Abstract
Electronic medical records are often de-identified before disseminated for secondary uses. However, unstructured natural language records are challenging to de-identify while utilizing a considerable amount of expensive human annotation. In this investigation, we incorporate active learning into the de-identification workflow to reduce annotation requirements. We apply this approach to a real clinical trials dataset and a publicly available i2b2 dataset to illustrate that, when the machine learning de-identification system can actively request information to help create a better model from beyond the system (e.g., a knowledgeable human assistant), less training data will be needed to maintain or improve the performance of trained models in comparison to the typical passive learning framework. Specifically, with a batch size of 10 documents, it requires only 40 documents for an active learning approach to reach an F-measure of 0.9, while passive learning needs at least 25% more data for training a comparable model.
Introduction
The significance of repurposing electronic medical records (EMR) for secondary usage1,2 has been increasingly acknowledged, driving the need for EMR data sharing3,4. In many instances, ensuring privacy for the patients to whom the data corresponds remains one of the primary challenges to disseminating such data. To achieve privacy protection, health care organizations often rely upon the de-identification standard of the Privacy Rule of the Health Insurance Portability and Accountability of 19965. Though it is straightforward to apply this standard to EMR data that is managed in a structured form6, where it is readily apparent where protected health information (PHI) including explicit (e.g., personal name) and quasi-identifiers (e.g., dates of birth and geocodes of residence), reside, it is more challenging to do so for clinical information communicated in a free or semi-structured form7,8. Yet a substantial amount of information exists in such a form9,10 and recent policies, such as those put forth by the European Medicines Agency11 require that narratives associated with clinical trials (e.g., clinical study reports) be made public. Given that modern EMR systems manage data on millions of patients, it is critical to develop de-identification routines for this data in a manner that are both effective and efficient12.
Machine learning methods can assist in realizing such goals, however, supporting state-of-the-art approaches requires the acquisition of sufficiently high-quality training data13,14. And, this must be accomplished under the limited budgets available to informatics teams running such systems. Thus, in this research, we specifically focus on incorporating an active learning in the process to reduce the overall cost for annotation and, thus, support the establishment of a more scalable de-identification pipeline. To do so, instead of randomly sampling a dataset for training purposes (or what is called passive learning), active learning works by allowing the machine learning system to select the data to be annotated by a human oracle and added to the set of training data iteratively15. The system is expected to learn and improve its accuracy throughout the process, while eventually fewer instances annotated by humans are required than passive learning16. In this work, we report on an active learning pipeline for de-identification based on machine learning. In doing so, we assess the extent to which active learning can lead to better results than passive learning (i.e., randomly selecting documents to be annotated by humans for the purposes of training a classifier). This research is based on the hypothesis that, actively requesting for more informative data can lead to a better model from human annotators with less training data.
This paper is organized as follows. We first review de-identification principles and systems for natural language EMR systems, as well as existing active learning applications in NLP (especially for named entity recognition tasks) and specifically with clinical documents. Next, we establish an active learning workflow for natural language de-identification and introduce several new heuristics for solving the key problem of active learning (i.e., choosing the most informative data for annotation), which is notable contribution of this work. We then conduct a series of controlled and systematic simulations on a real world dataset of CSRs for a clinical trial and a publicly accessible i2b2 dataset on the active learning pipeline, while evaluating the performance of the heuristics. We show that, in general, active learning can yield a comparable and, at times, better performance, with less training data than passive learning.
Background
Various machine learning methods have been applied for natural language de-identification17,18 by generalizing the problem to a named entity tagging task19. Specifically, conditional random fields20 (CRFs), a model with proven capability for solving NLP problems21,22, has been adopted by several software tools for de-identification23,24. For this paper, we work with the MITRE Identification Scrubber Toolkit23 (MIST). In this section, we review related research in 1) query strategies for active learning and 2) applications of active learning with clinical documents.
Active Learning Query Strategies
Several query strategies for active learning in sequence labeling tasks with CRFs were investigated and compared by Settles15, including uncertainty sampling, information density, Fisher information, and query-by-committee – some of which are more computationally costly than others in practice. A decision-theoretic active learning approach with Gaussian process classifiers was introduced and evaluated with voice messages classification25, utilizing the expected value of information (VOI) that accounts for the cost of misclassification. Also, the concept of return on investment (ROI) was implemented in active learning to account for the cost of annotation, which was assessed on a part of speech tagging task26. In our work, we introduce and assess query strategies based on uncertainty sampling and the notion of return on investment (ROI) for de-identification, as these approaches are more practical in real world.
Active Learning with Clinical Documents
Active learning has been shown to be an effective tool in named entity recognition tasks in clinical text27. These findings suggest that active learning is more efficient than passive learning in most cases. They further suggested that uncertainty sampling was the best strategy for reducing the annotation cost. The results implied that human annotation cost should be taken into account when evaluating the performance of active learning.
In the context of de-identification, Bostrom and colleagues28 proposed an active learning approach that relied upon a random forest classifier. They evaluated the approach with a dataset of 100 Swedish EMRs. In their framework, the query strategy to determine which documents humans should annotate next focused on entropy-based uncertainty sampling. However, this investigation was limited in several notable ways. First, entropy-based uncertainty sampling does not explicitly account for human annotation cost, such that we introduce and implement several query strategies in our system beyond. Second, we perform an expanded investigation and conduct controlled experiments using real, as well as a publicly accessible resynthesized, EMR data.
Methods
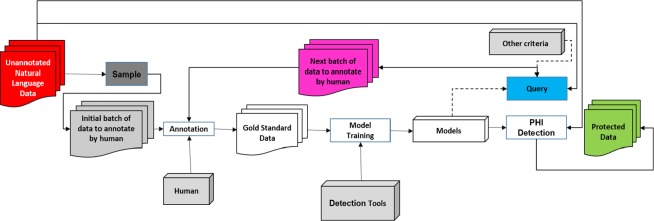
We adopt a pool-based active learning framework15. In our scenario, this means that there is a limited amount of annotated data to begin with and a large pool of unannotated data the framework could select from. The pipeline for the active learning framework for natural language de-identification is illustrated in Figure 1. Initially, we use a small batch of data that is selected randomly from the dataset as the starting point of the active learning. The human annotators then manually tag the PHI in the initial batch of data to create a gold standard for model training.
Figure 1.
The pipeline for the active learning framework for natural language EMR de-identification.
Since human annotation is costly, the goal of active learning is to reduce the total amount of human annotation needed in the process while maintaining (or even improving) the performance of de-identification model training. In reality, the human effort involved in the framework can be viewed as two parts: 1) the human annotation effort in gold standard creation and 2) the human correction effort that is needed to fix incorrect labels generated in the previous round when the de-identification model is applied to unannotated data (because no reasonable existing automated de-identification approaches yield a recall of 100%). After the first batch of gold standard data is created by human annotators, we train a de-identification model, which is applied to the remainder of the unannotated data. The active learning pipeline then queries for more informative data to be corrected by humans based on the performance of previous models and/or additional criteria. This information is expected to assist in better de-identification model development. Another way to view this strategy is, instead of randomly selecting a fixed amount of unannotated data for training data (passive learning), the system actively queries for the data that potentially contributes more information during model training.
This begs the question: how should we select the data that are more informative? Since the query for active learning is based on a heuristic, we propose several query strategies for our de-identification framework and compare the performance with our simulation. Before providing the details of each strategy, we formalize the problem statement.
Problem Formulation
Let D be a set of documents in the form of EMRs. Now, let DL and Du be the set of annotated and unannotated documents, where . DU consists of n documents, d1, d2, dn. Let Q(di) be the query strategy that the active learning framework utilizes to select additional documents for human annotation. Note that, we choose documents rather than tokens as the unit for selection for several reasons. First, it is impractical for a human to annotate tokens out of context. Second, selecting tokens could lead to a significant computational overhead since it takes time to retrain the model each time the training dataset is updated.
The goal of the query step is to choose the data dc that maximizes Q(di),
| (1) |
At this point, let Ds be the selected batch of k documents for a human to correct. In essence, this consists of the k documents that maximize Q(di). Note that the value of k could depend on the learning rate of the framework, as well as the time that it takes to retrain and reannotate.
Once corrected by the human, DS is removed from the unannotated document set DU, while DL is updated to include the annotated batch of documents:
| (2) |
Each time DS is annotated and added to training data, the de-identification model needs to be retrained using DL’. Next, we introduce several options for query strategies that we utilize in our system.
Uncertainty Sampling
One of the more prevalent query strategies for active learning is uncertainty sampling15. In this model, it is assumed that the active learning system picks the data that the current model is most uncertain of when making predictions.
Least confidence (LC)
For a CRF model, given a token x, let y be the most likely predicted label of x (e.g., a patient’s name or a date) and let P(y|x) be the posterior probability, or the confidence score of x given the current model. We define the uncertainty of token x as 1 – P(y|x). Note that we aim to find the document for which the current model has the least confidence. Upon doing so, we can use either 1) the summation of the LC-based uncertainty of all tokens in a document di:
| (3) |
or 2) the mean of all token uncertainty based on LC:
| (4) |
where li is the total number of tokens in di. The problem with adopting the mean of all token uncertainty is that it does not take into account the length of the documents in the selection process, which may not be optimal.
Our initial studies revealed several issues when employing the sum of tokens uncertainty approach. First, the predicted non-PHI tokens are more likely to produce a prediction confidence score of 0.95 or higher, while the confidence of the tokens that are predicted as PHI are, in most cases, lower. Also, since this strategy aims for documents with a higher sum of token uncertainty, it tends to be biased towards documents that contain a larger amount of tokens, even though the PHI density in the selected set of documents could be low (which is not desirable for model training).
To gain intuition into the problem, imagine we have two documents d1 and d2 as summarized in Table 1. Note that document d2 consists of a much higher PHI density than document d1 and might provide more information in de-identification model training. Nonetheless, uncertainty sampling will more likely choose document d1 over document d2 due to a higher sum of token uncertainty.
Table 1.
An example of how the number of tokens and PHI density influences the sum of token uncertainty.
| Feature | Document d1 | Document d2 |
|---|---|---|
| Total number of non-PHI tokens | 1000 | 100 |
| Average non-PHI token confidence | 0.99 | 0.99 |
| Total number of PHI tokens | 5 | 10 |
| Average PHI token confidence | 0.6 | 0.6 |
| PHI density | 0.5% | 9.1% |
| Sum of tokens uncertainty | 12 | 5 |
To mitigate this problem, we introduce a modified version of the least confidence approach, which we refer to as least confidence with upper bound (LCUB). Instead of summing the uncertainty of all tokens, the framework calculates the sum of uncertainty of tokens, when P(yt|xt) < θ, where θ is a cutoff value for uncertainty sampling. Based on this formulation, we say that Σ f(xt, θ) is the modified sum of token uncertainty with cutoff value θ:
| (5) |
Entropy
Entropy measures the potential amount of discriminative information available. Given a token x, the entropy H(x) is:
| (6) |
where m corresponds to the number of most probable labels of x, as predicted by the current classification model (e.g., the CRF). Here P(yj|x) is the probability that x’s label is yj. Again, for a document di that contains t tokens, the total entropy-based uncertainty of di can be calculated as:
| (7) |
Similar to the LC approach, entropy-based uncertainty sampling also tends to suffer from the problem of low PHI density documents. To mitigate this issue, we introduce an entropy with lower bound (ELB) approach. In this approach, we set a minimum threshold ρ for token entropy.
Thus, we define Σg(xt, ρ) as the modified sum of token entropy with minimum value ρ:
| (8) |
Return on Investment
The goal of active learning is to reduce the human effort needed for machine learning. Both the least confidence and the entropy-based uncertainty sampling methods seek to solve the problem by minimizing the amount of training data required. However, this implicitly assumes that the cost for human annotation is fixed and is not explicitly modeled during the query step. In reality, we need to acknowledge that the effort that a human annotator spends is more complex than the above assumption. Consider, it is likely that the cost varies based on PHI types, error types, human fatigue (due to the number, or length, of documents), among other factors. Additionally, the contribution of human correction towards a better model can also vary according to various factors, such as PHI types and error types. Thus, we designed a query strategy that accounts for both the cost and the contribution of human correction.
We assume there is a reading cost for the human annotator that is proportional to document length. The average reading cost per token is ctr, which implies the total reading cost for a document di of length li is ctr × li. Again, we formalize the problem as follows. Given token x with y as the most likely label, P(y|x) is the probability the active learning system assigns y as the label to x, while P’(y|x) is the true probability that x is of classy.
Without loss of generality, here, we consider only a two-class of the problem, PHI versus non-PHI. This indicates that we assume the annotation cost and the human contribution of correcting a PHI instance classified as the wrong PHI type cancel each other out. Now, let ctn and ctp be the human correction cost of correcting a false negative (FN) instance (i.e., a token mistakenly labeled by the current model as non-PHI) and a false positive (FP) instance (i.e., a non-PHI token mistakenly labeled by the current model as PHI), respectively. Similarly, cnn and cnp represent the human correction contribution of correcting an FN and FP instance, respectively. Thus, the expected total contribution of human correction for token x when y is a non-PHI instance can be defined as TCCN(x) and is calculated as:
| (9) |
The expected total cost of human correction for token x when y is a non-PHI instance is TCCT(x). Then
| (10) |
Note that cnn – ctn and cnp – ctp represent the net contribution of correcting an FN instance and an FP instance, respectively. At this point, let NCn and NCp denote the net contribution of an FN instance correction and an FP instance correction, respectively. Thus, the expected return on investment (ROI) of token x is:
| (11) |
Consequently, the total expected ROI of unannotated document di is:
| (12) |
The active learning approach will select documents that maximize ROI.
Experiments and Results
Datasets
After constructing the active learning pipeline, we utilized two datasets for simulation and evaluation: 1) a dataset drawn from a healthcare organization (anonymized due to contractual agreements), which we refer to as the clinical study report (CSR) dataset and 2) a publicly available dataset from the i2b2 de-identification challenge19.
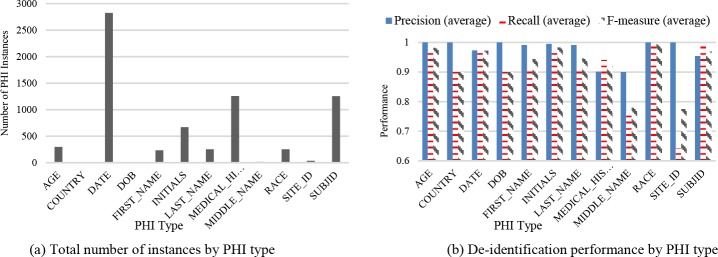
The CSR dataset consists of 370 documents partitioned from clinical study reports with a total number of 312,991 tokens, and 7,098 PHI instances across 12 PHI types. Figure 2(a) reports on the number of instances per PHI type in the CSR dataset. Note that certain PHI types contain no more than a couple of instances in this corpus (e.g., COUNTRY, DOB, MIDDLENAME, and SITE ID), which makes it challenging to train an effective model. As such we do not consider these further. To assess the de-identification performance of this dataset, we conducted a 10-fold cross-validation on this corpus before simulating the active learning process. Each time we trained a de-identification model with 90% of the documents and tested the model on the remaining 10% of the data. The average precision, recall and F-measure for the PHI types in this dataset are shown in Figure 2(b).
Figure 2.
CSR dataset statistics: (a) Number of PHI instances and (b) De-identification performance by PHI type.
Note that after excluding the aforementioned PHI types with very few instances, the FIRST_NAME, LAST_NAME and MEDICAL_HISTORY types appear to be the most difficult to detect among all types. Possible reasons behind this observation are that FIRST_NAME and LAST_NAME have only a few hundred instances, while MEDICAL_HISTORY lacks sufficient contextual cues in the documents. The i2b2 dataset contains 889 annotated discharge summaries from Partners Healthcare, with the real identifiers replaced by synthetic information19.
Experimental Design and Evaluation
We simulated the active learning query strategies with the CSR dataset. For uncertainty sampling strategies, namely least confidence and entropy, we varied the parameters to evaluate the original and modified models (i.e., LC with a cutoff value and entropy with a minimum value). Specifically, the cutoff value θ of LC ranged from 0.1 to 0.9, with a step size of 0.1 and the minimum entropy score was selected from a set of four values: {0.001, 0.01, 0.1, 1}.
The parameters of the ROI model include: 1) the costs of correcting an FP instance and an FN instance, 2) the contributions of the two types of correction towards creating a model with greater fidelity, and 3) P’(y|x), which, as noted earlier, is the true probability that token x is of class y. Since P’(y|x) is unknown in this scenario, we approximate P’(y|x) with P(y|x), the predicted probability based on the current model. Thus, the ROI reduces to:
| (13) |
We vary NCn, NCp and ctr to investigate how these parameters influence active learning performance.
Simulation Results and Analysis
We initially evaluated the approaches with a randomly selected initial batch of 10 documents. For each learning iteration, the simulation selects an additional batch of documents to add to the training set and learns a new de-identification model. The process proceeds for 10 iterations. For a baseline comparison, we use a random selection of the next batch of documents to be added to training, which we refer to as Random. All results are based on an average of 3 runs (as the variance was negligible). We evaluate the framework with batch sizes of 10, 5 and 1 document.
The CSR Dataset Results
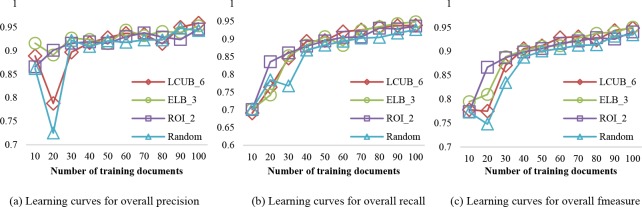
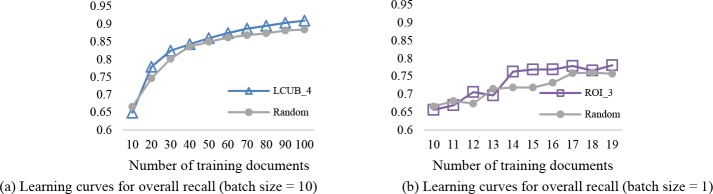
Batch of 10 documents. Figure 3 compares the learning curves of overall precision, recall and F-measure for the various query strategies. For brevity, we choose the setting with the best performance to plot for each strategy. Note that in LCUB 10, the upper bound of LC is set to 1, which implies that all confidence scores are taken into account. Also, ELB_1 is equal to using entropy without a minimum value.
Figure 3.
Learning curves for the active learning strategies and passive learning with a batch size of 10 documents for the CSR dataset. Each curve corresponds to the parameter setting that achieves the highest performance.
It can be seen that there is a general increasing trend for all query strategies (including random selection) as additional training data is added. This is particularly pronounced when there is less than 50 training documents, at which point most of the selection approaches yield an F-measure of over 0.9. The observed growth in performance slows down from training 50 to 100 documents. Additionally, the classification model generally favors precision over recall in all testing scenarios, especially at early training stages (i.e., when the number of training documents is smaller than 50).
In terms of recall, ELB_3 outperforms all other selection methods at the final training stages, achieving 0.949 at 100 training documents, while LCUB_6 achieves the best precision at the final training stage. However, ROI_2 learns faster at the beginning than all other strategies. Also, it provides the steadiest growth in all three performance measures among all participants in comparison, which is preferable in reducing human correction effort.
The previous observations are not guaranteed to hold true for each PHI type. More specifically, among the three types of PHI that we focus on (FIRST_NAME, LAST_NAME and MEDICAL_HISTORY), there is no obvious trend for precision for FIRST NAME or LAST NAME. By contrast, the precision for MEDICAL HISTORY increases with additional training data. Both the recall and F-measure exhibits more clear increasing trends for all three PHI types, in both the active and passive learning scenarios. For FIRST_NAME and LAST_NAME, the active learning approaches are more stable in growth than Random, but do not necessarily outperform Random in the final iterations. For MEDICAL_HISTORY, the selection based on active learning exceeds Random selection, especially for ROI_2.
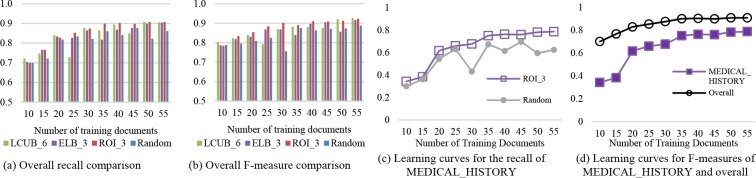
Batch of 5 documents. When the batch size reduces to 5, the advantage of active learning over passive learning (random selection) becomes more apparent (Figure 4(a-b)). Generally, ELB_3 and ROI_3 (the best performed settings of ELB and ROI) exceeds Random on all three performance measures (P, R, and F). Additionally, LCUB_6 (the best of LCUB) shows less stable growth than the other two active learning approaches, but is still better than Random most of the time.
Figure 4.
Overall and MEDICAL_HISTORY performance comparison for active and passive learning (with a batch size of 5 documents) for the CSR dataset.
Similar to a batch size of 10, the recall and F-measure improves with higher quantities of training documents for both of the active and passive approaches. The increasing trend does not hold for precision; it instead fluctuates around 0.9 for active learning and 0.85 for random. For recall, to yield a score 0.9, ROI_3 requires around 35 training documents, LCUB_6 and ELB_3 need 50 documents, and for Random the best score within 10 iterations is only 0.876. At a training level of 30 documents, ROI_3 reaches an F-measure over 90%, while the highest F-score for random is 0.887.
The advantage of adopting active learning over passive learning remains for ROI_3 in terms of performance for the three individual PHI types. Specifically for MEDICAL_HISTORY (as in Figure 4(c)), the recall of ROI_3 sees a steady growth and arrives at 0.787 after 10 iterations. By contrast, the random approach yields a much more unstable trend and never reaches above 0.7. In comparison to the overall F-measure (shown in Figure 4(d)), although the F-measure of MEDICAL HISTORY is generally worse, it shows much more drastic growth within the 10 iterations.
Batch of 1 document. The simulation with a batch size of 1 document leads to more complex results than the above scenarios. ROI_1 generally outperforms random selection as well as other active learning approaches within 10 iterations, reaching an F-measure of 0.87 with 19 training documents. LCUB_6 and ELB_3 lead to higher performance scores than random in most cases, but the learning is less stable than ROI_1. When considering the performance by PHI types, ROI_1 demonstrates a greater advantage than the other strategies, particularly in terms of recall and F-measure.
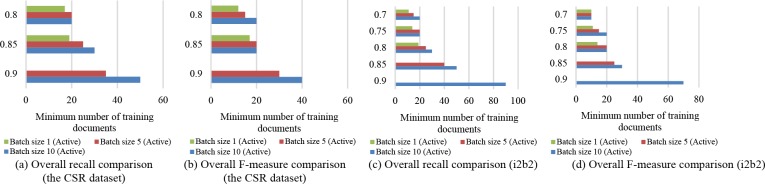
Batch Size. To compare the performance of models trained with different batch sizes, we plot the performance scores against the size of the training datasets needed for each of the batch sizes (i.e., the minimum number of training documents the active learning was provided to reach a level of performance). Since the precision across all PHI types could yield over 0.9 with the initial 10 (randomly selected) documents and does not exhibit a clear trend as more training data is supplied, we focus on the recall and F-measure for this comparison. Overall, when the active learning proceeds with smaller batch sizes, the performance scores, in terms of recall and F-measure, could attain a certain level with less training data involved.
As shown in Figure 6(a), a recall of 0.8 requires 20 training documents for batch sizes of 10 and 5, while only 17 documents are needed for a batch size of 1. A recall of 0.85 requires at least 30 documents for a batch size of 10, while 25 are needed for a batch size of 5, and only 19 for a batch size of 1. Finally, we observe that a batch size of 10 could reach a recall of 0.9 with 50 documents, while a batch size of 5 could yield the same level of recall with 40 documents in training. Since all batch sizes are tested with 10 iterations, the maximum training number for batch size of 1 is 19, the highest recall of which (0.86) is less than 0.9 and thus not included in the plot.
Figure 6.
Comparison of active learning batch sizes in terms of minimum number of training documents to reach a certain level of performance for the CSR dataset.
Generally, it takes less training data to achieve the same level of F-measure (Figure 6(b)). Starting from an F-measure of 0.8, a batch size of 10 requires 20 documents to build the model, and a batch size of 5 and 1 needs 15 and 12, respectively. When the goal of F-measure is set to 0.85, 20 documents are needed for both a batch size of 10 and 5, whereas 17 documents are required for a batch size of 1. Again, a batch size of 1 could never reach an F-measure of over 0.9 within 10 iterations, while a minimum of 30 to 40 documents are needed to train with a batch size of 5 or 10.
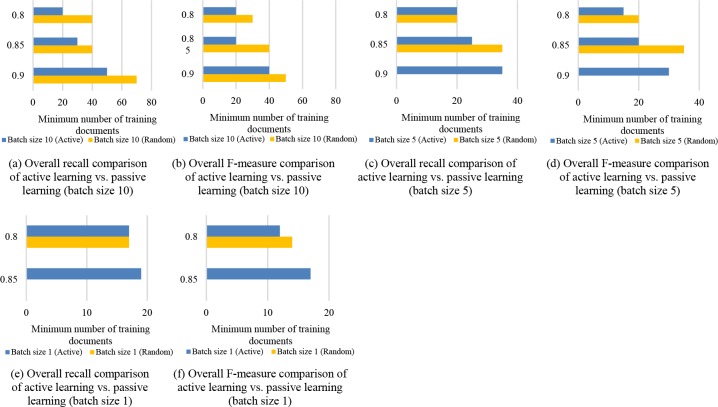
Minimum Number of Training Documents. To gain insight into training data reductions achieved by active learning, we compare the minimum number of training documents provided to active and passive learning given the same batch size (Figure 7). By comparing Figures 7(a) and 7(b) it can be seen that, at a batch size of 10 documents, to reach the same level of performance (over 0.8), passive learning requires 10 to 20 (which is 25% to 100%) more documents than active learning. When the batch size decreases to 5 (Figure 7(c-d)) or 1 (Figure 7(e-f)), active learning surpasses passive learning most of the time, while the latter fails to reach a recall or f-measure of 0.85 within the 10 iterations.
Figure 7.
Comparison of active learning vs. passive learning (random) in terms of minimum number of training documents to reach a certain level of performance for the CSR dataset.
i2b2 Results
When experimenting on the i2b2 dataset, the active learning approaches surpass passive learning for all three batch sizes (10, 5 and 1), especially in terms of recall and F-measure. As shown in Figure 5, similar to previous findings with the CSR dataset, smaller batch sizes lead to more significant difference between active learning and Random. Meanwhile, unlike in the CSR dataset, LCUB performs better than other active strategies with batch size of 1batch size of 10 and 5, and ROI is the best query strategy when the batch size is 1.
Figure 5.
Recall for active and passive learning (batch size of 10 documents or 1 document) for the i2b2 dataset.
Again, to investigate the impact of batch sizes on active learning, we plot the minimum number of documents that the system trains on to reach a certain level of recall or F-measure for the i2b2 dataset (Figure 6(c-d)). Generally, we observe a similar finding with the CSR dataset; that is, it requires less documents for the training to attain a certain level of performance when opting for a smaller batch size. For example, a batch size of 10 needs 30 documents for a recall of 0.8, while a batch size of 5 requires 25 documents, which is 16.7% less, and a bath size 1 needs only 19 documents to train, which is 36.7% less than a batch size of 10.
Discussion and Conclusions
There are several notable findings that we wish to highlight. First and foremost, the simulation results for both the CSR dataset and the i2b2 dataset lend credibility to the hypothesis that adopting active learning in training data selection for natural language de-identification could generally result in more efficient learning than selecting data randomly (passive learning). It is reasonable to conclude that, under the assumption that a CRF is used, active learning could lead to comparable or higher performance scores with less amount of training data needed than passive learning.
Additionally, it is worth mentioning that various query strategies in active learning exhibited different trends in learning. Depending on the specific learning goals (e.g., focusing on overall performance or an individual PHI type) the decision for which active learning query strategy should be adopted could vary. ROI often generated a more stable learning curve than other strategies for the CSR dataset, but the finding did not hold for the i2b2 dataset, which may be due to the fact that the i2b2 data is highly regularized and the results with it do not always transfer to real datasets29.
Last but not least, the choice of batch size could play a non-trivial part in the learning process, as smaller batch sizes could aid faster learning, yet might potentially result in more iterations and longer time in re-training. For the CSR dataset, when batch size is 1, the precision of random selection might beat active learning. Still, active learning retained the advantage over passive learning in recall and F-measure.
Despite the merits of our investigation, there is opportunity for improvement in active learning in this setting. First, in the ROI model, we provided results based on simulations, but the actual human annotation costs and contributions in real-world problems need to be measured through user studies. Second, instead of fixing a batch size over the entire active learning process, an adaptive batch sizing strategy might lead to better training performance. Lastly, deep learning methods (e.g., recurrent neural networks) might be considered for the active learning system, as they have recently shown promising results in de-identificaiton30. Note that, while the proposed pipeline will likely still be applicable by simply switching the machine learning basis, it is possible that doing so might influence the performance.
Finally, it should be recognized that the acceptability of machine learned de-identification models is currently left to the discretion of each healthcare organization. Based on the 2016 CEGS N-GRID shared tasks32, using off-the-shelf systems on new data led to F-scores in the range of 0.8, which is clearly not sufficient for deployment. However, appropriately trained systems can yield substantially better performance. For example, in the i2b2 2014 challenge31, the best system reached an F-measure of 0.964, which was improved to 0.979 via a deep neural network30. The latter system also achieved an F-measure of 0.99 on the MIMIC dataset. Still, there is no agreed upon standard F-measure threshold for natural language de-identification. So, while these are notable levels of performance, informatics teams need to work with their institutional review boards to ensure that they meet their requirements.
Acknowledgements
This research was sponsored in part by Privacy Analytics Inc. and grant U2COD023196 from the NIH.
References
- 1.Chapman WW, Dowling JN, Wagner MM. Fever detection from free-text clinical records for biosurveillance. J Biomed Inform. 2004;37(2):120–7. doi: 10.1016/j.jbi.2004.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newton KM, Peissig PL, Kho AN. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform. 2013;20(e1):e147–54. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarty CA, Chisholm RL, Chute CG. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Gen. 2011;4(1):13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Institutes of Health. Final NIH statement on sharing research data. NOT-OD-03-032. 2003. Feb 26,
- 5.Office for Civil Rights. U.S. Dept. of Health and Human Services. Standards for privacy of individually identifiable health information. Final rule. Federal Register. 2002 Aug 14;67(157):53181–273. [PubMed] [Google Scholar]
- 6.Van Vleck TT, Wilcox A, Stetson PD. 2008. Nov 6, Content and structure of clinical problem lists: a corpus analysis. AMIA Annu Symp Proc; pp. 753–7. [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson SB, Bakken S, Dine D. An electronic health record based on structured narrative. J Am Med Inform Assoc. 2008;15(1):54–64. doi: 10.1197/jamia.M2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman WW, Nadkarni PM, Hirschman L. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc. 2011 Sep;18(5) doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;42(5):760–72. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbloom ST, Denny JC, Xu H. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J Am Med Inform Assoc. 18(2):181–6. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Medicines Agency. External guidance on the implementation of the European Medicines Agency policy on the publication of clinical data for medicinal products for human use. 2017. Sep 22, Accessed 2018 Aug 7.
- 12.Dorr DA, Phillips WF, Phansalkar S, Sims SA, Hurdle JF. Assessing the difficulty and time cost of de-identification in clinical narratives. Meth Inf Med. 2006;45(3):246–52. [PubMed] [Google Scholar]
- 13.Hanauer D, Aberdeen J, Bayer S, Wellner B, Clark C, Zheng K, Hirschman L. Bootstrapping a de-identification system for narrative patient records: cost-performance tradeoffs. Int J Med Inform. 2013;82(9):821–31. doi: 10.1016/j.ijmedinf.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Douglass M, Clifford GD, Reisner A. Computer-assisted de-identification of free text in the MIMIC II database. In Computers in Cardiology. 2004. pp. 341–4.
- 15.Settles B. Active learning. Synthesis Lectures on Artificial Intelligence and Machine Learning. 2012;6(1):1–14. [Google Scholar]
- 16.Settles B, Craven M. An analysis of active learning strategies for sequence labeling tasks. Proc Conference on Empirical Methods in Natural Language Processing; 2008. pp. 1070–9. [Google Scholar]
- 17.Meystre SM, Friedlin FJ, South BR, Shen S, Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Metho. 2010 Dec;10(1):70. doi: 10.1186/1471-2288-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrández O, South BR, Shen S. BoB, a best-of-breed automated text de-identification system for VHA clinical documents. J Am Med Inform Assoc. 2012;20(1):77–83. doi: 10.1136/amiajnl-2012-001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uzuner Ö, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc. 2007;14(5):550–63. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafferty J, McCallum A, Pereira FC. 2001. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. Proc Int Conf Mach Learn; pp. 282–9. [Google Scholar]
- 21.Settles B. 2004. Biomedical named entity recognition using conditional random fields and rich feature sets. Proc International Joint Workshop on Natural Language Processing in Biomedicine and its Applications; pp. 104–7. [Google Scholar]
- 22.Sha F, Pereira F. Shallow parsing with conditional random fields. Proc Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology; 2003. pp. 134–41. [Google Scholar]
- 23.Aberdeen J, Bayer S, Yeniterzi R. The MITRE Identification Scrubber Toolkit: design, training, and assessment. Int J Med Inform. 2010;79(12):849–59. doi: 10.1016/j.ijmedinf.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Deleger L, Molnar K, Savova G. Large-scale evaluation of automated clinical note de-identification and its impact on information extraction. J Am Med Inform. 2013;20(1):84–94. doi: 10.1136/amiajnl-2012-001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapoor A, Horvitz E, Basu S. Selective supervision: guiding supervised learning with decision-theoretic active learning. Proc International Joint Conference on Artificial Intelligence; 2007. pp. 877–82. [Google Scholar]
- 26.Haertel RA, Seppi KD, Ringger EK, Carroll JL. Return on investment for active learning. Proc NIPS Workshop on Cost-Sensitive Learning. 2008.
- 27.Chen Y, Lasko TA, Mei Q, Denny JC, Xu H. A study of active learning methods for named entity recognition in clinical text. J Biomed Inform. 2015;58:11–8. doi: 10.1016/j.jbi.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boström H, Dalianis H. De-identifying health records by means of active learning. Proc International Conference on Machine Learning; 2012. pp. 90–7. [Google Scholar]
- 29.Yeniterzi R, Aberdeen J, Bayer S, Wellner B, Hirschman L, Malin B. Effects of personal identifier resynthesis on clinical text de-identification. J Am Med Inform Assoc. 2010 Mar 1;17(2):159–68. doi: 10.1136/jamia.2009.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dernoncourt F, Lee JY, Uzuner O, Szolovits P. De-identification of patient notes with recurrent neural networks. J Am Med Inform. 2017;24(3):596–606. doi: 10.1093/jamia/ocw156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stubbs A, Uzuner O. Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/UTHealth corpus. J Biomed Inform. 2015;58:S20–S29. doi: 10.1016/j.jbi.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stubbs A, Filannino M, Uzuner O. De-identification of psychiatric intake records: Overview of 2016 CEGS N-GRID Shared Tasks Track 1. J Biomed Inform. 2017;75:S4–S18. doi: 10.1016/j.jbi.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]