Abstract
Electronic health records (EHR) are valuable to define phenotype selection algorithms used to identify cohorts ofpatients for sequencing or genome wide association studies (GWAS). To date, the electronic medical records and genomics (eMERGE) network institutions have developed and applied such algorithms to identify cohorts with associated DNA samples used to discover new genetic associations. For complex diseases, there are benefits to stratifying cohorts using comorbidities in order to identify their genetic determinants. The objective of this study was to: (a) characterize comorbidities in a range of phenotype-selected cohorts using the Johns Hopkins Adjusted Clinical Groups® (ACG®) System, (b) assess the frequency of important comorbidities in three commonly studied GWAS phenotypes, and (c) compare the comorbidity characterization of cases and controls. Our analysis demonstrates a framework to characterize comorbidities using the ACG system and identified differences in mean chronic condition count among GWAS cases and controls. Thus, we believe there is great potential to use the ACG system to characterize comorbidities among genetic cohorts selected based on EHR phenotypes.
Introduction
Electronic health records (EHR) are rich resources to identify patients with specific conditions for inclusion in genetic studies. Within the NHGRI-funded electronic MEdical Records & GEnomics (eMERGE) Network1–3, for example, EHR phenotyping methods are used to identify cohorts with linked DNA samples used to discover new genetic associations. Given the variability in approaches to implement EHR phenotypes (e-phenotypes) among institutions, documentation is often shared as “pseudocode” and made accessible using the Phenotype KnowledgeBase4,5.
Several genome-wide association studies (GWAS) have been completed for a range of e-phenotypes defined by eMERGE institutions, such as dementia, cataracts, peripheral arterial disease, type 2 diabetes and cardiac conduction defects6–9. While GWAS are generally carried out for one phenotype at a time, for complex diseases, the existence of secondary (comorbid) phenotypes can influence results. For example, we can find significant overlap in genetic associations among related conditions10. One approach to consider comorbidities in GWAS is to stratify results by suspected or known comorbidities e.g., assessing whether common variants interact with hypertension to modify the risk of atrial fibrillation11. Comorbidity indices are often used in health research12, but GWAS analyses have not typically assessed comorbidities in ways that would distinguish whether observed variant-trait associations are with the primary phenotype or co-occurring “comorbid” phenotypes. Thus, the extent of the influence of comorbid phenotypes on GWAS findings is an area that often cannot be studied. This work proposes to comprehensively characterize comorbidities among GWAS cohorts to enable assessing the influence of those comorbidities on the GWAS results. The specific objectives of this study were to: (a) characterize comorbidities in a range of eMERGE phenotype-selected cohorts using the Johns Hopkins Adjusted Clinical Groups® (ACG®) system13, (b) assess the frequency of important comorbidities in three commonly studied GWAS phenotypes and (c) compare the comorbidity characterization of GWAS cases and controls. We also discuss the potential for sharing measures of comorbidity identified using the ACG software as part of genomic datasets.
Methods
Data source and preparation
De-identified EHR-derived electronic phenotype (e-phenotype) data and raw diagnostic codes were provided by the eMERGE Coordinating Center. The full dataset includes well-validated and published e-phenotypes4. For this analysis we used only the International Classification of Disease, Ninth Revision, Clinical Modification (ICD-9-CM), and International Classification of Disease, Tenth Revision, Clinical Modification (ICD-10-CM) codes for service dates ranging from 1978 to 2017 from the EHR of twelve eMERGE institutions.
We analyzed data for eMERGE Network study participants classified as a case or control for three eMERGE e-phenotypes including: Angiotensin converting enzyme (ACE)-inhibitor induced cough14, peripheral arterial disease (PAD)15 and heart failure (HF) (including both preserved and reduced ejection fraction subtypes)16. Two of the eMERGE e-phenotypes have led to published GWAS studies (ACE-inhibitor induced cough and peripheral arterial disease)6,7 We report the number of eMERGE institutions that implement each e-phenotype, the number of e-phenotype-selected cases and controls for GWAS, and the proportion of males and females among e-phenotype-selected cases and controls.
Analysis of comorbidities among phenotype-selected cohorts
Comorbidities were captured for eMERGE Network study participants using the Expanded Diagnosis Cluster (EDC) condition markers generated by the Johns Hopkins ACG system (version 11.2)13. For each study participant, overall ICD-9-CM, and ICD-10-CM codes from EHRs are used. The ACG system assigns all ICD codes to one or multiple of 282 EDCs. The ACG system also calculates the number of chronic condition comorbidities present for each individual (i.e., chronic condition count, CCC). For selected eMERGE phenotypes, we summarize the frequency of the top ten EDC chronic condition markers present in cases and controls. We also report the number of chronic conditions among cases and controls. In order to enable comparison of GWAS cases and controls for three eMERGE phenotypes, we report a t-test of the mean CCC among cases and controls. Statistical analyses were performed using SAS version 9.4.
Results
Study population
The representation of eMERGE institutions for each of the three selected e-phenotypes are summarized in Table 1. The case:control ratios were roughly 1:4 for ACE-inhibitor induced cough, 1:7 for PAD, and 1:3 for HF.
Table 1.
Study population
| ACE-inhibitor induced cough | Peripheral Arterial Disease | Heart Failure | |
|---|---|---|---|
| # eMERGE institutions | 7 | 5 | 8 |
| # Cases (% Female) | 1714 (62% female) | 2492 (37% female) | 3836 (46% female) |
| # Controls (% Female) | 7729 (47% female) | 18131 (61% female) | 13138 (60% female) |
Summary of comorbid conditions among eMERGE cohorts
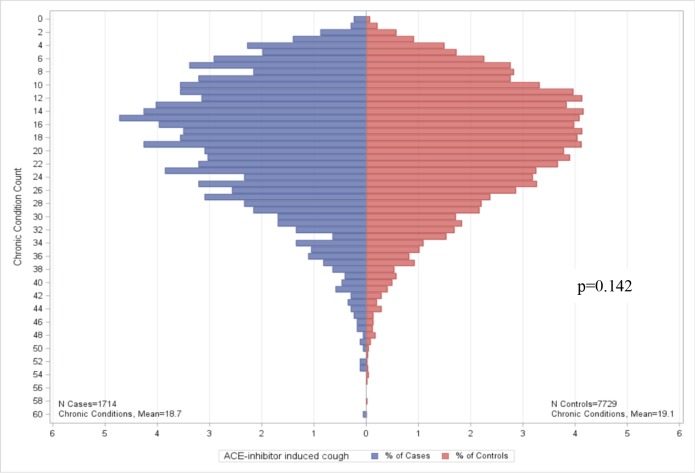
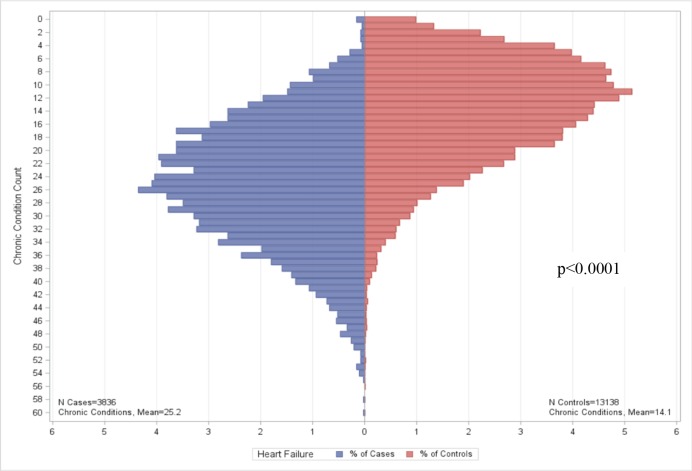
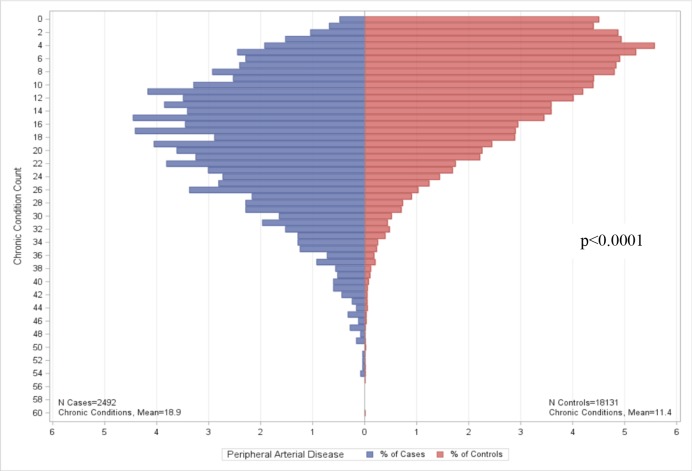
After filtering out non-disease conditions (e.g., preventive care), we reported the top ten comorbid conditions (see Table 2). Nine of ten, four of ten, and five of ten of the conditions for ACE-inhibitor induced cough, PAD and HF are the same between cases and controls, respectively. For all three conditions, the rank order of the top ten comorbid conditions differ between cases and controls. The number of chronic conditions identified by the ACG software for cases and controls are summarized in Figures 1–3. We found no significant differences in CCC for ACE-inhibitor induced cough cases and controls (p=0.1425). There are significant differences in CCC for PAD and HF cases and controls (p<0.0001), both of which CCC was lower in controls when compared to cases.
Table 2.
Top ten EDC condition markers among eMERGE e-phenotype cases and controls. NOTE: EDC condition markers that are not chronic conditions are excluded. Chronic conditions unique to cases or controls among the top ten EDC condition markers for the e-phenotype are bolded and italicized.
| ACE-inhibitor induced cough | Peripheral Arterial Disease | Heart Failure | |||
|---|---|---|---|---|---|
| Cases (%) (N=1,714) | Controls (%) (N=7,729) | Cases (%) (N=2,492) | Controls (%) (N=18,131) | Cases (%) (N=3,836) | Controls (%) (N=13,138) |
| Hypertension, w/o major complications (97%) | Hypertension, w/o major complications (96%) | Generalized atherosclerosis (86%) | Hypertension, w/o major complications (68%) | Congestive heart failure (98%) | Disorders of lipid metabolism (72%) |
| Disorders of lipid metabolism (84%) | Disorders of lipid metabolism (84%) | Disorders of lipid metabolism (85%) | Disorders of lipid metabolism (61%) | Hypertension, w/o major complications (93%) | Benign and unspecified neoplasm (66%) |
| Benign and unspecified neoplasm (69%) | Benign and unspecified neoplasm (65%) | Peripheral vascular disease (81%) | Benign and unspecified neoplasm (53%) | Cardiac arrhythmia (87%) | Hypertension, w/o major complications (66%) |
| Degenerative joint disease (59%) | Cardiac arrhythmia (60%) | Ischemic heart disease (excluding acute myocardial infarction) (71%) | Gastroesophageal reflux (42%) | Disorders of lipid metabolism (82%) | Musculoskeletal disorders, other (53%) |
| Musculoskeletal disorders, other (58%) | Iron deficiency, other deficiency anemias (59%) | Cardiac arrhythmia (65%) | Iron deficiency, other deficiency anemias (38%) | Ischemic heart disease (excluding acute myocardial infarction) (81%) | Bursitis, synovitis, tenosynovitis (53%) |
| Iron deficiency, other deficiency anemias (57%) | Degenerative joint disease (56%) | Cardiovascular disorders, other (63%) | Musculoskeletal disorders, other (38%) | Iron deficiency, other deficiency anemias (76%) | Degenerative joint disease (51%) |
| Bursitis, synovitis, tenosynovitis (55%) | Cataract, aphakia (55%) | Iron deficiency, other deficiency anemias (56%) | Degenerative joint disease (38%) | Respiratory disorders, other (74%) | Dermatitis and eczema (46%) |
| Gastroesophageal reflux (54%) | Musculoskeletal disorders, other (55%) | Cerebrovascular disease (55%) | Bursitis, synovitis, tenosynovitis (36%) | Cardiovascular disorders, other (71%) | Gastroesophageal reflux (44%) |
| Cardiac arrhythmia (53%) | Ischemic heart disease (excluding acute myocardial infarction) (54%) | Benign and unspecified neoplasm (54%) | Obesity (36%) | Benign and unspecified neoplasm (67%) | Peripheral neuropathy, neuritis (42%) |
| Ischemic heart disease (excluding acute myocardial infarction) (52%) | Bursitis, synovitis, tenosynovitis (52%) | Respiratory disorders, other (51%) | Other skin disorders (35%) | Musculoskeletal disorders, other (62%) | Iron deficiency, other deficiency anemias (40%) |
Figure 1.
Chronic conditions among eMERGE ACE-inhibitory induced cough cases and controls.
Figure 3.
Chronic conditions among eMERGE Heart Failure with differentiation between preserved and reduced ejection fraction cases and controls.
Discussion
Findings from this study demonstrate use of the Johns Hopkins ACG System to characterize comorbidities among GWAS cohorts. In summary, we show that use of the software enables comparing the prevalence of multiple comorbid chronic conditions (Figures 1–3) and of specific comorbid chronic conditions (Table 2). For two of the e-phenotypes, PAD and HF, the average number of chronic conditions present appears to be higher for cases when compared to controls. We also found differences in the ranking of four to nine of the top ten comorbid chronic conditions present in both cases and controls of selected eMERGE cohorts. Knowledge of such differences can help to inform unbiased control selection. It may also be possible to match cases and controls based on their overall burden of comorbidities for the purpose of genetic case-control analysis.
Bias and noise are well-known challenges to working with EHR data. Without understanding the complex processes under which the data were collected, incorrect conclusions can be drawn. For example, community-acquired pneumonia can simply be counted as healthy patients with high probability to have disease17. Some have explored approaches to factor out bias and noise18. The approach reported here comprehensively assessed e-phenotype-identified cohorts for a range of comorbid conditions using the ACG software. Providing summary data on the distribution of comorbid conditions has potential to guide actions for avoiding some forms of bias, and thus has implications for genomic data sharing. Currently eMERGE submits de-identified genetic and phenotype data to NIH’s database of Genotypes and Phenotypes (dbGap) for individual subjects. Measures of comorbidity identified using the ACG software has potential to be included as part of genomic datasets.
Limitations and future work
Our research has some limitations and areas for further investigation. First, there may be site bias due to our approach toward developing and validating eMERGE e-phenotype definitions used for case and control selection5. While e-phenotypes are often developed by one institution and used more broadly, there may be opportunities for approaches to select cases and controls in a way that is more tailored to each institution in order to optimize yield for the combined cohort. Second, more exploration is needed to understand where occurrences of gender and age imbalances exist and the impact on observed differences in CCC among cases and controls. In addition, differences in CCC among cases and controls for PAD and HF may be due in part to the major risk factors for the condition. For example, smoking, hypertension, diabetes and hyperlipidemia are risk factors for PAD. Third, within the top ranked comorbid chronic conditions, there is some confounding by the indication. For example, patients receiving ACE-inhibitors will almost always have hypertension. Chronic conditions identified may also be part of the definition itself. For example, PAD cases by definition have hypertension with major complications. Further investigation is needed to understand the extent to which differences between cases and controls remain after removing conditions known to be associated with the indication and e-phenotype definition. Fourth, the types of data we use influences our results. Administrative claims data are not currently included in the eMERGE datasets but could potentially augment the EHR-derived ICD data. For example, others have found that including claims data with EHR data rather than from the EHR alone has potential to improve sensitivity of detection19 and to improve the predictive power of risk stratification models20. In addition, laboratory data are captured for eMERGE cohorts but are not used by the ACG software in this analysis. There is work underway to expand the use of the ACG software to consider laboratory tests21, which may improve the identification of comorbidities. Last, further research is needed to compare the Johns Hopkins ACG software with other comorbidity definitions. While the specific focus of this work was on broad characterization of comorbid conditions using the Johns Hopkins ACG software, there are several other definitions of comorbidity that exist (e.g., Charlson comorbidity definitions22,23). Previous efforts comparing different definitions for chronic conditions found that the types of conditions included in different definitions may be important factors influencing analyses (e.g., estimating health care costs24, identifying individuals25, etc).
Conclusion
We characterized comorbidities in eMERGE datasets using the Johns Hopkins ACG system and compared the mean chronic condition count (CCC) among GWAS cases and controls. This study applied the ACG system to three eMERGE phenotype-selected cohorts: ACE-inhibitor induced cough, peripheral arterial disease (PAD) and heart failure (HF). Our analysis identified statistically significant differences in CCC among cases and controls for PAD and HF cohorts, suggesting that our framework for characterizing comorbidities among GWAS cohorts may enable improved selection of controls.
Figure 2.
Chronic conditions among eMERGE Peripheral Arterial Disease cases and controls.
References
- 1.Crawford DC, Crosslin DR, Tromp G, Kullo IJ, Kuivaniemi H, Hayes MG. eMERGEing progress in genomics-the first seven years. Front Genet. 2014;5:184. doi: 10.3389/fgene.2014.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15(10):761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirby JC, Speltz P, Rasmussen LV, Basford M, Gottesman O, Peissig PL. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc. 2016;23(6):1046–52. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20(e1):e147–54. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kullo IJ, Fan J, Pathak J, Savova GK, Ali Z, Chute CG. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc. 2010;17(5):568–74. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosley JD, Shaffer CM, Van Driest SL, Weeke PE, Wells QS, Karnes JH. A genome-wide association study identifies variants in KCNIP4 associated with ACE inhibitor-induced cough. Pharmacogenomics J. 2016;16(3):231–7. doi: 10.1038/tpj.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peissig PL, Rasmussen LV, Berg RL, Linneman JG, McCarty CA, Waudby C. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. J Am Med Inform Assoc. 2012;19(2):225–34. doi: 10.1136/amiajnl-2011-000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritchie MD, Verma SS, Hall MA, Goodloe RJ, Berg RL, Carrell DS. Electronic medical records and genomics (eMERGE) network exploration in cataract: several new potential susceptibility loci. Mol Vis. 2014;20:1281–95. [PMC free article] [PubMed] [Google Scholar]
- 10.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10(4):241–51. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 11.Weng LC, Lunetta KL, Muller-Nurasyid M, Smith AV, Theriault S, Weeke PE. Genetic Interactions with Age, Sex, Body Mass Index, and Hypertension in Relation to Atrial Fibrillation: The AFGen Consortium. Sci Rep. 2017;7(1):11303. doi: 10.1038/s41598-017-09396-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huntley AL, Johnson R, Purdy S, Valderas JM, Salisbury C. Measures of multimorbidity and morbidity burden for use in primary care and community settings: a systematic review and guide. Ann Fam Med. 2012;10(2):134–41. doi: 10.1370/afm.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Health Services Research & Development Center at the Johns Hopkins University Bloomberg School of Public Health. Baltimore, MD: The Johns Hopkins University Bloomberg School of Public Health; 2014. The Johns Hopkins ACG Case-Mix System Reference Manual Version 110 (Technical Reference Guide) [Google Scholar]
- 14.Mosley JD, Denny JC. ACE Inhibitor (ACE-I) induced cough. PheKB. Available from: https://phekb.org/phenotype/90 Vanderbilt University2012 [ [Google Scholar]
- 15.Kullo IJ. PheKB. Peripheral Arterial Disease - 2012. Available from: https://phekb.org/phenotype/16 Mayo Clinic2012 [ [Google Scholar]
- 16.Bielinski SJ. PheKB. Heart Failure (HF) with Differentiation between Preserved and Reduced Ejection Fraction. Available from: https://phekb.org/phenotype/147 Mayo Clinic2013 [ [Google Scholar]
- 17.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20(1):117–21. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hripcsak G, Albers DJ, Perotte A. Exploiting time in electronic health record correlations. J Am Med Inform Assoc. 2011;18(Suppl 1):i109–15. doi: 10.1136/amiajnl-2011-000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilchesky M, Tamblyn RM, Huang A. Validation of diagnostic codes within medical services claims. J Clin Epidemiol. 2004;57(2):131–41. doi: 10.1016/S0895-4356(03)00246-4. [DOI] [PubMed] [Google Scholar]
- 20.Kharrazi H, Weiner JP. A Practical Comparison Between the Predictive Power of Population-based Risk Stratification Models Using Data From Electronic Health Records Versus Administrative Claims: Setting a Baseline for Future EHR-derived Risk Stratification Models. Med Care. 2018;56(2):202–3. doi: 10.1097/MLR.0000000000000849. [DOI] [PubMed] [Google Scholar]
- 21.Lemke KW, Gudzune KA, Kharrazi H, Weiner JP. Assessing markers from ambulatory laboratory tests for predicting high-risk patients. Am J Manag Care. 2018;24(6):e190–e5. [PubMed] [Google Scholar]
- 22.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–83. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 23.Deyo RA, Cherkin DC, Ciol MA. Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J Clin Epidemiol. 1992;45(6):613–9. doi: 10.1016/0895-4356(92)90133-8. [DOI] [PubMed] [Google Scholar]
- 24.Brilleman SL, Gravelle H, Hollinghurst S, Purdy S, Salisbury C, Windmeijer F. Keep it simple? Predicting primary health care costs with clinical morbidity measures. J Health Econ. 2014;35:109–22. doi: 10.1016/j.jhealeco.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dattalo M, DuGoff E, Ronk K, Kennelty K, Gilmore-Bykovskyi A, Kind AJ. Apples and Oranges: Four Definitions of Multiple Chronic Conditions and their Relationship to 30-Day Hospital Readmission. J Am Geriatr Soc. 2017;65(4):712–20. doi: 10.1111/jgs.14539. [DOI] [PMC free article] [PubMed] [Google Scholar]