Abstract
Radiology reports contain descriptions of radiological observations followed by diagnosis and follow up recommendations, transcribed by radiologists while reading medical images. One of the most challenging tasks in a radiology workflow is to extract, characterize and structure such content to be able to pair each observation with an appropriate action. This requires classification of the findings based on the provided characterization. In most clinical setups, this is done manually, which is tedious, time-consuming and prone to human error yet of great importance as various types of findings in the reports require different follow-up decision supports and draw different levels of attention. In this work, we present a framework for detection and classification of change characteristics of pulmonary nodular findings in radiology reports. We combine a pre-trained word embedding model with a deep learning based sentence encoder. To overcome the challenge of access to limited labeled data for training, we apply Siamese network with pairwise inputs, which enforces the similarities between findings under the same category. The proposed multitask neural network classifier was evaluated and compared against state-of-the-art approaches and demonstrated promising performance.
1. Introduction
Radiology reports contain descriptions of imaging findings and the corresponding diagnosis transcribed by radiologists while reading radiology images. Criticality and significance of the findings vary based on the characterization of the observation in terms of size, shape, texture, etc. Considering pulmonary nodular findings, the significance is characterized by first determining whether the observation is a new finding or not. In the case of a prior finding follow-up, it is important to evaluate the interval change in terms of worsening, improving, or remaining unchanged. Radiologists typically provide a brief description of such characterization while transcribing a nodular finding. For example, “Ground-glass opacities are unchanged in the upper lobes”. Extraction and classification of the findings help determine and communicate the appropriate next care action more accurately and consistently. This is typically performed manually, which is tedious, time-consuming, and prone to human error. With the increase in the volume of imaging studies and consequently increase in the number of radiology reports, automatic structuring of radiology report content in the form of pairs of findings and the corresponding appropriate actions has gained significant interest during the last couple of decades. In this work, we propose a machine learning-based framework to classify findings in terms of characterization of nodular change. The task is defined as a sentence-level multi-class classification.
Sentence classification has previously been applied for different clinical applications including assessment detection1-3, automatic report summarization4, and incidental finding/follow-up recommendation extraction5-7. The state-of-the-art clinical sentence classification approaches use Natural Language Processing (NLP) techniques for parsing unstructured textual data. The computerized NLP techniques feature efficiency and scalability compared to the conventional approaches and are capable of processing large-scale data and obtaining results in near real-time, while the latter are time-consuming, labor-intensive, and require specific expertise.
Conventional sentence classification methods can be categorized into two groups8: rule-based/pattern matching2,3,6,9,10 and statistical machine learning-based approaches1,10-12. A rule-based approach is performed as a string-matching using a set of keywords pre-defined by experts or available through standard ontologies (such as SNOMED CT*). Rule-based approaches have been widely used for clinical tasks such as recommendation detection for incidental findings in radiology reports6, and acute bacterial Pneumonia detection and related concept extraction from chest X-ray reports9. A major drawback of rule-based approaches is the dependency of the performance on the completeness of the pre-defined keywords/patterns. On the other hand, machine learning-based approaches learn lexical and clinical features from a set of pre-labeled report contents to achieve the classification. Castro et al. proposed a machine learning-based classifier for automatic Breast Imaging Reporting and Data System (BI-RADS) categorization, using Bag-of-Words (BoW) and BI-RADS annotation occurrence as features to train Naive Bayes (NB) and Support Vector Machine (SVM) classifiers1. Solti et al. constructed a list of keywords and count their presences similar to BoW, and used maximum entropy for detection of acute Lung injury from radiology reports10. Lexical features such as term frequency - inverse document frequency (tf-idf)11,12 and n-grams13-15 have also been widely used as features for similar classification tasks. More recently, deep learning techniques, including Convolutional Neural Networks (CNN) and Recurrent Neural Networks (RNN), have been applied to general purpose word/sentence embedding16-18 and classification19-22 tasks such as sequence tagging and sentiment analysis, and demonstrated promising results compared with the conventional NLP approaches discussed above. More recently, the success of deep learning techniques in NLP-related tasks inspires the recent applications in the clinical domain such as clinical decision-making23,24, Name Entity Recognition (NER)25,26.
In this work, we present a multi-step framework for detection and classification of change characteristics in pulmonary nodular findings described in radiology reports using state-of-the-art deep learning. The proposed classification problem was motivated by a recent work by Hassanpour et al. who proposed an NLP-based pipeline to categorize the change and significance of clinical findings from radiology reports3, utilizing a keyword-matching algorithm. Nevertheless, this work suffers from similar shortcomings of keyword-matching approaches. Different from the conventional feature extraction steps, we combine a pre-trained word embedding model with a deep learning based sentence encoder to convert the text into dense vectors in a lower dimension. The word embedding (skip-gram) model is able to learn a distributed representation for each word and capture its semantics16,27. The obtained word embeddings are then fed into the sentence encoder to learn vector representations of the sentences that are used as features for the classification. The biggest challenge to apply deep learning approaches in clinical domain is the data scale as it requires large-scale labeled data to train deep learning models without overfitting. Therefore, we apply Siamese network, which is a dual network used to predict the similarity between a pair of input samples28,29, in addition to the sentence classification network to enforce the similarities between findings under the same category. We jointly learn the two neural networks in a multitask scheme. The proposed multitask neural network classifier was evaluated and compared against state-the-art approaches and demonstrated promising performance on pulmonary nodular finding classification.
The main contributions of this work can be summarized as following:
A multitask deep learning framework is proposed for classification of change characterization in pulmonary nodular findings described in radiology reports with a robust high-precision/recall. To the best of our knowledge, this is the first work utilizing multitask deep learning to tackle such problem and achieve promising performance.
We utilize Siamese network in a multitask scheme to overcome a typical challenge in deep learning solutions for healthcare tasks: lack of sufficient labeled data for training. This is achieved by considering all possible pairs of input sentences.
A thorough investigation and comparison is conducted and reported on an extensive selections of features and methods including conventional machine learning and state-of-the-art deep learning-based approaches, which resulted in insights and suggestions for future work.
2. Material and Methods
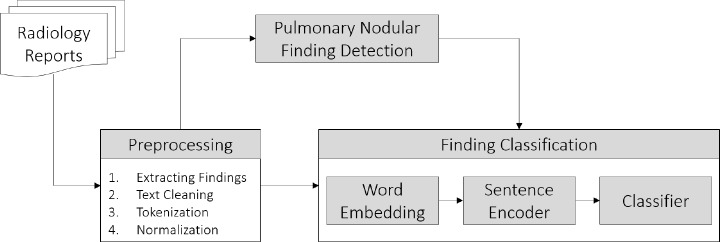
Figure 1 demonstrates an overview of the proposed framework. In summary, radiology reports are preprocessed to extract finding sentences. Next, sentences with an indication of pulmonary nodular findings are automatically identified. Finally, pulmonary nodular finding sentences are classified in terms of characterization of change. Details on each step are provided in the following sections.
Figure 1:
An overview of the proposed pulmonary nodular finding classification framework.
2.1. Preprocessing
Free-text radiology reports are processed to split into sentences using SpaCy*. The sentences are further processed to lower case, removing redundant spaces, correcting incidental sentence chunking caused by punctuations, and fixing special characters and encodings. In the end, we obtain the word tokens of the cleaned sentences using SpaCy. The tokens are further normalized based on the entity type such as measurements and dates: e.g., “3.0 × 2.0 × 3.0 mm” is normalized to “9 × 9 × 9 mm”, and time formats such as “01/01/2001” and “Jan 1st, 2001” are converted to “9-9-99”.
2.2. Pulmonary Nodular Finding Detection
Pulmonary nodular finding detection was achieved in two steps: First, a previously proposed anatomical phrase labeling approach was applied for pulmonary sentence classification30. Next, sentences with pulmonary nodular findings were automatically identified using a logistic regression-based binary classifier. The input feature vector consists of TFIDF features as well as binary features based on regular expression-based keyword matching. Here is a list of keywords extracted from SNOMED CT ontology: nodule, focus, mass, consolidation, lesion, focal opacity, cyst, ground-glass, tree-in-bud, abnormality, carcinoma, focal density, and tumor.
2.3. Pulmonary Nodular Finding Classification
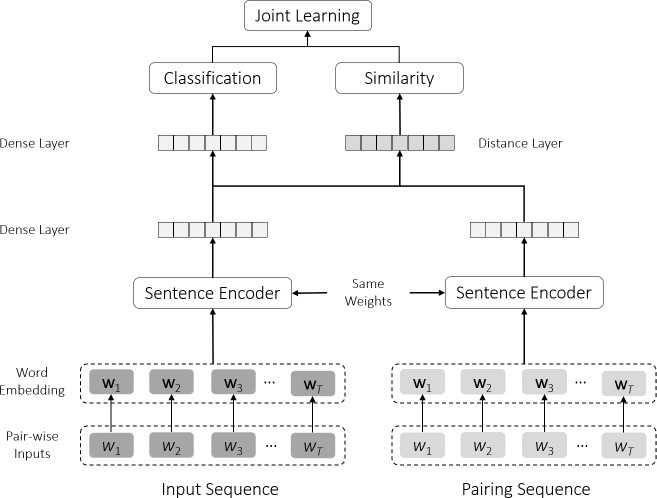
The basic finding classification model (denoted as encoder classifier) is a deep neural network including the following components: (1) word embedding which converts input words/tokens into dense vectors, (2) sentence encoder which reads embedding vectors and outputs highly abstracted features of the findings, and (3) fully-connected layers for further classification tasks based on the encoded feature. In addition, since we have a relatively small labeled dataset, we apply a Siamese network in addition to the encoder classifier in order to utilize pairwise data. The network configuration is shown in Figure 2.
Figure 2:
The proposed deep neural network for pulmonary nodular finding classification in a multitask scheme.
2.3.1. Word Embedding and the Pre-trained Model
We apply the skip-gram model for word embedding. The skip-gram works in an unsupervised fashion, and is proposed to learn semantic representations of the words16. The underlying assumption of skip-gram is that the semantics of the current word is distributed through the whole sequence or within a window of nearby words. For example, we have an input word sequence T = {w1, w2, wN} of N words where wt indicates the current word at step t, and let c denote the window size, the objective function of skip-gram model is to maximize the likelihood of the occurrences of words within the window range given the current word, as shown in Equation 1:
| (1) |
where the conditional probability is calculated from soft-max. A word wt in the sequence T is mapped into a word vector wt ∈ ℝn×1, and used to construct the vector sequence T ∈ ℝN×n to represent the input sequence where T = [w1, w2,…,wt]T. The number of rows is fixed to be the maximum length among all the input sequences, and zero-padding is performed for shorter sequences. The details on pre-training the skip-gram model is discussed in Section 2.4.
2.3.2. Sentence Encoder
Three one-layer architectures were considered for the sentence encoder: uni- and bi-directional Long Short Term Memory (LSTM)31 and one-dimensional CNN23,32. The learned features are fed into two layers of fully-connected layers where we use a soft-max function as the activation function in the last layer. Given N samples and C classes in total, let g(·) denote the encoder function including the first fully-connected layer, ŷi and yi denote the prediction and ground truth of sample xi, the prediction loss is thus defined in Equation 2.
| (2) |
2.3.3. Siamese Network
Deep learning techniques typically require large-scaled labeled data. Nevertheless, it is always a significant challenge to create large-scale labeled data. We utilize Siamese network in addition to the classification task in a multitask scheme in order to overcome the shortcoming of limited training data. Siamese network was initially proposed to tackle the one-shot and few-shot learning problem28,29. The inputs to the Siamese network are pairwise samples from different classes and the outputs are the probabilities of the input sample pairs belonging to the same class. The intuitions of applying the Siamese network is two folds: (1) by creating pairwise inputs we are able to upsample the data scale from O(N) to up to O(m • N), m ∈ [1, N/2] which is also controllable, and (2) the objective of the Siamese network is to enforce the within-class similarity while maximizing the inter-class dissimilarities. In our case, the Siamese network is constructed by the same sentence encoder as we introduced in the previous section. After we obtained the features for the two input samples xi and xj, a joint distance layer is used to compute the absolute distance per dimension between the two layers. Then, by adding a fully-connected layer and sigmoid activation σ(·), we obtain the final prediction , and the similarity loss following binary cross entropy in Equation 3. Output labels for the Siamese network are created based on the following rule: output label is set to “1” if the input sample pair belong to the same class, and “0” otherwise.
| (3) |
2.3.4. Multitask Learning
In order to achieve an end-to-end training where we can perform both the sentence classification and similarity prediction at the same time, we construct a joint neural network in a multitask fashion as shown in Figure 2. The two components of the multitask neural network share the same sentence encoder with the same weights. Therefore, during the training process, the encoder learns to encode the findings under the same class close to each other as well as to push the findings from different classes away from each other. The total loss function of the multitask neural network is calculated as a weighted average of the classification loss and the similarity loss as defined in Equation 4, where α is a scaler. We run validation after training for 10 batches each time and adjust the scaler α based on the loss values obtained on the validation set.
| (4) |
2.4. Clinical Data
Radiology reports from two different clinical sites, the University of Washington (UW) and the University of Chicago (UC) were used in this study. Radiology reports were collected with Institutional Review Board (IRB) approvals. All reports were de-identified by offsetting dates with randomly generated numbers. All other HIPAA patient health information including name, date of birth and address were removed. The following section describes the break down of the data:
Word embedding: 1,566,921 reports from the UW dataset as well as 334,486 unique terms extracted from SNOMED CT ontology (version 20150731) were used for training the word embedding model. After applying preprocessing, the training corpus contained 418,761,995 tokens with a vocabulary size of 270,015. Details are given in30.
Pulmonary nodular finding corpus: 2,000 radiology reports were randomly selected from the UC dataset and manually labeled for pulmonary nodular finding types: New/Indeterminate, Worsening, Unchanged, and Improving. Such choice of categorization is adopted from a previous work3 with a couple of differences: New and Indeterminate are combined as one class and Worsening is considered as a separate class. Definitions of the classes and the corresponding data distributions are provided in Table 1. The suggested class choice differences is based on recommendations by radiologists from the collaborating sites to adapt to their current care workflow. From the selected 2,000 reports, 438 radiology reports contain sentences of pulmonary nodular findings (918 sentences in total) are detected by the pulmonary nodular finding detection phase as discussed in Section 2.2. We further split the pulmonary nodular finding sentences to 80% and 20% as the training/validation and testing sets.
Table 1:
Definition of pulmonary nodular finding type based on characterization of change and sample size.
| Class Type | Sample Size (no. of sentences) | Definition |
|---|---|---|
| New/Indeterminate | 421 | Nodular finding was not present in a prior study and/or cannot be assessed as any of the other types |
| Worsening | 87 | Nodular finding has progressed |
| Unchanged | 340 | Nodular finding has not changed |
| Improving | 90 | Nodular finding has been partially or completely resolved |
3. Results
Since our task is a multi-class classification and the classes are imbalanced, we apply the weighted average of the precision, recall, and F1-score by the number of instances of each class33 to calculate the overall performances for evaluating and comparing the classification approaches. In this case, the F1-score can be not in the range of the precision and recall because it is a weighted average over the classes instead of a weighted average over the precision and recall per class.
The proposed classifier is compared against a number of conventional and state-of-the-art approaches. In order to conduct a fair comparison between our proposed approach and other methods, we incorporate the commonly used hand-crafted features including BoW, tf-idf, n-grams (including bi-grams and tri-grams), and all possible combinations, with various classifiers including NB, SVM, Logistic Regression (LR), and Random Forest (RF). All combinations of the aforementioned features were considered for both training and testing. In order to obtain the optimal performance for each feature set and classifier combination, we carefully run grid search within the training set on the hyper-parameters and choose the best performing settings. The options of L1 and L2 regularization are also included in the grid search to deal with the high dimensionality of the inputs. L1 regularization enforces the sparsity in the coefficients and tends to perform better on high dimensional tasks, while L2 regularization is more commonly used to prevent overfitting34. Additionally, we compare the proposed multitask finding classification model with Siamese network and joint loss with the state-of-the-art deep learning models based on different sentence encoders but without the Siamese network: Bi-directional LSTM (denoted as BiLSTM)35, uni-directional LSTM (denoted as LSTM)31,36, one-dimensional CNN29,32, and the Deep Averaging Network (DAN), which is a deep neural network consist of two fully-connected layers on top of the averaged word2vec features obtained from all the words in the input sequence37. The proposed multitask neural networks with different encoders are denoted as Multitask CNN, Multitask LSTM, and Multitask BiLSTM. All deep learning models are trained on batches with 64 samples. We also implemented the rule-based approach proposed by Hassanpour et al.3 to solve a similar finding classification problem. Nevertheless, we were unable to replicate the results on our test set using the provided keyword set3. The is due to the fact that: 1) creating a comprehensive list of keywords is always a challenge; and 2) determining the class label solely based on keywords may not be sufficient as in some scenarios, the hint to determine the appropriate class label is in the context of the sentence1. Consider the following example: “The other nodule previously noted on the prior study cannot be identified on the current examination”. By considering the context, one can simply conclude that the appropriate class label for the nodule is Improving. However, there is no specific keyword that highlights the class label.
Table 2 summarizes the performance of different approaches in terms of precision, recall, and F1-score. From Table 2, we can observe that the proposed Siamese network-based multitask CNN yields the best precision, recall, and F1-score outperforming the other state-of-the-art deep learning approaches by about 2%. Table 3 summarizes the performance of the proposed multitask neural network with the CNN encoder on each class separately. As can be observed from the table, the worst precision and recall belong to Worsening and Improving classes. We also present the confusion matrix among different classes in Figure 3. As can be observed from Figure 3, class New/Indeterminate is confused the most with other classes, specifically, the most confusion being with class Improving.
Table 2:
Results on pulmonary nodular finding classification. The best performing results are shown as bold. NB: Naive Bayes; SVM: Support Vector Machine; LR: Logistic Regression; RF: Random Forest; DAN: Deep Averaging Network; CNN: Convolutional Neural Network; LSTM: Long Short Term Memory; BiLSTM: Bi-directional LSTM.
| Models | Evaluation | ||
|---|---|---|---|
| Precision | Recall | F1-score | |
| NB + tf-idf | 75.50% | 75.54% | 75.21% |
| SVM + All Features | 88.65% | 86.96% | 86.65% |
| LR + All Featuers | 88.24% | 87.50% | 87.47% |
| RF + tf-idf | 82.86% | 80.43% | 81.10% |
| DAN | 82.97% | 81.52% | 81.87% |
| CNN | 88.93% | 86.96% | 86.34% |
| LSTM | 88.37% | 88.04% | 87.97% |
| BiLSTM | 87.24% | 86.96% | 86.72% |
| Our proposed Multitask CNN | 90.65% | 89.67% | 89.69% |
| Our proposed Multitask LSTM | 89.35% | 88.59% | 88.36% |
| Our proposed Multitask BiLSTM | 90.64% | 89.67% | 89.44% |
Table 3:
Per-class Precision/Recall/F1-score using the proposed CNN-based Multitask Neural Network.
| Class Names | Precision | Recall | F1-score |
|---|---|---|---|
| New/Indeterminate | 86.08% | 97.14% | 91.28% |
| Worsening | 70.59% | 92.31% | 80.00% |
| Unchanged | 98.61% | 87.65% | 92.81% |
| Improving | 87.50% | 70.00% | 77.78% |
Figure 3:
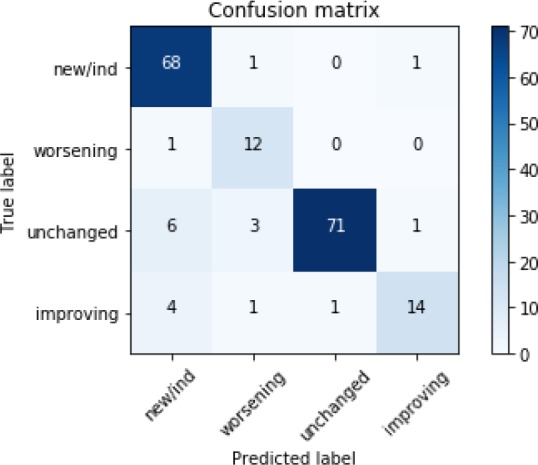
The confusion matrix of the proposed multitask neural network with CNN-based sentence encoder.
4. Discussion
Automatic identification and extraction of actionable information from unstructured radiology report content is a challenging task due to the significant variation in radiologists’ language in transcribing such information. Consider the following examples: “Previously described nodular opacity in the left upper lobe less conspicuous on current examination.”; “Marked interval decrease in size of the nodular opacity within the left upper lobe”; “left upper lobe nodular opacity has resolved since the prior examination.” All these sentences refer to an improving nodular finding yet in quite different languages.
While achieving a high recall is desired for guaranteeing the detection of all actionable sentences within the report, reaching a high precision is quite important for finding type classification to avoid confusions and mistakes in the workflow and care management. The proposed classifier yields relatively high precisions and recalls for three out of four class. Additionally, class confusion is quite an important factor in the evaluation of a classifier. However, not all confusions will have the same impact on the overall performance which is special in the healthcare domain. In other words, confusions between certain classes may result in higher costs in downstream applications. For example, for the current use case, confusing worsening with improving or unchanged classes could have a dramatic impact on the decision on the appropriate care action and as a result, pose a significant risk on patient’s health. Considering the confusion matrix provided in Figure 3, the most confusion is between new/indeterminate and other classes. This is an expected error considering a current limitation of the proposed framework. Consider the following example: “Small, noncalcified nodules are seen in the right upper lobe on image 18/102. These are unchanged in the interim.” The first sentence was labeled as new/indeterminate by the algorithm; however, by considering the following sentence, the appropriate label seems to be unchanged. This is due to the fact that the proposed framework does not take any extra context beyond the target sentence into account for the classification. The most worrisome confusion is between worsening and other three classes, as a patient falling under unchanged or improving classes may not require any follow-up action, whereas a patient under worsening class most definitely requires short-term or immediate attention. Fortunately, the proposed framework demonstrated no such confusion as can be seen in Figure 3.
Building a classifier that could reliably distinguish between different classes requires a significant amount of training labeled data to capture as much of the variation in the data as possible. Nevertheless, creating such size of labeled data is costly and requires a significant amount of effort, and time. To overcome such challenges, we incorporated Siamese network in a multitask and end-to-end scheme to tackle the limitation of data scale, which often occurs in deep learning applications. Such incorporation benefits the overall performance by constructing pairwise samples in a large-scale and enforce the differences between findings from different categories. The proposed multitask neural network framework can be potentially applied to other similar classification tasks in the clinical domain, where labeled data is usually limited in scales and very expensive to obtain.
5. Conclusions
In this work, we present a deep learning approach to tackle the pulmonary nodular finding classification in terms of characterization of change. To overcome the challenge of access to limited labeled data, we take advantage of the Siamese network in a multitask scheme. The proposed architecture takes pairwise samples as inputs to increase the number of training samples. Furthermore, the Siamese network is able to capture the similarities and subtle differences for samples under the same and different classes. We compared the proposed framework against a number of conventional machine learning approaches as well as a few state-of-the-art deep learning methods and demonstrated that the proposed approach outperforms the other methods.
Footnotes
https://www.snomed.org/snomed-ct
References
- 1.Sergio M. Castro, Eugene Tseytlin, Olga Medvedeva, Kevin J. Mitchell, Shyam Visweswaran, Tanja Bekhuis, Rebecca S. Jacobson. Automated annotation and classification of BI-RADS assessment from radiology reports. Journal of Biomedical Informatics. 2017;69:177–187. doi: 10.1016/j.jbi.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorothy A. Sippo, Graham I. Warden, Katherine P. Andriole, Ronilda Lacson, Ichiro Ikuta, Robyn L. Birdwell, Ramin Khorasani. Automated extraction of BI-RADS final assessment categories from radiology reports with natural language processing. J. Digital Imaging. 2013;26(5):989–994. doi: 10.1007/s10278-013-9616-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saeed Hassanpour, Graham Bay, Curtis P. Langlotz. Characterization of change and significance for clinical findings in radiology reports through natural language processing. J. Digital Imaging. 2017;30(3):314–322. doi: 10.1007/s10278-016-9931-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniel J. Goff, Thomas W. Loehfelm. Automated radiology report summarization using an open-source natural language processing pipeline. J. Digital Imaging. 2018;31(2):185–192. doi: 10.1007/s10278-017-0030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meliha Yetisgen-Yildiz, Martin L. Gunn, Fei Xia, Thomas H. Payne. A text processing pipeline to extract recommendations from radiology reports. Journal of Biomedical Informatics. 2013;46(2):354–362. doi: 10.1016/j.jbi.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Sayon Dutta, William J Long, David FM Brown, Andrew T Reisner. Automated detection using natural language processing of radiologists recommendations for additional imaging of incidental findings. Annals of emergency medicine. 2013;62(2):162–169. doi: 10.1016/j.annemergmed.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Anne-Dominique Pham, Aurélie Névéol, Thomas Lavergne, Daisuke Yasunaga, Olivier Clément, Guy Meyer, Rémy Morello, Anita Burgun. Natural language processing of radiology reports for the detection of thromboembolic diseases and clinically relevant incidental findings. BMC Bioinformatics. 2014;15:266. doi: 10.1186/1471-2105-15-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tianrun Cai, Andreas A Giannopoulos, Sheng Yu, Tatiana Kelil, Beth Ripley, Kanako K Kumamaru, Frank J Rybicki, Dimitrios Mitsouras. Natural language processing technologies in radiology research and clinical applications. Radiographics. 2016;36(1):176–191. doi: 10.1148/rg.2016150080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcelo Fiszman, Wendy Webber Chapman, Dominik Aronsky, Scott Evans R, Peter J. Haug. Research paper: Automatic detection of acute bacterial pneumonia from chest x-ray reports. JAMIA. 2000;7(6):593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imre Solti, Colin R Cooke, Fei Xia, Mark M Wurfel. Automated classification of radiology reports for acute lung injury: comparison of keyword and machine learning based natural language processing approaches. In Bioinformatics and Biomedicine Workshop, 2009. BIBMW 2009. IEEE International Conference on; IEEE; 2009. pp. 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brian E. Chapman, Sean Lee, Hyunseok Peter Kang, Wendy Webber Chapman. Document-level classification of CT pulmonary angiography reports based on an extension of the context algorithm. Journal of Biomedical Informatics. 2011;44(5):728–737. doi: 10.1016/j.jbi.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hari Trivedi, Joseph Mesterhazy, Benjamin Laguna, Thienkhai Vu, Jae Ho Sohn. Automatic determination of the need for intravenous contrast in musculoskeletal MRI examinations using IBM watson’s natural language processing algorithm. J. Digital Imaging. 2018;31(2):245–251. doi: 10.1007/s10278-017-0021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imre Solti, Cooke C, Fei Xia, Wurfel M. Peeling away the black box label: clinical validation of a maxent machine learning character n-gram feature set for acute lung injury. AMIA Summit on Translational Bioinformatics. 2010.
- 14.Meliha Yetisgen-Yildiz, Brad J Glavan, Fei Xia, Lucy Vanderwende, Mark M Wurfel. Identifying patients with pneumonia from free-text intensive care unit reports. In Proceedings of Learning from Unstructured Clinical Text Workshop of the International Conference on Machine Learning.2011. [Google Scholar]
- 15.Eamon Johnson, Baughman W Christopher, Gultekin Ozsoyoglu. Mixing domain rules with machine learning for radiology text classification. In Proceedings of the ACM SIGKDD Workshop on Health Informatics (HI-KDD 2014).2014. [Google Scholar]
- 16.Tomas Mikolov, Ilya Sutskever, Kai Chen, Gregory S. Corrado, Jeffrey Dean. Distributed representations of words and phrases and their compositionality. In Advances in Neural Information Processing Systems 26: 27th Annual Conference on Neural Information Processing Systems 2013. Proceedings of a meeting held December 5-8, 2013, Lake Tahoe, Nevada, United States; 2013. pp. 3111–3119. [Google Scholar]
- 17.Ryan Kiros, Yukun Zhu, Ruslan Salakhutdinov, Richard S. Zemel, Raquel Urtasun, Antonio Torralba, Sanja Fidler. Skip-thought vectors. In Advances in Neural Information Processing Systems 28: Annual Conference on Neural Information Processing Systems 2015; Canada: Montreal, Quebec; 2015 2015. Dec 7-12, pp. 3294–3302. [Google Scholar]
- 18.Edouard Grave, Tomas Mikolov, Armand Joulin, Piotr Bojanowski. Bag of tricks for efficient text classification. In Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics, EACL 2017; Spain: Valencia; 2017 2017. Apr 3-7, pp. 427–431. [Google Scholar]
- 19.Yoon Kim. Convolutional neural networks for sentence classification. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing, EMNLP 2014; Doha, Qatar: A meeting ofSIGDAT, a Special Interest Group of the ACL; 2014 2014. Oct 25-29, pp. 1746–1751. [Google Scholar]
- 20.Mohit Iyyer, Varun Manjunatha, Jordan L. Boyd-Graber, Hal Daume., III Deep unordered composition rivals syntactic methods for text classification. In Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing of the Asian Federation ofNatural Language Processing, ACL 2015; Beijing, China: 2015 2015. Jul 26-31, pp. 1681–1691. [Google Scholar]
- 21.Yafeng Ren, Yue Zhang, Meishan Zhang, Donghong Ji. Context-sensitive twitter sentiment classification using neural network. In Proceedings of the Thirtieth AAAAI Conference on Artificial Intelligence; Phoenix, Arizona, USA: 2016 2016. Feb 12-17, pp. 215–221. [Google Scholar]
- 22.Zhengqiu He, Wenliang Chen, Zhenghua Li, Meishan Zhang, Wei Zhang, Min Zhang. SEE: syntax-aware entity embedding for neural relation extraction. In Proceedings of the Thirty-Second AAAI Conference on Artificial Intelligence; New Orleans, Louisiana, USA: 2018 2018. Feb 2-7, [Google Scholar]
- 23.Jianbo Yuan, Chester Holtz, Tristram H. Smith, Jiebo Luo. Autism spectrum disorder detection from semi-structured and unstructured medical data. EURASIP J. Bioinformatics and Systems Biology. 2017 2017;3 doi: 10.1186/s13637-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenglong Ma, Radha Chitta, Jing Zhou, Quanzeng You, Tong Sun, Jing Gao. Dipole: Diagnosis prediction in healthcare via attention-based bidirectional recurrent neural networks. In Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; Halifax, NS, Canada: 2017 2017. Aug 13-17, pp. 1903–1911. [Google Scholar]
- 25.Fei Li, Meishan Zhang, Guohong Fu, Donghong Ji. A neural joint model for entity and relation extraction from biomedical text. BMC Bioinformatics. 2017;18(1):198:1–198:11. doi: 10.1186/s12859-017-1609-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xuan Wang, Yu Zhang, Xiang Ren, Yuhao Zhang, Marinka Zitnik, Jingbo Shang, Curtis Langlotz, Jiawei Han. Cross-type biomedical named entity recognition with deep multi-task learning. CoRR. 2018. (abs/1801.09851).
- 27.Yaoyun Zhang, Hee-Jin Li, Jingqi Wang, Trevor Cohen, Kirk Roberts, Hua Xu. Adapting word embeddings from multiple domains to symptom recognition from psychiatric notes. AMIA Summits on Translational Science Proceedings. 2017 2018;281 [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory Koch, Richard Zemel, Ruslan Salakhutdinov. Siamese neural networks for one-shot image recognition. In ICML Deep Learning Workshop. 2015.
- 29.Jianbo Yuan, Han Guo, Zhiwei Jin, Hongxia Jin, Xianchao Zhang, Jiebo Luo. One-shot learning for finegrained relation extraction via convolutional siamese neural network. In 2017 IEEE International Conference on Big Data, BigData 2017; Boston, MA, USA: 2017 2017. Dec 11-14, pp. 2194–2199. [Google Scholar]
- 30.Amir M. Tahmasebi, Henghui Zhu, Gabriel Mankovich, Peter Prinsen, Prescott Klassen, Sam Pilato, Rob van Ommering, Pritesh Patel, Martin L. Gunn, Paul Chang. Journal of Digital Imaging. 2018. Aug, Automatic normalization of anatomical phrases in radiology reports using unsupervised learning. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sepp Hochreiter, Jurgen Schmidhuber. Long short-term memory. Neural computation. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 32.Daojian Zeng, Kang Liu, Yubo Chen, Jun Zhao. Distant supervision for relation extraction via piecewise convolutional neural networks. In Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, EMNLP 2015; Lisbon, Portugal: 2015 2015. Sep 17-21, pp. 1753–1762. [Google Scholar]
- 33.Shantanu Godbole, Sunita Sarawagi. Discriminative methods for multi-labeled classification. In Advances in Knowledge Discovery and Data Mining, 8th Pacific-Asia Conference, PAKDD 2004; Sydney, Australia: 2004 2004. May 26-28, pp. 22–30. [Google Scholar]
- 34.Rong-En Fan, Kai-Wei Chang, Cho-Jui Hsieh, Xiang-Rui Wang, Chih-Jen Lin. LIBLINEAR: A library for large linear classification. Journal of Machine Learning Research. 2008;9:1871–1874. [Google Scholar]
- 35.Alex Graves, Santiago Fernandez, Jurgen Schmidhuber. Bidirectional LSTM networks for improved phoneme classification and recognition. In Artificial Neural Networks: Formal Models and Their Applications - ICANN 2005, 15th International Conference; Warsaw, Poland: 2005 2005. Sep 11-15, pp. 799–804. [Google Scholar]
- 36.Martin Sundermeyer, Ralf Schluter, Hermann Ney. LSTM neural networks for language modeling. In IN-TERSPEECH 2012, 13th Annual Conference of the International Speech Communication Association; Portland, Oregon, USA. 2012 2012. Sep 9-13, pp. 194–197. [Google Scholar]
- 37.Mohit Iyyer, Varun Manjunatha, Jordan Boyd-Graber, Hal Daume., III Deep unordered composition rivals syntactic methods for text classification. In Association for Computational Linguistics. 2015.