Abstract
Epidemiological studies identifying biological markers of disease state are valuable, but can be time-consuming, expensive, and require extensive intuition and expertise. Furthermore, not all hypothesized markers will be borne out in a study, suggesting that higher quality initial hypotheses are crucial. In this work, we propose a high-throughput pipeline to produce a ranked list of high-quality hypothesized marker laboratory tests for diagnoses. Our pipeline generates a large number of candidate lab-diagnosis hypotheses derived from machine learning models, filters and ranks them according to their potential novelty using text mining, and corroborate final hypotheses with logistic regression analysis. We test our approach on a large electronic health record dataset and the PubMed corpus, and find several promising candidate hypotheses.
Introduction
Epidemiological studies associating changes in biological markers (often measured by laboratory tests) and disease states are an invaluable tool for better understanding disease mechanism, such as the Framingham heart study1. Moreover, many diagnostic criteria exploit such associations by testing for abnormal changes in the associated biological marker. For example, the diagnostic criteria for Type II diabetes (a metabolic disorder that impacts how the cells uptake glucose) includes a measurement of fasting blood glucose levels. The benefits of a well performed epidemiological study are clear, but such studies can be time-consuming, expensive, and like any scientific study they require some intuition of the link searched for. Further, not all hypothesized associations will be borne out in the data, and thus the higher the quality of initial hypothesis, the more likely the study yields valuable medical information. In this work, we attempt to generate a ranked set of high-quality candidate hypotheses through a combination of machine learning, text-mining based literature searches, and traditional logistic regression analysis.
Our method identifies “novel predictive lab tests,” which we define as a previously unknown association between a given disease state and a given biological marker (measurable by a laboratory test) that changes prior to diagnosis. Our approach hinges on the assumption that a novel diagnostic lab test is 1) useful for predicting a given diagnosis (via a machine learning model) and 2) not currently discussed in published medical literature on PubMed. Using these criteria we hypothesize that given a set of candidate diagnoses, we can generate a set of high-quality novel diagnostic lab tests in a five step procedure:
For each diagnosis, produce a machine learning model to predict it, and select the top k lab tests used for prediction by their feature importance.
Map the clinic used name for each diagnosis and lab test to the literature used name (clinic names in our data source commonly used names and abbreviations that may not have been found in literature).
Perform automated text-mining to search for literature associations between diagnoses and lab tests. This provides a pseudo knowledge-base of known diagnostic lab tests.
Rank novel diagnostic lab tests in a way that encourages high predictive usefulness and low literature presence.
Evaluate the top candidates with traditional logistic regression analysis to only retain hypotheses that are statistically significant both with and without inclusion of potential confounders.
The use of Electronic Health Records (EHRs) to digitally capture patient health encounters has grown substantially in recent years2. This has created unprecedented opportunity for secondary use of EHR data in combination with machine learning algorithms to build predictive models for critical patient health events such as breast cancer3 or heart attack1 risk. Machine learning algorithms flexibly learn relationships in a data set without the need for hard coded rules. In this work we first utilize a machine learning algorithm, specifically random forests4, to build predictive models of disease for which we are interested in finding novel diagnostic lab tests. Random forests work by forming an ensemble of multiple decision trees, each learned on a randomly bootstrapped sample of the original dataset. They are well known for their strong predictive performance5 and resilience to applications with very large numbers of features4 (which is true of EHR data).
While machine learning algorithms can give a quantitative prediction, or even a confidence of the prediction, one of their common critiques is interpretability. That is, machine learning algorithms do not offer reasoning along with their predictions. One way that data scientists can interpret machine learned models is by observing which features most impact their predictions. The random forest algorithm has support for “feature importances”4 which provide a window into the model by ranking the contribution of each feature to the construction of the model. In random forests, feature importances are non-negative values for which larger values suggest a greater contribution of that feature towards prediction. In this work, we use feature importances to discover potential novel predictive lab tests.
While a researcher might consider conducting manual literature searches to validate potential novel discoveries from a trained model, the size and exponential growth in scientific literature6,7 make this approach infeasible. The literature search space to validate tens or hundreds of lab tests across tens or hundreds of diagnoses is too large for an individual to reasonably explore. We therefore use a text mining approach to search for associations between diagnoses and lab tests. For this work, we rely on KinderMiner, a previously developed algorithm designed to filter and rank a list of target terms by their association in the literature with a key phrase of interest8. In our particular application, a diagnosis name is the key phrase of interest, and the names of important lab features are the target terms to be ranked by association with the diagnosis. In contrast to the original intent of KinderMiner, which is to rank the target terms by their positive association with the key phrase within the literature, we are looking for labs deemed useful for prediction, but which are not already well known in the literature. Thus, we modify KinderMiner to suit our needs by instead filtering and ranking terms by significant lack of association with the diagnosis in the literature.
In this work, we demonstrate our proposed method of identifying novel predictive lab tests by gathering a set important diagnoses and their most predictive lab tests from machine learning models. We then use text mining to filter and rank hypothesized predictive lab tests based on negative association within the literature. Finally, we evaluate the top hypotheses proposed by our method and find several to be promising candidates for further investigation. In the following sections we specifically describe the pipeline: the selection of diagnoses and labs, the construction of predictive models, text mining based filtration and ranking, and final evaluation of the hypotheses.
Selection of Diagnoses and Lab Tests
To select the set of important diagnoses to consider, we started with the 100 most common diagnoses by patient count in our EHR dataset. We then manually filtered out diagnoses that we considered unlikely to be diagnosed via lab tests or that were effectively a restatement of an abnormal lab value. For example, we removed ICD-9 code 719.46 (Pain in joint; lower leg) because it is likely a result of a mechanical ailment, and we removed ICD-9 code 272 (Pure hypercholesterolemia) because it is a diagnosis of an abnormal lab value. We also manually curated our diagnosis descriptions to better reflect what we would expect to find in the literature and to include synonyms. For example, “gout; unspecified” became just “gout” and “dysthymic disorder” became “dysthymic disorder” or “dysthymia.” See Table 1 for our final chosen list of 69 diagnoses and the search terms we used for each.
Table 1:
The 69 diagnoses that we considered along with all search terms we used for each. Alias search terms are separated by semicolons.
| ICD Code | Diagnosis Name | Curated Search Terms |
|---|---|---|
| 162.9 | Malignant neoplasm of bronchus and lung; unspecified site | malignant lung cancer |
| 174.9 | Malignant neoplasm of breast (female); unspecified site | malignant breast cancer |
| 274.9 | Gout; unspecified | gout |
| 300.01 | Anxiety state; unspecified | anxiety; gad; generalized anxiety disorder |
| 300.4 | Dysthymic disorder | dysthymic disorder; dysthymia |
| 305.1 | Tobacco use disorder | tobacco use |
| 309.28 | Adjustment disorder with mixed anxiety and depressed mood | adjustment disorder |
| 314.00 | Attention deficit disorder of childhood without mention of hyperactivity | attention deficit disorder |
| 314.01 | Attention deficit disorder of childhood with hyperactivity | attention deficit hyperactivity disorder; adhd |
| 327.23 | Obstructive sleep apnea (adult) (pediatric) | obstructive sleep apnea |
| 362.51 | Nonexudative senile macular degeneration of retina | senile macular degeneration |
| 366.10 | Unspecified senile cataract | senile cataract; senile cataracts |
| 366.16 | Nuclear sclerosis | nuclear sclerosis |
| 367.0 | Hypermetropia | hypermetropia; farsightedness; hyperopia; farsighted |
| 367.1 | Myopia | myopia |
| 367.4 | Presbyopia | presbyopia |
| 372.30 | Unspecified conjunctivitis | conjunctivitis |
| 379.21 | Vitreous degeneration | vitreous degeneration |
| 382.9 | Unspecified otitis media | otitis media |
| 388.70 | Unspecified otalgia | otalgia |
| 389.9 | Unspecified hearing loss | hearing loss |
| 410.71 | Acute myocardial infarction; subendocardial infarction; initial episode of care | acute myocardial infarction; heart attack; subendocardial infarction |
| 411.1 | Intermediate coronary syndrome | intermediate coronary syndrome |
| 413.9 | Other and unspecified angina pectoris | angina |
| 414 | Other forms of chronic ischemic heart disease | chronic ischemic heart disease |
| 424.1 | Aortic valve disorders | aortic valve disorder |
| 427.31 | Atrial fibrillation | atrial fibrillation; afib |
| 427.89 | Other specified cardiac dysrhythmias | cardiac dysrhythmia |
| 427.9 | Unspecified cardiac dysrhythmia | cardiac dysrhythmias |
| 428.0 | Congestive heart failure; unspecified | congestive heart failure |
| 434.91 | Unspecified cerebral artery occlusion with cerebral infarction | ischemic stroke |
| 440.9 | Generalized and unspecified atherosclerosis | atherosclerosis |
| 443.9 | Unspecified peripheral vascular disease | peripheral vascular disease |
| 461.9 | Acute sinusitis; unspecified | acute sinusitis |
| 462 | Acute pharyngitis | acute pharyngitis |
| 465.9 | Acute upper respiratory infections of unspecified site | acute upper respiratory infection |
| 466.0 | Acute bronchitis | acute bronchitis |
| 472.0 | Chronic rhinitis | chronic rhinitis |
| 473.9 | Unspecified sinusitis (chronic) | chronic sinusitis |
| 477.9 | Allergic rhinitis; cause unspecified | allergic rhinitis; hay fever; seasonal allergies |
| 486 | Pneumonia; organism unspecified | pneumonia |
| 490 | Bronchitis; not specified as acute or chronic | bronchitis |
| 493.90 | Asthma; unspecified; unspecified status | asthma |
| 496 | Chronic airway obstruction; not elsewhere classified | chronic airway obstruction |
| 521.00 | Unspecified dental caries | dental caries; dental cavity |
| 530.81 | Esophageal reflux | esophageal reflux; gerd |
| 558.9 | Other and unspecified noninfectious gastroenteritis and colitis | non-infectious gastroenteritis; noninfectious gastroenteritis; non-infectious colitis; noninfectious colitis |
| 562.10 | Diverticulosis of colon (without mention of hemorrhage) | colon diverticulosis |
| 564.0 | Unspecified constipation | constipation |
| 564.1 | Irritable bowel syndrome | irritable bowel syndrome |
| 574.20 | Calculus of gallbladder without mention of cholecystitis or obstruction | gallbladder calculus; gallstones; gallstone |
| 584.9 | Acute kidney failure; unspecified | acute kidney failure; acute renal failure |
| 585.3 | Chronic kidney disease; Stage III (moderate) | stage 3 chronic kidney disease; ckd stage 3 |
| 592.0 | Calculus of kidney | kidney calculus; kidney stone; nephrolithiasis |
| 593.9 | Unspecified disorder of kidney and ureter | kidney disorder; ureter disorder |
| 599.0 | Urinary tract infection; site not specified | urinary tract infection |
| 600.0 | Hypertrophy (benign) of prostate | benign prostate hypertrophy |
| 611.72 | Lump or mass in breast | breast mass; breast lump |
| 616.10 | Unspecified vaginitis and vulvovaginitis | vaginitis; vulvovaginitis |
| 625.3 | Dysmenorrhea | dysmenorrhea |
| 626.2 | Excessive or frequent menstruation | excessive menstruation; frequent menstruation; menorrhagia; polymenorrhea; hypermenorrhea |
| 627.2 | Symptomatic menopausal or female climacteric states | symptomatic menopause; symptomatic menopausal |
| 692.9 | Contact dermatitis and other eczema; due to unspecified cause | contact dermatitis; eczema |
| 702.0 | Actinic keratosis | actinic keratosis |
| 706.1 | Other acne | acne |
| 709.9 | Unspecified disorder of skin and subcutaneous tissue | skin disorder |
| 723.4 | Brachial neuritis or radiculitis NOS | brachial neuritis |
| 724.4 | Thoracic or lumbosacral neuritis or radiculitis; unspecified | thoracic neuritis; thoracic radiculitis; lumbosacral neuritis; lumbosacral radiculitis; thoracic radiculopathy; lumbosacral radiculopathy |
| 729.1 | Unspecified myalgia and myositis | myalgia; myositis |
To select the lab tests to consider, we assembled the union of the top 10 most important lab features (according to our random forest models) from each of our 69 chosen diagnoses. There was substantial overlap of important features between diagnoses, leaving us with a total of 52 different lab features from our EHR dataset. Just as with the diagnoses, we curated the lab test names to better reflect what we would expect to find in the literature and to include synonyms. See Table 2 for our list of 52 lab tests and the search terms we used for each.
Table 2:
The 52 lab tests that we considered along with all search terms we used for each. Alias search terms are separated by semicolons.
| Lab Name | Curated Search Terms |
|---|---|
| ALT (GPT) | alanine aminotransferase |
| AST (GOT) | aspartate aminotransferase test |
| Anion Gap | anion gap |
| Bacteriuria Screen (Esterase) | bacteriuria esterase; bacteriuria screen; bacteriuria test |
| Bacteriuria Screen (Nitrate) | bacteriuria nitrate; bacteriuria screen; bacteriuria test |
| Bicarbonate (CO2) | blood bicarbonate; blood co2; serum bicarbonate; serum co2 |
| Bilirubin, Total-Neonatal | neonatal bilirubin; neonatal bile |
| Calcium | blood calcium; serum calcium |
| Chloride (Cl) | blood chloride; serum chloride |
| Cholesterol | cholesterol blood; serum cholesterol |
| Creatinine, Blood | blood creatinine; serum creatinine |
| Culture Organism | culture organism |
| Differential Segment Neut-Segs | segmented neutrophils; segmented pmn |
| Direct Bilirubin | direct bilirubin; conjugated bilirubin |
| Glom Filter Rate (GFR), Est | estimated glomerular filtration rate; egfr |
| Glucose | blood glucose; serum glucose |
| HDL Cholesterol | high density lipoprotein; hdl cholesterol |
| Hematocrit (Hct) | hematocrit |
| Hemoglobin (Hgb) | hemoglobin |
| Low Density Lipoprotein(LDL-C) | low density lipoprotein; ldl cholesterol |
| MCH | mean corpuscular hemoglobin |
| MCHC | mean corpuscular hemoglobin concentration |
| Mean Corpuscular Volume (MCV) | mean corpuscular volume |
| Phosphorus | blood phosphorus; serum phosphorus |
| Platelet Count (Plt) | platelet count |
| Potassium (K) | blood potassium; serum potassium |
| Prothrombin Time (PT)-INR | prothrombin time; international normalized ratio |
| Rapid Strep Antigen | rapid strep test |
| Red Blood Cell (RBC) Count | red blood cell count |
| Red Cell Distribute Width (RDW) | red cell distribution width |
| Sodium, Bld (Na) | blood sodium; serum sodium |
| Thyroid Stimul Hormone-Mfld | thyroid stimulating hormone |
| Total Cholesterol/HDL Ratio | cholesterol ratio |
| Triglycerides | triglycerides blood; triglycerides serum |
| Unconjugated Bilirubin | unconjugated bilirubin |
| Urea Nitrogen,Bld | urea nitrogen blood; serum urea nitrogen |
| Uric Acid,Bld | uric acid blood; uric acid serum |
| Urinalysis-Coarse Gran | urine coarse granular casts |
| Urinalysis-Color | urine color |
| Urinalysis-Fine Gran | urinary cast fine |
| Urinalysis-Hyaline | urine hyaline |
| Urinalysis-RBC | urine red blood cell |
| Urinalysis-Renal Epi | renal epithelial cells urine |
| Urinalysis-Spec Type | urinalysis specimen |
| Urinalysis-Specific Gravity | urine specific gravity |
| Urinalysis-Turbidity | urine turbidity |
| Urine Bile | urine bile; urine bilirubin |
| Urine Blood | urine blood |
| Urine Ketones | urine ketones |
| Urine Urobilinogen | urine urobilinogen |
| Urine pH | urine ph |
| White Blood Cell Count (WBC) | white blood cell count |
Predictive Models and Feature Importance
For each of the 69 diagnoses of interest we constructed a random forest model using case-control matched patient EHR data from Marshfield Clinic in Wisconsin. We phenotyped cases and controls from the EHR data using the “rule of 2” with cases having 2 or more entries of the diagnosis on their record and controls having no entries. We matched cases and controls based on age and date of birth (within 30 days) and we truncated all data for a case-control pair following 30 days prior to the case patient’s first entry of the diagnosis of interest. In this fashion we generated 5,000 case-control pairs (a total of 10,000 patients). Our patient data included demographics, diagnoses, labs, vitals, and procedures. Demographic features included age, sex, and date of birth. We summarize this information in Table 3. For all features except demographics, we extracted the features as counts in the time windows: 1-year, 3-years, 5-years, and ever. In this manner, our features were of the form “4 influenza diagnoses in the last 3-years”, or “2 high blood glucose labs in the last 1-year”. We used the random forest implementation from the Python package scikit-learn9 version 0.15. Each forest was trained with 500 trees and 10% of the features randomly selected at each split. We chose these setting a priori, as our prior research has performed well with these choices. All other settings for the forest used default parameters. We extracted feature importance values using scikit-learn’s built in functionality which uses the standard random forest feature importance calculation method4.
Table 3:
Demographic summary for the Marshfield electronic health record population.
| Characteristic | Women | Men | Total |
|---|---|---|---|
| n | 565,011 (51.5%) | 532,083 (48.5%) | 1,097,094 |
| Mean age, yrs | 46.7 ± 25.7 | 44.9 ± 25.5 | 45.8 ± 25.6 |
| < 18 y.o | 84,917 (15.0%) | 98,183 (18.5%) | 183,100 (16.7%) |
| 18-39 y.o. | 157,827 (27.9%) | 137,967 (25.9%) | 295,794 (27.0%) |
| 40-59 y.o. | 132,280 (23.4%) | 123,642 (23.2%) | 255,922 (23.3%) |
| ≥ 60 y.o. | 189,987 (33.6%) | 172,291 (32.4%) | 362,278 (33.0%) |
Text-Mining
We modify the KinderMiner algorithm for the text mining portion of this work to determine which diagnostic lab tests are likely novel in the literature. KinderMiner filters and ranks a list of target terms by their association with a key phrase of interest. It accomplishes this through simple string matching and document counting within a given text corpus. For each search, the user must specify the key phrase representing a concept of interest along with the list of target terms to be filtered and ranked by their association with the key phrase. KinderMiner then searches a given text corpus for article counts matching the target terms and key phrase. Specifically, it computes a contingency table of counts for each target term. For each target term, KinderMiner computes the number of articles containing both, either, and neither of the target term and key phrase. The result of this procedure is a list of contingency tables of document counts, one table for each target term. KinderMiner then performs a one-sided Fisher’s exact test on each contingency table, filtering out target terms that do not demonstrate statistically-significant co-occurrence with the key phrase according to a specified p-value threshold. Finally, the remaining target terms are ranked by the co-occurrence ratio, which is the number of articles in which a target term co-occurs with the key-phrase divided by the total number of articles in which the target term appears.
In this work, we make four modifications to the original KinderMiner algorithm (see Figure 1 for a visual representation). First, while the original KinderMiner algorithm finds exact string matches for target terms and key phrases, we extend this by breaking target terms and key phrases into their constituent tokens and matching on all tokens in any order or location within the document. For example, in the original KinderMiner a key phrase like “stage 3 chronic kidney disease” would need to match that string exactly to be counted and would not match the similar phrase “chronic kidney disease, stage 3.” Our modification breaks this key phrase into five tokens (“stage”, “3”, “chronic”, “kidney”, and “disease”) which must all be present in the document, but which do not need to match exactly in the original phrasing order.
Figure 1:
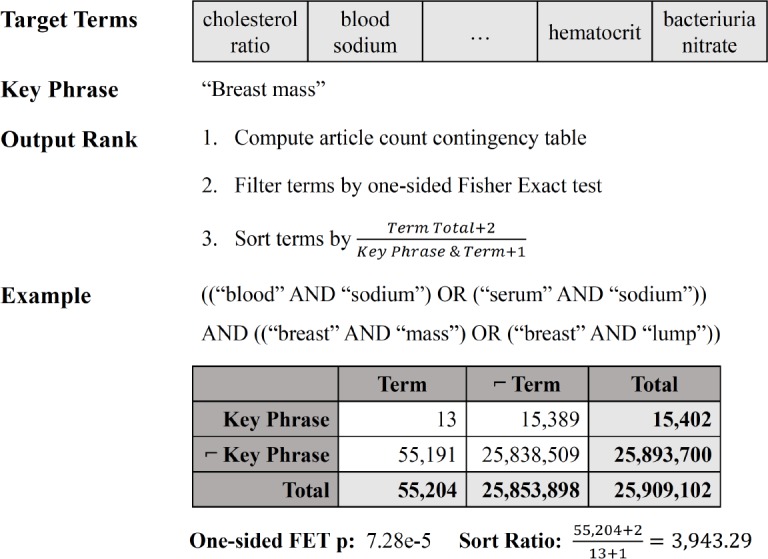
Visual example of our modified KinderMiner, with contingency table and disassociation Fisher’s Exact Test (FET) analysis of the diagnosis key phrase “breast mass” and the lab target term “blood sodium.” Target terms are filtered by significance of disassociation with the key phrase and then sorted by the inverted co-occurrence ratio.
Second, we extend KinderMiner to accommodate alias matching for target terms and key phrases. For example, we may expand a target term like “blood sodium” with an alias like “serum sodium.” Similarly, we can expand a key phrase like “breast mass” with an alias like “breast lump.” This is important for key phrases and target terms that may be referred to in multiple ways within the literature.
Third, in contrast to the original goal of KinderMiner, we wish to identify targets that are negatively associated with the key phrase in the literature. To accomplish this, we modify KinderMiner’s filtration step by changing the one-sided Fisher’s exact test to the opposite side test, thereby testing for significant negative association between each target term and key phrase.
Fourth, we also change how KinderMiner ranks the final filtered set of associations. KinderMiner typically ranks results by the co-occurrence ratio, the proportion of articles in which both the key phrase and target term occur over all articles in which the target term occurs. This is useful because it gives a rough estimate of the magnitude of association between the key phrase and the target term. In this work, we instead use the inverted co-occurrence ratio because it gives a rough estimate of the disassociation. When computing this ratio, we also add a pseudo-count of one to each of the article counts for when the key phrase and term co-occur, and when the target term appears without the key phrase. We add these pseudo-counts because it is not rare to find a significant disassociation when there are zero articles in which the term and key phrase co-occur.
As part of the filtration step, KinderMiner requires a p-value threshold for the Fisher’s exact test. While the original paper used 0.00001 in all cases, we loosen that threshold to 0.05 for our work. Because there are already few candidate lab-diagnosis pairs that appear unexpectedly disassociated in the literature, we care more about getting sufficient candidate discoveries than filtering out false positives.
KinderMiner also requires a text corpus to search. We constructed our text corpus from the National Library of Medicine’s MEDLINE/PubMed publicly available citation records10. We downloaded the annual baselines in XML format, parsed, and then ingested them into an Elasticsearch index (version 2.4.6). Our initial ingest of the 2017 annual baseline was performed in June and July 2017, and we updated to the 2018 baseline in November 2017. The dataset contains 27,947,480 citation records, with the abstracts indexed by Elasticsearch using two analysis chains. The default analysis that we use for all of the searches in our work is Elasticsearch’s standard analyzer, which applies a grammar-based tokenizer and lowercase filter to the text. We then use the Elasticsearch Query Domain Specific Language to construct each of our queries in JSON. Altogether, a search for the key phrase “breast mass” (with alias “breast lump”) and target term “blood sodium” (with alias “serum sodium”) would be equivalent to the following:
((“breast” AND “mass”) OR (“breast” AND “lump”)) AND ((“blood” AND “sodium”) OR (‘‘serum” AND “sodium”))
Hypothesis Ranking and Evaluation
Once we have gathered a set of hypothesized lab-diagnosis pairs to consider, we must rank them to help prioritize the best candidates for further investigation. Recall that we initially rank lab-diagnosis pairs by the inverted co-occurrence ratio. While this provides a lab test ranking for a particular diagnosis, we have several hypotheses from different diagnoses and we want to identify the most promising hypotheses overall. To do this, we construct a combined rank score defined as the product of a literature score and a feature score. For the literature score, we simply use the inverted co-occurrence ratio, which takes on large values when a lab is infrequently mentioned with a diagnosis. For the feature score, we use the feature importance of the lab value in the diagnosis model multiplied by the number of features in that diagnosis model (as to not bias models with small numbers of features). The product of the literature score and feature score thus gives a combined estimate of the novelty and the diagnostic importance of the lab.
With all the lab-diagnosis pairs ranked, we consider the top 20 hypotheses in more detail. To evaluate each candidate, we perform logistic regression analyses on the same dataset that was used to train the random forest. First, for each hypothesis, we perform a logistic regression with the diagnosis as the response variable and the laboratory test as the sole covariate. We use this to calculate the odds ratio of the lab in question. Second, we assess the odds ratio of the lab test in the presence of potential confounders. Finally, we perform manual literature search for important findings and decide if each is in fact novel.
To select potential confounders for a given diagnosis, we perform L1-regularized logistic regression on a feature set containing demographics, laboratory tests, and the top-level, whole integer ICD-9 codes. Moreover, our features for laboratory tests and demographics are binary features capturing if the patient did or did not have an entry of a particular health event in the last one year. We perform the L1-regularized logistic regression with scikit-learn9 and choose the minimum number of covariates greater than or equal to five by slowly increasing the regularization parameter. Note that as the cases and controls for this data were already age and sex matched we do not see these as discovered confounders. We then use these five (or more) selected features as confounders in logistic regression analysis (with R version 3.3.1) where we compute the odds ratio (and 95% confidence interval) of the lab in question both with an without the identified confounders. If the lab in question has an odds ratio that both maintains the same sign in both evaluations, and if the 95% confidence interval for both odds ratios exclude 1.0 (no change in odds), then we consider the hypothesis to be corroborated by the logistic regression analysis.
Results and Discussion
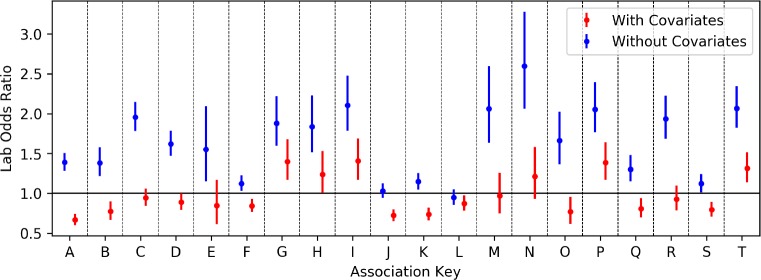
In Table 4 we present, in rank order, the top 20 hypotheses found between labs and diagnoses. In Figure 2 we plot the odds ratios of all 20 hypotheses both with and without potential confounders. We find that four of the 20 hypotheses passed the secondary logistic regression analysis and maintained an odds ratio 95% confidence interval above 1.0 both with and without potential confounders. In all hypotheses except one, hypothesis L, we see that the inclusion of confounders either diminishes or even reverses the trend found without confounders. For the four bolded hypotheses that passed our logistic regression analysis, we present in Table 5 the covariates selected by the L1-regularized logistic regression.
Table 4:
Summary and logistic regression odds ratios for the top 20 hypotheses. We compute the odds ratio of the lab test for a given hypothesis in two ways: as the sole covariate, “Odds”, and including potential confounders, “Adjusted Odds”. For both odds ratio calculations, we present the 95% confidence interval and bold 4 of the top 20 hypotheses whose 95% confidence intervals exclude 1.0 and whose odds are in the same direction, both before and after including confounders.
| Key | ICD-9 | Diagnosis | Lab Test | Odds | Adjusted Odds |
|---|---|---|---|---|---|
| A | 702.0 | Actinic Keratosis | Glucose | 1.4 [1.3, 1.5] | 0.67 [0.6, 0.74] |
| B | 702.0 | Actinic Keratosis | Creatinine, Blood | 1.4 [1.2, 1.6] | 0.78 [0.67, 0.9] |
| C | 367.4 | Presbyopia | Glucose | 2.0 [1.8, 2.1] | 0.95 [0.84, 1.1] |
| D | 600.0 | Benign Prostate Hypertrophy | LDL Cholesterol | 1.6 [1.5, 1.8] | 0.89 [0.79, 1.0] |
| E | 702.0 | Actinic Keratosis | Sodium, Bld (Na) | 1.6 [1.2, 2.1] | 0.85 [0.61, 1.2] |
| F | 162.9 | Malignant Lung Cancer | LDL Cholesterol | 1.1 [1.0, 1.2] | 0.84 [0.76, 0.93] |
| G | 461.9 | Acute Sinusitis | HDL Cholesterol | 1.9 [1.6, 2.2] | 1.4 [1.2, 1.7] |
| H | 461.9 | Acute Sinusitis | LDL Cholesterol | 1.8 [1.5, 2.2] | 1.2 [1.0, 1.5] |
| I | 472.0 | Chronic Rhinitis | Glucose | 2.1 [1.8, 2.5] | 1.4 [1.2, 1.7] |
| J | 162.9 | Malignant Lung Cancer | HDL Cholesterol | 1.0 [0.94, 1.1] | 0.72 [0.65, 0.8] |
| K | 496 | Chronic Airway Obstruction | LDL Cholesterol | 1.1 [1.0, 1.3] | 0.74 [0.66, 0.82] |
| L | 521.00 | Dental caries | LDL Cholesterol | 0.95 [0.85, 1.1] | 0.87 [0.78, 0.97] |
| M | 461.9 | Acute Sinusitis | Hemoglobin (Hgb) | 2.1 [1.6, 2.6] | 0.97 [0.75, 1.3] |
| N | 473.9 | Chronic Sinusitis | Hemoglobin (Hgb) | 2.6 [2.1, 3.3] | 1.2 [0.93, 1.6] |
| O | 367.0 | Hypermetropia | Hemoglobin (Hgb) | 1.7 [1.4, 2.0] | 0.77 [0.62, 0.96] |
| P | 461.9 | Acute Sinusitis | Cholesterol | 2.1 [1.8, 2.4] | 1.4 [1.2, 1.6] |
| Q | 496 | Chronic Airway Obstruction | Triglycerides | 1.3 [1.1, 1.5] | 0.81 [0.7, 0.94] |
| R | 530.81 | Esophageal Reflux-Gerd | WBC Count | 1.9 [1.7, 2.2] | 0.93 [0.79, 1.1] |
| S | 162.9 | Malignant Lung Cancer | Cholesterol | 1.1 [1.0, 1.2] | 0.8 [0.71, 0.89] |
| T | 466.0 | Acute Bronchitis | Cholesterol | 2.1 [1.8, 2.3] | 1.3 [1.1, 1.5] |
Figure 2:
The odds ratios and confidence intervals of all 20 lab-diagnosis hypotheses. Includes odds ratios for both with (right, red) and without (left, blue) covariates.
Table 5:
Covariates included as potential confounders for the 3 diagnoses included in the 4 hypotheses that passed the regression analysis. A minimum of 5 covariates were identified for each diagnosis, with Chronic Rhinitis including a 6th covariate as there was no L1 penalty that achieved 5.
| Confounder | Acute Sinusitis | Chronic Rhinitis | Acute Bronchitis |
|---|---|---|---|
| 1 | V72: Examination | 461: Acute Sinusitus | V72: Examination |
| 2 | Lab: Hemoglobin | 473: Chronic Sinusitis | Lab: MCHC |
| 3 | 786: Respiratory Symptoms | V72: Examination | Lab: Hemoglobin |
| 4 | 465: Acute URI | 465: Acute URI | 786: Respiratory Symptoms |
| 5 | 462: Acute Pharyngitis | 493: Asthma | 465: Acute URI |
| 6 | 786: Respiratory Symptoms |
The four hypotheses that met our odds ratio criteria for further consideration effectively represented three distinct hypotheses: cholesterol for acute sinusitis, cholesterol for acute bronchitis, and blood glucose for chronic rhinitis. While we expected to find few hits by design, we manually searched PubMed for articles related to these findings.
First, manual literature search for an association between cholesterol and acute sinusitis did not turn up direct associations. It did, however, turn up several hits for cholesterol granuloma of the maxillary sinus, which describes cysts containing cholesterol crystals and other fluids surrounded by fibrous tissue11. Symptoms are vague, and there are only two noted specific symptoms: clear golden yellow antral washout fluid, and washout containing cholesterol crystals. A family history of hypercholesterolemia was noted in one study12. This tangential association between cholesterol and sinus ailments within the literature, suggests to us that this discovered hypothesis is a promising lead for further investigation.
Second, literature search for an association between cholesterol and chronic bronchitis turned up two relevant studies. One study notes that low plasma lipid levels, particularly HDL cholesterol, is indicative of bacterial infection, and that low total cholesterol is predictive of adverse outcomes in patients with lower respiratory infections13. Another study suggests that lipid levels in airway mucus may be diagnostic for infection14. While the presence of these studies suggests prior awareness of an association, the literature is limited and can be viewed as confirmatory of our approach.
Third, literature search for an association between blood glucose and chronic rhinitis turned up one study. The study suggests that some antihistamine medications may affect blood glucose levels15. If true, this indicates that our hypothesis may instead be a confounded result of patients with chronic rhinitis having abnormal blood glucose as a result of antihistamine prescription, rather than blood glucose being predictive of rhinitis. Given the limited literature, however, the hypothesis may still warrant further investigation.
Conclusion
In this work, we propose a high-throughput pipeline for generating high-quality hypotheses for novel lab tests to predict diagnoses. We test our pipeline on a large electronic health record dataset and the PubMed corpus and, after manual evaluation, find several promising hypotheses in the top candidates.
One limitation of this work is the current need for manual mapping of diagnosis and lab terms to curated search terms. We expect that future work incorporating standardized naming and coding schemes in EHR datasets may obviate this need or that the process may be automated more completely in the future. This would facilitate higher throughput of diagnostic lab discovery by allowing us to run our pipeline on all diagnoses and all laboratory tests rather than a subset.
Our pipeline leverages the latent knowledge and patterns present in electronic health record data and the PubMed corpus to identify potentially interesting epidemiological findings. However, our proposed method does not eliminate the need for experimental design and further investigation of findings. On the contrary, it augments this process, and we argue that it represents a valuable addition by assisting with the prioritization of experiments when identifying biomarkers of disease.
Acknowledgments
The authors acknowledge support from the National Institutes of Health (NIH) grant U54-AI117924 and the National Library of Medicine (NLM) grant 5T15LM007359. The authors also thank Marv Conney for a grant to Ron Stewart and Finn Kuusisto.
References
- [1].Dawber TR, Meadors GF, Moore FE. Epidemiological approaches to heart disease: the Framingham Study. American journal of public health and the nation’s health. 1951 Mar;41(3):279–81. doi: 10.2105/ajph.41.3.279. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14819398http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC1525365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hsiao CJ, Hing E, Ashman J. National health statistics reports. 75. 2014. Trends in electronic health record system use among office-based physicians: United States, 2007-2012; pp. 1–18. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24844589. [PubMed] [Google Scholar]
- [3].Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. Journal of the National Cancer Institute. 1989 Dec;81(24):1879–86. doi: 10.1093/jnci/81.24.1879. Available from: http://www.ncbi.nlm.nih.gov/pubmed/2593165. [DOI] [PubMed] [Google Scholar]
- [4].Breiman L. Random forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- [5].Caruana R, Niculescu-Mizil A. An empirical comparison of supervised learning algorithms. Proceedings of the 23rd international conference on Machine learning; 2006. pp. 161–168. Available from: http://portal.acm.org/citation.cfm?doid=1143844.1143865. [Google Scholar]
- [6].Pautasso M. Publication growth in biological sub-fields: patterns, predictability and sustainability. Sustainability. 2012;4(12):3234–3247. [Google Scholar]
- [7].Bornmann L, Mutz R. Growth rates of modern science: a bibliometric analysis based on the number of publications and cited references. Journal of the Association for Information Science and Technology. 2015;66(11):2215–2222. [Google Scholar]
- [8].Kuusisto F, Steill J, Kuang Z, Thomson J, Page D, Stewart R. A simple text mining approach for ranking pairwise associations in biomedical applications. AMIA Summits on Translational Science Proceedings. 2017;2017:166–174. [PMC free article] [PubMed] [Google Scholar]
- [9].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2012;12:2825–2830. Available from: http://dl.acm.org/citation.cfm?id=2 078195{\%}5Cnhttp://arxiv.org/abs/1201.0490. [Google Scholar]
- [10].US National Library of Medicine. MEDLINE/PubMed citation records. https://www.nlm.nih.gov/databases/download/pubmed_medline.html. [Google Scholar]
- [11].Chao TK. Cholesterol granuloma of the maxillary sinus. European Archives of Oto-Rhino-Laryngology and Head & Neck. 2006 Jun;263(6):592–597. doi: 10.1007/s00405-006-0015-0. [DOI] [PubMed] [Google Scholar]
- [12].Dilek FH, Kiris M, Ugras S. Cholesterol granuloma of the maxillary sinus. Rhinology. 1997;35:140–141. [PubMed] [Google Scholar]
- [13].Gruber M, Christ-Crain M, Stolz D, Keller U, Mller C, Bingisser R. Prognostic impact of plasma lipids in patients with lower respiratory tract infections-An observational study. Swiss Medical Weekly. 2009;139:166–72. doi: 10.4414/smw.2009.12494. [DOI] [PubMed] [Google Scholar]
- [14].Bhaskar K, O’Sullivan D, Opaskar-Hincman H, Reid L. Lipids in airway secretions. European Journal of Respiratory Diseases. 1987;153:215–221. [PubMed] [Google Scholar]
- [15].Lal A. Effect of a few histamine(1)-antagonists on blood glucose in patients of allergic rhinitis. Indian Journal of Otolaryn-gology and Head and Neck Surgery. 2000;52(2):193–195. doi: 10.1007/BF03000353. [DOI] [PMC free article] [PubMed] [Google Scholar]