Abstract
Characterization of a patient’s clinical phenotype is central to biomedical informatics. ICD codes, assigned to inpatient encounters by coders, is important for population health and cohort discovery when clinical information is limited. While ICD codes are assigned to patients by professionals trained and certified in coding there is substantial variability in coding. We present a methodology that uses deep learning methods to model coder decision making and that predicts ICD codes. Our approach predicts codes based on demographics, lab results, and medications, as well as codes from previous encounters. We are able to predict existing codes with high accuracy for all three of the test cases we investigated: diabetes, acute renal failure, and chronic kidney disease. We employed a panel of clinicians, in a blinded manner, to assess ground truth and compared the predictions of coders, model and clinicians. When disparities between the model prediction and coder assigned codes were reviewed, our model outperformed coder assigned ICD codes.
Introduction
Accurate identification, documentation, and coding of disease is important to health care, relating directly to patient care, revenue, and performance evaluation. ICD (International Classification of Disease) codes are used to classify mortality, define cohorts, evaluate health care policy, and drive health care finance, yet there is considerable inaccuracy and variability in these assigned codes1. Codes associated with acute care hospital stays in the United States are determined by certified and trained coders2. A study in 2014 from the Brigham & Women’s Hospital evaluating patients undergoing back surgery, showed that in those patients with degenerative disease, the primary ICD-9-CM diagnosis matched the surgeon’s diagnosis in only 48% of cases3. Thus, improving the accuracy of coded diagnoses will have a significant effect on many aspects of health care.
The use of electronic health records (EHRs) has increased dramatically with the introduction of the Health Information Technology for Economic and Clinical Health (HITECH) Act of 2009; from 9.4% in 2008 to 83.8% in 20154. Understanding the patient through a data driven approach is now possible by using EHR data to generate derived phenotypes5,6which include efforts, such as, eMERGE7,8. From data in the EHR, much can be learned about the patient both to more completely characterize their state of health, both past and present as well as ultimately predict who may be at risk for specific diseases going forward. While considerable material in the EHR is locked in text, critical information can be found in the discrete data elements, such as, diagnosis and procedure codes, medication lists and lab values. The ICD codes reflecting the underlying disease state of the patient is determined by trained coders who review the chart for relevant documented information. By presenting more complete information both to the care providers during the hospitalization, as well as to the coders at the time of chart review, potential gaps in documentation and care can be closed. This process is sometimes as simple as noting chronic conditions coded from previous encounters that are unlikely to have been resolved, for example, end stage renal disease or diabetes. In a more complex scenario, a series of lab measurements can be used to infer a state which has not been explicitly noted, for example a spike in creatinine levels in blood suggesting acute renal failure.
In this work, we analyzed 83,987 inpatient encounters including more than 5,000 features. A feature is an attribute or value that is used by the model to learn from, most of which are results of lab tests. We applied machine learning methods to three test cases to see if we could identify patients with three common diseases during a hospitalization: diabetes, chronic kidney disease and acute renal failure. These three disease states were chosen as they are common conditions that can be implied from a record of laboratory test values and medications. As input, our models used only demographic, laboratory and medication data plus past diagnoses if available. This approach, using data easily available at the time of discharge, allows us to identify patients who may have a diagnosis of diabetes, chronic kidney disease or acute renal failure, but who were not recognized as such at the time of discharge. This will drive improved ICD coding and allow a better view of an individual, of the population, and permit creation of more robust derived clinical phenotypes, based on more accurate ICD codes.
There have been many previous works discussing use of machine learning methods on EHR data, and there are comprehensive review papers on the topic9,10. Previous attempts to predict chronic kidney disease or failure, for example, have relied on a single relatively small well-curated data set11,12. These studies reported very high predictive accuracy, however do not address the overwhelming amount of heterogeneity and lack of structure in real-world prediction scenarios. More recent works have explored deep learning methods and showed that they work better than simpler methods with real EHR datasets. Some studies have focused on patients’ background information for predicting clinical events using recurrent neural networks (RNN)13,14,15,16. Others have employed convolutional neural networks for discovering patterns17or finding diagnosis codes18. Another study applied deep learning on EHR data to suggest candidates for palliative care through a fully connected deep neural network19. In our study, we avoid limitations of previous works by completely avoiding manual curation of data or features, while working towards an approach which we expect to generalize well in the prediction of a wide range of disease codes.
Methods
Data Source
Inpatient encounter data for adults age greater than equal to 50 years was extracted from the Cerner HealthFacts database, a large multi-institutional de-identified database derived from EHRs and administrative systems20. The HealthFacts database was determined by the Stony Brook University Institutional Review Board to be non-human subjects research. Chosen inpatient encounters needed to have at least one diagnosis code and at least one laboratory test. Extracted data was mapped to the OHDSI Common Data Model (version 5.3) and vocabulary release (2/10/2018). The OHDSI common data model is becoming the standard data model for observational health studies21,22,23. Laboratory tests were mapped directly using LOINC codes, diagnoses and procedures were mapped from ICD-9-CM. Inpatient administered medication were mapped from NDC (National Drug Code) to RxNorm RxCUI, and if the NDC did not match then full name of the drug was matched to RxNorm drug names.
The HealthFacts database contains data from 599 facilities, however many of the facilities are community hospitals with a small number of beds and contain a mix of inpatient and outpatient encounters. We started by selecting acute-care medical facilities which have laboratory tests and coded diagnoses in the database. We selected the facilities with the largest volumes of inpatient encounters. For the bulk of the results discussed here we focused on a single 500+ bed urban academic medical facility located in the south of the United States which is identified as hospital 143. We also tested the model building process on another facility (67) and on subset of patients from the 10 highest volume facilities. This acute-care facility (143) was selected randomly from among the top 10 inpatient volume facilities in the HealthFacts data. The data extracted were for inpatient encounters starting from 8/24/2006 and ending in 12/31/2013. It is important to note that we do not necessarily have any previous information for all of these patients, and we did not intentionally select a cohort of patients with rich historical data. By considering this, we are solving a more difficult and generalizable problem. Also, this strategy gives us flexibility to cover more input features and more disease codes without any specific restrictions. In this manner, the strategy for using RNNs does not apply, as a rich background is required for RNN such as in a previous study15which had 54 visits per patient on average. Here, for more than 2/3 of patients, we do not have any background information at all.
Features Description
Demographic Information: Age, gender, race and ethnicity of the patient are added to feature space, as this information are important factors in disease development, severity, and prevalence. For categorical features such as gender, race and ethnicity, we took advantage of one-hot encoding scheme for converting these into vector format.
Laboratory Tests: Results from labs prescribed for the patient; commonly, blood tests, urine tests, or blood pressure. For instance, a blood test could contain information such as Sodium [Moles/volume] in Blood or Glucose [Mass/volume] in Blood. How features are built from laboratory tests will be expanded in detail in later sections.
Medications: We take advantage of the number of times each medicine is ordered for the patient during their inpatient stay. Currently, we are not considering any information related to dose of medications in our model. Since this information are subject to highly vary among patients, we automatically filter medications based on their relation to the disease, and get rid of unrelated medications to each disease, explained further in details.
Past History: In this facility, two-thirds of the patients did not have any prior visits which makes incorporating this information in a model challenging. When this information is available it can be extremely useful for making predictions. For example, a patient who has previously been assigned a diagnosis of end stage renal disease almost certainly has chronic kidney disease. Data elements, such as demographics, are more or less static through time, as a result we do not consider their historical pattern. Past information can become outdated, for example, a blood glucose test from 2 years ago is less relevant than if the test took place a day ago. Discrete data elements including demographics, medications and laboratory data from the current encounter were analyzed, as well as past medical conditions, when available. For this purpose, for each encounter of the patient, we aggregate all previous encounters diagnosis codes into one vector, showing whether the patient ever had a previous ICD code. This simplifies the temporal dimension and allows the model training process to incorporate this vital information.
Constructing a Dataset for Supervised Learning
Combining all information described earlier is extremely valuable, but there are two main challenges. First, the amount of available information for a single encounter can be highly variable. Even when patients are diagnosed with the same disease the hospital course can be variable. Second, a patient can have multiple lab tests during a single encounter. As neither the tests or the number of repeats is fixed, it is not possible to directly code the laboratory test results in the matrix.
For overcoming these two issues we came up with two strategies. First, we simply filter out information unrelated to each disease without any background knowledge about the disease and only based on training data. We select common features between patients diagnosed with that disease. Although with deep learning algorithms having an enormous feature set is not necessarily prohibitive, for speed and memory considerations, features that obviously have nothing to do with the outcome are removed when possible. To accomplish this, we use a rule of thumb: for each disease we considered patients who had that disease, and then selected only features that are common between at least 5% of these patients. By common features, we mean there is a non-empty value for that feature in 5% of encounters with positive outcome. This simple but effective technique helps us to get rid of irrelevant features including labs and medications for unrelated diseases. For instance, by applying this technique, the number of features reduced from 5,347 to 880 for predicting the diagnosis of diabetes.
Second, to manage the variety of times a patient took a test, instead of retaining exact values of each lab, we aggregate these results into summary statistics. These statistics include: the count, median, minimum, maximum, and delta (most recent value minus the first value) of each lab value. Additionally, each lab value was mapped to a semantical category based on each facility’s own standards, i.e. the number of times a test was low, within range, high, normal, abnormal or other. For example, if patient’s glucose in blood was measured 20 times during the encounter, these 20 values might be summarized to the following aggregated values: count (20), min (86), max (231), median (135), high values (15), normal values (5), low values (0), and a delta (72). For a patient who took the test two times we again map that to these features, with a count of 2 retaining the information that the test was only performed twice. By doing this, we reduce the variable feature set to a fixed number of values for each lab. Now two patients are represented by identically structured data, and are straightforward to compare with each other programmatically, regardless of the number of times a lab was ordered. Previous studies have used a similar strategy19,24, which aims at maintaining salient information while easing computability.
In this effort, we focus on prediction of ICD-9-CM codes associated with three diseases. The disease categories include: Diabetes mellitus, acute renal failure, and chronic kidney disease. The ICD-9-CM codes for diabetes were defined using the ICD-9-CM codes associated with CCS (Clinical Classification Software25) codes (49, 50). The codes for acute renal failure (ICD-9-CM: 584.5-584.9, 958.5) and chronic kidney disease (ICD-9-CM: 585.3-585.9, 586, 403.01, 403.11, 403.91, 404.02, 404.03, 404.12, 404.13, 404.92, 404.93, V42.0, V45.11, V45.12, V56.0-V56.8) are from code mappings developed by Vizient, Inc. for their risk adjustment models.
In summary, we created our feature set by combining all information and features available, aggregating each lab test into summary statistics, filtering relevant information to the disease and ignoring the rest (explained in detail in the next section). We face this problem as a binary classification problem and grouped all ICD codes for each disease into one binary value. In the next sections, we introduce our proposed method and measure its functionality and accuracy in detail.
Model Methodology
We explored multiple models, including logistic regression and random forests. However, as there are many features that could be used to predict any disease, we decided to take advantage of deep learning methods as well. Deep Learning is well suited to handle huge amounts of data as input without an explicit feature selection step. Our deep learning approach was based on a multilayer perceptron with fully connected layers. In this manuscript, we compared results of deep learning models with machine learning methods, but for each machine learning method, we used the same data preprocessing procedure.
The first step was to create the feature space from available data. For each disease we retained only common labs, medications, and past diseases-- only features present in more than 5% of cases with the disease. We added demographic information to this list and created a feature vector for each encounter. This process retained approximately 900 features for each disease from a pool of over 5,000 features.
After construction of the feature matrix for our training cases, we imputed any missing data. Even after filtering out rare laboratory tests, a majority of the data set had missing values that needed to be filled. We started by filling these missing values using the median of available lab values. This gave us the capability of running learning methods on the dataset, however the median test result of all patients likely represents a ‘normal’ value, thus our replacement procedure was tantamount to assuming any test that was not ordered would return a normal result. We also tried more complicated imputation methods like MICE26, Soft-Impute27and SVD-Impute28but these did not improve performance enough to justify their added complexity. In our investigations, we noticed that many key features which would typically be correlated and thus helpful for imputation were measured in groups, e.g. the features representing a complete blood count all occur together or not at all. Therefore, even a complex imputation algorithm has a hard time inferring informative values for those.
After preparing the dataset by filtering out unrelated features and filling missing values by imputation, the dataset was then normalized. We kept 20% of patient encounters untouched for testing and evaluating the model, and the rest were used for training and validation purposes.
For training the deep learning model the Python programming language (2.7), Keras framework29with underlying Tensorflow30, and scikit learn library31were used for developing and testing this research project. The training was performed on an NVIDIA Tesla V100 (16GB RAM).
Results
Model Architecture
We employed a Multilayer Perceptron, Deep Neural Network32. The input layer, depending on the disease, has approximately 900 features, and the output has a single node with a sigmoid activation function. After hyperparameter searching, we determined that a network with 8 hidden layers worked well. Of the activation functions we considered (tanh, ReLU, and SeLU), tanh performed better than others. The network was optimized with the Adam optimizer33. We added regularization and dropout to each hidden layer to avoid overfitting to the training set. For the loss function, mean squared error was used.
Since the input dataset was imbalanced in terms of patients with disease, versus patients without the disease, we specified class weights of input labels for calculating loss and added more importance to positive cases. In the predicted use case of our model, i.e. detecting existing diseases that have not yet been coded, false negatives were more important than false positives, and thus our loss function reflected this.
Deep Learning VS Baseline Methods
First, we compared the results of the fully connected, optimized, deep learning model to logistic regression and random forests (Table 1). For these machine learning methods, we tried both knowledge-based features selected from a team of experts as well as automatic feature selection. A gridsearch was applied to optimize both the number of features to be extracted and the max depth of the random forest model. Only the best-case result is reported below in Table 1 when comparing to deep learning. Results of the automatic feature selection algorithm surpassed all knowledge-based feature sets (result not shown).
Table 1.
Deep Learning vs. Baseline Methods Results
| Learning Method | Disease | Accuracy | Precision | Recall | F1-Score | AUC ROC |
|---|---|---|---|---|---|---|
| Random Forest | Diabetes | 84.59 | 72.21 | 80.44 | 76.10 | 90.53 |
| Logistic Regression | Diabetes | 85.63 | 75.22 | 78.86 | 77.00 | 89.94 |
| Deep Learning | Diabetes | 87.12 | 75.93 | 84.60 | 80.04 | 91.53 |
| Random Forest | Chronic Kidney Disease | 87.37 | 61.32 | 85.02 | 71.25 | 93.62 |
| Logistic Regression | Chronic Kidney Disease | 87.77 | 62.20 | 85.51 | 72.02 | 93.88 |
| Deep Learning | Chronic Kidney Disease | 90.91 | 74.35 | 77.26 | 75.77 | 94.24 |
| Random Forest | Acute Renal Failure | 83.59 | 46.91 | 85.41 | 63.01 | 91.50 |
| Logistic Regression | Acute Renal Failure | 84.31 | 51.27 | 82.93 | 63.37 | 91.23 |
| Deep Learning | Acute Renal Failure | 89.06 | 66.27 | 67.43 | 66.86 | 91.94 |
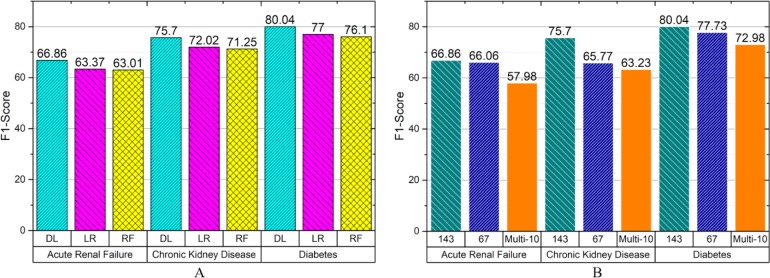
Since the disease prevalence is typically far from 50%, for instance in acute renal failure and chronic kidney disease there are only 16% positive cases, and for diabetes about 30%, accuracy would be a poor measurement for comparing results with each other. For this purpose, recall, precision, F1 and AUC-ROC are calculated, however, AUC-ROC also could be misleading when the dataset is imbalanced19,34,35. Because detecting rarer positive cases are defined as more important in this problem, F1 alone is a good summary measurement for comparing different algorithms with each other (Figure 1 A).
Figure 1.
(A) F1 comparison of different learning methods across three different diseases. Deep Learning (DL), Logistic Regression (LR) and Random Forest (RF) are shown. (B) Performance of deep learning prediction in other data sets, for facilities with identifier 143, 67, or the multi-10 dataset consisting of 10 different facilities.
To estimate the generalizability of this process, we applied the same training and testing process to data from one other facility (67), as well as a pool of 10 other facilities (Figure 1 B). In diabetes, the F1 score was 80.0 or 77.7 when training and testing on independent subsets of encounters from a single facility, but the F1 score dropped to 73.0 when data from 10 different facilities were included in the training and testing sets. In the most difficult challenge, we trained on data from one facility, and tested on data from other facilities, resulting in an F1 score of 57.9.
Validation with Review by Experts
Having established that a deep learning model could reproduce existing coder-generated ICD-9 codes, we investigated false positive and false negative cases where the model contradicted the documented diagnosis. We sampled a set of encounters from the test set where the model predicted with high confidence that the existing code was incorrect (>85% for positive cases, and <15% for negative cases). For three diseases, we considered 20 false positives and 20 false negatives. This resulted in a total of 120 cases for further review. We then asked 3 board-certified practicing physicians to review these cases ("experts"). We asked them two questions: (1) Whether the patient does or does not have the disease, and (2) Whether the experts are answering with high or low confidence. In addition to these 120 cases of disagreement between the model and the coders, we also reviewed 40 cases where the model and coders agreed on diabetes status, and the experts concurred in all 40 cases.
Importantly, the group of experts as well as the test administrator were blind to the model predictions and current diagnosis codes. Experts were provided with all information available to the model regarding the patient, including demographic information (age, race, gender, ethnicity), lab results summary statistics as described in data section, history of past diagnosis (if available), medications prescribed for the same encounter. Furthermore, detailed individual lab results with relative time stamps were prepared for this group (i.e. all tests, not just the summary statistics) to provide as complete a picture as possible.
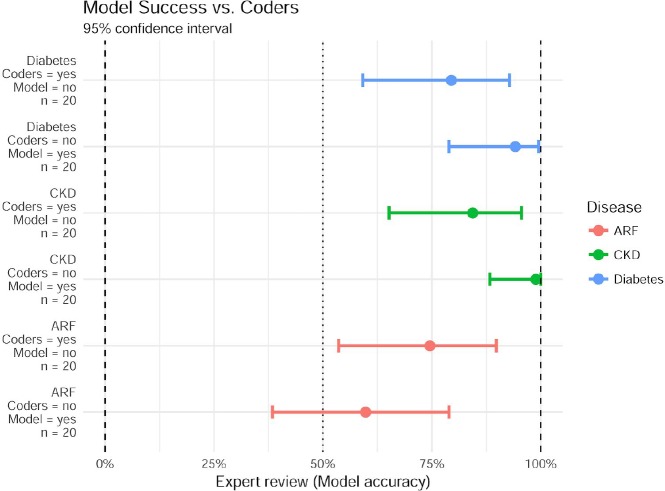
For cases in which the model predicted presence of diabetes, but the code was not documented, the experts overwhelmingly agreed with the model prediction (19/20). For cases in which the model predicts no disease, but a code indicates diabetes was present, the experts still agreed with the model in (16/20) cases. Results for diabetes and both types of kidney failure are summarized in figure 2. For all 3 diseases surveyed, when the model challenges the result of a coded disease, the model is correct significantly more than 50% of the time (p<0.025). The model was also exceptionally good at recognizing false negative (missed codes) for diabetes and chronic kidney disease but was not significantly better than a coin flip at identifying new uncoded cases of Acute Renal Failure. Nevertheless, a system that succeeds at even 50% of challenged diagnosis codes has the potential for enormous impact.
Figure 2.
Using expert review as ground truth, data shows the success of model predictions in cases where they disagree with existing codes. Model accuracy is shown as median (dot) and the 95% confidence interval (bar) using a beta-binomial model with Jeffreys prior. CKD - Chronic Kidney Disease, ARF - Acute Renal Failure.
Top Features
A deep learning method, unlike a logistic regression method for instance, does not provide transparent outputs of exact importance for each feature. The model is composed of multiple operations in each layer, including an activation function which makes sensitivity analysis complicated. However, it is important to see whether the model is actually deciding based on reasonable features, and which features are the most important to the model. We therefore generated a heuristic function to get a better look inside this complex model.
In this heuristic function, we multiplied weight matrices used to compute each node of the the network and cascaded this process until reaching a single vector the same size of input features. This methodology, although imperfect, provides good approximation since activation functions (tanh specifically) at least do not change the order of inputs. For instance, if inputs w1> w2, the respective outputs of tanh activation function are a1> a2. An example of these results is shown in Table 2.
Table 2.
Top 5 features for Diabetes detected by deep learning algorithm for different feature categories.
| Category | Important Features | Magnitude |
|---|---|---|
| Measurements/Observation | Glucose finger stick - High Count | 3.10 |
| Glucose finger stick - Total Count | 2.94 | |
| Lipase [Enzymatic activity/volume] in Serum or Plasma - Min | 1.53 | |
| Lipase [Enzymatic activity/volume] in Serum or Plasma - Median | 1.53 | |
| Glucose finger stick - Within reference range Count | 1.40 | |
| Medications | Humalog | 2.55 |
| Novolin R | 2.35 | |
| NovoLog | 2.22 | |
| Lantus | 2.11 | |
| Glucophage | 2.07 | |
| Patient Background Information (0/1) | Previously had Diabetes | 1.93 |
| Past Information available | -0.97 | |
| ICD 250.00, Diabetes mellitus without mention of complication | 0.91 | |
| ICD 250.60, Diabetes with neurological manifestations | 0.51 | |
| ICD 401.9, Unspecified essential hypertension | -0.48 |
The two highest scored features in the prediction of diabetes were the total count of glucose finger sticks as well as the number of glucose measurements that were high. We believe that this was evidence of the model not only capturing patients who had repeatedly high glucose levels (indicative of uncontrolled diabetes) but also those patients which were being constantly monitored for glucose but did not necessarily have abnormal results (indicative of controlled diabetes). In this way, the count of tests is often an indicator of what the physician was thinking and knowing what they were testing was independently important with respect to the results of the test.
When the magnitude is negative, it suggests that an increase in the feature relates to not having the disease. For instance, the “Past Information available” coefficient is negative, at first glance suggesting that among patients with background information, they were less likely to have diabetes. However, having previous information with diabetes coded had an even larger coefficient which appears to make up for the negative impact of the generic “Past Information available” feature. In other words, if the patient had information from previous encounters available, but diabetes was not coded previously, they were less likely to have diabetes according to the model.
Discussion
ICD codes are commonly used to classify the disease states present during a hospital encounter and are used to characterize the patient, generate derived phenotypes, populate disease registries, and drive public health decisions. Today, while many clinical health terminologies are used, including the more comprehensive and precise SNOMED CT, in the US the most available information is available in ICD codes, assigned by trained coders upon discharge.
These ICD codes, as generated today, have limited reliability1. Improving the quality of the ICD codes would allow a more accurate representation of the patient, of the population and allow for more robust derived phenotypes. Leveraging deep learning methods, we have developed a methodology that employs readily available discrete data elements to train a model capable of improving ICD code accuracy. We expect our model could be used to flag encounters for which the existing coding data are suspect. Flagged cases could be reviewed, either by humans or through an informatics process able to leverage additional data sources such as text notes.
One goals of this project was to create a “disease-agnostic” system which required no domain knowledge in the development of each model. An important caveat here is that the conditions we targeted are common conditions that can be associated with patterns of discrete data elements. The case study of acute renal failure also highlighted a shortcoming of our model that may be improved in future versions. Notably, the criteria for defining acute failure which was used by our panel of experts included evaluating the maximal change in serum creatinine levels over a 48-hour period. While this time scale information was available initially, it was lost to the model during the aggregation of the serum creatinine feature into summary statistics which no longer included time scale, and thus may be partially responsible for the relatively lower predictive performance for ARF.
The diseases we chose to focus on in this paper are relatively predictable, and may represent an optimistic assessment of the potential of models to predict other disease codes. Codes that are more rare or for which there are no lab-based diagnostic tests should prove to be harder to predict. However, we expect future versions to be able to include features from additional data sources such as text notes and images. Furthermore, the difficulty of coding a disease may correlate between models and coders. Aside from accuracy, we also reviewed the experts’ confidence level in their answers. Of the 35/40 disputed diabetes encounters where the experts agreed with the model, 29 (82.8%) of the times, the experts responded with high confidence. However, for the 5 cases in which the experts disagreed with the model, only 3 (60%) were highly confident. While these data are not statistically conclusive (due to the small number of times that the model was incorrect), they still represent a comforting trend that suggests our model struggles at the same times that the experts themselves are less confident.
There are a variety of limitations to this study. Primarily, our methods were thoroughly evaluated and validated on only a single deidentified acute care facility. This approach shows that models can be built and tested on a single acute care facility, but that more work will need to be done on testing and training across multiple acute care facilities. One likely limiting factor in cross-facility success is the difference between the way different facilities record data. For example, the lab values for “glomerular filtration rate” are recorded by one facility as a continuous variable, whereas another facility reported continuous values only from 0-60, and report “60+” for any value greater than 60. In another example, a single facility had no values reported whatsoever for the feature “glucose finger stick”. We expect that this limitation is not insurmountable, given the depth of training data available.
Another limitation of our work is that we did not predict to the level of the individual ICD-9-CM code but rather to a cluster of codes that are used in Vizient risk models and AHRQ’s CCS coding. While not specifically useful for predicting billable codes yet, our model can currently predict phenotypes sufficiently for population health applications.
In conclusion, the results suggest that deep learning models have the potential to provide ICD coding critiques that can be used in a pipeline to improve coding accuracy. While our initial results target three common conditions, the methodology should be applicable to many other common conditions that deep learning algorithms can characterize by patterns of discrete data elements.
Acknowledgements
We thank the Stony Brook Medicine Health System for discussions with the team on the coding and clinical documentation improvement process.
Figures & Table
References
- 1.O'malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, Ashton CM. Measuring diagnoses: ICD code accuracy. Health services research. 2005 Oct;40(5p2):1620–39. doi: 10.1111/j.1475-6773.2005.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Health Information Management Association [Internet] 2018 [cited 2018 Aug]. Available at: http://www.ahima.org/
- 3.Gologorsky Y, Knightly JJ, Lu Y, Chi JH, Groff MW. Improving discharge data fidelity for use in large administrative databases. Neurosurgical focus. 2014 Jun;36(6):E2. doi: 10.3171/2014.3.FOCUS1459. [DOI] [PubMed] [Google Scholar]
- 4.Henry J, Pylypchuk Y, Searcy T, Patel V. Adoption of electronic health record systems among US non-federal acute care hospitals: 2008–2015. ONC Data Brief. 2016 May;35:1–9. [Google Scholar]
- 5.Post AR, Kurc T, Cholleti S, Gao J, Lin X, Bornstein W, Cantrell D, Levine D, Hohmann S, Saltz JH. The Analytic Information Warehouse (AIW): A platform for analytics using electronic health record data. Journal of biomedical informatics. 2013 Jun 1;46(3):410–24. doi: 10.1016/j.jbi.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cholleti S, Post A, Gao J, Lin X, Bornstein W, Cantrell D, Saltz J. Leveraging derived data elements in data analytic models for understanding and predicting hospital readmissions; AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2012. p. p. 103. [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford DC, Crosslin DR, Tromp G, Kullo IJ, Kuivaniemi H, Hayes MG, Denny JC, Bush WS, Haines JL, Roden DM, McCarty CA. eMERGEing progress in genomics—the first seven years. Frontiers in genetics. 2014 Jun 17;5:184. doi: 10.3389/fgene.2014.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarty C. A., Chisholm R. L., Chute C. G., Kullo I. J., Jarvik G. P., Larson E. B., Wolf W. A. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Medical Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shickel B, Tighe P, Bihorac A, Rashidi P. Deep EHR: a survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. arXiv preprint arXiv:1706.03446. 2017 Jun 12; doi: 10.1109/JBHI.2017.2767063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miotto R, Wang F, Wang S, Jiang X, Dudley JT. Deep learning for healthcare: review, opportunities and challenges. Briefings in bioinformatics. 2017 May 6; doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salekin A, Stankovic J. Detection of chronic kidney disease and selecting important predictive attributes.. In Healthcare Informatics (ICHI), 2016 IEEE International Conference on 2016 Oct 4; IEEE; pp. 262–270. [Google Scholar]
- 12.Gunarathne WH, Perera KD, Kahandawaarachchi KA. Performance Evaluation on Machine Learning Classification Techniques for Disease Classification and Forecasting through Data Analytics for Chronic Kidney Disease (CKD). Bioinformatics and Bioengineering (BIBE), 2017 IEEE 17th International Conference on 2017 Oct 23; IEEE; pp. 291–296. [Google Scholar]
- 13.Lipton ZC, Kale DC, Elkan C, Wetzel R. Learning to diagnose with LSTM recurrent neural networks. arXiv preprint arXiv:1511.03677. 2015 Nov 11; [Google Scholar]
- 14.Esteban C, Staeck O, Baier S, Yang Y, Tresp V. Predicting clinical events by combining static and dynamic information using recurrent neural networks. Healthcare Informatics (ICHI), 2016 IEEE International Conference on 2016 Oct 4; Ieee; pp. 93–101. [Google Scholar]
- 15.Choi E, Bahadori MT, Schuetz A, Stewart WF, Sun J. Doctor ai: Predicting clinical events via recurrent neural networks; InMachine Learning for Healthcare Conference; 2016. Dec 10, pp. 301–318. [PMC free article] [PubMed] [Google Scholar]
- 16.Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, Liu PJ, Liu X, Marcus J, Sun M, Sundberg P. Scalable and accurate deep learning with electronic health records. npj Digital Medicine. 2018 May 8;1(1):18. doi: 10.1038/s41746-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehrabi S, Sohn S, Li D, Pankratz JJ, Therneau T, Sauver JL, Liu H, Palakal M. Temporal pattern and association discovery of diagnosis codes using deep learning. Healthcare Informatics (ICHI), 2015 International Conference on 2015 Oct 21; IEEE; pp. 408–416. [Google Scholar]
- 18.Razavian N, Sontag D. Temporal convolutional neural networks for diagnosis from lab tests. arXiv preprint arXiv:1511.07938. 2015 Nov 25; [Google Scholar]
- 19.Avati A, Jung K, Harman S, Downing L, Ng A, Shah NH. Improving palliative care with deep learning. Bioinformatics and Biomedicine (BIBM), 2017 IEEE International Conference on 2017 Nov 13; IEEE; pp. 311–316. [Google Scholar]
- 20.Choudhry S. A., Li J, Davis D, Erdmann C, Sikka R, Sutariya B. A public-private partnership develops and externally validates a 30-day hospital readmission risk prediction model. Online Journal of Public Health Informatics. 2013;5(2):219. doi: 10.5210/ojphi.v5i2.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hripcsak G, Ryan P. B., Duke J. D., Shah N. H., Park R. W., Huser V, Madigan D. Characterizing treatment pathways at scale using the OHDSI network. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(27):7329–7336. doi: 10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park R. W. Sharing Clinical Big Data While Protecting Confidentiality and Security: Observational Health Data Sciences and Informatics. Healthcare Informatics Research. 2017;23(1):1–3. doi: 10.4258/hir.2017.23.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuemie M. J., Hripcsak G, Ryan P. B., Madigan D, Suchard M. A. Empirical confidence interval calibration for population-level effect estimation studies in observational healthcare data. Proceedings of the National Academy of Sciences. 2018;115(11):2571–2577. doi: 10.1073/pnas.1708282114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Scientific reports. 2016 May 17;6:26094. doi: 10.1038/srep26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Healthcare Cost and Utilization Project (HCUP) Clinical Classifications Software (CCS) for ICD-9-CM [Internet]. 2008 [updated 2018 July; cited 2018 Aug]. Available at: www.hcup-us.ahrq.gov/toolssoftware/ccs/ccs.jsp.
- 26.Azur MJ, Stuart EA, Frangakis C, Leaf PJ. Multiple imputation by chained equations: what is it and how does it work? International journal of methods in psychiatric research. 2011 Mar;20(1):40–9. doi: 10.1002/mpr.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazumder R, Hastie T, Tibshirani R. Spectral regularization algorithms for learning large incomplete matrices. Journal of machine learning research. 2010;11(Aug):2287–322. [PMC free article] [PubMed] [Google Scholar]
- 28.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001 Jun 1;17(6):520–5. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 29.Chollet F. others. keras [Internet]. 2015 [cited 2018 July 20]. Available from: https://keras.io.
- 30.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M. Tensorflow: a system for large-scale machine learning. OSDI. 2016 Nov 2;Vol. 16:265–283. [Google Scholar]
- 31.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J. Scikit-learn: Machine learning in Python. Journal of machine learning research. 2011;12(Oct):2825–30. [Google Scholar]
- 32.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015 May;521(7553):436. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 33.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. 2014 Dec 22; [Google Scholar]
- 34.Davis J, Goadrich M. The relationship between Precision-Recall and ROC curves. Proceedings of the 23rd international conference on Machine learning 2006 Jun 25; ACM; pp. 233–240. [Google Scholar]
- 35.Boyd K, Costa VS, Davis J, Page CD. Unachievable region in precision-recall space and its effect on empirical evaluation. Proceedings of the… International Conference on Machine Learning. International Conference on Machine Learning 2012 Dec 1; NIH Public Access; p. 349. [PMC free article] [PubMed] [Google Scholar]