduct AKI prediction. In this section we investigate the prediction performance of GDBT with different type of features with 24 hours of data collection window. The results are shown in Table 2. From the table we can observe that the laboratory features can achieve better performance than other feature groups. The comorbidity and medication feature groups demonstrated similar performance, and the demographic feature group contributed the least to AKI risk prediction. This is likely because ICU stay-level data rather than patient-level cohort were used for prediction. For example, if a patient had several hospital ICU stays and he/she might satisfy AKI criteria in one ICU stay and not in another. For this case, the demographic-only classifier would produce worse results, because it was difficult to distinguish this patient who belong to AKI and non-AKI class by just demographic features. Lab covariates provide specificity to each individual patient that can further classify their risk beyond traditional demographic and past medical history information.
Table 2.
The performance of different group features based on 24 hours data.
| Demog | Med | Comm | Chart | Lab | |
|---|---|---|---|---|---|
| Auc | 0.68 ±0.007 | 0.693±0.007 | 0.683±0.008 | 0.694±0.006 | 0.703±0.006 |
| Recall | 0.57±0.014 | 0.65±0.015 | 0.621±0.013 | 0.651±0.012 | 0.678±0.014 |
| Precision | 0.243±0.011 | 0.271±0.012 | 0.265±0.01 | 0.312±0.012 | 0.356±0.011 |
| F-score | 0.341±0.012 | 0.383±0.013 | 0.371±0.011 | 0.422±0.013 | 0.467±0.012 |
The Important Features Selected from all Feature Groups
There are 147 features in total used for AKI prediction. The importance of each factor in AKI prediction was further explored in experiment. The GBDT model was used to obtain the important score of each feature. Table 3 showed the most important features based on their importance scores and correlations with label, which were obtained by using the spearman correlation coefficient. The positive and negative signs indicated positive and negative correlation, respectively. From the table, we can find that CREATININE, CHLORIDE, and urine are more important, because they have strong correlation with AKI label. These results could corroborate well with some previous reports. For example, the decrease in urine output and the magnitude of increase in serum creatinine level can be used to determine the severity of AKI40. We also found that the minimum of eGFR value was informative, because eGFR is a number obtained by testing creatinine in blood, which can tell how well your kidneys are working. Besides, Chloride levels are associated with the severity of AKI41. And Ishikawa et al.42 found the percentage of patients with intraoperative hypoxemia (SpO2 < 90%) was significantly different between the AKI groups and none-AKI groups.
Table 3.
The top 10 features selected from all feature groups based on importance score. The value nearby name of feature is spearman correlation coefficient with label.
| 24h | 48h | 72h | ||||
|---|---|---|---|---|---|---|
| 1 | CREATININE slope | 0.0724 | eGFR min | -0.0431 | eGFR min | 0.004 |
| 2 | MeanBP slope | -0.0192 | RespRate slope | -0.0132 | SpO2 slope | 0.0769 |
| 3 | avg urine | -0.0457 | BICARBONATE slope | -0.0501 | DiasBP max | 0.0694 |
| 4 | eGFR min | -0.1129 | MeanBP slope | 0.0224 | avg urine | 0.1628 |
| 5 | RespRate avg | 0.0161 | Glucose slope | 0.0148 | MeanBP slope | 0.0451 |
| 6 | Glucose slope | -0.0329 | SpO2 slope | 0.054 | CHLORIDE count | 0.1585 |
| 7 | CREATININE last | 0.1423 | CALCIUM slope | -0.0051 | Temp slope | -0.0137 |
| 8 | CHLORIDE count | 0.1241 | PLATELET slope | 0.0183 | PTT last | 0.0793 |
| 9 | HeartRate slope | 0.0399 | CREATININE slope | -0.0046 | Glucose slope | 0.0337 |
| 10 | HeartRate avg | 0.0263 | age | 0.0605 | RespRate slope | -0.0294 |
| 96h | 120h | 144h | ||||
| 1 | avg urine | 0.2205 | SpO2 slope | 0.0995 | avg urine | 0.2448 |
| 2 | Glucose slope | 0.0436 | Glucose slope | 0.0473 | HeartRate slope | 0.0302 |
| 3 | Temp slope | -0.0239 | avg urine | 0.0243 | Glucose slope | 0.0451 |
| 4 | HeartRate slope | 0.0285 | Temp slope | -0.0239 | Temp max | 0.1655 |
| 5 | CHLORIDE count | 0.1817 | DiasBP max | 0.1194 | SpO2 slope | 0.1 |
| 6 | HeartRate max | 0.1401 | HeartRate max | 0.1146 | RespRate slope | -0.027 |
| 7 | eGFR min | 0.0404 | Temp max | 0.1578 | DiasBP max | 0.1244 |
| 8 | SpO2 slope | 0.0919 | MeanBP slope | 0.0408 | HeartRate max | 0.1445 |
| 9 | RespRate slope | -0.1315 | HeartRate slope | 0.0306 | Glucose max | 0.1112 |
| 10 | DiasBP max | 0.098 | Temp min | -0.0781 | DiasBP slope | 0.0292 |
The Comparisons of Different Size of Time Slot during Sampling Temporal Variable
We investigated the effect of time slots size during extracting feature for temporal variables on prediction. During fea- ture engineering, each temporal variable was compressed into a statistical vector calculated on observations recorded during overall observational window. The observational window could be split into several sub-windows, the time slots size was set to 2h, 4h, 6h, 8h, 12h and 24h in experiments. Within each time-slot, statistics for a temporal vari- able were got. Then we can concatenate these fine-grained statistics from all time slots together as a feature vector for a temporal variable.
The performance of different size of time slots were shown in Figure 3. This experiment were done based on 24 hours data using GBDT classification model and individualization predictive modeling. From these result, we could find that when the size of time slot was set to 8h, better results could been obtained. There were the worst results when the time slot was set to 2h, which might be because there were more missing values during sampling the variable values with high frequency, and simple imputation for missing values might distort the original distribution. And setting time slot to the observational window may lose time-dependency within a temporal variable.
Figure 3.
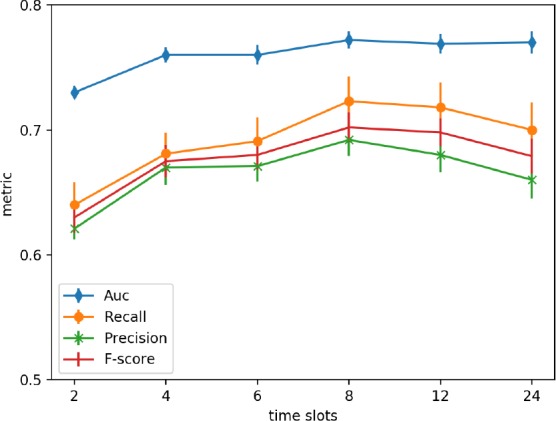
The performance of different time slots during sampling variable values.
Conclusion and Discussion
Predicting AKI accurately is helpful for clinicians to take properly measures for patients. The imbalance of case and control can produce worse results for the AKI prediction. This study investigated the impact of data imbalance on the performance of AKI prediction. Two strategies (case-control matching and individualized predictive modeling) were proposed to address class imbalance problem. These two strategies could construct balanced cohort by finding similar background cases and controls, which reduced the interference of noise data and captured important information for the prediction of AKI. In addition, we investigated most of class-balancing strategies. Some popular machine learning models (e.g., logistic regression, RF, and GBDT) were integrated with these class-balancing strategies for the AKI prediction. GBDT showed better performance than other methods for the AKI prediction in this study.
Prior models of risk prediction of AKI in critically ill patients typically use static clinical parameters43, 44. The utiliza- tion of real-time data as performed in this manuscript provides an opportunity to predict AKI with features specific to each individual patient. Dynamic real-time data may allow for clinical monitoring of AKI risk beyond solely static prediction upon ICU arrival. The use of dynamic clinical monitoring algorithms for AKI prediction may allow for incorporation into the electronic medical record11. The ability to identify patients at high risk of AKI using real-time data may allow for initiation of earlier intervention to prevent or reduce the deterioration of kidney function45. Further optimization of dynamic clinical risk prediction models can be further improved with the incorporation of novel blood, urine, imaging, and genomic biomarkers to allow for individualized precise detection of AKI risk.
Acknowledgement
This work was supported in part by NIH Grants 2R01GM105688-06 and 1R21LM012618-01.
Figures & Table
References
- [1].Peng Cheng, Lemuel R Waitman, Yong Hu, Mei Liu. Predicting inpatient acute kidney injury over different time horizons: How early and accurate? 2017;2017:565. [PMC free article] [PubMed] [Google Scholar]
- [2].Andrew JP Lewington, Jorge Cerdá, Ravindra L Mehta. Raising awareness of acute kidney injury: a global perspective of a silent killer. Kidney international. 2013;84(3):457–467. doi: 10.1038/ki.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jeremiah R Brown, Michael E Rezaee, Emily J Marshall, Michael E Matheny. Hospital mortality in the united states following acute kidney injury. BioMed research international. 2016;2016 doi: 10.1155/2016/4278579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lakhmir S Chawla, Paul W Eggers, Robert A Star, Paul L Kimmel. Acute kidney injury and chronic kidney disease as interconnected syndromes. New England Journal of Medicine. 2014;371(1):58–66. doi: 10.1056/NEJMra1214243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brian F Gage, Amy D Waterman, William Shannon, Michael Boechler, Michael W Rich, Martha J Radford. Validation of clinical classification schemes for predicting stroke: results from the national registry of atrial fibrillation. Jama. 2001;285(22):2864–2870. doi: 10.1001/jama.285.22.2864. [DOI] [PubMed] [Google Scholar]
- [6].Adil Ahmed, Srinivasan Vairavan, Abbasali Akhoundi, Gregory Wilson, Caitlyn Chiofolo, Nicolas Chbat, Ro-drigo Cartin-Ceba, Guangxi Li, Kianoush Kashani. Development and validation of electronic surveillance tool for acute kidney injury: A retrospective analysis. Journal of critical care. 2015;30(5):988–993. doi: 10.1016/j.jcrc.2015.05.007. [DOI] [PubMed] [Google Scholar]
- [7].Hilde RH de Geus, Michiel G Betjes, Jan Bakker. Biomarkers for the prediction of acute kidney injury: a narrative review on current status and future challenges. Clinical kidney journal. 2012;5(2):102–108. doi: 10.1093/ckj/sfs008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shigehiko Uchino, Rinaldo Bellomo, Hiroshi Morimatsu, Stanislao Morgera, Miet Schetz, Ian Tan, Catherine Bouman, Ettiene Macedo, Noel Gibney, Ashita Tolwani, et al. External validation of severity scoring systems for acute renal failure using a multinational database. Critical care medicine. 2005;33(9):1961–1967. doi: 10.1097/01.ccm.0000172279.66229.07. [DOI] [PubMed] [Google Scholar]
- [9].Christine J Porter, Irene Juurlink, Linda H Bisset, Riaz Bavakunji, Rajnikant L Mehta, Mark AJ Devonald. A real-time electronic alert to improve detection of acute kidney injury in a large teaching hospital. Nephrology Dialysis Transplantation. 2014;29(10):1888–1893. doi: 10.1093/ndt/gfu082. [DOI] [PubMed] [Google Scholar]
- [10].Stuart L Goldstein. Automated/integrated real-time clinical decision support in acute kidney injury. Current opinion in critical care. 2015;21(6):485. doi: 10.1097/MCC.0000000000000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jay L Koyner, Richa Adhikari, Dana P Edelson, Matthew M Churpek. Development of a multicenter ward- based aki prediction model. Clinical Journal of the American Society of Nephrology. 2016:1935–1943. doi: 10.2215/CJN.00280116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rohit J Kate, Ruth M Perez, Debesh Mazumdar, Kalyan S Pasupathy, Vani Nilakantan. Prediction and detection models for acute kidney injury in hospitalized older adults. BMC medical informatics and decision making. 2016;16(1):39. doi: 10.1186/s12911-016-0277-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li Zhi Zhou, Xiao Bing Yang, Ying Guan, Xing Xu, Ming T Tan, Fan Fan Hou, Ping Yan Chen. Development and validation of a risk score for prediction of acute kidney injury in patients with acute decompensated heart failure: A prospective cohort study in china. Journal of the American Heart Association. 2016;5(11):e004035. doi: 10.1161/JAHA.116.004035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Todd Wilson, Samuel Quan, Kim Cheema, Kelly Zarnke, Rob Quinn, Lawrence de Koning, Elijah Dixon, Neesh Pannu, Matthew T James. Risk prediction models for acute kidney injury following major noncardiac surgery: systematic review. Nephrology Dialysis Transplantation. 2015;31(2):231–240. doi: 10.1093/ndt/gfv415. [DOI] [PubMed] [Google Scholar]
- [15].Hamid Mohamadlou, Anna Lynn-Palevsky, Christopher Barton, Uli Chettipally, Lisa Shieh, Jacob Calvert, Nicholas R Saber, Ritankar Das. Prediction of acute kidney injury with a machine learning algorithm using electronic health record data. Canadian Journal of Kidney Health and Disease. 2018;5:2054358118776326. doi: 10.1177/2054358118776326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Scott M Sutherland, Lakhmir S Chawla, Sandra L Kane-Gill, et al. Utilizing electronic health records to pre- dict acute kidney injury risk and outcomes: workgroup statements from the 15 th adqi consensus conference; Canadian journal of kidney health and disease; 2016. p. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nathalie Japkowicz, Shaju Stephen. The class imbalance problem: A systematic study. Intelligent data analysis. 2002;6(5):429–449. [Google Scholar]
- [18].Alistair EW Johnson, Tom J Pollard, Lu Shen, H Lehman Li-wei, Mengling Feng, Mohammad Ghassemi, Benjamin Moody, Peter Szolovits, Leo Anthony Celi, Roger G Mark. Mimic-iii, a freely accessible critical care database. Scientific data. 2016;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Paul M Palevsky, Kathleen D Liu, Patrick D Brophy, et al. Kdoqi us commentary on the 2012 kdigo clinical practice guideline for acute kidney injury. American Journal of Kidney Diseases. 2013;61(5):649–672. doi: 10.1053/j.ajkd.2013.02.349. [DOI] [PubMed] [Google Scholar]
- [20].Yuan Luo, Yun Li. Medications documents. https://docs.google.com/spreadsheets/d/1G3Jj33m1BlfdOpaGMojEkSrDtNu3iBnORtYcmVTEX78/edit?usp=sharing.
- [21].Michael R Smith, Tony Martinez, Christophe Giraud-Carrier. An instance level analysis of data complexity. Machine learning. 2014;95(2):225–256. [Google Scholar]
- [22].Miroslav Kubat, Stan Matwin, et al. Icml. volume 97. USA: Nashville; 1997. Addressing the curse of imbalanced training sets: one-sided selection; pp. 179–186. [Google Scholar]
- [23].Ivan Tomek. Two modifications of cnn. IEEE Trans. Systems, Man and Cybernetics. 1976;6:769–772. [Google Scholar]
- [24].Dennis L Wilson. Asymptotic properties of nearest neighbor rules using edited data. IEEE Transactions on Systems, Man, and Cybernetics. 3(1972):408–421. [Google Scholar]
- [25].Nitesh V Chawla, Kevin W Bowyer, Lawrence O Hall, W Philip Kegelmeyer. Smote: synthetic minority over-sampling technique. Journal of artificial intelligence research. 2002;16:321–357. [Google Scholar]
- [26].Gustavo EAPA Batista, Ronaldo C Prati, Maria Carolina Monard. A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD explorations newsletter. 2004;6(1):20–29. [Google Scholar]
- [27].William A Knaus, Elizabeth A Draper, Douglas P Wagner, Jack E Zimmerman. Apache ii: a severity of disease classification system. Critical care medicine. 1985;13(10):818–829. [PubMed] [Google Scholar]
- [28].Mary E Charlson, Peter Pompei, Kathy L Ales, C Ronald MacKenzie. A new method of classifying prog- nostic comorbidity in longitudinal studies. Journal of chronic diseases. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- [29].John C Gower. A general coefficient of similarity and some of its properties. Biometrics. 1971:857–871. [Google Scholar]
- [30]. https://www.scribd.com/doc/7343061/gower-s-similarity-coefficient.
- [31].Saskia Le Cessie, Johannes C Van Houwelingen. Ridge estimators in logistic regression. Applied statistics. 1992:191–201. [Google Scholar]
- [32].Hui Zou, Trevor Hastie. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67(2):301–320. [Google Scholar]
- [33].Leo Breiman. Random forests. Machine learning. 2001;45(1):5–32. [Google Scholar]
- [34].Jerome H Friedman. Stochastic gradient boosting. Computational Statistics & Data Analysis. 2002;38(4):367–378. [Google Scholar]
- [35].Fabian Pedregosa, Gaël Varoquaux, Alexandre Gramfort, et al. Scikit-learn: Machine learning in python. Journal of machine learning research. 2011;12(Oct):2825–2830. [Google Scholar]
- [36].Tianqi Chen, Carlos Guestrin. Xgboost: A scalable tree boosting system. 2016:785–794. [Google Scholar]
- [37].Guillaume Lemaître, Fernando Nogueira, Christos K Aridas. Imbalanced-learn: A python toolbox to tackle the curse of imbalanced datasets in machine learning. Journal of Machine Learning Research. 2017;18(17):1–5. [Google Scholar]
- [38]. https://sourceforge.net/projects/gower-distance-4python/files/
- [39].Léon Bottou, Vladimir Vapnik. Local learning algorithms. Neural computation. 1992;4(6):888–900. [Google Scholar]
- [40].John A Kellum, Florentina E Sileanu, Raghavan Murugan, Nicole Lucko, Andrew D Shaw, Gilles Clermont. Classifying aki by urine output versus serum creatinine level. Journal of the American Society of Nephrology. 2015;26(9):2231–2238. doi: 10.1681/ASN.2014070724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hyung Jung Oh, Sungwon Kim, Jung Tak Park, et al. Baseline chloride levels are associated with the incidence of contrast-associated acute kidney injury. Scientific reports. 2017;7(1):17431. doi: 10.1038/s41598-017-17763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Seiji Ishikawa, Donald EG Griesdale, Jens Lohser. Acute kidney injury within 72 hours after lung transplan- tation: incidence and perioperative risk factors. Journal of cardiothoracic and vascular anesthesia. 2014;28(4):931–935. doi: 10.1053/j.jvca.2013.08.013. [DOI] [PubMed] [Google Scholar]
- [43].Kianoush Kashani, Ali Al-Khafaji, Thomas Ardiles, et al. Discovery and validation of cell cycle arrest biomark- ers in human acute kidney injury. Critical care. 2013;17(1):R25. doi: 10.1186/cc12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Marine Flechet, Fabian Güiza, Miet Schetz, et al. Akipredictor, an online prognostic calculator for acute kidney injury in adult critically ill patients: development, validation and comparison to serum neutrophil gelatinase- associated lipocalin. Intensive care medicine. 2017;43(6):764–773. doi: 10.1007/s00134-017-4678-3. [DOI] [PubMed] [Google Scholar]
- [45].Sehoon Park, Seon Ha Baek, Soyeon Ahn, et al. Impact of electronic acute kidney injury (aki) alerts with automated nephrologist consultation on detection and severity of aki: a quality improvement study. American Journal of Kidney Diseases. 2018;71(1):9–19. doi: 10.1053/j.ajkd.2017.06.008. [DOI] [PubMed] [Google Scholar]